Abstract
Analysis of Caenorhabditis elegans syp-1 mutants reveals that both synapsis-dependent and -independent mechanisms contribute to stable, productive alignment of homologous chromosomes during meiotic prophase. Early prophase nuclei undergo normal reorganization in syp-1 mutants, and chromosomes initially pair. However, the polarized nuclear organization characteristic of early prophase persists for a prolonged period, and homologs dissociate prematurely; furthermore, the synaptonemal complex (SC) is absent. The predicted structure of SYP-1, its localization at the interface between intimately paired, lengthwise-aligned pachytene homologs, and its kinetics of localization with chromosomes indicate that SYP-1 is an SC structural component. A severe reduction in crossing over together with evidence for accumulated recombination intermediates in syp-1 mutants indicate that initial pairing is not sufficient for completion of exchange and implicates the SC in promoting crossover recombination. Persistence of polarized nuclear organization in syp-1 mutants suggests that SC polymerization may provide a motive force or signal that drives redispersal of chromosomes. Whereas our analysis suggests that the SC is required to stabilize pairing along the entire lengths of chromosomes, striking differences in peak pairing levels for opposite ends of chromosomes in syp-1 mutants reveal the existence of an additional mechanism that can promote local stabilization of pairing, independent of synapsis.
Keywords: Meiosis, chromosome pairing, synaptonemal complex, C. elegans, syp-1
At the onset of meiosis, an extensive spatial reorganization of chromosomes within the nucleus culminates in an arrangement in which homologous chromosomes are lengthwise-aligned, intimately paired, and capable of undergoing crossover recombination. For nearly all sexually reproducing organisms, the accuracy of chromosome segregation during meiosis I depends on the physical exchange between DNA molecules of homologous chromosomes provided by crossover recombination events that are completed in this context. In conjunction with sister-chromatid cohesion, crossing over results in the formation of chiasmata, which serve as mechanical connections that facilitate proper orientation and subsequent segregation of homologs toward opposite poles of the meiosis I spindle (Roeder 1997; Zickler and Kleckner 1999).
Specific associations between homologs are established early in meiotic prophase and are maintained prior to and during completion of crossover recombination and chiasma formation. Initial pairing events occur soon after premeiotic S phase, and are typically accompanied by an overall spatial reorganization of the nucleus that leads to a striking polarized distribution of chromosomes and other nuclear contents (for reviews, see Zickler and Kleckner 1998; Scherthan 2001). Many or all aspects of early prophase polarization are lost upon entry into the pachytene stage of meiotic prophase, as homologous chromosomes achieve full intimate alignment with one another along their entire lengths. A hallmark feature of pachytene chromosome organization is the synaptonemal complex (SC), a highly ordered protein structure positioned at the interface of lengthwise-aligned paired homologs (von Wettstein et al. 1984; Roeder 1997; Zickler and Kleckner 1999); most meiotic crossover events are thought to be completed within the context of the SC. Following exit from the pachytene stage, the SC disassembles and side-by-side alignment is lost, leaving homologs associated only if they have successfully completed crossing over and formed chiasmata.
Despite decades of cytological observation of meiosis, how homologous chromosomes recognize and stably associate with one another in a manner that is productive for crossover recombination is still not well understood. The early meiotic events that lead to initial establishment of homologous associations between chromosomes are particularly mysterious, as there exists little functional evidence from any system regarding the molecular mechanisms by which initial homolog recognition is achieved.
There has been considerable speculation, however, regarding possible roles that certain chromosome-associated structures and subchromosomal domains might play in the pairing process. Based on widespread observation of the SC as a prominent ultrastructural feature of meiosis, it had been a popular assumption that this highly ordered protein structure must play a crucial role in some aspect of homolog pairing (although cytological and genetic evidence from multiple systems suggests that the SC is not essential for initial homolog recognition; Roeder 1997). A prominent role for the SC in homolog pairing had been called into question, however, by the finding that in the absence of the Saccharomyces cerevisiae major SC central region component Zip1, pachytene-stage homologous chromosomes still maintain close parallel alignment (Sym et al. 1993; Nag et al. 1995). Moreover, the fact that the frequency of crossing over was only reduced by 50%–70% in zip1 mutants had additionally called into question the assumption that the SC has a conserved essential role in promoting crossover formation (Sym and Roeder 1994).
It has also been postulated that cis-acting chromosomal sites or domains may play important roles in promoting productive homolog alignment. Evidence for such sites comes from experiments examining the meiotic behavior of chromosome rearrangements in several different systems (e.g., Hawley 1980; Maguire 1985, 1986; McKim et al. 1988, 1993; Villeneuve 1994). Specifically, these experiments found that the presence of particular chromosomal domains in cis influenced the likelihood of crossover or chiasma formation over larger chromosome segments or whole chromosomes. A role for these domains in homolog pairing was inferred from the chromosome-wide nature of their influence on crossing over and/or chiasma formation; however, pairing per se was never examined in these experiments.
Clearly, longstanding questions remain regarding the molecular mechanisms that underlie meiotic chromosome pairing. The nematode Caenorhabditis elegans is emerging as a powerful experimental system for addressing these questions. The C. elegans gonad contains hundreds of syncitial germ-line nuclei whose spatial arrangement follows a temporal progression through meiotic prophase (Schedl 1997); each germ line contains many nuclei that are establishing pairing, as well as hundreds of nuclei that exhibit full alignment and synapsis between homologs. Moreover, the arrangement of chromosomes within germ-line nuclei exhibits distinguishing characteristics, readily detectable at the light-microscopic level, that reflect hallmark features of different meiotic prophase substages (Albertson et al. 1997). Finally, cytological tools such as fluorescence in situ hybridization (FISH) and immunolocalization can be used to further probe the organization and morphogenesis of meiotic chromosomes in detail within three-dimensionally preserved germ lines (Dernburg et al. 1998).
Previous work established that stable side-by-side alignment of homologous chromosomes and SC formation in C. elegans do not depend on prior initiation of meiotic recombination or the presence of recombination machinery components. Meiotic nuclei in mutants lacking components of the core meiotic recombination machinery exhibit normal chromosome organization until the last stage in prophase, diakinesis, when SC disassembly and loss of side-by-side alignment reveal a lack of chiasmata between homologous chromosomes (Dernburg et al. 1998; Zalevsky et al. 1999; Kelly et al. 2000; Chin and Villeneuve 2001). This property of C. elegans meiosis has enabled us to identify genes involved in meiotic homolog alignment per se by screening among mutants containing achiasmate chromosomes at diakinesis for those with disrupted chromosome morphology and/or organization at earlier prophase stages (MacQueen and Villeneuve 2001).
Our approach previously identified a role for C. elegans chk-2 in promoting both the initial establishment of homolog pairing and the extensive nuclear reorganization that normally accompanies initial pairing at the onset of meiotic prophase. Here we present analysis of a phenotypically distinct pairing mutant, which has revealed essential roles for a C. elegans SC component in crossover recombination and in maintaining stable alignment of homologs subsequent to initial establishment of pairing. Furthermore, our time-course analysis of pairing in syp-1 mutants has additionally revealed a capacity of chromosome ends that harbor postulated cis-acting pairing centers to achieve local stabilization of pairing even in the absence of synapsis.
