Abstract
Cell division in bacteria is mediated by the tubulin-like protein FtsZ, which assembles into a structure known as the Z ring at the future site of cytokinesis. We report the discovery of a Z-ring-associated protein in Bacillus subtilis called ZapA. ZapA was found to colocalize with the Z ring in vivo and was capable of binding to FtsZ and stimulating the formation of higher-order assemblies of the cytokinetic protein in vitro. The absence of ZapA alone did not impair cell viability, but the absence of ZapA in combination with the absence of a second, dispensable division protein EzrA caused a severe block in cytokinesis. The absence of ZapA also caused lethality in cells producing lower than normal levels of FtsZ or lacking the division-site-selection protein DivIVA. Conversely, overproduction of ZapA reversed the toxicity of excess levels of the division inhibitor MinD. In toto, the evidence indicates that ZapA is part of the cytokinetic machinery of the cell and acts by promoting Z-ring formation. Finally, ZapA is widely conserved among bacteria with apparent orthologs in many species, including Escherichia coli, in which the orthologous protein exhibited a strikingly similar pattern of subcellular localization to that of ZapA. Members of the ZapA family of proteins are likely to be a common feature of the cytokinetic machinery in bacteria.
Keywords: FtsZ, cytokinesis, Bacillus subtilis, protein localization
Binary fission in rod-shaped bacteria entails the formation of a transverse septum that divides a progenitor cell into two equal-sized daughter cells. Despite its apparent simplicity, cell division in bacteria is subject to tight spatiotemporal control. Septum formation is faithfully coordinated with chromosome replication and segregation, as illustrated by the very low frequency of anucleated cells normally found in bacterial cultures (Hiraga et al. 1989; Ireton et al. 1994). Spatial control, on the other hand, is evident in the stereotypical placement of the newly formed septum at precisely the middle of the progenitor cell (Trueba 1982), which ensures that the resulting daughter cells are equivalent both genetically and morphologically.
Centrally important to cytokinesis in bacteria is the GTPase FtsZ, which represents an ancestral form of the eukaryotic cytoskeletal protein tubulin (Erickson 1995; van den Ent et al. 2001). FtsZ is capable of GTP-induced self-assembly into higher-order polymers structurally analogous to tubulin polymers under a variety of in vitro conditions (Mukherjee and Lutkenhaus 1994; Erickson et al. 1996; Lowe and Amos 1999; Lu et al. 2000). These polymers exhibit dynamic behavior, which depends on the ability of FtsZ to hydrolyze its bound GTP (Mukherjee and Lutkenhaus 1998; Stricker et al. 2002).
The earliest event in bacterial cytokinesis is the localization of FtsZ to the future division site, where it assembles into a ring-like structure called the Z ring (Bi and Lutkenhaus 1991). The Z ring then serves as a scaffold for the recruitment of several other proteins into a complex that constitutes the cytokinetic machinery. In Bacillus subtilis, the Z ring directly or indirectly recruits at least nine proteins to the division site (Errington and Daniel 2002). The roles of these proteins in septum formation remains unknown with the exception of Pbp2B (FtsI in Escherichia coli) and FtsW, which participate in the synthesis of the peptidoglycan cross-wall of the septum.
The polymerization of FtsZ into the Z ring at the right place and time in the cell should reflect both intrinsic self-assembly properties of FtsZ as well as the effects of positively and negatively acting factors. There are several known inhibitors of Z-ring formation. The best understood is the MinCD complex (de Boer et al. 1989), which consists of MinC, an inhibitor of FtsZ polymerization (Hu et al. 1999), and MinD, an ATPase that recruits MinC to the membrane (Hu and Lutkenhaus 1999; Marston and Errington 1999; Raskin and de Boer 1999a). The Min complex is a site-specific inhibitor of Z-ring formation and its absence leads to a high frequency of septum formation near the cell poles. The topological specificity proteins MinE in E. coli (de Boer et al. 1989) and DivIVA in B. subtilis (Cha and Stewart 1997; Edwards and Errington 1997) bias the localization of MinCD to the ends of the cell, thereby preventing septum formation near the poles but not at the cell middle (Marston et al. 1998; Hu and Lutkenhaus 1999; Raskin and de Boer 1999a,b; Rowland et al. 2000).
A second, negatively acting factor is the bacterial chromosome or nucleoid, which inhibits Z-ring formation in the region of the cell that it occupies (Woldringh et al. 1990; Sun et al. 1998; Harry et al. 1999; Yu and Margolin 1999). The molecular basis for nucleoid occlusion is unclear, but it is generally accepted that the phenomenon plays a role in the medial placement of the septum and in coordinating cytokinesis with chromosome replication and segregation (Harry 2001; Margolin 2001). A third negatively acting factor in Z-ring formation is the B. subtilis membrane protein EzrA (Levin et al. 1999). The absence of EzrA leads to the formation of extra Z rings and may facilitate Z-ring formation when the cellular concentration of FtsZ is low (Levin et al. 1999). EzrA is homogeneously distributed in the cell membrane and localizes to the division site once the cytokinetic ring is assembled, presumably via an interaction with the FtsZ polymer (Levin et al. 1999). Thus, in contrast with the Min system, EzrA seems to lack site specificity and acts by destabilizing FtsZ polymers throughout the cell.
In comparison, less is known about proteins that promote Z-ring formation. Components of the cytokinetic machinery that interact directly with FtsZ, such as FtsA and the E. coli protein ZipA, are likely to stabilize Z rings. ZipA has been shown to promote Z-ring formation in vivo (RayChaudhuri 1999) and to stabilize FtsZ polymers in vitro (RayChaudhuri 1999; Hale et al. 2000). Nevertheless, Z rings still form in the absence of ZipA (Hale and de Boer 1999; Liu et al. 1999; Pichoff and Lutkenhaus 2002) or FtsA (Addinall et al. 1996; Pichoff and Lutkenhaus 2002), suggesting either that FtsZ is intrinsically capable of forming Z rings or that additional proteins exist that promote Z-ring formation. The purpose of the present work was to attempt to identify such proteins in B. subtilis. Our strategy was to search for proteins that, when overproduced, would promote Z-ring formation. Here, we report the discovery and characterization of a Z-ring-associated protein called ZapA. ZapA is a component of the division machinery that is capable of binding to FtsZ and stimulating the formation of higher-order FtsZ structures in vitro. ZapA is not essential for septum formation, but several lines of evidence indicate that ZapA is important for Z-ring formation and septation in vivo. In particular, the absence of ZapA has a strong synergistic effect in combination with the absence of the otherwise dispensable division protein EzrA, resulting in a severe block in cell division. The absence of ZapA also exacerbates the block in division caused by the absence of the division site-selection protein DivIVA. Finally, ZapA is a widely conserved protein with orthologs in many species, including bacteria that are distantly related to B. subtilis, such as E. coli. The E. coli ortholog of ZapA exhibits a strikingly similar pattern of subcellular localization to that of ZapA in B. subtilis. Thus, members of the ZapA family of proteins are a common feature of the cytokinetic machinery of bacteria.
