Abstract
Mutations in Caenorhabditis elegans gene mab-23 cause abnormal male tail morphology and abolish male fecundity but have no obvious effect in the hermaphrodite. Here we show that mab-23 encodes a DM (Doublesex/MAB-3) domain transcription factor necessary for specific aspects of differentiation in sex-specific tissues of the male. mab-23 is required for the patterning of posterior sensory neurons in the male nervous system, sex muscle differentiation, and morphogenesis of the posterior hypodermis, spicules, and proctodeum. Failure of mab-23 mutant males to sire progeny is due primarily to defective sex muscle-mediated turning during copulatory behavior and likely compounded by impairment of sperm passage through the proctodeum. In the male nervous system, mab-23 refines ray neuron subtype distribution by restricting expression of dopaminergic neurotransmitter identity through interactions with the Hox gene egl-5 and a TGF-β-related signaling pathway. mab-23 has distinct roles and functions independent of mab-3, indicating different aspects of C. elegans male sexual differentiation are coordinated among DM domain family members. Our results support the hypothesis that DM domain genes derive from an ancestral male sexual regulator and suggest how regulation of sexual development has evolved in distinct ways in different phyla.
Keywords: mab-23, DM domain, neuron, behavior, dopamine, C. elegans
Most multicellular organisms have evolved sexually dimorphic characteristics that serve in conspecific mate recognition, copulation, or fertilization. However, the mechanisms that direct the development of sex-specific features are not well understood. Unlike many other regulatory pathways that govern patterning of the body and tissue differentiation, the regulatory pathways that initiate sexual differentiation are not conserved among the species in which they have been studied in detail. This raises the question as to the nature of an ancestral sex determination system and whether any remnant of this is retained in sexual differentiation pathways downstream of the nonconserved regulators.
Two downstream genes, Drosophila doublesex (dsx) and Caenorhabiditis elegans mab-3, appear to function exclusively in differentiation of sexually dimorphic characteristics and share common features, suggesting that they have descended from an ancestral sex-determining gene (Raymond et al. 1998; Zarkower 2001). Both dsx and mab-3 encode transcription factors with DM (DSX/MAB-3) DNA-binding domains. Both repress yolk synthesis in males and both direct similar aspects of sensory organ differentiation such that dsx can functionally replace mab-3 in vivo (Baker and Ridge 1980; Belote et al. 1985; Shen and Hodgkin 1988; Raymond et al. 1998). Whereas mab-3 appears to function exclusively in male differentiation, dsx is required for both male and female development (Baker and Ridge 1980; Shen and Hodgkin 1988). In vertebrates, at least one DM domain gene family member, Dmrt1, is known to be required for testis differentiation (Raymond et al. 2000), whereas another, zebrafish terra, does not appear to function in sexual development and is involved in somitogenesis (Meng et al. 1999). Phylogenetic analyses, however, have been unable to clarify whether sexual differentiation function was the ancestral function of this gene family and has been lost by some family members during evolution, or whether it is a characteristic acquired independently in separate branches by convergent evolution (Ottolenghi et al. 2002).
In only a few cases is it understood how the activities of DM domain proteins and other sexual regulators promote the establishment of sex-specific characteristics at precise axial locations in the body (Conradt and Horvitz 1999; Yi and Zarkower 1999; Kopp et al. 2000; Estrada and Sánchez-Herrero 2001; Keisman and Baker 2001; Keisman et al. 2001; Sánchez et al. 2001). Understanding the mechanisms by which sexual regulators promote sexual dimorphism in the nervous system poses a particular challenge because of the complexity and cellular heterogeneity of this tissue. Whereas fru (fruitless), dsx, and dsf (dissatisfaction) are required in Drosophila for sex-specific aspects of nervous system development (see Goodwin 1999), little is known of their transcriptional targets in the nervous system or of their relationship to the lineal or spatial patterning mechanisms that shape this tissue. Almost nothing is known about the genetic origins of sexual dimorphism within vertebrate nervous systems. Questions of sex-specific nervous system development may be addressed in C. elegans, which possesses a simple nervous system with a high degree of sexual dimorphism. The male nervous system contains 381 neurons, 87 of which are male specific, whereas the hermaphrodite nervous system contains 302 neurons, eight of which are specific to this sex (Hodgkin 1988). These sex-specific differences have a correlate in behavioral complexity. Male copulatory behavior consists of a sequence of sub-behaviors, whereas hermaphrodites have a relatively passive role in mating (Loer and Kenyon 1993; Liu and Sternberg 1995). Most sex-specific neurons are generated by differential numbers of cell divisions or by differential cell fates, as are other sex-specific cell types in this species (Sulston and Horvitz 1977; Sulston et al. 1980).
To identify the genetic programs that drive sexual dimorphism in the C. elegans nervous system, we performed a forward genetic screen for mutants defective in male nervous system patterning. We identified a second member of the C. elegans DM domain gene family, mab-23, required for the differentiation of multiple sex-specific characteristics of the male. Our findings expand the known role of the DM domain transcription factor family in male sexual development in C. elegans and reinforce the notion of an ancestral function for this family in male development. Our results suggest both similarities and differences in the way nematodes and insects have solved the problem of establishing sexual dimorphism through regulation of this gene family.
Results
mab-23 is required for the development of male sexual structures
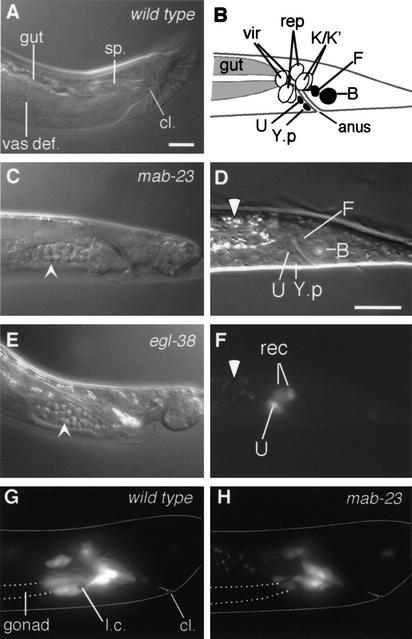
The mab-23 locus is defined by two recessive putative loss of function mutations, e2518 and bx118, which are similar with respect to the spectrum and penetrance of defects they cause. We identified mab-23(bx118) in a forward genetic screen for mutants affected in dopaminergic ray neuron patterning. mab-23(e2518) was identified in the wild isolate strain KR314 on the basis of abnormal male morphology (Hodgkin and Doniach 1997). mab-23(bx118) and (e2518) males are unable to sire progeny, ∼85% are Morpho-Mab (Morphologically Male abnormal) due to abnormal tail and gonad morphology (Fig. 2E, below), 30% have a mild crumpled spicule defect, and all display abnormal neurotransmitter patterning and axon guidance in a subset of male ray sensory neurons. In striking contrast to mab-23 males, mab-23 hermaphrodites have no obvious phenotypic defects. This observation, together with the existence of a wild population carrying a mutation in mab-23 that likely propagated only by hermaphrodite self-fertilization, suggests that mab-23 is dispensable in the hermaphrodite and that it might encode a male-specific developmental factor.
Figure 2.