Results
syp-1 mutants exhibit defective chromosome organization within meiotic prophase nuclei
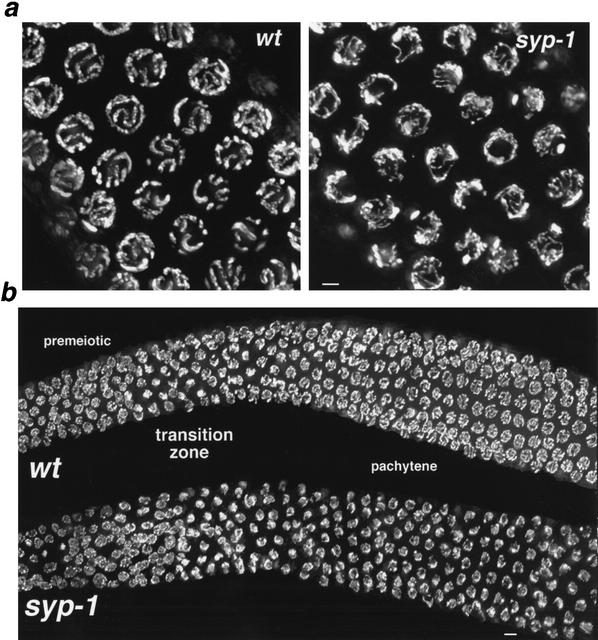
DAPI staining and high resolution imaging of three-dimensionally preserved syp-1 mutant germ-line nuclei revealed defects in chromosome organization soon after the onset of meiotic prophase (Fig. 1). In wild-type germ lines, nuclei entering meiotic prophase undergo a dramatic spatial reorganization in which chromosomes cluster toward one side of the nucleus; this polarized nuclear organization imparts a crescent-shaped appearance to the DAPI-stained chromatin in nuclei within the transition-zone region of the germ line (Fig. 1b). Nuclear polarization is normally lost during the transition into the pachytene stage, as aligned and synapsed chromosomes redisperse about the periphery of the nucleus. In syp-1 mutant germ lines, nuclei within the transition-zone region exhibit polarized organization (Fig. 1b), indicating that early meiotic reorganization has taken place. However, the vast majority of nuclei within the pachytene region of syp-1 germ lines also exhibit an asymmetric distribution of chromosomes, indicating that aspects of nuclear polarization persist in the absence of syp-1 function. Occasional nuclei with a widely dispersed spatial distribution of chromosomes can be detected only at the very end of the pachytene region of syp-1 mutant germ lines.
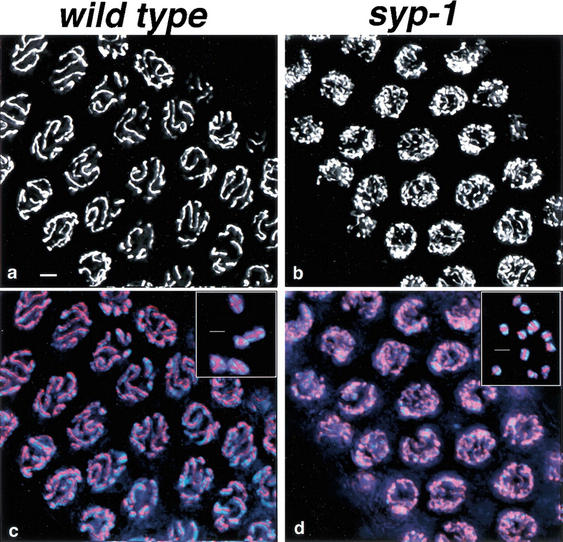
Figure 1.
Defective chromosome organization in syp-1 meiotic prophase nuclei. (a) High-magnification images of DAPI-stained nuclei from the pachytene region of wild-type and syp-1 mutant germ lines. Widely distributed about the periphery of wild-type pachytene nuclei are parallel pairs of DAPI-stained tracks, corresponding to lengthwise-aligned and synapsed homologous chromosomes. In the syp-1 mutant, pachytene region nuclei exhibit disorganized DAPI-stained tracks not aligned in parallel pairs, and chromatin is distributed asymmetrically within the nuclei. Images are projections approximately halfway through 3D data stacks of whole nuclei. (b) Low-magnification images of DAPI-stained germ lines. In both wild-type and syp-1 mutant germ lines, DAPI-stained chromatin is widely dispersed in premeiotic nuclei, whereas the polarized organization of nuclei in the transition zone (entering meiotic prophase) imparts a crescent-shaped appearance to DAPI-stained chromatin. In the pachytene region of the wild-type germ line, DAPI signals are again widely dispersed; early prophase nuclear polarization persists in the pachytene region of the syp-1 mutant germ line, with most nuclei in this region exhibiting an asymmetric distribution of chromatin. Images are projections of 3D data stacks that encompass entire nuclei (except at the edges, where only a portion of some nuclei are projected). Bars, a, 2 μm; b, 4 μm.
Chromosomes within pachytene region nuclei in the syp-1 mutant are not only asymmetrically distributed, but they also fail to achieve normal pachytene alignment. Wild-type DAPI-stained pachytene nuclei exhibit closely aligned, parallel tracks that correspond to pairs of homologous chromosomes fully synapsed along their entire lengths (Fig. 1a). In contrast, thin DAPI-stained tracks, unaligned with any other chromosome, are frequently observed in pachytene-region nuclei of syp-1 mutant germ lines, indicative of a defect in homologous synapsis.
syp-1 is required to stabilize homologous associations between meiotic prophase chromosomes
We used FISH to assess the relationships between specific regions of homologous chromosomes during meiotic prophase in syp-1 mutants (Fig. 2). In C. elegans, homologs are unpaired prior to entry into meiosis (Dernburg et al. 1998); therefore, most premeiotic nuclei, located in the distal region of wild-type germ lines, exhibit two separated FISH signals for each probe. Initial pairing occurs within the transition zone, which contains a mixture of nuclei with separated signals and nuclei with closely associated or completely overlapping hybridization signals. Closely juxtaposed or overlapping FISH signals are detected in all nuclei throughout the entire pachytene region of wild-type germ lines, where chromosomes are fully synapsed.
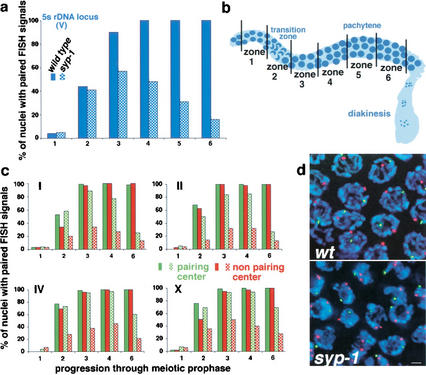
Figure 2.
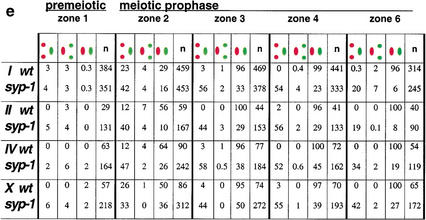
Homolog pairing is established but not maintained in syp-1 mutants. (a) Graph exhibiting the percent of nuclei with paired FISH signals at the 5S rDNA locus on Chromosome V. The numbers of nuclei scored per zone for wild-type and syp-1 (respectively) are as follows: zone 1: 103, 191; zone 2: 133, 242; zone 3: 125, 235; zone 4: 120, 219; zone 5: 102, 286; zone 6: 84, 148. In a and c, solid bars correspond to wild-type pairing frequencies, whereas hatched bars represent pairing frequencies in the syp-1 mutant. (b) Diagram of the germ line, indicating the positions of zones used in time-course analysis of homolog pairing. Zone 1, which begins approximately three nuclear diameters from the distal tip, contains exclusively premeiotic nuclei; zone 2 includes some premeiotic nuclei, but contains predominantly nuclei at the leptotene/zygotene stages of meiotic prophase; zones 3 and 4 contain mainly early- to mid-pachytene stages (zone 3 may also include a few leptotene/zygotene nuclei); zones 5 and 6 contain late-pachytene nuclei (zone 5 was used only for the experiment shown in a). (c) Graphs exhibiting data from double-label experiments in which pairing at the pairing-center end (green) and the non-pairing-center end (red) of a single chromosome were quantified simultaneously. For Chromosome I, the non-pairing-center end locus is ∼15% of the physical chromosome length from the left end of I; in all other cases, probes targeted loci <3% of chromosome length from the nearest chromosome end. Numbers of nuclei scored for each zone are exhibited in e; see Materials and Methods for statistical analysis. (d) High-magnification images of zone 4 nuclei hybridized with FISH probes targeting opposite ends of Chromosome IV (scale bar, 2 μm). FISH signals for both probes are paired in all wild-type nuclei. In the syp-1 mutant, the non-pairing-center end (red) is frequently unpaired, whereas the pairing-center end (green) is usually paired. (e) Percent of nuclei with distinct pairing configurations (e.g., green paired, red unpaired) within zones 1–6 for both wild-type and syp-1 mutant germ lines; the numbers of nuclei scored are in parentheses.