Results
A screen for genes that promote cell division
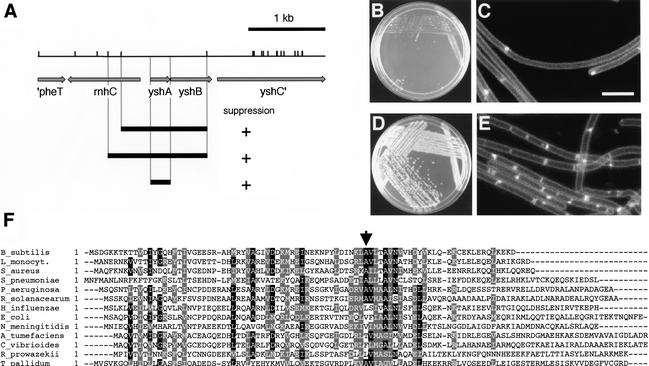
Overexpression of minD disrupts the spatially restricted action of the MinCD division inhibitor, thereby blocking FtsZ polymerization throughout the cell and causing filamentation and lethality (Marston and Errington 1999; F.J. Gueiros-Filho, unpubl.). We exploited this phenotype in a screen for genes that would stimulate FtsZ polymerization. Our strategy was to seek genes whose overexpression would suppress the division inhibition caused by excess MinD. A strain (FG247) was constructed that contained an extra copy of minD under the control of the IPTG-inducible promoter Pspac-hy. This strain was unable to grow in the presence of IPTG due to a block in division. Next, an overexpression library was constructed by cloning B. subtilis DNA fragments downstream of the xylose-inducible promoter Pxyl in an integrational vector. FG247 was transformed with the overexpression library and transformants were sought that were able to grow into colonies on medium containing IPTG and xylose. Transformants that were capable of growth on IPTG-containing medium arose at a frequency of ∼3 × 10−4. Sequencing of the inserts in several of these transformants revealed overlapping segments of DNA corresponding to the previously uncharacterized yshAB operon (Fig. 1A). The only complete ORF in these inserts was yshA, suggesting that this gene was responsible for suppressing the lethal effect of overexpression of minD. Indeed, subcloning and expression of yshA alone (to create strain FG332) under the control of Pxyl was sufficient to suppress the lethality of minD overexpression (Fig. 1B,D).
Figure 1.
Identification of the yshA (zapA) gene. (A) Sau3AI restriction map of the yshAB region of the B. subtilis chromosome. Arrows correspond to predicted genes as annotated by the B. subtilis genome consortium (http://genolist.pasteur.fr/SubtiList). Black bars correspond to Sau3AI fragments present in library clones that were capable of suppressing MinD toxicity or to the yshA (zapA) ORF. (B–E) Colony formation (B,D) and division phenotype (C,E) of cells overexpressing minD alone (B,C) or both minD and yshA (zapA; D,E). Strain FG332 was streaked on LB plates containing 250 μM IPTG (B) or 250 μM IPTG plus 1% xylose (D) and grown overnight at 37°C. For microscopy, the cells were grown in liquid LB medium to the mid-exponential phase of growth and treated with IPTG (C) or with IPTG and xylose (E) for 2 h before being harvested and stained with the membrane dye TMA-DPH. For details see Materials and Methods. (F) Multiple sequence alignment of representative ZapA orthologs. Arrow points to the start of the predicted coiled-coil region in the B. subtilis sequence, as determined by the program Coils. Alignment was made with Clustal-W and BoxShade programs. Sequence accession nos.: Agrobacterium tumefaciens, NP_356887; Bacillus subtilis, NP_390739; Caulobacter vibrioides, NP_422041; Escherichia coli, NP_289478; Haemophilus influenzae, NP_439017; Listeria monocytogenes, NP_464754; Neisseria meningitides, NP_275048; Pseudomonas aeruginosa, NP_253914; Ralstonia solanacearum, NP_520523; Rickettsia prowazekii, NP_221108; Treponema pallidum, NP_219282; Staphylococcus aureus, NP_371665; Streptococcus pneumoniae, NP_344927. The A. tumefaciens sequence has been truncated and only the first 108 amino acids of 125 are shown. Bar, 5 μm.
To determine whether yshA restored the capacity of minD-overexpressing cells to divide, we examined cells of FG332 that had been stained with the membrane dye trimethylammonium diphenylhexatriene (TMA-DPH) by fluorescence microscopy. When grown in the presence of IPTG, but in the absence of xylose, FG332 formed long, septumless filaments (Fig. 1C). However, when grown in the presence of both IPTG and xylose, FG332 formed septa at almost the same frequency as when the cells were grown in the absence of IPTG (Fig. 1E).
The yshA gene is predicted to encode a protein of 85 amino acids with a neutral isoelectric point (pH 7.17). Hydrophobicity analysis suggests that the gene product is a cytoplasmic protein and a structural prediction reveals a possible coiled-coil region in the C-terminal region of the protein. As we will show below, the yshA gene product is a Z ring-associated protein, and we henceforth refer to the gene as zapA and to its product as ZapA.