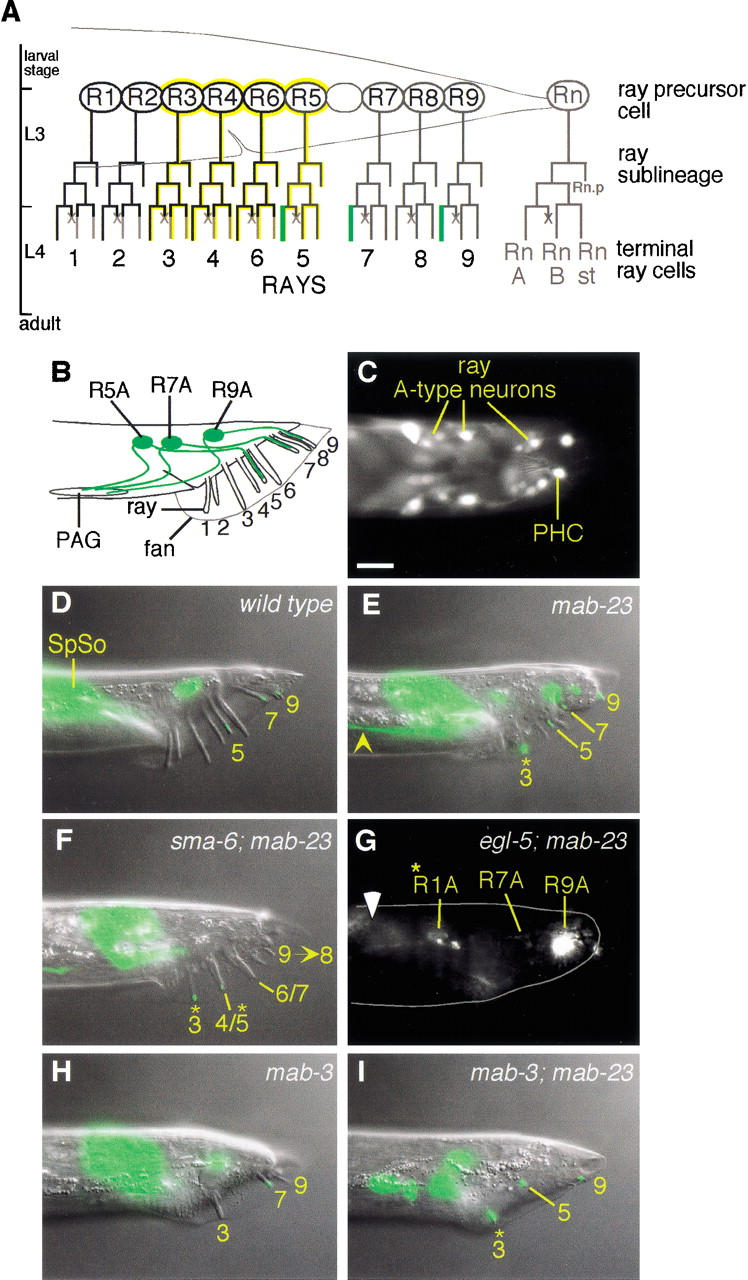
mab-23 is required for ray dopaminergic patterning and axon pathfinding. (A) Postembryonic cell lineages leading to the ray neurons (Sulston and Horvitz 1977). RnA, A-type neuron; RnB, B-type neuron; Rnst, structural cell; ×, programmed cell death. Rn.p is a hypodermal cell. Dopaminergic A-type neurons (green; Sulston and Horvitz 1977), EGL-5-expressing cells (yellow; Ferreira et al. 1999), MAB-23 ∷ GFP-expressing cells (black). (B) Adult male tail (lateral view; anterior, left; dorsal, up) showing the location of dopaminergic ray neurons (green). PAG, preanal ganglion. (C) mab-23 expression in wild-type adult male rays. Fluorescent micrograph of a transgenic adult male (dorsal view) expressing EM#308 reporter. MAB-23 ∷ GFP localizes predominantly to cell nuclei in the A-type neurons of rays 1–4 and 6 and the nonsex-specific PHCL/R neurons. (D–I) Dopaminergic fate expression in wild-type and mutant males (lateral view; anterior, left; dorsal, up). The dopaminergic fate marker is CAT-2 ∷ GFP (Lints and Emmons 1999). Nomarski and fluorescent micrographs of the same animal are superimposed. (D) Wild type. CAT-2 ∷ GFP is visible in ray 5, 7, and 9 A-type neurons. CAT-2 ∷ GFP signal more anteriorly in the tail corresponds to male spicule socket cell (SpSo) expression. (E) mab-23(bx118). The ray 3 A-type neuron expresses CAT-2 ∷ GFP ectopically (*) and its axon projects abnormally (yellow arrowhead). The Morpho-Mab tail defect characteristic of mab-23 males is also apparent in this animal. (F) sma-6(wk7); mab-23(bx118). Ray abnormalities typical of DBL-1 pathway single mutants are apparent: fusion of rays 4 and 5 (4/5) and rays 6 and 7 (6/7), and ray 9 has been transformed to ray 8 (9→8). CAT-2 ∷ GFP is ectopically expressed in rays 3 and 4/5. (G) egl-5(u202); mab-23(bx118). Fluorescent micrograph [the presence of egl-5(u202) disrupts male tail retraction and, therefore, rays and fan are embedded in the tail]; body outline (gray solid line); gut autofluorescence (white arrowhead). CAT-2 ∷ GFP is absent form the A-type neurons of egl-5-dependent rays 3 and 5. CAT-2 ∷ GFP is present in the A-type neurons of non-egl-5-dependent rays 7 and 9 (R7A and R9A, respectively) and ectopically in ray 1 (R1A*). (H) mab-3 (e1240). Only rays 3, 7, and 9 are generated in this animal. CAT-2 ∷ GFP expression is observed in ray 7, one of the rays that express this fate in wild type. No ectopic expression of CAT-2 ∷ GFP is observed in the ray 3 that is present. (I) mab-3 (e1240); mab-23 (bx118). Double mutant shows both mab-3 and mab-23 phenotypes: missing rays and ectopic expression of CAT-2 ∷ GFP in ray 3, respectively. Magnification, 1000×. Bar, 10 μm.
mab-23 encodes a DM domain transcription factor
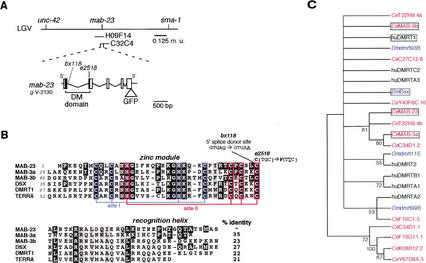
We cloned mab-23 by genetic mapping and complementation rescue of the Morpho-Mab and ray neuron dopaminergic patterning defect (Fig. 1A). The minimal mab-23 rescuing region contains a small ORF (identified by the Genie gene prediction program and designated g-V-2130) predicted to encode a 175 amino acid protein with a DNA-binding domain of the DM (DSX/MAB-3) transcription factor class (Fig. 1B; Erdman and Burtis 1993; Raymond et al. 1998). The corresponding region of the Caenorhabditis briggsae genome encodes a potential homolog that shares 78% amino acid identity with C. elegans MAB-23. In addition, 5′ noncoding regions and introns of the gene contain multiple blocks of sequence identity (50–200 bp in length) that may correspond to conserved regulatory elements (data not shown). Both mab-23(bx118) and mab-23(e2518) mutations alter nucleotides within the predicted DM domain region, and so are likely to disrupt DNA-binding function (Fig. 1A,B). e2518 converts the last conserved cysteine of the DM motif to phenylalanine. bx118 alters the fifth position of a conserved 5′ donor splice site in intron 1 and appears to affect splicing (see Materials and Methods). Mutations affecting the fifth position of a 5′ donor splice site are also associated with loss-of-function in two other C. elegans genes, egl-46 and unc-52, indicating that the identity of this position can be functionally important (Rogalski et al. 1995; Wu et al. 2001). mab-23(RNAi) phenocopies the mab-23 mutants inducing the full spectrum of defects observed in mab-23 males and no apparent defects in hermaphrodites. mab-23(RNAi) did not cause embryonic or early larval lethality (see Materials and Methods). Taken together, these data suggest that mab-23(bx118) and mab-23(e2518) are simple loss-of-function mutations and that reduction of mab-23 function may affect only males. However, as bx118 and e2518 may not represent null mutations and because RNAi may not be fully penetrant, we cannot rule out the possiblity that mab-23 could have functions in early development or in the hermaphrodite.
Figure 1.
mab-23 encodes a DM domain protein. (A) Genetic map of a portion of linkage group V (top) and the corresponding physical map showing the minimal 9.5-kb mab-23 rescuing region (middle), common to cosmids C32C4 and H09F14, and the predicted mab-23 ORF, g-V-2130 (bottom). The positions of mab-23 mutations and site of the GFP ORF insertion in mab-23 reporter EM#308 is indicated. (B) Sequence alignment of DM domains. Cysteines and histidines that form the Zn2+-binding sites are indicated (boxed red or blue; for review, see Zhu et al. 2000). Percentage identity with MAB-23 is shown; identical amino acids are indicated in black; similar amino acids in grey. MAB-23 (GenBank accession no. AF535153); MAB-3a, and MAB-3b, the two DM domains of C. elegans MAB-3 (accession no. O18214); Drosophila DSX (accession no. P23022/P23023); human DMRT1 (accession no. AF130728); zebrafish TERRA (accession no. AF080622). e2518 converts the last conserved cysteine of the DM motif to phenylalanine. bx118 alters the fifth position of the 5′ donor splice site within intron 1. (C) Unrooted phylogenetic tree of DM domains encoded by predicted genes in human (hu, black), Drosophila melanogaster (Dm, blue), and C. elegans (Ce, red) genomes. The DM domains (zinc module plus recognition helix) were aligned and subjected to phylogenetic analysis by maximum parsimony using the Protpars algorithm of PHYLIP v. 3.573c (Felsenstein 1989). The tree presented is a consensus of the most parsimonious trees derived from 100 bootstrap resamplings of the original data. Bootstrap support values over 50% are shown; nodes with values of <50% were collapsed. DM domains of known sexual regulator genes are boxed. Accession numbers are as follows: human DMRTA1 (AJ290954), DMRTA3 (AAF78891), DMRTA2 (AJ301580), DMRTC2 (AJ291669), DMRTB1 (AJ291671), DMRT2 (AF130729); Drosophila dmrt93B (AAF55843), dmrt11E (AAF48261), dmrt99B (AAF56919); C. elegans C27C12.6 (CAA93739), C34D1.1 (CAB01491), C34D1.2 (CAB01490), F10C1.5 (AAA93409), F13G11.1 (CAB05898/CAB05899), K08B12.2 (AAB52258), T22H9.4a and T22H9.4b, the two DM domains of T22H9.4 (AAC69225), Y43F8C.10 (CAA21612), and Y67D8A.3 (AAK68545).