Nuclei in the pachytene region of syp-1 mutant germ lines frequently contained widely separated FISH signals, indicative of a defect in homolog pairing. However, in our initial experiments using the 5S rDNA probe (Fig. 2a), we also noted that FISH signals were paired in a significant fraction of nuclei. This pairing defect contrasts with that seen in chk-2 mutants, in which pairing never rose above the premeiotic baseline in any region of the germ line (MacQueen and Villeneuve 2001). Furthermore, nuclei containing paired FISH signals appeared to be more frequent in the transition zone compared with late-pachytene regions of syp-1 mutant germ lines, raising the possibility that syp-1 function is required for stabilizing paired associations between chromosomes.
To explore whether the status of pairing differs between earlier and later stages of prophase in syp-1 mutants, we conducted a time-course analysis of meiotic pairing. The architecture of the C. elegans germ line facilitates this analysis, because the spatial arrangement of nuclei from distal to proximal regions of the germ line corresponds to the temporal progression into and through meiotic prophase. For most of the time-course experiments, the frequency of paired FISH signals was assessed in five equal-sized zones (36 × 36 μm) distributed along the distal–proximal axis of the germ line (Fig. 2b). Four of the five zones were contiguous, with the first (most distal) domain containing exclusively premeiotic nuclei. The three subsequent zones spanned meiotic prophase entry through mid-pachytene stages, and the final zone began roughly one zone-length from the end of zone 4, corresponding to late pachytene. (Our first time-course experiment, using the 5S rDNA probe, was performed similarly except that it included one additional zone between zone 4 and the final zone in the time course.)
Figure 2 illustrates the pairing profiles observed for each of nine loci, representing five of the six C. elegans chromosomes. In wild-type germ lines, paired signals were very infrequent (<6%) in premeiotic nuclei (zone 1); the frequency of paired FISH signals increased dramatically in zones 2 and 3, which span the transition-zone region of the germ line, where meiotic entry occurs. Full pairing was achieved by the pachytene stage, and was maintained throughout pachytene (zones 4–6).
In syp-1 mutant germ lines, homologous associations initially formed upon entry into meiotic prophase (zone 2), but homologs dissociated prematurely. For each of the nine probes analyzed, the frequency of nuclei with paired FISH signals increased above premeiotic levels in zones 2 and 3 in syp-1 mutants and peaked within zone 3 or 4. Furthermore, for each probe the final zone displayed a significantly reduced frequency of nuclei with paired FISH signals compared with the peak value (Fig. 2; Materials and Methods). The pairing profiles exhibited by syp-1 mutants indicate that meiotic chromosome pairing in C. elegans involves genetically separable steps: a syp-1-independent process that establishes initial homolog pairing during early prophase and a syp-1-dependent process that stabilizes pairing through later prophase stages.
Strikingly, opposite ends of a single chromosome exhibited drastically different peak pairing levels in syp-1 mutants (Fig. 2c). In experiments where pairing profiles of opposite ends of a single chromosome (I, II, III, or X) were analyzed simultaneously, one chromosome end exhibited a substantially higher frequency of homologous association than the other end. These experiments revealed a requirement for syp-1 in maintaining parallel alignment between homologs during prophase, as many nuclei in mid- to late-pachytene regions of syp-1 germ lines exhibited paired FISH signals at one end of a homolog pair and unpaired signals, ranging from 0.8 to 4.0 μm apart, at the opposite end of the same pair. Interestingly, in all cases the chromosome end that exhibited a relatively high degree of syp-1-independent pairing corresponded to the end containing the pairing center, a cis-acting domain genetically defined by its ability to permit and/or promote crossover recombination along the entire length of a given chromosome (Albertson et al. 1997). These data imply the existence of a second, syp-1-independent mechanism that promotes stabilization of pairing locally within a specific domain of each C. elegans chromosome. Notably, prior to achieving full pairing, even in wild-type germ lines chromosome I exhibited a significantly higher frequency of meiotic paired associations at the pairing center end compared with the opposite end (zone 2; Fig. 2c,e).
SC is not detected in syp-1 mutant prophase nuclei
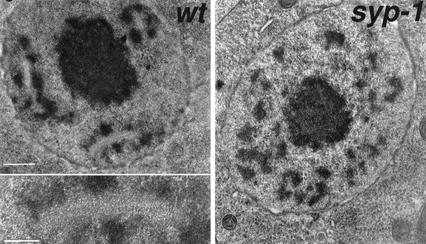
As the defects in chromosome organization in syp-1 mutants first become apparent during the period of prophase when the SC would normally assemble, we asked whether the SC is formed in syp-1 mutants. We used transmission electron microscopy (TEM) to evaluate SC organization in wild-type and syp-1 mutant germ-line nuclei in prophase stages ranging from late leptotene/zygotene through late pachytene (>85% of nuclei scored were in the pachytene stage; see Materials and Methods). In wild-type nuclei, 0.3–1.8-μm-long stretches of a zipper-like structure, corresponding to an ordered array of transverse filaments that make up the SC central region, were readily detected. These structures were typically flanked along their lengths by densely stained chromatin. An average of 2.5 SC stretches per nucleus was observed in the scored regions of wild-type germ lines (n = 195), whereas the SC was never detected in nuclei from similar regions of syp-1 mutant germ lines (n = 212). Patches of electron-dense chromatin were clearly visible in syp-1 mutant germ lines, but were never associated with an ordered zipper-like structure (Fig. 3).
Figure 3.
The SC is absent in the syp-1 mutant. TEM images of 100-nm-thick equatorial sections of nuclei from the late-pachytene regions of wild-type and syp-1 mutant germ lines. In the wild-type nucleus, three extensive stretches of zipper-like tracks (corresponding to transverse filaments that comprise the SC central region) are flanked by densely stained patches of chromatin; the inset shows a higher-magnification view of one SC stretch. The centrally located, densely stained entity is the nucleolus. The syp-1 nucleus contains numerous patches of densely stained chromatin, but no zipper-like structures. Bar, 500 nm; inset, 200 nm.
Meiotic chromosome axis component HIM-3 assembles onto chromosomes in the absence of SYP-1
Just prior to or at the onset of meiotic prophase, chromosomes develop specialized axial structures along their lengths, built on a foundation of sister-chromatid cohesion proteins. These meiotic chromosome axes, or cores, become incorporated into the mature SC during the process of synapsis and are known as lateral elements within the context of the mature SC (Moses 1969; Rufas et al. 1992; Zickler and Kleckner 1999). C. elegans HIM-3, a meiosis-specific, axis-associated protein related to S. cerevisiae Hop1p, localizes at the interface between aligned homologs during pachytene, when chromosome axes are fully synapsed, and remains at the interface between sister chromatids through diakinesis and metaphase I (Zetka et al. 1999).
When we assessed HIM-3 localization in syp-1 mutant germ lines, we observed loading of HIM-3 onto chromosomes beginning early in prophase, demonstrating that chromosomal localization of HIM-3 does not require syp-1 function (Fig. 4). Furthermore, HIM-3 remained localized to chromosomes throughout the diakinesis stage in a narrow stripe that presumably corresponds to the interface between sister chromatids. Thus, at least some aspects of meiotic chromosome axis morphogenesis are unaffected by the syp-1 mutation, raising the possibility that syp-1 plays a role in organization of the SC central region.
Figure 4.
HIM-3 localizes to meiotic chromosomes in syp-1 mutants. Anti-HIM-3 staining is exhibited in white (a,b) or red (c,d), and DAPI-stained chromosomes are exhibited in blue (c,d). HIM-3 is visible as a single, continuous line at the interface between synapsed homologous chromosomes in wild-type pachytene nuclei (a,c), and remains associated with chromosome axes during diakinesis, where it is detected as a single line or a cross-shaped pattern on homolog pairs attached by a chiasma (c, inset). Although chromosomes are asynapsed in pachytene-region nuclei in the syp-1 mutant, they nevertheless exhibit extensive continuous stretches of anti-HIM-3 staining. Likewise, HIM-3 remains associated with diakinesis chromosomes in syp-1 mutants; each univalent chromosome exhibits one or two discrete tracks of anti-HIM-3 staining, presumably corresponding to chromosome axes (d, inset). Bars, 2 μm.
syp-1 encodes a coiled-coil protein that localizes to the interface of synapsed chromosomes
We initially mapped syp-1(me17) to a 1-Mb region between stP18 and the 5S rDNA locus (rrs-1) on Chromosome V (see Materials and Methods). In parallel, our laboratory had initiated a functional genomics approach to assess potential meiotic roles for a defined set of candidate genes (Colaiácovo et al. 2002). This approach used RNA interference (RNAi) to deplete the products of a subset of genes exhibiting germ-line-enriched expression in microarray analyses conducted by Reinke et al. (2000). One gene whose expression was strongly induced in germ lines, F26D2.2, was located in the interval to which syp-1 had been mapped. When F26D2.2 was targeted by RNAi, affected worms exhibited meiotic defects similar to those seen in syp-1(me17) mutants: Late-prophase nuclei contained achiasmate chromosomes, whereas nuclei in the pachytene region exhibited both disrupted chromosome organization and a persistence of polarized nuclear organization.