ZapA is a component of the division machinery
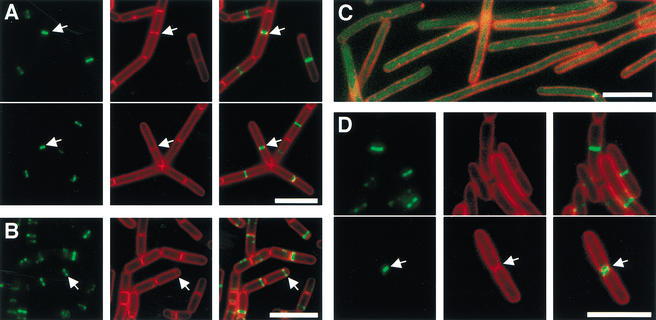
One model for how zapA acts was that its overexpression stimulates the synthesis of FtsZ, thereby compensating for the inhibitory effect of MinD. However, immunoblot analysis with anti-FtsZ antibodies revealed no increase in the levels of the cell division protein in a strain overexpressing zapA (data not shown). Another possibility was that ZapA is itself a component of the division machinery. To investigate this possibility, we fused the coding sequence for the Green Fluorescent Protein in-frame to the 5′ end of zapA. The resulting gfp–zapA gene fusion was functional as judged by its ability when overexpressed to reverse the lethality of minD overexpression (data not shown) and to complement the phenotypes associated with a zapA-null mutation (see below). Fluorescence microscopy of a GFP–ZapA-producing strain revealed transverse bands at the middle of cells, a localization pattern characteristic of division proteins, such as FtsZ (Fig. 2A). Comparison of the pattern of fluorescence from GFP–ZapA with that from the membrane dye TMA-DPH showed that transverse bands of the fusion protein were often present at the midcell position in cells in which no septum could be detected. This finding is consistent with the idea that ZapA localization precedes septum formation, as is true for FtsZ (Fig. 2A). We also observed cells in which fluorescence from GFP–ZapA coincided with a transverse band of staining from the membrane dye (Fig. 2A, arrow). In these cells, GFP–ZapA localized as a narrow band or a dot. We interpret these images as representing cells in which a septum had formed and in which the ZapA-containing, cytokinetic ring had constricted but had not entirely disassembled.
Figure 2.
Subcellular localization of GFP–ZapA and GFP–YgfE. Cells were grown, stained, and subjected to fluorescence microscopy as described in Materials and Methods. Fluorescence from GFP is in green and that from TMA-DPH is false colored in red. (A) Growing cells of the GFP–ZapA-producing B. subtilis strain FG347. The arrows point to cells in which the cytokinetic ring is constricting. (B) Sporulating cells of the GFP–ZapA-producing strain FG347. The arrows point to a polar GFP–ZapA band. GFP–ZapA can also be seen in V-like structures and membrane-associated dots, which we interpret as indicating an association of ZapA–GFP with the FtsZ spiral-like structures observed in cells undergoing sporulation (Ben Yehuda and Losick 2002). (C) ZapA localization in cells depleted of FtsZ. Cells of strain FG369 were grown to OD ∼0.6 in the presence of 1 mM IPTG, harvested, suspended in medium without IPTG, and incubated for 1.5 h at 37°C. (D) GFP–YgfE-producing cells of E. coli strain DH5α harboring pFG53 and growing in LB medium with ampicillin. The arrows point to a cell in which the cytokinetic ring is constricting. Bars, 5 μm.
We also investigated the localization of GFP–ZapA in cells undergoing sporulation. Entry into sporulation leads to a switch in the positioning of the Z ring from the midcell to sites near both the poles of the cell and the formation of a septum near one pole (Levin and Losick 1996). GFP–ZapA localized similarly in transverse bands near the cell poles in sporulating cells (Fig. 2B), and thus seems to be recruited to the Z ring during sporulation. The majority of such cells exhibited a single polar band of GFP–ZapA (Fig. 2B), but cells were also detected that displayed the bipolar localization pattern described for FtsZ (Levin and Losick 1996) and SpoIIE (Arigoni et al. 1995).
To confirm that GFP–ZapA was being recruited to the Z ring, we determined whether ZapA localization depends on FtsZ. To do this, we introduced the GFP–ZapA fusion into a strain in which synthesis of FtsZ is controlled by the IPTG-inducible Pspac promoter to create strain FG369. Depletion of FtsZ by growth in the absence of IPTG caused filamentation and led to delocalization of GFP–ZapA, which became evenly distributed in the cytoplasm (Fig. 2C). Similar experiments showed that ZapA localization was not dependent upon the late-acting division proteins DivIB and DivIC (data not shown). We conclude that GFP–ZapA is incorporated into the cytokinetic ring at an early stage of its assembly, possibly interacting directly with FtsZ.
ZapA is not essential for division
To investigate the role of ZapA in septum formation, we created a deletion of the zapA–yshB operon by replacing the two ORFs with a tetracycline-resistance cassette to create strain FG343. Next, we constructed a strain that lacked zapA alone by introducing a copy of yshB at the thrC locus of FG343. The resulting strain (FG356) exhibited no significant defect in division under a variety of conditions, including growth in minimal medium and growth at 16, 37, and 45°C (Fig. 3C,D; data not shown). FG356 also exhibited no recognizable defect in septum formation during sporulation or in the capacity of the cells to form spores. Thus, ZapA is not essential for septum formation during growth or sporulation under normal laboratory conditions. No defect in growth or division were observed for FG343, which lacks both zapA and yshB, as well as for a strain that lacked only yshB (FG345).
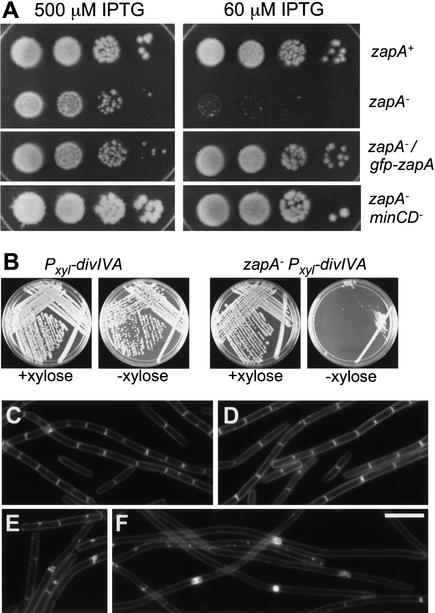
Figure 3.