Phylogenetic analysis of the DM domains in genes predicted in C. elegans, Drosophila, and human genomes places MAB-23 in a well-supported nematode-specific clade with one of the two DM domains of MAB-3 and two other nematode genes (Fig. 1C). These four DM domains, therefore, may have arisen by duplication events in the nematode lineage. Two other well-supported clades, one containing vertebrate and Drosophila members, and the other vertebrate, Drosophila and C. elegans members, support the conclusion that the common ancestor of all three groups contained multiple genes of this family. However, this analysis could not resolve the relationship of the four known sexual regulators (mab-3, mab-23, dsx, and DMRT1) to each other or to ancestral family members.
mab-23 is expressed in both sexes
To understand the basis of the pleiotropic mab-23 mutant phenotype and its male specificity, we analyzed the mab-23 expression pattern by means of a functional mab-23 ∷ gfp reporter (Fig. 1A). In males, MAB-23 ∷ GFP is observed in several sex-specific cell types during larval development and in the adult, including the A-type ray sensory neurons, ventral male-specific muscles, and unidentified neurons of the male posterior ventral nerve cord (Figs. 2C and 5A,B, below). MAB-23 ∷ GFP is also detected in a limited number of non-sex-specific tissues in the adult male, including 6–8 unidentified neurons of the head, ventral body wall muscle (Fig. 5A, below), and the PHCL/R neurons (Fig. 2C). It is transiently expressed during larval development in the hindgut (Fig. 6F, below) and in the tail spike (data not shown), two tissues that undergo significant remodeling in the male. Many of these MAB-23 ∷ GFP-positive tissues have identifiable defects in mab-23 males (ray neurons, tail hypodermis, sex muscles, and hindgut).
Figure 5.
mab-23 is required for male-specific muscle cell differentiation. (A,B) MAB-23 ∷ GFP is expressed in ventral posterior muscles of the male. Fluorescent micrographs of MAB-23 ∷ GFP in non-sex-specific ventral body wall (b.w.) muscle, male-specific outer longitudinal (o.l.), and diagonal muscles (lateral view, A) and diagonal muscles (ventral view, B). (C,D) Affect of exogenous 20 mM 5HT on wild-type and mab-23 males (lateral view). (C) Wild type. Note tight ventral flexure of the tail (white arrowhead) and altered posture affecting more anterior regions of the body. (D) mab-23. Ventral flexure of the tail is weak and more anterior regions of the body also differ in posture from wild type. (E,F) SER-2 ∷ GFP expression in the diagonal muscles. Fluorescent micrographs showing strong expression of SER-2 ∷ GFP in the diagonal muscles (arrowheads) of a wild-type male (E; ventral view) and weak expression in a mab-23 male (F; ventral view). v.n.c., ventral nerve cord. (G) Frequency of 5HT-induced male tail curling. The percentage of males that displayed a tight ventral flexure of the tail (as shown in C) after exposure to 20 mM 5HT is shown. Number of males scored were as follows: wild-type, 70; mab-23(bx118), 43; mab-23(e2518), 50; and mab-3(e1240), 40. (H) Mean number of diagonal muscles per male side that were SER-2 ∷ GFP-positive and have wild-type morphology (revealed by UNC-27 ∷ GFP labeling) for the genotype indicated is shown, as well as the range observed in that population. In both mab-23 and mab-3 males, SER-2 ∷ GFP signal is also weak (as shown in F) compared with wild-type. Number of male sides scored (SER-2 ∷ GFP, morphology) were as follows: wild-type (49, 30); mab-23(bx118) (48, 39); mab-23(e2518) (48, 43); and mab-3(e1240) (30, 49). Magnification: A, 400×; B,E,F, 1000×. Bar, 10 μM.
Figure 6.
mab-23 is required for development of the male proctodeum. (A,C,E) Proctodeum of wild-type, mab-23 and egl-38 adult males. Nomarski micrographs (lateral view; anterior, left; dorsal, up). (A) In wild-type males the vas deferens (vas def.) is devoid of sperm. cl., cloaca (the anus in larval animals); sp., spicules. In mab-23 (C) and egl-38 (E) males, sperm (arrowhead) accumulates in the vas def. (B) Schematic of L1 male hindgut (lateral view). Black, blast cells; vir, rectal valve cells; rep, rectal epithelial cells (Sulston et al. 1980). (D) Nomarski micrograph of L1 male hindgut (lateral view; anterior, left; dorsal, up). (F) Fluorescent micrograph of D showing MAB-23 ∷ GFP in U and two rectal epithelial cells (rec), either K, K.a, K‘, or repD; gut autofluorescence (white arrowhead). (G,H) Fluorescent micrograph showing plin-48 ∷ gfp-expressing hindgut cells in late L4 males (lateral view). U descendents and K‘/K.a are located over the posterior end of the gonad (outlined with broken line) in the ventral half of the animal. In wild-type (G), these cells extend anteriorly over the gonad; in mab-23 males (H), these cells show only limited extension. Body outline (solid line), engulfed linker cell corpse inside U.l/rp (l.c). Magnification, 1000×. Bar, 10 μM.
The male specificity of mab-23 function does not appear to be due to sex-specific expression. In adult hermaphrodites, the same set of non-sex-specific tissues are MAB-23 ∷ GFP positive as in the male (data not shown). In addition, MAB-23 ∷ GFP is expressed in several hermaphrodite-specific tissues that contribute to the egg-laying apparatus, namely the ventral uterus and spermatheca of the oviduct and the Hermaphrodite Specific Neurons (HSNs; data not shown). However, mab-23 hermaphrodites have no obvious defects in the hindgut or the tail and do not differ significantly from wild-type animals in either egg-laying rate or brood sizes generated (data not shown; Hodgkin and Doniach 1997). Thus, mab-23 may be either dispensable or have subtle or redundant functions in these tissues in the hermaphrodite.
Regulation of axial patterning in the nervous system by mab-23
To understand how mab-23 affects differentiation of the male nervous system, we examined its role in ray neuron development. The male tail bears nine bilateral pairs of sensory rays aligned along its anterior-posterior axis (Fig. 2A,B). Each ray contains two neurons, an A-type neuron and a B-type neuron, that are thought to register chemo- and mechanosensory cues used to guide mating (Sulston and Horvitz 1977; Liu and Sternberg 1995; Barr and Sternberg 1999). The cells of a single ray derive from a common precursor cell, Rn (in which n stands for rays 1–9). MAB-23 ∷ GFP is expressed from the Rn cell stage onward in the lineages that give rise to rays 1–4 and 6, and in the A-type neurons of these rays in the adult (Fig. 2A,C).
mab-23 loss-of-function affects at least two aspects of ray neuron differentiation, neurotransmitter expression and axon guidance. In wild-type males, expression of dopamine biosynthesis gene cat-2 (tyrosine hydroxylase) and dopamine are restricted to the A-type neurons of rays 5, 7, and 9 (Figs. 2A,B,D, 3A) (Sulston and Horvitz 1977; Lints and Emmons 1999). In mab-23 males, the A-type neurons of three additional rays, rays 1, 3, and 6, ectopically express cat-2 reporters and synthesize dopamine (Figs. 2E, 3B). In addition, the axons of all ectopically expressing neurons and 80% of ray 1 B-type neurons (see Materials and Methods) project anteriorly, instead of ventrally, and fail to synapse with cognate targets in the preanal ganglion (Fig. 2E).
Figure 3.