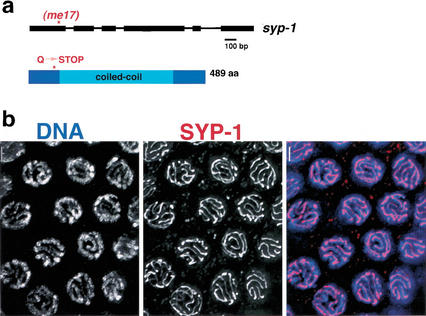
Sequence analysis exhibited that syp-1(me17) mutant worms contain a nonsense mutation in the F26D2.2 coding region, which results in a stop codon inserted at position 74 of the 489-amino-acid F26D2.2 protein (Fig. 5). The size of the predicted SYP-1 protein (GenBank accession no. AF515883) is based on a revised gene structure derived from sequencing cDNAs obtained by RT-PCR and 5′ RACE; this revised structure lacks three upstream exons that had been predicted by GeneFinder, but is consistent with the structure suggested by EST alignment data presented in WormBase (http://www.wormbase.org).
Figure 5.
SYP-1 is a coiled-coil protein that localizes to the interface of synapsed pachytene chromosomes. (a) Schematic representation of syp-1 gene structure and the predicted SYP-1 protein, indicating the location of the early stop caused by the me17 mutation. (b) Wild-type pachytene nuclei stained with an antibody raised against the SYP-1 C terminus. The images shown are projections approximately halfway through 3D data stacks that encompass entire nuclei. Bars, 2 μm.
The region between residues 48 and 402 of the 489-amino-acid SYP-1 protein is predicted to contain α-helical coiled coils, based on analysis using the MultiCoil, COILS, and Paircoil programs (Berger et al. 1995; Lupas 1997; Wolf et al. 1997); outside of the coiled-coil domain, SYP-1 is not similar to other known proteins. In light of the requirement for syp-1 function in SC morphogenesis, it is interesting to note that coiled-coil domains are the only described structural characteristics shared in common between proteins comprising the central region of the SC in diverged organisms (Roeder 1997; Page and Hawley 2001).
Immunolocalization of SYP-1 protein provides further evidence for a structural role for SYP-1 in SC assembly. Using two independent anti-SYP-1 antibodies raised against N-terminal and C-terminal peptides, we observed SYP-1 protein along the length of synapsed chromosomes during meiotic prophase (Fig. 5b). Within the premeiotic region of the germ line, SYP-1 protein is not detectable on chromosomes. At or just prior to the beginning of the transition zone, a small number of nuclei are closely associated with one or two SYP-1 foci (Fig. 6b, arrows), while the majority of transition-zone nuclei exhibit several continuous tracks of SYP-1 staining that localize to a portion of the total DAPI-stained chromatin (Fig. 6). In the pachytene region, nuclei exhibit highly ordered SYP-1 staining: Thick, continuous lines of SYP-1 lie at the interface between synapsed homologs along their full lengths (Fig. 5b). Finally, SYP-1 disassembles from the majority of the length of each chromosome pair by diakinesis, and is undetectable on chromosomes by the end of prophase (see below).
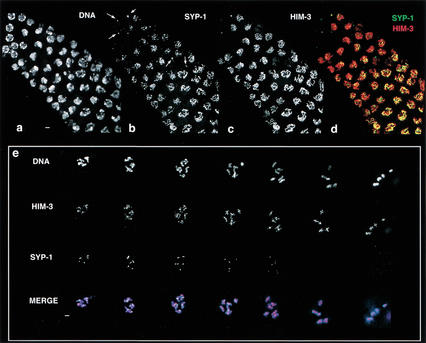
Figure 6.
SYP-1 and HIM-3 localize to chromosomes and disassemble from chromosomes with different kinetics. (a–d) Region of the germ line flanking the transition zone, where nuclei enter meiotic prophase; earlier stages are at the upper left. One or two SYP-1 foci are associated with nuclei just prior to or in earliest meiotic prophase (arrows in b). Within the first early prophase nuclei to exhibit short, continuous stretches of SYP-1 staining, SYP-1 localizes over a limited portion of the chromatin. At earliest prophase stages, HIM-3 is sometimes found as a single focus but more typically exhibits a uniform distribution along chromosomes (c). Numerous early prophase nuclei exhibit robust HIM-3 localization uniformly along chromosomes but only partial or no chromosomal localization of SYP-1 (b–d). The images in e represent all diakinesis nuclei contained within a single wild-type germ line: Left to right arrangement of nuclei corresponds to a temporal progression through diakinesis, with latest at right. Robust α-HIM-3 staining is present on all bivalents in all diakinesis nuclei. In contrast, anti-SYP-1 staining appears as a single focus associated with each bivalent in early diakinesis nuclei, and is virtually absent from all bivalents by the end of diakinesis. In merged images, α-HIM-3 is shown in red, α-SYP-2 in green (d,e), and DAPI in blue (e). Bars, 2 μm.
Antibodies that recognize either the N or C terminus of SYP-1 do not detect SYP-1 protein on chromosomes in germ-line nuclei from syp-1(me17) mutants (data not shown), further confirming the specificity of the antibody and suggesting that syp-1(me17) is likely a null allele.
Dynamics of SYP-1 localization during meiotic prophase
Further evidence that SYP-1 comprises part of the SC central region comes from simultaneous analysis of HIM-3 and SYP-1 localization (Fig. 6). Although HIM-3 and SYP-1 exhibit indistinguishable patterns of localization on chromosomes in pachytene nuclei, their kinetics of assembly and disassembly differ substantially. Within the transition zone, a uniform distribution of chromosome-associated HIM-3 is faintly detectable in a few nuclei that are presumably just entering meiotic prophase or are in late-premeiotic S phase. Most other nuclei in this region exhibit the polarized organization characteristic of the onset of meiotic prophase and also display robust HIM-3 uniformly distributed along the chromosomes. A small number of nuclei with either undetectable or very faint levels of HIM-3 contain a strong focus of SYP-1 positioned adjacent to the nuclear boundary, which sometimes overlaps with a similarly positioned HIM-3 focus. This SYP-1 focus is found in nuclei that have not yet achieved the early prophase polarized organization, and is not apparent in nuclei with chromosomal localization of SYP-1, suggesting that it may correspond to a pool of protein that will later be assembled onto chromosomes. In contrast to HIM-3, early chromosomal localization of SYP-1 does not occur uniformly along all chromosomes: In some early prophase nuclei, SYP-1 is assembled in robust stretches over several limited chromosomal segments while remaining undetectable on other chromosomal regions. In addition, chromosomal SYP-1 is detected only in nuclei that already have robust, widespread chromosomal localization of HIM-3; thus, many nuclei in this region exhibit uniform chromosomal localization of HIM-3 with only partial chromosomal localization of SYP-1. In more proximal regions of the germ line, where nuclei are entering early pachytene, both HIM-3 and SYP-1 exhibit maximal chromosomal localization, and the distribution patterns of the two proteins are indistinguishable at the light-microscopic level. The differences in localization dynamics between SYP-1 and HIM-3 in early prophase are consistent with a model in which SYP-1 loads onto chromosomes subsequent to the morphogenesis of chromosome axes.