Division phenotypes of zapA mutation. (A) Failure of a zapA mutant to form colonies at lowered FtsZ levels. All of the strains harbored a construct in which ftsZ was under the control of an IPTG-inducible promoter and, in addition, were zapA+ (RL861), mutant for zapA (FG367), mutant for zapA, and harboring a Pxyl–gfp–zapA construct (FG371), or mutant for zapA and mutant for minCD (FG373). The cells were grown to stationary phase in the presence of 500 μM IPTG, serially diluted (10-fold steps), and then 10 μl of each dilution was spotted onto LB plates containing the indicated concentrations of IPTG. For strain FG371 (mutant for zapA and containing Pxyl–gfp–zapA), the plates additionally contained 0.1% xylose. The plates were incubated ∼12 h at 37°C before being photographed. (B–F) Synergy of the zapA mutation with mutations of diviIVA and ezrA. (B) Colonies of strain FG449, which is mutant for zapA and divIVA and contains a Pxyl–divIVA construct, and the congenic zapA+ strain FG94, grown in LB medium containing 1% xylose or lacking xylose. (C) Wild-type strain PY79. (D) zapA mutant strain FG356. (E,F) Strain FG381, which is mutant for ezrA and zapA and contains a Pxyl–zapA construct. In E, cells of strain FG381 were grown in the presence of 0.1% xylose, whereas in F, the cells were depleted for ZapA by growth in LB medium in the absence of xylose. Cells in C–F were stained with TMA-DPH.
Reducing cellular FtsZ levels reveals a dependence of division on ZapA
In an attempt to uncover a role for ZapA in division, we investigated the effect of the absence of ZapA on division under conditions in which the cellular level of FtsZ was reduced. This was accomplished by introducing the zapA–yshB deletion into a strain in which ftsZ expression was under the control of the IPTG-inducible promoter Pspac and growing the cells at varying concentrations of inducer. An otherwise wild-type strain (RL861) harboring the Pspac–ftsZ construct was able to grow and form colonies on solid medium containing IPTG at concentrations ranging from 1 mM to as low as 60 μM. A strain (FG367) deleted for the zapA–yshB operon and harboring the Pspac–ftsZ construct was similarly able to grow and form colonies when the concentration of inducer was high (Fig. 3A). However, at low concentrations (60 μM) of inducer, colony formation by FG367 was strongly reduced (Fig. 3A). Microscopic examination of RL861 and FG367 cells grown in medium containing 60 μM IPTG showed that the difference in viability between the two strains correlated with a more pronounced inability of the zapA–yshB deletion mutant to divide (data not shown). That this division defect was due to the absence of ZapA was shown by use of a strain (FG371) that contained a copy of gfp–zapA at amyE in addition to the deletion of the zapA–yshB operon. Colony formation by strain FG371 was similar to that of RL861, even at low concentrations of inducer (Fig. 3A).
The identification of zapA as a gene whose overexpression compensates for the inhibitory effect of minD overexpression suggested that ZapA and MinCD have antagonistic effects. To investigate this possibility, we asked whether the division defect caused by the absence of ZapA under conditions of low levels of FtsZ could be suppressed by a deletion of the min operon. Accordingly, we introduced a deletion of minCD into FG367 and tested the ability of the resulting strain (FG373) to form colonies on solid medium containing 60 μM IPTG. As shown in Figure 3A, FG373 was able to form colonies at low inducer concentration as efficiently as a zapA–yshB+ strain (RL861) harboring the Pspac–ftsZ construct. Thus, the absence of minCD had reversed the growth-inhibitory effect of the absence of zapA. These results are consistent with the idea that ZapA and MinCD have opposite effects on division.
The absence of ZapA enhances the division defect of a divIVA mutant
Even though the absence of ZapA caused no recognizable phenotype on its own, we wondered whether a synergistic effect would be observed in cells mutant for the cell division gene divIVA. DivIVA is responsible for the polar localization of the MinCD inhibitor (Marston et al. 1998), and its absence leads to MinCD distribution throughout the cell membrane and inhibition of Z-ring formation at the midcell position. Cells mutant for divIVA are impaired in division but are nonetheless capable of growth and colony formation (Cha and Stewart 1997; F.J. Gueiros-Filho, unpubl). However, when we attempted to introduce a divIVA mutation into cells deleted for the zapA–yshB operon, few transformants were recovered, and these were capable of producing only small colonies that were composed of highly filamentous cells. The synergistic effect was a consequence of the absence of zapA (and not yshB), because transformants were readily recovered when the deletion-mutated recipient harbored a copy of zapA in trans. Control experiments showed that cells lacking zapA exhibited no defect in genetic competence and could be transformed readily with divIVA+ DNA.
As an alternative strategy for testing synergism between mutations in the two genes, we constructed a strain (FG449) that was doubly mutant for zapA and divIVA and that also harbored an inducible copy of divIVA. Thus, FG449 was conditionally mutant for divIVA. Whereas a zapA+ strain (FG94) that was conditionally mutant for divIVA formed approximately normal-sized colonies in the absence and presence of inducer, FG449 formed much smaller colonies in the absence of inducer than in its presence (Fig. 3B). In other experiments (data not shown), we established that the zapA mutation exacerbated the filamentous phenotype of cells depleted of DivIVA. We interpret these results as follows. In the absence of DivIVA, MinCD is free to impair cell division at the midcell position, but because ZapA counteracts the effect of MinCD, divIVA mutant cells are only partially impaired in growth and division. However, in the absence of both DivIVA and ZapA, the inhibitory effect of MinCD is unopposed, resulting in a severe division defect. We conclude that ZapA's role in cell division is masked by the action of DivIVA. Conversely, the role of DivIVA in opposing the effect of MinCD is partially compensated for by the action of ZapA.
The absence of ZapA and EzrA results in a synthetic defect in division
Next, we investigated the dependence on ZapA in a mutant that exhibits an especially mild defect in division. For this purpose, we used a mutant of ezrA, which encodes a membrane protein that is a negative regulator of Z-ring formation (Levin et al. 1999). A mutant of ezrA forms extra Z rings, but exhibits little or no defect in growth, although the mutant cells are slightly longer than the wild type and occasionally produce minicells (Levin et al. 1999; Fig. 3E). As in the case of the divIVA mutation, attempts to create an ezrA zapA double mutant led to the production of colonies that were too unhealthy for routine maintenance. Accordingly, we created a conditional mutant (strain FG381) that contained a deletion of ezrA and a copy of zapA under the control of an inducible promoter. Cells of strain FG381 were able to grow normally in the presence of inducer, but exhibited a strong block in division and filamentation in the absence of inducer (Fig. 3E,F). Thus, even though the absence of ZapA or EzrA alone has little effect on viability, the absence of both proteins severely impairs division.
We conclude that ZapA and EzrA both play important roles in cell division, but that the contribution of each is masked by the contribution of the other. Nonetheless, the observation that a ezrA zapA double mutant exhibits a synthetic phenotype is, as discussed below, surprising in light of the fact that EzrA is a negative regulator of FtsZ polymerization and ZapA, as we shall argue, is a positive regulator of FtsZ polymerization.