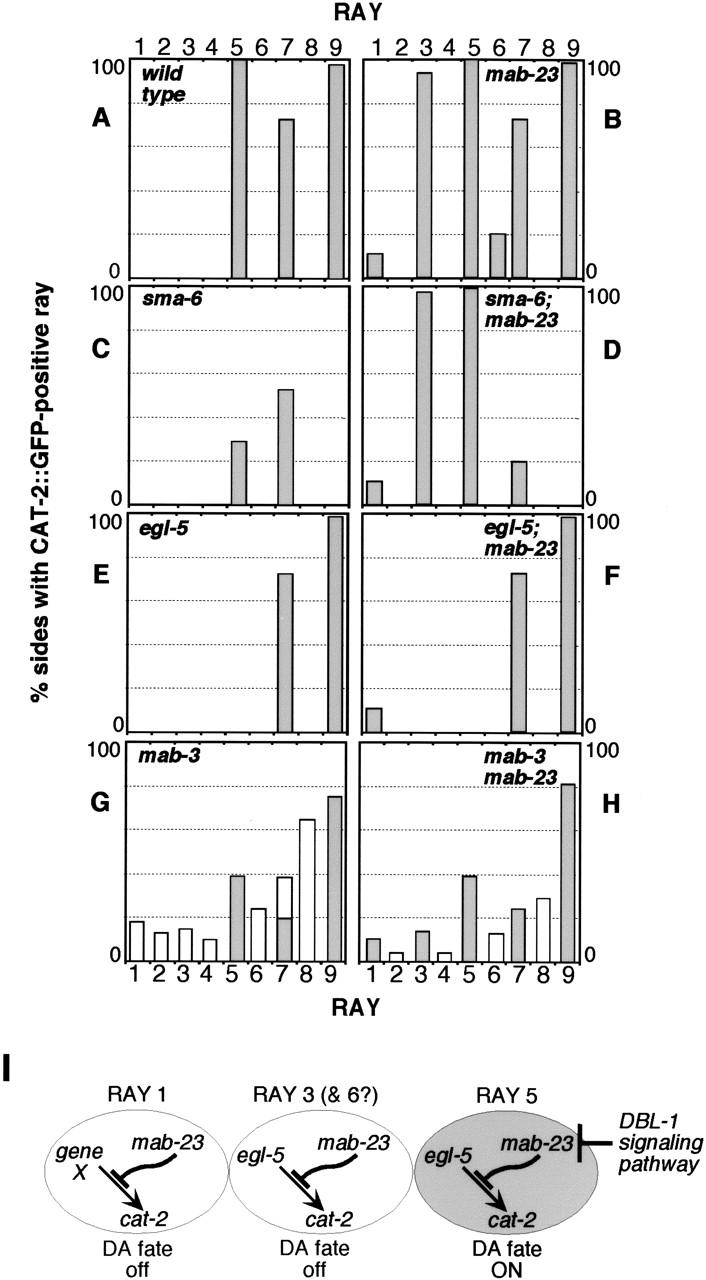
Genetic interactions between mab-23, egl-5, and the DBL-1 pathway. (A–H) Frequency and distribution of CAT-2 ∷ GFP-positive rays in wild-type and mutant males. Single, double, and triple mutant strains were generated by crossing either wild-type, mab-23(bx118), or mab-23(e2518);nIs118 animals with DBL-1 pathway, egl-5, or mab-3 mutants. The DBL-1 pathway is defined by the six genes, dbl-1, daf-4, sma-6, sma-2, sma-3, and sma-4 (Patterson and Padgett 2000); for mutations used in this study, see Materials and Methods. sma-6(wk7), egl-5(u202), and mab-3(e1240) are presumptive null mutations (Wang et al. 1993; Raymond et al. 1998; Krishna et al. 1999). CAT-2 ∷ GFP-expressing rays were identified as described in Lints and Emmons (1999). mab-23(bx118) and mab-23(e2518) mutant combinations gave similar results, as did mutations in all genes of the DBL-1 pathway. Tail sides develop independently and are, therefore, scored separately. The number of sides scored in a given mutant background ranged from 60 to 144. For each genetic background, the pattern of CAT-2 ∷ GFP observed was found to be consistent with the dopamine staining pattern obtained using FIF (Sawin et al. 2000; data not shown). In DBL-1 pathway mutant backgrounds, fusions involving rays 5, 7, or 9 have been grouped with unfused ray 5, 7, or 9 identities, respectively, as fusions contain a single CAT-2 ∷ GFP-positive neuron likely to correspond to the neuron that normally expresses CAT-2 ∷ GFP in wild type, namely the A-type neuron of ray 5, 7, or 9 (Lints and Emmons 1999). In mab-3 single and mab-3; mab-23 double mutants, rays 1–6 are variably missing so that the frequency with which a particular ray identity was generated is indicated (unshaded), in addition to the frequency with which it was CAT-2 ∷ GFP-positive (shaded). (I) Genetic relationship between mab-23, the DBL-1 pathway, and egl-5 in dopaminergic (DA) patterning (see text).
Previously, we have shown that expression of dopaminergic fate in rays 5, 7, and 9 requires the activity of a TGF-β-like signaling pathway, the DBL-1 pathway (Fig. 3C; Lints and Emmons 1999). The pathway ligand, DBL-1, is expressed in several tail cells, but the source relevant to ray development is not known (Suzuki et al. 1999). Downstream components of the pathway act cell autonomously in the ray cells (Savage et al. 1996). Regulated expression of DBL-1 is required to ensure that expression of dopaminergic fate is restricted to rays 5, 7, and 9, because ubiquitous expression of dbl-1 (driven by a heat-shock promoter) can induce ectopic expression of dopaminergic fate in rays 3, 4, 6, and 8 (Lints and Emmons 1999). The mab-23 mutant phenotype indicates that mab-23 normally functions to repress inappropriate expression of dopaminergic fate in rays 1, 3, and 6. However, mab-23 does not act by restricting DBL-1 availability or by repressing pathway activation in rays 1, 3, and 6. In mab-23;DBL-1 pathway double-mutants, rays 1, 3, and 6 ectopically express dopaminergic fate as in mab-23 single mutants (Figs. 2F, 3D). In addition, ray 5 expresses dopaminergic fate at a frequency of 100% in double mutants, instead of 25% as in DBL-1 pathway single mutants (Fig. 3C). mab-23 therefore appears to repress an activity that confers dopaminergic fate potential in rays 1, 3, 5, and 6 independently of DBL-1 pathway function.
Expression of MAB-23 ∷ GFP in rays 1, 3, and 6 suggests that mab-23 acts cell autonomously in these rays. The absence of MAB-23 ∷ GFP from ray 5 raises the possibility that DBL-1 signaling might induce dopaminergic fate in this ray by repressing mab-23 expression. However, in DBL-1 pathway mutants, we observed no significant change in the MAB-23 ∷ GFP expression pattern; in particular, there was no obvious expression in ray 5 (data not shown). It is possible that the mab-23 reporter gene lacks promoter elements necessary for expression in ray 5. Alternatively, mab-23 expression might be reduced in ray 5 by a DBL-1-independent mechanism. This could account for the continued expression of dopamine at decreased frequency in ray 5 in DBL-1 pathway mutants. Finally, mab-23 could have a cell nonautonomous effect on ray 5.
C. elegans AbdominalB-like Hox gene egl-5 is expressed in the lineages of rays 3–6 (Ferreira et al. 1999) and is required for competence to adopt dopaminergic fate in ray 5, either in the absence or presence of DBL-1 signaling (Fig. 3E), and for heat-shock: ∷ dbl-1-induced ectopic dopaminergic fate in rays 3, 4, and 6 (Lints and Emmons 1999). We found that egl-5 is also necessary for ectopic dopaminergic fate expression in ray 3 in mab-23 mutants (Figs. 2G, 3F; ray 6 could not be tested because it is not generated in an egl-5(u202) background).
Taken together, the results suggest a model for restricted expression of dopaminergic fate in wild type (Fig. 3I). In rays 3, 5, and possibly 6, egl-5 activity confers a state of competence to adopt dopaminergic fate that is repressed by mab-23. In ray 1, mab-23 is presumed to similarly affect the activity of a gene analogous to egl-5. As a consequence of differential exposure to the ligand, DBL-1 pathway activation occurs preferentially in ray 5, abrogating mab-23 repression in this ray and thereby allowing dopaminergic fate to be expressed.
mab-23 males are defective in specific steps of male mating behavior
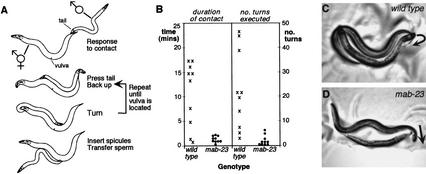
Defective development of a subset of ray neurons is unlikely to underlie the complete inability of mab-23 males to sire progeny, as there appears to be some functional redundancy among rays (Liu and Sternberg 1995). To determine whether mab-23 males are affected in the execution of steps in male mating behavior that do not involve ray function, we examined their performance in mating assays. We found that mab-23 males were unable to complete the stereotyped sequence of copulatory behaviors, regardless of whether they had distorted tail morphology (data not shown) or near wild-type tail morphology (Fig. 4A,B; Loer and Kenyon 1993; Liu and Sternberg 1995). Their primary defect was an inability to adopt the appropriate posture for performing a systematic vulva search or for turning (Fig. 4C,D). As a consequence, males frequently lost contact with the hermaphrodite, failed to locate the vulva, and did not copulate. The inability of mab-23 males to turn and their anomalous body posture may be due to abnormalities in sex-specific muscles (see below) and possibly to additional defects in the male-specific nervous system.