At the end of prophase, SYP-1 and HIM-3 exhibit distinct localization patterns (Fig. 6e). During SC disassembly, chromosomes lose side-by-side alignment and continue to condense, giving rise to six compact bivalents in diakinesis nuclei. Axis association of HIM-3 is retained through diakinesis and persists until it is lost at anaphase of meiosis I (Zetka et al. 1999). In contrast, SYP-1 disassembles from the majority of the length of chromosomes concurrent with loss of side-by-side alignment, and is undetectable on chromosomes by late diakinesis (Fig. 6e). These localization dynamics during meiotic prophase entry and exit most likely reflect a structural role for SYP-1 as a building block of the SC central region.
syp-1 function is required for crossover recombination
In syp-1 mutants, 12 univalent chromosomes are observed in diakinesis nuclei (Fig. 4, inset), reflecting a failure in chiasma formation. Because lack of chiasmata leads to homolog missegregation and consequent aneuploid gametes, the vast majority of embryos produced by syp-1 mutant hermaphrodites are inviable (95%, n = 982). Moreover, a high incidence of males (Him phenotype) is found among the rare surviving progeny (38% male self-progeny, n = 1669 compared with 0.2% for wild type; Hodgkin et al. 1979); this Him phenotype is diagnostic of X chromosome missegregation, because X chromosome ploidy determines sex in an otherwise diploid C. elegans animal.
Absence of chiasmata at diakinesis in syp-1 mutants could indicate either a failure to complete crossover recombination or a defect in deriving functional chiasmata from meiotic recombination events. Assessment of crossover frequencies for two intervals spanning most of the X chromosome genetic map demonstrated a severe reduction or absence of crossing over in syp-1 mutant germ lines (Table 1). For a 40-cM interval encompassing the majority of X, crossover frequency was measured using a pair of morphological markers. Crossover frequency was also measured for a 3-cM interval corresponding to the pairing center region at the left end of X that exhibited substantially higher peak pairing levels than the opposite end of the X chromosome in FISH analysis; crossing over in this interval was assessed using codominant single nucleotide polymorphism (SNP) markers (Wicks et al. 2001). No crossovers were detected in either interval in syp-1 mutants, indicating that initial pairing of homologs is not sufficient to allow exchange.
Table 1.
Absence of Crossover Recombination in syp-l Mutants
| A. Genotype
|
|
Recombinant progeny
|
Total progeny
|
Map distance (cM)
|
|---|---|---|---|---|
| +/(syp-l or +); dpy-3 unc-3/++ | 1103 | 3420 hermaphrodites | 40 | |
| syp-l/syp-l; dpy-3 unc-3/++ | 0 | 286 hermaphrodites | <0.2 | |
| 186 males | ||||
| B. Genotypea | SNP markers assayed | Recombinant chromosomes | Total chromosomes | Map distance (cM) |
| +/(syp-l or +); X(CB4856)/X(N2) | pkP6100, pkP6118 | 17 | 560 | 3 |
| syp-l/syp-1; X(CB4856)/X(N2) | pkP6100, pkP6118 | 0 | 710 | <0.2 |
Assayed worms were heterozygous for an X chromosome derived from the Bristol N2 strain background and an X chromosome derived from the CB4856 mapping strain.
Positions of markers on the genetic map are as follows: pkP6100 (−19); dpy-3 (−16.4); pkP6118 (−14.6); unc-3 (21.2).
syp-1 mutants exhibit spo-11-dependent activation of the pachytene checkpoint
Absence of crossing over in syp-1 mutants could reflect a failure to initiate meiotic recombination in the first place, or an inability to complete initiated recombination events as crossovers. To distinguish between these possibilities, we sought evidence of unrepaired meiotic recombination intermediates in syp-1 mutant germ-line nuclei. Gartner et. al (2000) previously showed that unrepaired meiotic double-strand breaks (DSBs) can trigger a pachytene DNA-damage checkpoint in C. elegans, resulting in apoptosis of meiocytes in the late-pachytene region of the germ line. In the late-pachytene region of syp-1 mutant germ lines, we observed an elevated number of apoptotic nuclei compared with that seen in age-matched wild-type controls (Fig. 7). Moreover, we found that elevated germ cell death in syp-1 mutants was largely suppressed by a mutation in spo-11, which encodes a protein responsible for making the DSBs that initiate meiotic recombination. These data imply that SPO-11-induced DSBs do form in the absence of syp-1 function, and further implicate the SC in promoting the completion of crossover recombination events. Finally, the fact that the checkpoint response triggered in syp-1 mutants is largely spo-11-dependent suggests that a defect in synapsis per se may not elicit a robust pachytene checkpoint response in C. elegans.
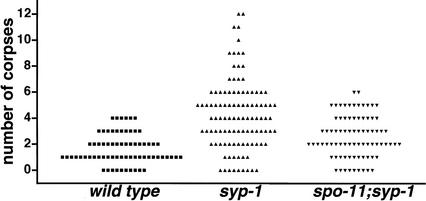
Figure 7.
spo-11-dependent elevation of germ cell apoptosis in syp-1 mutant germ lines. Scatterplot depicting the number of apoptotic corpses detected in germ lines of wild-type, syp-1, or syp-1; spo-11 worms. Germ cell corpses were scored in adult hermaphrodites 22–24 h post-L4 as in Gartner et al. (2000). The Y-axis value for each point represents the number of corpses observed in a single gonad arm. The differences between wild-type and syp-1 data sets and between spo-11; syp-1 and syp-1 data sets are extremely significant (two-tailed Mann-Whitney p < 0.0001; this test also indicated a modestly significant difference between wild type and spo-11; syp-1: p = 0.024).
Persistent polarized organization of prophase nuclei in syp-1 mutants is not a consequence of incomplete meiotic recombination
The asymmetric distribution of chromosomes exhibited by nuclei in the pachytene region of syp-1 mutant germ lines is distinct from the chromosome distribution normally found in C. elegans pachytene nuclei, and is reminiscent of the polarized organization that is usually restricted to nuclei at the onset of meiotic prophase. We found that nuclei in the pachytene regions of spo-11; syp-1 double-mutant germ lines exhibited persistent polarized organization indistinguishable from that seen in syp-1 single mutants (data not shown). This argues against the possibility that failure in redispersal of chromosomes in syp-1 mutants represents an arrest response elicited by incomplete recombination events and further suggests that the role of SYP-1 in driving large-scale chromosome redispersal may be through the assembly or function of the SC itself.
Discussion
SYP-1 and SC structure
The experimental evidence presented above supports the conclusion that SYP-1 is an SC structural component and likely assembles within the central region. Zip1 and SCP1 are structural components of the central region of budding yeast and mammalian SC, respectively (Meuwissen et al. 1992; Sym et al. 1993; Dong and Roeder 2000). Although these proteins are otherwise dissimilar, both contain extensive coiled-coil domains, and there is evidence that they form rod-shaped dimers that orient head-to-head to build the transverse filaments that span the central region of the SC (Meuwissen et al. 1992; Sym et al. 1993; Tung and Roeder 1998; Dong and Roeder 2000). Based on its size and the fact that it also contains an extensive central coiled-coil region, the product of the c(3)G gene has been proposed to serve a structural role similar to that of Zip1 or SCP1 within the Drosophila SC (Page and Hawley 2001). C. elegans syp-1 encodes a protein that, like Zip1, SCP1, and C(3)G, contains an extended coiled-coil motif. However, SYP-1 is approximately half the size of Zip1 or SCP1; given that the width of the C. elegans SC is comparable to that of mammals, yeast, and flies (Goldstein and Slaton 1982; Dernburg et al. 1998), we propose that SYP-1 exhibits an organization within the SC central region that is distinct from that of Zip1 or SCP1, and/or that additional structural proteins work in conjunction with SYP-1 to form the mature central region of the C. elegans SC.
A role for the C. elegans SC in the global organization of chromosomes within meiotic prophase nuclei
As a rule, progression through meiotic prophase entails extensive changes in the global organization and arrangement of chromosomes within the nucleus (Scherthan 2001). Immediately upon entry into meiosis, a dramatic polarization event occurs in which entire chromosomes or chromosome ends become clustered at a limited sector of the nuclear periphery. Accompanying this early prophase reorganization is the initial establishment of homologous associations between chromosomes; a mechanistic coupling between nuclear reorganization and initial pairing is evidenced by several C. elegans meiotic mutants in which both of these major early prophase events are abolished (MacQueen and Villeneuve 2001; A.J. MacQueen, K. Nabeshima, N. Miley, and A.M. Villeneuve, unpubl.). Polarized chromosome organization within the nucleus is often lost as the pairing process culminates in an intimate parallel alignment between homologs; this organizational change is temporally coupled with completion of assembly of the SC (Roeder 1997; Zickler and Kleckner 1998, 1999; MacQueen and Villeneuve 2001). In C. elegans, loss of nuclear polarization and completion of SC assembly result in a nuclear organization in which lengthwise-aligned, synapsed chromosomes are widely dispersed about the periphery of pachytene nuclei.