ZapA interacts with FtsZ
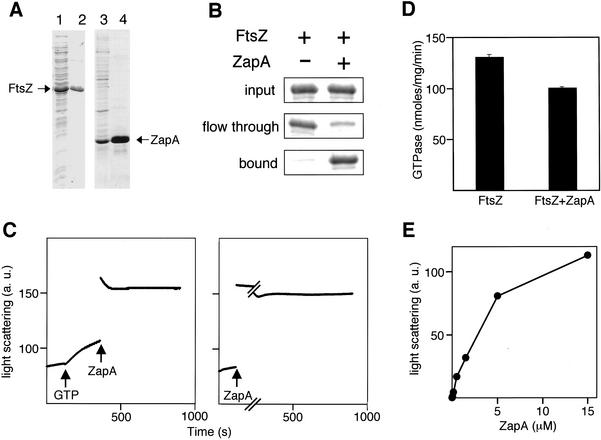
The simplest interpretation of the results presented so far is that ZapA is a component of the division machinery and that it has a positive effect on FtsZ polymerization. Such a positive effect could be the consequence of a direct interaction between ZapA and FtsZ. To investigate this possibility, we purified both proteins from strains of E. coli that had been engineered to produce the B. subtilis proteins at high levels (Fig. 4A). FtsZ was purified in its native form, whereas ZapA was produced as a fusion protein (His6–ZapA) that contained six histidine residues at its N terminus. Affinity chromatography experiments carried out using Ni++-agarose revealed that FtsZ bound quantitatively to the resin and did so in a manner that was dependent upon the presence of the histidine-tagged fusion protein (Fig. 4B). Binding of FtsZ to His6–ZapA was observed both in the presence of added GTP and in its absence (a condition under which FtsZ is presumed to be associated with GDP; Fig. 4B; data not shown).
Figure 4.
Biochemical properties of ZapA. (A) SDS-PAGE of FtsZ and His6–ZapA. The following were subjected to SDS-PAGE: an extract from cells of the FtsZ-overproducing strain W3110(pBS58)(pCXZ) (lane 1); purified FtsZ (lane 2); an extract from cells of the His6–ZapA-overproducing strain FG443 (lane 3); and purified His6–ZapA (lane 4). (B) ZapA binds to FtsZ. Ni++–NTA affinity chromatography of FtsZ in the presence or absence of His6–ZapA. Bands correspond to FtsZ and are from a Coomasie stained SDS–polyacrylamide gel. Note that the eluate fractions were more concentrated than the input or the flowthrough (see the Materials and Methods). (C) ZapA promotes assembly of FtsZ in the presence of GTP (left) or in the absence of added nucleotide (right). Light scattering traces of FtsZ polymerization reactions containing 5 μM FtsZ in PEM buffer (50 mM PIPES at pH 6.5, 1 mM EDTA, 5 mM MgCl2). The arrows indicate the time of addition of GTP (1 mM) or ZapA (5 μM). Light scattering is expressed in arbitrary units. (D) Effect of ZapA on the GTPase activity of FtsZ. GTPase activity was measured as described in Materials and Methods. FtsZ concentration was 5 μM and ZapA, when present, was at 5 μM. Error bars, the standard deviation of three replicates. Similar results were obtained in three experiments. (E) FtsZ polymerization as a function of ZapA concentration. The maximum light scattering signal of individual reactions containing 5 μM FtsZ in PEM buffer and various concentrations of ZapA is plotted. Light scattering expressed in arbitrary units.
ZapA promotes FtsZ polymerization in vitro
Two alternative models for how ZapA acts are that it promotes the assembly of FtsZ into polymers (stimulating either the formation of the single-stranded polymers known as protofilaments or the bundling of filaments), or that it protects FtsZ from polymerization inhibitors, such as MinCD. To investigate whether ZapA can stimulate FtsZ polymerization, we used light scattering to monitor the assembly of FtsZ into large structures (Mukherjee and Lutkenhaus 1999). Purified B. subtilis FtsZ behaved similarly in its polymerization properties to that observed previously for E. coli FtsZ (Mukherjee and Lutkenhaus 1998): Assembly occurred readily in the presence of Mg++ and GTP, but not when GTP was substituted with GDP (data not shown). Polymers formed with GTP were dynamic and disassembled as GTP was converted to GDP (data not shown).
The effect of ZapA was determined in reactions containing 5 μM FtsZ and either 1 mM GTP or in the absence of added nucleotide. FtsZ was diluted in polymerization buffer and a scattered light baseline was acquired for 2 min before the reaction was started with nucleotide. Addition of GTP to FtsZ led to polymer formation as indicated by an increase in light scattering (Fig. 4C, left). The further introduction of ZapA at 5 μM into the GTP-containing reaction mixture caused a dramatic additional increase in light scattering (Fig. 4C, left). Addition of ZapA to a reaction lacking GTP also led to a strong increase in light scattering (Fig. 4C, right), suggesting that ZapA can promote the polymerization of GDP-bound FtsZ as well as GTP-containing FtsZ. The ZapA-mediated increase in light scattering was not due to oligomerization or precipitation of ZapA itself, because ZapA had little effect on light scattering in the absence of FtsZ (data not shown).