Figure 4.
mab-23 males are defective in specific steps of male mating behavior. (A) Wild-type C. elegans male mating behavior. The male responds to contact with the hermaphrodite by halting forward locomotion, placing the ventral side of his tail against the hermaphrodite's body and swimming backward in search of the vulva. If the male reaches the end of her body without finding the vulva he executes a turn to continue the search on the other side. After locating the vulva, he inserts anchoring spicules and transfers sperm. (B) Performance of wild-type (✖) and mab-23 (•) males in mating assays. Ten adult virgin males of each genotype were individually observed and scored for the amount of time spent in direct contact with the hermaphrodite (left) and for the number of successful turns executed (right) during the first 30 min of a mating assay (see Materials and Methods). (C,D) Examples of posture adopted by wild-type (C) and mab-23 (D) males during execution of the turn. Trajectory of the turn is indicated by the direction of the arrow. The posture adopted by wild-type males ensures that the ventral side of the male tail is pressed against the hermaphrodite throughout the turn. mab-23 males do not adopt this posture and execute loose or missed turns (shown here), during which contact with the hermaphrodite is lost.
mab-23 is required for full differentiation of male-specific sex muscles
The posture required for performing the vulval search and for turning during mating is controlled in part by male-specific muscles located in the ventral posterior (Loer and Kenyon 1993). MAB-23 ∷ GFP expression is observed in three muscles of this class, the diagonal muscles and the posterior inner and outer longitudinal muscles (Fig. 5A,B) as well as in their precursors during development (data not shown). Exposure of wild-type males to exogenous serotonin (5HT) can induce a posture analogous to that observed during turning, namely, a tight ventral flexure of the tail (Fig. 5C,G; Loer and Kenyon 1993). This is likely due to 5HT stimulation of the diagonal muscles, which normally receive inputs from the serotonergic CP motor neurons of the ventral cord (White 1988), and possibly of other male-specific muscles. Exposure of mab-23 males to exogenous 5HT, in contrast, cannot induce this posture and males produce, at best, a weak ventral tail flexure, suggesting a defect in diagonal muscle function (Fig. 5D,G). In mab-23 males, a reporter for muscle troponin I gene unc-27 (T. Allen, pers. comm.; L. Jia and S.W. Emmons, unpubl.) was strongly expressed in diagonal muscles, as in wild type (data not shown). However, in mab-23 males, diagonal muscle cells appeared slightly narrower in width than in wild type, and in 30% of sides scored, two to three cells had abnormal morphology (disorganized filaments or ectopic projections) compared with 6% in wild type. In addition, we found that expression of the putative 5HT/octopamine receptor SER-2 (T. Niacaris and L. Avery, pers. comm.) was significantly reduced in mab-23 males compared with wild type (Fig. 5, cf. E,F and H). Taken together, these data suggest that mab-23 is required for specific aspects of diagonal muscle differentiation and possibly differentiation of other muscles, and that loss of its function in these tissues contributes to the abnormal mating behavior of mab-23 males.
mab-23 is necessary for development of the male proctodeum
In addition to deficits in mating behavior, 85% of mab-23 males have an abnormal gonad morphology that suggests that they may be impaired in their ability to transfer sperm during copulation. In mab-23 males, the lumen of the vas deferens is distended and often contains sperm, whereas in wild-type males, the vas deferens lumen is generally devoid of sperm except during ejaculation (Fig. 6A,C). The mab-23 phenotype is reminiscent of certain mutants defective in male hindgut/proctodeum development. The proctodeum is a male-specific structure that consists of the junction between the alimentary canal and the genital tract together with the copulatory spicules and the spicule channels (Fig. 6A; Sulston et al. 1980). This structure is generated from the progeny of hindgut blast cells F, U, B, Y, and nonblast cells K.a, K‘, and rep(V/D) (Fig. 6B). MAB-23 ∷ GFP is apparent in U and K cells in both sexes at L1 (Fig. 6D,F) with occasional expression in other hindgut cells. During male development, expression is strong throughout the U lineage and transient and weak in F and Y.p lineages (H.M. Chamberlin, pers. comm.). After formation of the adult proctodeum, MAB-23 ∷ GFP is no longer apparent.
The mab-23 gonad phenotype resembles that of two mutants affected in K, K‘, and U fate, egl-38 and lin-48 (Fig. 6, cf. C and E; Chamberlin et al. 1997, 1999). K.a, K‘, and descendents of U play a role in establishing a connection between the gonad and the developing proctodeum. During L4, the vas deferens of the male gonad grows anteriorly along the ventral side of the body cavity. In late L4, U.lp or U.rp (or their fusion products) engulf the posterior cell of the gonad, the linker cell, whereas K.a and K‘ grow anteriorly along the outside of the advancing gonad and establish contact with it (Sulston et al. 1980). To determine whether cells K.a, K‘, or descendents of U were affected in mab-23 males, we visualized these cells with GFP generated under control of the lin-48 promoter (Johnson et al. 2001) and compared their behavior in wild-type and mab-23 animals. We observed that in all wild-type males examined (n = 12), cells likely to correspond to K.a or K‘, and U descendents extended anteriorly over the advancing gonad during L4. In contrast, in all mab-23 males examined (n = 15), these cells showed no, or limited, extension and remained in close proximity to the rectum (Fig. 6G,H). No significant differences were observed between mab-23 and wild-type hermaphrodites in reporter expression pattern or morphology of GFP-positive hindgut cells (n = 24 per genotype). This indicated that mab-23 function is required in hindgut cells of the male, in which it may confer cell adhesion or migratory properties that promote the formation of a functional gonad-proctodeum junction.
Two DM domain genes are required for male sexual differentiation in C. elegans
The DM domain gene mab-3 is necessary for male ray and diagonal muscle development, spicule and tail-tip morphogenesis, repression of yolk protein synthesis in the gut, and mating behavior (Shen and Hodgkin 1988). mab-23 affects some, but not all of these processes, and, with the exception of tail morphogenesis, its loss-of-function has less severe consequences. This raises the possibility that, in tissues that require both gene functions, mab-23 might be subordinate to mab-3 and regulated by it. Our assessment of mab-3 and mab-23 function in the rays and the diagonal muscles, however, suggests that this is not the case. In the ray lineages, mab-3 is expressed in all Rn cells and their descendents (Yi et al. 2000) and is required for generation of rays 1–6; in mab-3 null mutants, rays 1–6 are variably missing due to defects in ray lineage execution from the Rn stage onward (Shen and Hodgkin 1988). In a mab-3(e1240) null mutant background, as in wild type, mab-23 reporter expression initiates in the Rn cell stage and in developing diagonal muscles, indicating that mab-23 does not require mab-3 for its expression. In addition, in mab-3(e1240) males, we did not observe ectopic expression of dopaminergic fate in rays 1, 3, or 6 when these were generated (Fig. 2, cf. H and I; Fig. 3, cf. G and H). The absence of ectopic dopaminergic fate in mab-3 males argues that mab-23 still functions in a mab-3 null mutant background. Taken together, these data suggest that mab-3 and mab-23 are independently regulated and that they have distinct roles in sex-specific tissue differentiation.
A striking feature of mab-3;mab-23 double mutants is that considerable male development still occurs in these animals, including development of a thin body, fan, rays, spicules, hook, sex muscles, and male gonad and germ line (data not shown). This observation indicates that although mab-3 and mab-23 are required for the specification of many male-specific characteristics, additional genes are involved in directing male differentiation.
Discussion
The results presented here extend our understanding of the role of the DM domain gene family in bringing about sexual dimorphism in C. elegans. Existing mab-23 mutants and mab-23(RNAi) suggest that like mab-3, mab-23 is required for male, but not hermaphrodite development. mab-23 loss-of-function affects the differentiation of diverse sex-specific tissues necessary for male copulatory behavior, but has no apparent effect in the hermaphrodite. Substantive support for this conclusion comes from the isolation of a wild nematode population homozygous for a mab-23 loss-of-function mutation that likely propagated by hermaphrodite self-fertilization (Hodgkin and Doniach 1997). A male-specific function for mab-23 is consistent with that of DM family members from other species and with the proposed ancestral function of the DM transcription factor class in the implementation of sexual development (Zarkower 2001).
Regulation of axial patterning by DM domain proteins
In the developing nervous system of the C. elegans male, mab-23 and mab-3 promote the expression of sex-specific characteristics at precise axial locations by modulating the activities of general patterning factors. By this means, mab-3 and mab-23 integrate sex-specific information with appropriate spatial and temporal coordinates. Studies of Drosophila DM domain gene dsx reveal that a similar strategy has been used to establish sex-specific characteristics of non-neuronal tissues. Drosophila genital discs are specified in segments A8 and A9 through the combined activities of DSX and the posterior Hox genes abdominal-A (abd-A) and Abdominal-B (Abd-B). The disc then generates either male or female genitalia as a consequence of differential regulation of wg and dpp by sex-specific isoforms of DSX (Estrada and Sánchez-Herrero 2001; Keisman and Baker 2001; Keisman et al. 2001; Sánchez et al. 2001). Elsewhere in the animal, dual regulation of the gene bric-a-brac (bab) by Abd-B and sex-specific isoforms of DSX induces sex-specific abdominal pigmentation in segments A5 and A6 (Kopp et al. 2000). In the posterior nervous system of the C. elegans male, the combined activities of mab-3 and the proneural gene lin-32 confer neuronal competence in the male-specific Rn cells, leading to the generation of ray neurons (Yi et al. 2000). mab-23 ensures that dopaminergic identity is appropriately restricted among these neurons by suppressing expression of an egl-5(Abd-B-like)-dependent dopaminergic potential and rendering induction of dopaminergic fate dependent on the DBL-1 (DPP-like)-signaling pathway. These data show how DM domain proteins participate in sex-specific patterning of the nervous system by modulating the activities of non-sex-specific developmental genes and suggest the possibility that, despite phylogenetic differences in upstream sex determination mechanisms, DM domain genes could be similarly used to elaborate aspects of sexual dimorphism in the nervous systems of other animals, perhaps including vertebrates.