In syp-1 mutants, chromosomes fail to disperse from their early prophase configuration, resulting in a persistence of polarized nuclear organization into late-pachytene stages. Because persistence of polarized organization is not a consequence of unrepaired recombination intermediates, this observation suggests that SC morphogenesis and early pachytene chromosome redispersal may be mechanistically coupled. Perhaps the SC assembly process itself provides a direct motive force that drives chromosomes out of their polarized distribution within the nucleus, possibly as a result of a rigidity imposed on chromosome pairs by the SC itself. An alternative possibility is that the progression of synapsis might produce a signal that promotes either an active (e.g., driven by molecular motors) chromosome redispersal process, or the release of chromosomes from a higher level of organization, enabling their passive dispersal. This model fits well with the idea that chromosome redispersal might be functionally important for facilitating completion of synapsis in a manner that avoids interlocking of chromosomes (Zickler and Kleckner 1999).
The role of the SC in homolog pairing throughout meiotic prophase
The SC structure assembles at the interface between homologs as they achieve an intimate lengthwise alignment, and its prominent presence at this stage has fueled speculation that it must play some role in homolog pairing. However, there is presently little direct evidence that speaks to an essential function for the SC in mediating chromosome pairing. In fact, the major SC central region component Zip1 is dispensable for stable lengthwise alignment between prophase chromosomes in budding yeast: In zip1 mutants, homolog axes lie farther apart than normal in spread preparations, but nonetheless are aligned along their entire lengths (Sym et al. 1993). In fact, pachytene-stage homologs in zip1 mutants lie sufficiently close to one another for homologous FISH signals to appear paired (Nag et al. 1995). In contrast, our TEM and light-microscopic analyses of germ-line nuclei from syp-1 mutants indicate that the C. elegans SC plays a key role in maintaining stable, parallel alignment between homologs throughout the majority of meiotic prophase.
This difference suggests that C. elegans and S. cerevisiae may rely predominantly on different mechanisms to stabilize initial homolog pairing events. In zip1 mutants, places of close contact between chromosome axes are observed at intervals between aligned but unsynapsed homologs, and these axial associations are dependent on recombination machinery components Dmc1 and Rad51 (Rockmill et al. 1995). Thus, in S. cerevisiae, it appears that initial pairing is stabilized by nascent recombination intermediates that can form even in the absence of Zip1 (and presumably the SC). However, synapsis may play a partially redundant role in stabilizing pairing in S. cerevisiae, because dmc1 and rad51 mutants, in which synapsis does occur with delayed kinetics but axial associations are presumably severely reduced or abolished, nevertheless exhibit substantial levels of paired homologous associations during the pachytene stage (Rockmill et al. 1995).
In contrast, prior work has demonstrated that stable homolog alignment and synapsis in C. elegans do not depend on initiating meiotic recombination events (Dernburg et al. 1998). Thus, the requirement for SYP-1 in stabilizing parallel alignment between homologs does not simply reflect a requirement for this protein in promoting recombination; instead, the SC structure itself appears to play a predominant role in stabilizing lengthwise alignment between homologs in C. elegans.
The role of cis-acting chromosomal domains in homolog pairing during C. elegans meiosis
Genetic analysis of C. elegans chromosome rearrangements has defined a cis-acting region (the pairing center) at one end of each C. elegans chromosome that promotes normal levels of crossover recombination along the length of the chromosome pair (Albertson et al. 1997). These pairing centers have been proposed to play roles either in homolog recognition or in promoting directional assembly of the SC between homologous chromosomes, initiating at the pairing-center end. When we examined pairing profiles for opposite ends of four different C. elegans chromosomes, we observed that the pairing center ends exhibited a robust maintenance of homologous associations even in the absence of syp-1 function and, by extension, normal SC. These data imply that pairing-center domains play an SC-independent role in maintaining homolog pairing locally. This role does not preclude the possibility that pairing centers might also function in a second capacity to promote the initiation or progression of synapsis.
The discovery that pairing-center regions of chromosomes can promote local, SC-independent pairing stabilization raises the possibility that homotypic interactions between pairing centers might serve to coordinate initial homolog pairing with synapsis progression. This idea may initially seem at odds with earlier assertions regarding pairing-center function (Villeneuve 1994). Specifically, Villeneuve argued against a model in which the information content for homolog recognition could be restricted to pairing-center ends of chromosomes, based on the observation that in worms heterozygous for a normal X chromosome and a pairing-center-deletion X chromosome, the X chromosome pair formed chiasmata and carried out successful disjunction most of the time. One implication of this result was that a homotypic association between the pairing-center regions of the two X chromosomes is not absolutely essential for a substantial level of chiasma formation between homologs. We can reconcile this earlier observation with our present findings by proposing the existence of two separable roles for pairing-center domains: one role in mediating local, SC-independent stabilization of pairing, which perhaps does rely on homotypic associations between pairing centers, and a second role in promoting SC assembly, a function that can be accomplished by a single pairing center (albeit likely with reduced efficiency) despite the fact that homotypic interactions in the pairing-center region are precluded.
In this way, C. elegans pairing-center associations may fulfill a role analogous to that proposed for axial associations in budding yeast, which appear to both stabilize pairing between homologs and serve as sites for synapsis initiation (Rockmill et al. 1995). However, whereas the formation of axial associations in S. cerevisiae is rooted in recombination between homologs, synapsis in C. elegans does not depend on meiotic recombination. Moreover, meiotic recombination initiation is dispensable for SYP-1-independent stabilization of pairing at the pairing-center region of the X chromosome (A.J. MacQueen, unpubl.). Perhaps the worm uses a distinct molecular mechanism, which may involve homotypic interactions at pairing centers but is independent of meiotic recombination, for mediating the coordination of chromosome pairing with synapsis progression.
Moreover, it is tempting to suggest that the synapsis-independent associations at pairing centers might additionally ensure that SC assembly occurs exclusively between homologous chromosomes. By fortifying early pairing events between single, defined regions on each homolog, pairing-center activity could coordinate an enforcement of chromosome partner choice with the progression of synapsis along the remainder of the chromosome pair. The S. cerevisiae Hop2 protein and the recently identified interacting protein Mnd1 have been proposed to play an analogous role in regulating chromosome partner choice during the meiotic pairing process in budding yeast (Leu et al. 1998; Tsubouchi and Roeder 2002). In hop2 mutants, most of the DSBs that initiate meiotic recombination are not repaired, and single-stranded DNA tails accumulate, similar to the consequences of lacking proteins involved in the strand-invasion step of meiotic recombination, such as Dmc1 and Rad51 (Bishop et al. 1992; Shinohara et al. 1992). In addition, as in rad51 and dmc1 mutants, synapsis occurs but axial associations are not observed in the absence of Hop2 function (Rockmill et al. 1995; Leu et al. 1998). However, unlike dmc1 and rad51 mutants, the SC frequently occurs between nonhomologous chromosome regions in hop2 mutants. These data raise the possibility that Hop2 might regulate chromosome partner choice by participating with Dmc1 and Rad51 in the formation of axial associations; perhaps Hop2 can promote the selective stabilization of axial associations between bona fide homologs. The notion that chromosome partner choice in yeast might be enforced in conjunction with axial association formation may provide an explanation for why Hop2, Mnd1, and Dmc1 proteins are conserved among diverse organisms including yeast, plants, and mammals but are apparently absent from the genomes of organisms such as Drosophila and C. elegans. Perhaps Hop2, Mnd1, and Dmc1 belong to a cassette of proteins that together coordinate the meiotic recombination process with the fortification of proper pairing events, and this cassette has been lost from organisms that do not rely on a recombination-based mechanism to achieve stable pairing and synapsis.