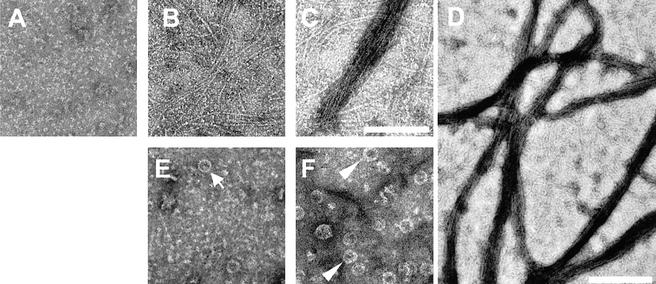
We used electron microscopy of negatively stained samples to characterize the FtsZ polymers formed in the presence of ZapA. B. subtilis FtsZ is monomeric or present as small oligomers at 5 μM in the absence of added nucleotide (Fig. 5A). Similar to E. coli FtsZ (Erickson et al. 1996; RayChaudhuri 1999; Romberg et al. 2001), B. subtilis FtsZ formed mostly straight, single-stranded, protofilaments, or lateral associations of a few protofilaments, when polymerization was induced by GTP in the absence of ZapA (Fig. 5B). The addition of ZapA at a 1:20 molar ratio to FtsZ caused strong bundling of the GTP-induced FtsZ protofilaments into large branched networks (Fig. 5C,D). Formation of such bulky polymer networks is reminiscent of the effects of agents known to cause bundling of FtsZ, such as Ca++ (Yu and Margolin 1997; Mukherjee and Lutkenhaus 1999) and the E. coli protein ZipA (RayChaudhuri 1999; Hale et al. 2000). A different type of polymer was detected in the reactions carried out in the absence of GTP. Under such conditions, ZapA promoted the appearance of FtsZ mini-rings (Fig. 5E,F), which measured ∼25 nm in diameter and tended to be more frequent in reactions with high ZapA concentrations. Some of the mini-rings seemed to be formed by a single curved protofilament and were similar to the mini-rings detected when E. coli FtsZ is assembled onto cationic phospholipid monolayers (Erickson et al. 1996). Other rings seemed to be formed by more than one protofilament and likely represented segments of the helical tubes similar to those that form when FtsZ is polymerized in the presence of GDP and DEAE-dextran (Lu et al. 2000; Fig. 5F). All of the structures detected by electron microscopy represent polymers of FtsZ alone or FtsZ and ZapA, because no structures could be detected in reaction mixtures containing ZapA alone. We conclude that ZapA promotes the assembly of FtsZ into complex polymeric structures.
Figure 5.
Electron microscopy of the products of FtsZ polymerization. (A) Unpolymerized FtsZ (5 μM FtsZ in polymerization buffer); (B) 5 μM FtsZ in the presence of 1 mM GTP; (C) 5 μM FtsZ in the presence of 1 mM GTP; and 0.25 μM His6–ZapA. (D) Low magnification of reaction products similar to those of C to show the extent of FtsZ bundling; (E) 5 μM FtsZ in the presence of 0.25 μM His6–ZapA. The arrow points to a mini-ring formed by a single protofilament; (F) 5 μM FtsZ in the presence of 1.25 μM His6–ZapA. The arrowheads point to mini-rings formed by more than one protofilament. Bars, A–C,E,F, 100 nm; D, 200 nm.
Bundling of FtsZ polymers via agents such as Ca++ or DEAE-dextran is accompanied by a decrease in the rate of GTP hydrolysis by FtsZ (Yu and Margolin 1997; Lu et al. 1998). This inhibition has been interpreted as being due to the stabilization of FtsZ polymers when assembled into bundles. Stabilization is believed to reduce the recycling of subunits and, hence, the rate of nucleotide hydrolysis (Lu et al. 1998). We measured the relative levels of GTPase activity of FtsZ in the presence and absence of ZapA and found that ZapA caused a modest (20%–30%), but reproducible reduction in the rate of GTP hydrolysis (Fig. 4D). This result is consistent with the idea that polymers formed in the presence of ZapA and GTP are more stable than polymers formed in the presence of nucleotide alone.
To determine whether ZapA acts catalytically or stoichiometrically, we investigated the rate of polymer formation and the amount of polymer at equilibrium as a function of ZapA concentration. If ZapA acts as an enzyme, then the rate of ZapA-promoted FtsZ polymerization should increase with increasing ZapA concentration, whereas the total amount of polymer at equilibrium should not vary. We found, instead, that ZapA-promoted polymerization was similarly rapid in reactions containing from 0.15 to 15 μM ZapA, and that the amount of FtsZ polymer at equilibrium increased as more ZapA was present, reaching a plateau at ∼5 μM ZapA. This represents a stoichiometry of approximately one molecule of ZapA for each molecule of FtsZ (data not shown; Fig. 4E). We conclude that ZapA acts stoichiometrically in the polymerization of FtsZ.
In light of these results, we attempted to investigate the abundance of ZapA in vivo. To do this, we created a strain that harbored GFP fusions to both ZapA and FtsZ, and determined the relative amounts of the two fusion proteins by Western blot analysis using anti-GFP antibodies. Preliminary results indicate that ZapA is a low-abundance protein, being present at 5% the level of FtsZ or less.
ZapA is widely conserved in bacteria
Finally, we investigated the presence of orthologs of ZapA in other bacteria. Using the program PSI-Blast (http://www.ncbi.nlm.nih.gov/BLAST), we found that zapA is widely conserved in eubacteria. There are 35 obvious orthologs, including 1 in E. coli, among the 60 complete genome sequences available in the data bases, and a multiple sequence alignment with representative sequences is shown in Figure 1F. The E. coli ortholog is ygfE, whose function was unknown previously . The predicted product of ygfE is similar to ZapA along its length, exhibiting identity at 9 of 79 positions (11%) and similarity (conserved substitutions) at 27 of 79 positions (34%). Like ZapA, YgfE displays a predicted coiled-coil region near its C terminus.
That ZapA and YgfE are true orthologs is supported by a subcellular localization experiment based on the use of a fusion of gfp to ygfE. Like GFP–ZapA in B. subtilis, GFP–YgfE localized to the division site in E. coli cells, suggesting that YgfE is a bonafide division protein and the counterpart of B. subtilis ZapA (Fig. 2D).
Discussion
We have discovered a novel Z-ring-associated protein called ZapA in B. subtilis that both binds to FtsZ and stimulates its assembly into bundles of protofilaments. ZapA is widely conserved in bacteria. Orthologs of ZapA exist in many species, including bacteria that are distantly related to B. subtilis. Among these is the gram-negative bacterium E. coli, in which the orthologous protein (YgfE) exhibits a strikingly similar pattern of subcellular localization to that of ZapA itself in B. subtilis. We therefore presume that members of the ZapA family of proteins are a general (or at least a common) feature of the cytokinetic machinery of bacteria.