The role of DM domain transcription factors in C. elegans sexual development
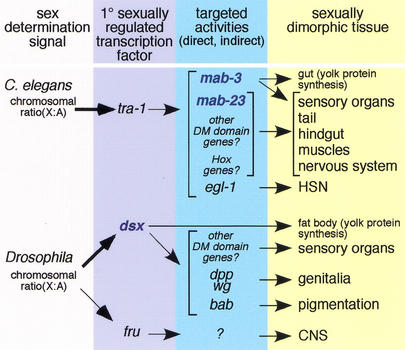
Although mab-23 and mab-3 loss-of-function affect essentially the same sets of male tissues, their phenotypes are additive in animals carrying mutations in both genes, indicating that they have distinct and independent functions. Interestingly, a considerable amount of characteristic male development occurs in the mab-23;mab-3 double mutant, suggesting that additional genes of the male differentiation pathway remain to be described. This remaining male differentiation may be accounted for by some of the nine additional DM domain gene family members predicted in the C. elegans genome, or by sex-specific regulation of other developmental activities by the sex determination pathway (Fig. 7). In either case, it is likely that sexual differentiation involves the activities of multiple genes, with each gene defining limited aspects of sexual development.
Figure 7.
Hierarchy of sexual differentiation genes in C. elegans and Drosophila. In both C. elegans and Drosophila, the primary sex determining signal, the X chromosome to autosome ratio (X : A), sets the activity of the globally acting sex determination pathway. DM domain genes (blue) in C. elegans and Drosophila differ with respect to their relationship to this pathway. In C. elegans, the primary sexually regulated transcription factor is a Gli-related Zn finger protein, TRA-1A, encoded by the terminal gene of the sex determination pathway, tra-1. tra-1 activity affects the development of most sexually dimorphic tissues, acting through non-sex-specific developmental gene targets (e.g., cell death gene egl-1), as well as dedicated sexual regulators (mab-3). mab-3 (in tissues other than the gut) and mab-23 are not directly targeted by TRA-1A. The sex specificity of their activity derives from the cellular context in which they are expressed. In Drosophila, the primary sexually regulated transcription factor is the DM domain gene dsx. In contrast to C. elegans DM domain genes, dsx is a direct target of the sex-determination pathway in most somatic tissues. Like tra-1, dsx is broadly acting and functions early in sexual differentiation to sex-specifically regulate multiple developmental genes. In the CNS, fru appears to be the primary sexually regulated transcription factor.
The role of DM domain genes and the dispersed regulation of sexual differentiation in C. elegans are in striking contrast to sexual differentiation pathways of Drosophila. In Drosophila, a single DM domain gene, dsx, appears to control sex-specific development of most tissues in the soma, with the exception of the CNS (Baker and Ridge 1980). In addition, dsx directs female as well as male development; the dsx gene generates either male- or female-promoting protein isoforms, depending on whether the sex determination pathway is inactive or active, respectively (Burtis and Baker 1989). In a dsx null mutant, tissues develop with a variable mixture of male and female features. Thus, dsx acts as a switch gene that establishes sexual identity by directing the activities of developmental factors into one of the two alternative developmental pathways. It is unlikely that C. elegans encodes such a broadly acting DM domain factor, because extensive genetic analysis has failed to identify such a gene. Functional characterization of other DM domain genes predicted in the Drosophila genome should reveal whether dsx is the only DM domain sexual differentiation gene in this organism, or whether multiple DM domain family members contribute as in C. elegans.
Regulation of sexual development by the sex-determination pathway
Drosophila and C. elegans also differ in the way the activities of DM domain genes are regulated by globally acting sex-determination pathways. In Drosophila females, the splicing machinery of the sex-determination pathway directly targets the dsx transcript generating a female-specific splice and protein isoform (Burtis and Baker 1989). In C. elegans hermaphrodites, the product of tra-1, TRA-1A, promotes female sexual fate by repressing transcription of male developmental genes (Goodwin and Ellis 2002). mab-23 and mab-3, however, are generally not targeted, and both genes, although apparently necessary only for male development, are also expressed in the hermaphrodite. mab-3 is expressed in the lateral seam of both sexes, but generates rays from the posterior cells of this tissue only in males. This is due to overlapping expression of coactivator lin-32 in male, but not hermaphrodite, posterior seam (Yi et al. 2000), presumably as a consequence of TRA-1A activity. mab-23 is expressed in the hindgut of both sexes, but appears to function only in males, in which it contributes to formation of the proctodeum. It is possible that mab-23 may be inactive in the hermaphrodite hindgut, because TRA-1A represses expression of a necessary coactivator, analogous to lin-32, or because TRA-1A blocks mab-23 target promoters in this tissue. Direct regulation of C. elegans DM domain gene expression does occur in one tissue, the hermaphrodite gut, in which TRA-1A directly represses mab-3 transcription (Yi and Zarkower 1999). Preliminary experiments suggest that this mechanism of regulation is unlikely to play a major role in restricting mab-23 activity in hermaphrodites, as mutagenesis of the only candidate TRA-1A DNA-binding site identified in the mab-23 promoter (−2048 to −2061 bp from the predicted translational initiation site) that is conserved in C. briggsae, does not alter the mab-23 ∷ gfp expression pattern in this sex (R. Lints and S.W. Emmons, unpubl.). Other tissues that require mab-3 and mab-23 function, such as the male sex muscles, are not present in the hermaphrodite. Together, these observations suggest that in C. elegans, DM domain gene activity may be controlled primarily through sex-specific regulation of other developmental genes by the sex determination pathway.
Evolution of DM domain family function in sexual differentiation
In spite of the foregoing differences in the way sex determination pathways regulate DM domain genes in Drosophila and C. elegans, and the differing levels in the hierarchy at which DM domain genes act, there remains one significant similarity – in both organisms, the default sexual pathway is male, that is, in the absence of upstream sex-determination pathway activity, the male-specific functions of DM domain genes are expressed. This similarity might derive from the ancestral function of these genes as dominant male-determining factors in a primordial sex-determination system (Hodgkin 1992; Zarkower 2001). According to this hypothesis for the origin of sex determination pathways, subsequent evolution has resulted in the activity of this male factor being brought under genetic control. Subsequent to this, a female function, such as that of the DSX female isoform, might evolve as a reinforcement mechanism.
The present-day differences in the respective roles of the DM domain proteins in male sexual differentiation in nematodes and flies may be a consequence of the different ways that genetic control of sexual development evolved in these two organisms. In Drosophila, the sex-determination signal regulates a splicing activity (Burtis and Baker 1989), whereas in C. elegans, it regulates the activity of a Gli-related Zn finger transcription factor (Zarkower and Hodgkin 1992). In both systems, for increased efficiency, the activity of the first transcription factor in the pathway may have evolved to target ever-higher levels of the developmental hierarchy. Because this transcription factor was TRA-1A in C. elegans rather than a transcription factor of the DM domain family, a greater number of general regulatory genes may be targeted directly by the sex-determination pathway in C. elegans than in Drosophila, and in C. elegans, the DM domain proteins act later in developmental programs (Fig. 7).
Materials and methods
Strains and cultures
Nematodes were grown and maintained as described in Brenner (1974). Bristol(N2)-derived strains him-8(e1489) and him-5(e1490), which generate a high incidence of males, were used as reference wild-type strains. Unless otherwise stated, strains were maintained at 20–25°C. The following alleles were used. LG II: mab-3 (mu15) [courtesy of C. Kenyon (University of California, San Francisco)], (e1240), and (e2093), sma-6(wk7); LG III: egl-5(u202), lin-48(sy234), pha-1(e2123ts) maintained at 15°C, sma-2(e502)sma-3(e491), and sma-2(e502)sma-3(e491)sma-4(e729) [courtesy of C. Savage-Dunn (Queens College, CUNY, Flushing, NY)], sma-2(e502), sma-3(e491), sma-4(e729); LGIV: egl-38(sy294), him-8(e1489); LGV: him-5(e1490)V, mab-23(e2518), and (bx118), unc-42(e270)sma-1(e30). Except for mab-23(bx118), all mutations have been described previously (Brenner 1974; Hodgkin et al. 1979; Shen and Hodgkin 1988; Wang et al. 1993; Granato et al. 1994; Hodgkin and Doniach 1997; Raymond et al. 1998; Chamberlin et al. 1999; Krishna et al. 1999).
Transgenic reporter gene arrays used are as follows: nIs118X, an integrated array of Ex[cat-2 ∷ gfp (EM#282) + lin-15(+)] [courtesy of H. Schwartz and H.R. Horvitz (Massachusetts Institute of Technology, Cambridge, MA); Lints and Emmons 1999]; otIs107, an integrated derivative of Ex[ser-2 ∷ gfp + lin-15(+)] [courtesy of T. Niacaris and L. Avery (University of Texas Southwestern Medical Center, Dallas, TX); Altun-Gultekin et al. 2001]; saIs14, an integrated derivative of Ex[plin-48 ∷ gfp + unc-119(+)] [Johnson et al. 2001; courtesy of H.M. Chamberlin (Ohio State University, Columbus, OH)]; bxIs14V, an integrated derivative of Ex[pkd-2 ∷ gfp + pha-1(+)] (Barr and Sternberg 1999; L. Jia and S.W. Emmons, unpubl.); bxEx97 = Ex[unc-27 ∷ gfp + pha-1(+)] (L. Jia and S.W. Emmons, unpubl.); bxEx83 and bxEx84, independently generated versions of complex extrachromosomal array Ex[mab-23 ∷ gfp (EM#308) + N2 + pha-1(+)].