Synapsis and crossover recombination
The idea that the SC might play a crucial role in promoting crossover recombination is an attractive one and had been a prevailing view for decades. However, this view was called into question by the finding that S. cerevisiae is still able to form 30%–50% of its normal number of crossovers even in the absence of Zip1, the major central-region building block (Sym and Roeder 1994). Furthermore, based on the lack of interference between residual crossover events in zip1 mutants, Sym and Roeder (1994) had suggested that a primary role of Zip1, and by extension the SC central region, was in mediating crossover interference.
An alternative interpretation of the modest crossover deficit and lack of interference in S. cerevisiae zip1 mutants postulates that two different recombination pathways can give rise to crossovers during meiosis. One pathway uses conserved core meiotic recombination-machinery components Msh4 and Msh5, together with the SC central region structural components, to promote the regulated formation of crossovers within the context of constraints imposed by meiotic chromosome structure (discussed in Zalevsky et al. 1999; Villeneuve and Hillers 2001); one manifestation of constraints governing these Msh4/Msh5/SC-component-dependent “regulated crossovers” is crossover interference (Sym and Roeder 1994; Novak et al. 2001; N. Hunter and N. Kleckner, pers. comm.). Whereas an alternative recombination pathway can generate a reduced number of crossovers in an unregulated fashion (i.e., not governed by interference) in yeast cells lacking Msh4, Msh5, or Zip1, the fact that meiotic crossovers are absent in C. elegans him-14 (msh-4) and msh-5 mutants (Zalevsky et al. 1999; Kelly et al. 2000) indicates that this “unregulated” alternative crossover pathway does not operate in worms.
According to this view, SC central region components would, indeed, have a conserved role in crossover recombination, in promoting formation of the set of regulated meiotic crossovers that are essential for ensuring the proper segregation of all chromosome pairs at the meiosis I division. Our finding that crossovers are absent in C. elegans mutants that lack a putative SC central region structural component supplies an important missing piece to this argument, and bolsters the idea that during C. elegans meiosis, crossovers are generated exclusively by this regulated pathway. The requirement for SYP-1 for crossover formation, together with our evidence suggesting that recombination is initiated in syp-1 mutants, strongly supports the reemerging view that structural components of the SC central region play a conserved and essential role in crossover formation, in promoting the crossover outcome of initiated recombination events. Although it has been shown that S. cerevisiae Zip1 retains a capacity to promote crossover formation under conditions where the normal SC is not elaborated (Storlazzi et al. 1996), it remains an open question whether local recruitment of Zip1 at crossover sites is sufficient for its crossover-promoting role or whether some extent of Zip1 polymerization is required. Because synapsis may initiate at sites distant from eventual crossovers in C. elegans, we favor the idea that SC polymerization may be required for the crossover-promoting role of SYP-1.
Materials and methods
Genetics
The following mutations and chromosome rearrangements (background Bristol N2) were used: LGIV: spo-11(ok79), nT1[unc-?(n754) let-?(m435)](IV,V); LGV: syp-1(me17); LGX: dpy-3(e27), unc-3(e151) (Riddle et al. 1997; Dernburg et al. 1998; this work). The me17 allele was generated by EMS mutagenesis as in Villeneuve (1994). Wild-type controls were N2 unless otherwise noted. Additional wild-type isolates used included CB4856 (for recombination analysis using SNP markers; Wicks et al. 2001) and RW7000 (Williams et al. 1992).
me17 was mapped to the right of stP18 (V) as in Williams et al. (1992). FISH experiments showed that the 5S rDNA locus (rrs-1) is paired in heterozygotes carrying the nT1 reciprocal translocation (IV, V), indicating that rrs-1 is located in the portion of Chromosome V that is not balanced by nT1; me17 is stably balanced by nT1, placing it within the ∼1-Mb region between stP18 and rrs-1. Recombination analysis using visible markers was performed as in Kelly et al. (2000), and recombination frequency (p) was calculated as p = 1 − (1 − 2R)1/2, where R is the frequency of phenotypically recombinant progeny (Brenner 1974). Recombination analysis using SNP markers was performed as follows: Males heterozygous for syp-1(me17) and carrying an X chromosome derived from the CB4856 strain were crossed with syp-1/+ hermaphrodites. Outcross F1 progeny were plated individually, and 300 F2 hermaphrodites from non-Syp F1s and 447 hermaphrodites or males from syp-1/ syp-1 F1s were collected. Individual F2 worms were lysed in 10 μL of worm lysis buffer (Williams et al. 1992) and were typed for the presence of N2 and CB4856 alleles for SNP markers pkP610 and pkP6118 as in Wicks et al. (2001).
DAPI analysis and immunohistochemistry
DAPI staining, anti-HIM-3 immunostaining, and analysis of stained meiotic nuclei were carried out as in MacQueen and Villeneuve (2001). Anti-SYP-1 immunostaining was performed as for anti-HIM-3, except the primary antibody incubation step was performed overnight at room temperature. Double-labeling of SYP-1 and HIM-3 was performed as for anti-SYP-1 single-antibody experiments, with both rabbit anti-HIM-3 (1:200) and guinea pig anti-SYP-1 (1:50) primary antibodies present simultaneously during the first incubation step. Similarly, the secondary incubation contained a mixture of appropriate secondary antibodies (FITC anti-rabbit and Cy3 anti-guinea pig; Jackson Immunochemicals), each at 1:200.
FISH and time-course analysis of chromosome pairing
With the exception of the 5S rDNA probe, all probes were derived from yeast artificial chromosome (YAC) clones carrying C. elegans genomic DNA. The following YAC clones were used: Y68A3 (X, extreme right); Y51E2 (X, left); Y40H8 (IV, right); Y44F12 (IV, left); Y25B10 (II, left); Y26G1 (II, right), Y13H5 (I, ∼15% of total chromosome length from left end); Y48E9 (I, right). Labeled probes were generated as in Zalevsky et al. (1999) and Dernburg et al. (1998). Quantitation of pairing was carried out as in MacQueen and Villeneuve (2001), using germ lines from age-matched adult worms 14–16 h post-L4 larval stage. Distances between peak intensities of FISH signals were measured, and considered paired if ≤0.75 μm.
Fisher's Exact Test was used for statistical analysis of pairing data. First, each chromosome region examined was found to achieve a statistically significant peak in homolog pairing in syp-1 mutants: For each probe in all experiments, premeiotic pairing data (zone 1) were compared with pairing data for the zone corresponding to peak pairing levels (typically zone 3). In addition, peak pairing data for each probe were compared with pairing data for zone 6. In every case, pairing levels in the peak zone differed significantly from those within either zone 1 or zone 6 (two-sided p values were <0.0001 for all comparisons, with the exception of the comparison between peak and zone 6 for the non-pairing-center end of Chromosome II, for which p < 0.0012). Second, we showed that the modest level of peak pairing exhibited by the non-pairing-center chromosome ends in syp-1 mutants differs significantly from the incidence of nonhomologous associations. For experiments in which opposite ends of a single chromosome were examined, nonhomologous associations were assessed using distance measurements between nonhomologous signals within the same experimental data sets; two-sided p values were <0.0001 in every case. Finally, comparisons of pairing data for opposite ends of Chromosome I within zone 2 of wild-type germ lines indicated that the level of pairing at the pairing-center end of Chromosome I was significantly higher than pairing at the opposite end of the chromosome (two-sided p value <0.0001). Analyses were performed using InStat3 software (http://www.Graphpad.com).
Gene structure
First-strand cDNA corresponding to F26D2.2 was generated by reverse-transcription of adult poly(A) RNA, using reverse primer: 5′-TTCCCTCCTCTCTTTCGGCG-3′ (located in the final predicted exon of F26D2.2). This cDNA pool was used as template in PCR reactions to amplify distinct regions of the F26D2.2 predicted transcript. PCR products generated by 5′ RACE or using SL1 forward primers and reverse primer 5′-AACTTTTGCAGTCTCCGC-3′ were ∼900 bp shorter than expected based on the GeneFinder prediction for F26D2.2. Sequence analysis showed that the SL1 transsplice leader is spliced to an exon beginning at position 1996 of the F26D2 sequence (corresponding to exon 4 of the previous GeneFinder prediction). The initiation codon for the 489-amino-acid predicted protein is 10 nt downstream from the SL1 leader. Splice junctions for all other predicted exons were confirmed, in agreement with analysis of EST cDNA clones by the C. elegans Transcriptome project (http://www.wormbase.org). Furthermore, the absence of any ESTs corresponding to the upstream exons predicted by GeneFinder, despite an abundance of EST cDNA clones derived from this gene, supports our revised gene structure for F26D2.2.