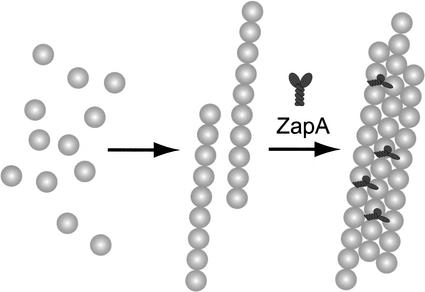
Like certain microtubule-associated proteins (MAPs) in eukaryotes, ZapA promotes the assembly of FtsZ, the bacterial equivalent of tubulin, into a higher-order structure. MAPs often achieve their effect by cross-linking adjacent tubulin monomers within the polymer lattice (Hirokawa 1994). Cross-linking requires multivalent interactions between the MAPs and tubulin, and this is accomplished by the presence of multiple tubulin-binding sites within a single molecule or by the oligomerization of the microtubule-associated protein (Hirokawa 1994). ZapA has a coiled-coil motif in its C-terminal region, and preliminary evidence suggests that ZapA interacts with itself and exists as a dimer in solution (A. Handler and R.L., unpubl.; S. Robson and G. King, pers. comm.). Thus, the ability of ZapA to promote the bundling of protofilaments could be a consequence of the bridging of FtsZ monomers between adjacent protofilaments. According to this model, ZapA stabilizes the relatively weak lateral bonds between FtsZ monomers in adjacent protofilaments (Romberg et al. 2001; Fig. 6). In light of the low abundance of ZapA, the cross bridges would be present at a frequency of about 1 in every 20 FtsZ molecules or less. An alternative possibility to explain the bundling activity of ZapA is that dimers of ZapA interact with adjacent FtsZ monomers along the axis of individual protofilaments and thus promote bundling indirectly by helping to stabilize individual protofilaments. Stabilization of longitudinal bonds could explain the formation of single protofilament minirings (Fig. 5E).
Figure 6.
Model for the function of ZapA in Z-ring formation. For simplicity, the figure depicts FtsZ polymerization as occurring by the assembly of FtsZ monomers (gray spheres) into protofilaments, which then associate with each other laterally to form bundles. We do not mean to exclude the possibility that monomers are also directly added to bundles. The model proposes that dimers of ZapA (black Y-shapes) cross-link FtsZ monomers in adjacent protofilaments, thereby stabilizing the lateral interactions. Alternatively, ZapA could promote bundling indirectly by stabilizing the longitudinal bonds along a protofilament. See Discussion for details.
Two other cytokinetic proteins that are known to interact directly with FtsZ are ZipA (which is present in E. coli but not B. subtilis) and FtsA. Like ZapA, ZipA and FtsA help to promote Z-ring formation. In the absence of either protein, Z rings form with reduced frequency (Addinall et al. 1996; Hale and de Boer 1999; Liu et al. 1999), and when both FtsA and ZipA are absent, FtsZ forms irregular assemblies at potential division sites that differ significantly from normal-looking Z rings (Pichoff and Lutkenhaus 2002). Moreover, ZipA is known to stabilize Z rings formed by a mutant FtsZ protein (RayChaudhuri 1999) and to promote the bundling of FtsZ protofilaments in vitro (RayChaudhuri 1999; Hale et al. 2000). An important difference between ZipA and FtsA, on the one hand, and ZapA, on the other hand, however, is that the former are essential for viability, whereas the latter is not. A likely explanation for this difference is that the essentiality of FtsA and ZipA is not due to their role in FtsZ polymerization. Rather, cells depleted of ZipA or FtsA, although capable of forming Z rings, are blocked in cytokinesis because they fail to recruit other division proteins to the cytokinetic ring (Wang and Lutkenhaus 1998; Pichoff and Lutkenhaus 2002). ZapA, in contrast, appears to have no such downstream function in the recruitment of essential cytokinetic proteins. We therefore propose that ZapA represents a hitherto unrecognized class of prokaryotic cell division proteins that functions exclusively in the assembly of FtsZ into higher-order structures. We also propose that ZapA and FtsA in B. subtilis and YgfE (the ZapA ortholog), ZipA and FtsA in E. coli play overlapping and perhaps partially redundant roles in promoting Z-ring formation.
Z ring assembly in vivo displays the properties of a cooperative process as follows: assembly of Z rings happens abruptly (Addinall et al. 1997; Sun and Margolin 1998), and a relatively modest reduction of the intracellular FtsZ concentration prevents division, probably due to a block in Z-ring formation (Dai and Lutkenhaus 1991; Garrido et al. 1993). Interestingly, recent studies have shown that FtsZ can assemble into single-stranded protofilaments in vitro in a noncooperative manner (Romberg et al. 2001). Assembly of multistranded polymers, which should display cooperativity, does not seem to be kinetically favored unless stabilizing agents are present (Mukherjee and Lutkenhaus 1994; Lu et al. 1998). This raises the possibility that the cooperative behavior of Z-ring formation is not solely a consequence of intrinsic properties of FtsZ, but requires cellular factors. We propose that ZapA, and other Z ring-interacting proteins like ZipA and FtsA, could promote the cooperative assembly of FtsZ in the cell by increasing the tendency of protofilaments to associate laterally (Fig. 6).
We discovered ZapA in our overexpression screen on the basis of its ability to antagonize the effect of the division inhibitor MinCD. How does MinCD act in blocking Z-ring formation? Evidence indicates that MinCD is unlikely to inhibit protofilament formation. For example, MinC does not inhibit the GTPase activity of FtsZ (Hu et al. 1999), and because GTP hydrolysis depends on FtsZ monomer–monomer association (Wang and Lutkenhaus 1993; Sossong et al. 1999; Scheffers et al. 2002), this finding implies that monomer association is not severely affected by MinC. An alternative possibility is that MinC inhibits Z-ring formation by inhibiting the bundling of protofilaments into larger structures. Because ZapA promotes bundling, the antagonism between ZapA and MinCD could be understood as the opposing effects of the two factors on the stability of lateral associations between protofilaments. In keeping with this idea, FtsA, which is also likely to be involved in cross-linking FtsZ protofilaments, is capable of antagonizing the inhibitory effect of MinCD when both FtsA and MinCD are overproduced in E. coli (Justice et al. 2000) or in B. subtilis (F. J. Gueiros-Filho, unpubl.).
Despite its striking pattern of subcellular localization, capacity to antagonize the effects of the MinCD division inhibitor, and ability to promote bundling of FtsZ protofilaments in vitro, ZapA is not, as we have noted, required for normal growth and cell division. Nonetheless, we believe that ZapA does play a central role in cytokinesis because of the strong synergistic effect of combining a zapA mutation with a mutation in the gene for another Z-ring-associated protein, EzrA. An ezrA-null mutation has little effect on viability by itself, although the mutant cells are longer and divide less frequently than do wild-type cells. In contrast, a zapA ezrA double mutant is severely impaired in division, forming long aseptate filaments. The synergism between a zapA mutation and an ezrA mutation shows that both otherwise dispensable genes do play an important role in cytokinesis, but that each partially or completely masks the contribution of the other.