Isolation of mab-23(bx118)
F2 or F3 male progeny of EMS-treated EM641 [him-5(e1490)V; nIs118X)] hermaphrodites were screened for altered expression of dopaminergic fate marker CAT-2 ∷ GFP and mutants were recovered from sibling hermaphrodites. bx118 was mapped to LGV near to unc-42 (0 map units, m.u.) using standard procedures. bx118 males displayed a Morpho-Mab phenotype similar to that of mab-23(e2518)V (2.44 m.u.) (Hodgkin and Doniach 1997) and failed to complement this mutation, indicating that bx118 is an allele of mab-23. Three factor mapping data (this study; Hodgkin and Doniach 1997) placed mab-23 between 2.56 and 2.83 m.u. (http://www.wormbase.org).
Cloning of mab-23
Cosmids from the mab-23 genetic region [provided by A. Coulson (Sanger Center, Cambridge, UK)] and pha-1(+) transformation plasmid pBX-1 (Granato et al. 1994), were coinjected into mab-23(e2518); pha-1(e2123ts); him-8(e1489) hermaphrodites at concentrations of 2–10 ng/μL and 100 ng/μL, respectively (Mello et al. 1991). A 9.5-kb region within C32C4 (nucleotides 5071–14476) strongly rescued both the Morpho-Mab and abnormal dopaminergic patterning phenotype (assayed by Formaldehyde Induced Fluorescence, FIF; Sawin et al. 2000). According to gene prediction program Genie (Kulp et al. 1996; Kent and Zahler 2000), but not Gene Finder (C. elegans Genome Sequencing Project), this region contains a single ORF, g-V-2130. One species of mab-23 (g-V-2130) cDNA, identical to the g-V-2130 prediction, was recovered from him-5 mixed-stage total RNA with the RLM kit (Ambion) and Thermoscript Reverse Transcriptase (Invitrogen). Mutations in bx118 and e2518 were identified by sequencing all mab-23 exons and introns in three independently generated PCR products derived from the relevant worm strains. The only consistent sequence change identified in mab-23(bx118)-derived PCR products was G→A at the fifth position of intron 1. To determine whether intron 1 splicing was affected in mab-23(bx118) animals, RT–PCR experiments were performed using a 5′ and 3′ primer pair based on exon 1 and 3 sequences, respectively. RNA samples from mab-23(bx118) animals generated a 1064-bp product, likely to correspond to an incompletely processed product containing intron 1, and a 235-bp product corresponding to the wild-type, fully spliced mab-23 product. Wild-type control RNA generated only the 235-bp product (data not shown).
RNAi experiments
RNA was synthesized using the MEGAscript T3 and T7 kit (Ambion) from a mab-23 cDNA PCR product generated with primer pairs based on positions 9585–11934 of C32C4, which contained T3 and T7 promoter sequences. Approximately 200 ng/μL dsRNA was introduced into EM641 by microinjection (Fire et al. 1998) or by soaking L2–L3 stage larvae for 20 h at 25°C (Petcherski and Kimble 2000). In the F1 progeny of injected hermaphrodites, no significant difference was observed between mock (M9) and mab-23 dsRNA treatments in the number of eggs layed, animals hatched, or ratio of male:hermaphrodite progeny (data not shown). F1 mab-23(RNAi) males generated after injection or males subjected to dsRNA soaking showed similar results. Ectopic expression of CAT-2 ∷ GFP: 12/92 and 67/92 male sides showed ectopic expression in rays 1 and 3, respectively (c.f. M9 control males in which 0/92 sides scored for either ray). Abnormal gonad morphology: 33/55 (c.f. M9 control, 0/89). Abnormal tail morphology: 35/55 males (c.f. M9 control, 0/89). Defective 5HT-induced tail curling: 10/37 males (c.f. M9 control, 24/33).
mab-23 ∷ gfp reporter construction and transgenic lines
mab-23 ∷ gfp reporter (EM#308) is based on the minimal 9.5-kb rescuing region in C32C4 and consists of mab-23-coding sequence flanked by 5.3 kb of 5′ promoter and 2.8 kb of 3′ noncoding sequence with GFP inserted in frame immediately 5′ of the mab-23 termination codon. EM#308 was generated by inserting a 7-kb fragment (position 4938–11939 in C32C4) and a 2.8-kb fragment (position 11939–14494 in C32C4) into the BssH–KpnI and NheI–StuI sites of pPD117.75 [a gift from A. Fire (Carnegie Institution of Washington, Baltimore, MD)], respectively. bxEx83 and bxEx84 were generated by coinjection of linearized EM#308 (1 ng/μL), N2 (50 ng/μL) and pBX-1[pha-1(+)] (1 ng/μL) DNA into a pha-1(e2123ts); him-5(e1490) strain. Low levels of reporter expression, generated by noncomplex EM#308-containing arrays, rescue the mab-23 phenotypes described. However, high levels of reporter expression, generated by complex arrays bxEx83 or bxEx84, resulted in partial rescue of gonad and ray phenotypes. As the presence of bxEx83 or bxEx84 in a wild-type background also caused a low frequency of mab-23 gonad phenotype (20% of males) and ray 3 loss (15% of male tail sides, compared with 5% in nontransgenic males), incomplete rescue by these arrays may be due to sensitivity to elevated levels of mab-23 expression in these tissues.
Cell identification
GFP-positive cells were identified according to their position and morphology (Sulston and Horvitz 1977; Sulston et al. 1980). The A-type neuron identity of ray cells expressing MAB-23 ∷ GFP in wild-type animals and ectopic CAT-2 ∷ GFP in mab-23 mutants was deduced by staining relevant reporter lines with 5HT antisera (purchased from H.W.M. Steinbusch, Maastricht University), which labels ray neurons R1B, R3B, and R9B (R. Lints, L. Jia, and S.W. Emmons, unpubl.). GFP and antisera label did not colocalize, indicating that the GFP-positive cells were A-type neurons. B-type neurons were visualized with PKD-2 ∷ GFP (Barr and Sternberg 1999).
Behavioral assays
Mating behavior assays (Liu and Sternberg 1995) are as follows: ∼20 L4 mab-23 or wild-type males were allowed to mature overnight at 20°C on a plate seeded with bacteria. Individual adult males were placed on the 3-mm bacterial lawn of a mating plate containing three L4 paralyzed (unc-51) hermaphrodites. Animals were observed at 50× magnification on a Wild Makroskop M420, and the following variables were recorded during the first 30 min of the assay: time of initial contact, time of contact response, number of turns executed, and time spent in direct contact with the hermaphrodite. Data shown (Fig. 4B) corresponds to mating assays performed on four separate occasions for each genotype. mab-23 males used had near to wild-type tail morphology.
The 5HT-induced tail curling assays (Loer and Kenyon 1993) were performed as follows: 5 to 10 adult males were placed in 75 μL of 20 mM 5HT (creatinine sulfate complex) in a sterile 96-well culture plate, and the percentage of animals with tightly curled tails was recorded after 10 min. Data shown (Fig. 5G) were collected from experiments performed on three separate occasions for each genotype.
Acknowledgments
We thank Helen Chamberlin for her insights into mab-23 expression in the hindgut, Ricardo Azevedo for the phylogenetic analysis, H. Schwartz and H.R. Horvitz, C. Savage-Dunn, T. Niacaris, L. Avery, O. Hobert, C. Kenyon, D. Zarkower, and A. Coulson for kindly providing strains and reagents. We also thank H. Zhang, D. Zarkower, T. Lints, D. Portman, and R. Azevedo for helpful discussions. Some nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. R.L. is a NARSAD Young Investigator. This work was supported by NIH grant R01 NS30986. S.W.E. is the Siegfried Ullmann Professor of Molecular Genetics.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
EMAIL emmons@aecom.yu.edu; FAX (718) 430-8778.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1012602.