Antibodies
A peptide corresponding to the final 20 amino acids of SYP-1 (generated by Biosynthesis Inc.) was used to generate rabbit anti-SYP-1 C-terminal polyclonal antibodies. A peptide corresponding to the first 20 amino acids was used to generate guinea pig anti-N-terminal antibodies. Animals were immunized and bled at Covance Research Products.
Electron microscopy
Adult hermaphrodites (17–20 h post-L4) were prepared for high-pressure freezing (HPF; McDonald 1999) as in Dernburg et al. (1998), except 50–100 worms were processed per 100-μm specimen carrier. Freeze substitution and poststaining were as in Dernburg et al. (1998). Longitudinal sections 100 nm thick were cut through three wild-type and three syp-1 mutant worms. One or two sections per gonad arm were scored for the presence of SC stretches within late-leptotene/zygotene through late-pachytene stages; when two sections were scored in the same worm they were derived from regions separated by more than two nuclear diameters and thus represented two different populations of nuclei (one population residing above and one below the central rachis of the germ line). Nuclei were scored from late in the transition-zone region (just prior to entry into pachytene) through the bend of the germ line, where nuclei are nearly exiting pachytene. Based on wild-type sections, >85% of nuclei scored were in early through late pachytene (leptotene/zygotene nuclei could be identified in wild-type germ lines based on their characteristic polarized chromosome organization). Three wild-type germ lines (195 nuclei total) and three syp-1 mutant germ lines (212 nuclei total) were analyzed.
Acknowledgments
A.J.M. and M.P.C. are grateful to R. Zalpuri and the EM lab at UC Berkeley for help and training in using the TEM. We thank the Caenorhabditis Genetics Center, the National Institute of Genetics (Japan), and the Sanger Center for strains and clones, and M. Zetka for the anti-HIM-3 antibody. We are grateful to K. Reddy for sharing her initial analysis of F26D2.2 RNAi, V. Reinke and S. Kim for early access to microarray data, and J. Engebrecht for critical reading of the manuscript. This work was supported by NIH grant R01GM53804 to A.M.V, by a Stanford Graduate Fellowship to A.J.M., and by Individual NRSA fellowship F32HD41329 to M.P.C.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL: villen@cmgm.stanford.edu, FAX: (650) 725-7739.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1011602.
References
- Albertson DG, Rose AM, Villeneuve AM. Chromosome organization, mitosis, and meiosis. In: Riddle DL, et al., editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 47–78. [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin GM, Villeneuve AM. C. elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G2 DNA damage checkpoint. Genes & Dev. 2001;15:522–534. doi: 10.1101/gad.864101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiácovo MP, Stanfield GM, Reddy KC, Reinke V, Kim SK, Villeneuve AM. A targeted RNAi screen for genes involved in chromosome morphogenesis and nuclear organization in the C. elegans germ line. Genetics. 2002;162:777–792. doi: 10.1093/genetics/162.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald J, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Dong H, Roeder GS. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J Cell Biol. 2000;148:417–426. doi: 10.1083/jcb.148.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- Goldstein P, Slaton DE. The synaptonemal complexes of Caenorhabditis elegans: Comparison of wild-type and mutant strains and pachytene karyotype analysis of wild-type. Chromosoma. 1982;84:585–597. doi: 10.1007/BF00292857. [DOI] [PubMed] [Google Scholar]
- Hawley RS. Chromosomal sites necessary for normal levels of meiotic recombination in Drosophila melanogaster. I. Evidence for and mapping of the sites. Genetics. 1980;94:625–646. doi: 10.1093/genetics/94.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly KO, Dernburg AF, Stanfield GM, Villeneuve AM. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics. 2000;156:617–630. doi: 10.1093/genetics/156.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu JY, Chua PR, Roeder GS. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- Lupas A. Predicting coiled-coil regions in proteins. Curr Opin Struct Biol. 1997;7:388–393. doi: 10.1016/s0959-440x(97)80056-5. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Villeneuve AM. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes & Dev. 2001;15:1674–1687. doi: 10.1101/gad.902601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire MP. The pattern of pairing that is effective for crossing over in complex B-A chromosome rearrangements in maize. II. Chromosoma. 1985;91:101–107. doi: 10.1007/BF00294052. [DOI] [PubMed] [Google Scholar]
- ————— The pattern of pairing that is effective for crossing over in complex B-A chromosome rearrangements in maize. III. Possible evidence for pairing centers. Chromosoma. 1986;94:71–85. doi: 10.1007/BF00293532. [DOI] [PubMed] [Google Scholar]
- McDonald K. High-pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol Biol. 1999;117:77–97. doi: 10.1385/1-59259-201-5:77. [DOI] [PubMed] [Google Scholar]
- McKim KS, Howell AM, Rose AM. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics. 1988;120:987–1001. doi: 10.1093/genetics/120.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Peters K, Rose AM. Two types of sites required for meiotic chromosome pairing in Caenorhabditis elegans. Genetics. 1993;134:749–768. doi: 10.1093/genetics/134.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen RL, Offenberg HH, Dietrich AJ, Riesewijk A, van Iersel M, Heyting C. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J. 1992;11:5091–5100. doi: 10.1002/j.1460-2075.1992.tb05616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses MJ. Structure and function of the synaptonemal complex. Genetics. 1969;61 Suppl.:41–51. [PubMed] [Google Scholar]
- Nag DK, Scherthan H, Rockmill B, Bhargava J, Roeder GS. Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics. 1995;141:75–86. doi: 10.1093/genetics/141.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak JE, Ross-Macdonald PB, Roeder GS. The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics. 2001;158:1013–1025. doi: 10.1093/genetics/158.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page SL, Hawley RS. c(3)G encodes a Drosophila synaptonemal complex protein. Genes & Dev. 2001;15:3130–3143. doi: 10.1101/gad.935001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, et al. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- Rockmill B, Sym M, Scherthan H, Roeder GS. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes & Dev. 1995;9:2684–2695. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- Roeder GS. Meiotic chromosomes: It takes two to tango. Genes & Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Rufas JS, Santos JL, Diez M, Suja JA. Meiotic chromosome structure: Relationship between the synaptonemal complex and the chromatid cores. Genome. 1992;35:1054–1061. [Google Scholar]
- Schedl T. Developmental genetics of the germ line. In: Riddle DL, et al., editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 241–269. [PubMed] [Google Scholar]
- Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Biol. 2001;2:621–627. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Storlazzi A, Xu L, Schwacha A, Kleckner N. Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc Natl Acad Sci. 1996;93:9043–9048. doi: 10.1073/pnas.93.17.9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M, Roeder GS. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- Sym M, Engebrecht JA, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS. The Mnd1 protein forms a complex with Hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol Cell Biol. 2002;22:3078–3088. doi: 10.1128/MCB.22.9.3078-3088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung KS, Roeder GS. Meiotic chromosome morphology and behavior in zip1 mutants of Saccharomyces cerevisiae. Genetics. 1998;149:817–832. doi: 10.1093/genetics/149.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve AM. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics. 1994;136:887–902. doi: 10.1093/genetics/136.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve AM, Hillers KJ. Whence meiosis? Cell. 2001;106:647–650. doi: 10.1016/s0092-8674(01)00500-1. [DOI] [PubMed] [Google Scholar]
- von Wettstein D, Rasmussen SW, Holm PB. The synaptonemal complex in genetic segregation. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Williams BD, Schrank B, Huynh C, Shownkeen R, Waterston RH. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf E, Kim PS, Berger B. MultiCoil: A program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J, MacQueen AJ, Duffy JB, Kemphues KJ, Villeneuve AM. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics. 1999;153:1271–1283. doi: 10.1093/genetics/153.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes & Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. The leptotene–zygotene transition of meiosis. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- ————— Meiotic chromosomes: Integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]