References
- Altun-Gultekin Z, Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, Hobert O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, control cell fate specification of a defined interneuron class in C. elegans. Development. 2001;128:1951–1969. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- Baker BS, Ridge K. Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics. 1980;94:383–423. doi: 10.1093/genetics/94.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401:386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- Belote JM, Handler AM, Wolfner MF, Livak KJ, Baker BS. Sex-specific regulation of yolk protein gene expression in Drosophila. Cell. 1985;40:339–348. doi: 10.1016/0092-8674(85)90148-5. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis KC, Baker BS. Drosophila doublesexgene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- Chamberlin HM, Palmer RE, Newman AP, Sternberg PW, Baillie DL, Thomas JH. The PAX gene egl-38 mediates developmental patterning in Caenorhabditis elegans. Development. 1997;124:3919–3928. doi: 10.1242/dev.124.20.3919. [DOI] [PubMed] [Google Scholar]
- Chamberlin HM, Brown KB, Sternberg PW, Thomas JH. Characterization of seven genes affecting Caenorhabditis eleganshindgut development. Genetics. 1999;153:731–742. doi: 10.1093/genetics/153.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1cell death activator gene. Cell. 1999;98:317–327. doi: 10.1016/s0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- Erdman SE, Burtis KC. The Drosophiladoublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 1993;12:527–535. doi: 10.1002/j.1460-2075.1993.tb05684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B, Sánchez-Herrero E. The Hox gene Abdominal-B antagonizes appendage development in the genital disc of Drosophila. Development. 2001;128:331–339. doi: 10.1242/dev.128.3.331. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Ferreira HB, Zhang Y, Zhao C, Emmons SW. Patterning of Caenorhabditis elegans posterior structures by the Abdominal-B homolog, egl-5. Dev Biol. 1999;207:215–228. doi: 10.1006/dbio.1998.9124. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Goodwin EB, Ellis RE. Turning clustering loops: Sex determination in Caenorhabditis elegans. Curr Biol. 2002;12:R111–R120. doi: 10.1016/s0960-9822(02)00675-9. [DOI] [PubMed] [Google Scholar]
- Goodwin SF. Molecular neurogenetics of sexual differentiation and behaviour. Curr Opin Neurobiol. 1999;9:759–765. doi: 10.1016/s0959-4388(99)00030-6. [DOI] [PubMed] [Google Scholar]
- Granato M, Schnabel H, Schnabel R. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 1994;22:1762–1763. doi: 10.1093/nar/22.9.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. Sexual dimorphism and sex determination. In: In: Wood WB, editor. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 243–279. [Google Scholar]
- ————— Genetic sex determination mechanisms and evolution. BioEssays. 1992;14:253–261. doi: 10.1002/bies.950140409. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Doniach T. Natural variation and copulatory plug formation in Caernorhabiditis elegans. Genetics. 1997;146:149. doi: 10.1093/genetics/146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Fitzsimmons D, Hagman J, Chamberlin HM. EGL-38 Pax regulates the ovo-related gene lin-48 during Caenorhabditis elegansorgan development. Development. 2001;128:2857–2865. doi: 10.1242/dev.128.15.2857. [DOI] [PubMed] [Google Scholar]
- Keisman EL, Baker BS. The Drosophila sex determination hierarchy modulates wingless and decapentaplegic signaling to deploy dachshundsex–specifically in the genital imaginal disc. Development. 2001;128:1643–1656. doi: 10.1242/dev.128.9.1643. [DOI] [PubMed] [Google Scholar]
- Keisman EL, Christiansen AE, Baker BS. The sex determination gene doublesex regulates the A/P organizer to direct sex-specific patterns of growth in the Drosophilagenital imaginal disc. Dev Cell. 2001;1:215–225. doi: 10.1016/s1534-5807(01)00027-2. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Zahler AM. The Intronerator: Exploring introns and alternative splicing in C. elegans. Nucleic Acids Res. 2000;28:91–93. doi: 10.1093/nar/28.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A, Duncan I, Carroll SB. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature. 2000;408:553–559. doi: 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- Krishna S, Maduzia LL, Padgett RW. Specificity of TGFβ signaling is conferred by distinct type I receptors and their associated SMAD proteins in Caenorhabditis elegans. Development. 1999;126:251–260. doi: 10.1242/dev.126.2.251. [DOI] [PubMed] [Google Scholar]
- Kulp DD, Haussler D, Reese MG, Eeckman FH. A generalized Hidden Markov Model for the recognition of human genes in DNA. ISMB-96. St. Louis, MO: AAAI/MIT Press; 1996. [PubMed] [Google Scholar]
- Lints R, Emmons SW. Patterning of dopaminergic neurotransmitter identity among Caenorhabditis elegans ray sensory neurons by a TGFβ family signaling pathway and a Hox gene. Development. 1999;126:5819–5831. doi: 10.1242/dev.126.24.5819. [DOI] [PubMed] [Google Scholar]
- Liu KS, Sternberg PW. Sensory regulation of male mating behavior in Caenorhabditis elegans. Neuron. 1995;14:79–89. doi: 10.1016/0896-6273(95)90242-2. [DOI] [PubMed] [Google Scholar]
- Loer CM, Kenyon CJ. Serotonin-deficient mutants and male mating behavior in the nematode Caenorhabditis elegans. J Neurosci. 1993;13:5407–5417. doi: 10.1523/JNEUROSCI.13-12-05407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng A, Moore B, Tang H, Yuan B, Lin S. A Drosophila doublesex-related gene, terra, is involved in somitogenesis in vertebrates. Development. 1999;126:1259–1268. doi: 10.1242/dev.126.6.1259. [DOI] [PubMed] [Google Scholar]
- Ottolenghi C, Fellous M, Barbieri M, McElreavey K. Novel paralogy relations among human chromosomes support a link between the phylogeny of doublesex-related genes and the evolution of sex determination. Genomics. 2002;79:333–343. doi: 10.1006/geno.2002.6711. [DOI] [PubMed] [Google Scholar]
- Patterson GI, Padgett RW. TGFβ-related pathways. Roles in Caenorhabditis elegans development. Trends Genet. 2000;16:27–33. doi: 10.1016/s0168-9525(99)01916-2. [DOI] [PubMed] [Google Scholar]
- Petcherski AG, Kimble J. LAG-3 is a putative transcriptional activator in the C. elegansNotch pathway. Nature. 2000;405:364–368. doi: 10.1038/35012645. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–694. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Murphy MW, O’Sullivan MG, Bardwell VJ, Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes & Dev. 2000;14:2587–2595. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski TM, Gilchrist EJ, Mullen GP, Moerman DG. Mutations in the unc-52 gene responsible for body wall muscle defects in adult Caenorhabditis elegansare located in alternatively spliced exons. Genetics. 1995;139:159–169. doi: 10.1093/genetics/139.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez L, Gorfinkiel N, Guerrero I. Sex determination genes control the development of the Drosophilagenital disc, modulating the response to Hedgehog, Wingless and Decapentaplegic signals. Development. 2001;128:1033–1043. doi: 10.1242/dev.128.7.1033. [DOI] [PubMed] [Google Scholar]
- Savage C, Das P, Finelli AL, Townsend SR, Sun C-Y, Baird SE, Padgett RW. Caenorhabditis elegans genes sma-2, sma-3, and sma-4define a conserved family of transforming growth factor β pathway components. Proc Natl Acad Sci. 1996;93:790–794. doi: 10.1073/pnas.93.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin ER, Ranganathan R, Horvitz HR. C. eleganslocomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Shen MM, Hodgkin J. mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell. 1988;54:1019–1031. doi: 10.1016/0092-8674(88)90117-1. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Albertson DG, Thomson JN. The Caenorhabditis elegans male: Postembryonic development of nongonadal structures. Dev Biol. 1980;78:542–576. doi: 10.1016/0012-1606(80)90352-8. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Yandell MD, Roy PJ, Krishna S, Savage-Dunn C, Ross RM, Padgett RW, Wood WB. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development. 1999;126:241–250. doi: 10.1242/dev.126.2.241. [DOI] [PubMed] [Google Scholar]
- Wang BB, Müller-Immergluck MM, Austin J, Robinson NT, Chisholm A, Kenyon C. A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell. 1993;74:29–42. doi: 10.1016/0092-8674(93)90292-x. [DOI] [PubMed] [Google Scholar]
- White J. The anatomy. In: Wood WB, editor. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. pp. 81–122. [Google Scholar]
- Wu J, Duggan A, Chalfie M. Inhibition of touch cell fate by egl-44 and egl-46 in C. elegans. Genes & Dev. 2001;15:789–802. doi: 10.1101/gad.857401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Zarkower D. Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogasterDSX suggests conservation of sex determining mechanisms. Development. 1999;126:873–881. doi: 10.1242/dev.126.5.873. [DOI] [PubMed] [Google Scholar]
- Yi W, Ross JM, Zarkower D. mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegansmale sexual development and behavior. Development. 2000;127:4469–4480. doi: 10.1242/dev.127.20.4469. [DOI] [PubMed] [Google Scholar]
- Zarkower D. Establishing sexual dimorphism: Conservation amidst diversity? Nat Rev Genet. 2001;2:175–185. doi: 10.1038/35056032. [DOI] [PubMed] [Google Scholar]
- Zarkower D, Hodgkin J. Molecular analysis of the C. elegans sex-determining gene tra-1: A gene encoding two zinc finger proteins. Cell. 1992;70:237–249. doi: 10.1016/0092-8674(92)90099-x. [DOI] [PubMed] [Google Scholar]
- Zhu L, Wilken J, Phillips NB, Narendra U, Chan G, Stratton SM, Kent SB, Weiss MA. Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers. Genes & Dev. 2000;14:1750–1764. [PMC free article] [PubMed] [Google Scholar]