Nonetheless, the synergism between mutations of zapA and ezrA is surprising from a mechanistic point of view. EzrA is believed to be an inhibitor of Z-ring formation, and its effect in cells is to increase the concentration of FtsZ required for Z-ring formation [in its absence, cells form extra Z rings (Levin et al. 1999); the absence of EzrA was also shown recently to suppress the division inhibition caused by MinCD overexpression (Levin et al. 2001)]. In contrast, ZapA acts positively on Z-ring formation and decreases the concentration of FtsZ needed for Z-ring formation. Thus, the absence of one protein might have been expected to cancel the effect of the absence of the other protein. One way to resolve the paradox is to think of EzrA as a protein whose predominant role is to disassemble the Z ring as cytokinesis progresses. Thus, in the absence of EzrA, the Z ring would not be entirely disassembled (or would disassemble more slowly). As a result, the cytoplasmic pool of free FtsZ monomers or protofilaments would be reduced and, hence, less would be available for the next round of Z-ring formation. This interpretation of EzrA function is consistent with the observation that cells mutant for EzrA are longer and seem to divide less frequently than do wild-type cells (Levin et al. 1999). Thus, in doubly mutant cells, the pool of cytoplasmic FtsZ would be low due to the absence of EzrA, and the concentration of FtsZ required for Z-ring formation would be high due to the absence of ZapA. Under such circumstances, Z-ring formation could be expected to be severely impaired.
Finally, we raise the possibility that ZapA is part of a system that imparts dynamism on the Z ring. Recent fluorescence photobleaching studies in E. coli reveal that the Z ring is undergoing constant remodeling on a time scale of seconds (Stricker et al. 2002). The authors propose that such remodeling could generate the force for Z-ring constriction during cytokinesis. The remodeling of the Z ring reflects, at least in part, intrinsic properties of FtsZ polymerization because the turnover rate of rings made up of different FtsZ variants correlates well with their GTPase activity. It is possible, however, that accessory proteins also help promote rapid turnover of the ring. This situation would be analogous to the case of microtubules in which stabilizing and destabilizing proteins are responsible for the dynamic behavior of the polymer in vivo (Tournebize et al. 2000; Kinoshita et al. 2001). Thus, FtsZ-interacting proteins, depending on how they affect the rate of assembly and disassembly of the Z ring, could significantly influence the rate of Z-ring remodeling. Assuming that ZapA and EzrA contribute to Z-ring remodeling by accelerating assembly and disassembly, respectively, this hypothesis offers another possible explanation for the phenotype of the zapA ezrA double mutant. According to this view, the simultaneous absence of both proteins, although not having a large effect on the monomer/polymer equilibrium, would slow down Z-ring remodeling sufficiently so as to impair the formation of the septum.
Materials and methods
General methods
All B. subtilis strains were derived from the prototrophic strain PY79 (Youngman et al. 1983) and are listed in Supplemental Table 1 (see Supplementary Material at http://www.genesdev.org). B. subtilis was grown in LB medium at 37°C, unless indicated otherwise. Preparation of competent cells and transformations were carried out essentially as described by Kunst and Rapoport (1995). Antibiotics were used at the following concentrations: chloramphenicol, 5 μg/mL; erythromycin plus lincomycin, 1 μg/mL and 25 μg/mL; spectinomycin, 100 μg/mL; tetracycline, 10 μg/mL; phleomycin, 8 μg/mL.
DNA methods
DNA manipulations and cloning were carried out according to standard methods (Sambrook and Russel 2001). Pfu DNA polymerase (Stratagene) was used in PCR reactions. Oligonucleotide primers were purchased from IDT (Coralville) and are listed in Supplemental Table 2 (see Supplementary Material at http://www.genesdev.org). Sequencing was carried out in the Molecular and Cellular Biology departmental facility.
Plasmids and their relevant characteristics are listed in Supplemental Table 1 (see Supplementary Material at http://www.genesdev.org). Descriptions of the plasmids, the construction of the expression library, and the construction of the zapA–yshB deletion are in the Supplemental Material (see Supplementary Material at http://www.genesdev.org).
Fluorescence microscopy
The equipment and general conditions used for microscopy were as described by King et al. (1999), and a detailed description can be found in the Supplemental Material (see Supplementary Material at http://www.genesdev.org).
Electron microscopy
A total of 5 μL of polymerization reactions were spotted on glow-discharged, carbon-coated 400-mesh grids (Electron Microscopy Sciences). After 30 sec, excess liquid was removed by blotting and the grid was stained with 5 μL of 1% uranyl acetate for 30 sec. Microscopy was carried out in the Harvard Medical School Electron Microscope Facility using a JEOL 1200EX microscope.
Protein purification
The purification of FtsZ and His6–ZapA is described in the Supplemental Material (see Supplementary Material at http://www.genesdev.org).
Ni++–NTA-agarose affinity chromatography
One hundred microliter reactions were assembled containing FtsZ (0.2 mg/mL) with or without His6–ZapA (0.1 mg/mL) in binding buffer (PEM plus 0.5 mg/mL BSA) and incubated at room temperature for 15 min. Twenty microliters of Ni++–NTA-agarose were added, binding was allowed to proceed for 5 min, and the resin was collected by low-speed centrifugation. The supernatant fluid (flowthrough) was saved, and the resin was washed twice in 100 μL of binding buffer. Proteins were eluted in two 20-μL steps with PEM containing 250 mM imidazole (pH 7.0). Thus, the eluate was in effect 2.5 more concentrated than the material that was applied to the column and the flowthrough. Twenty microliters of the input and 20 μL of each fraction were subject to SDS-PAGE in an 18% gel.
FtsZ polymerization assays
Light-scattering measurements of FtsZ polymerization were carried out essentially as described by Romberg et al. (2001) using a SPEX FluroMax fluorimeter. Polymerization buffer was PEM (50 mM PIPES at pH 6.5, 1 mM EDTA, 5 mM MgCl2), and reactions were carried out a room temperature.
GTPase assay
The GTPase activity of FtsZ was determined by measuring 32Pi released in reactions containing [γ-32P]GTP (NEN). Briefly, 200-μL reactions were assembled on ice in PEM buffer plus 50 mM KCl, 2 mM GTP, 5 μCi of 30 Ci/mmole [γ32P]GTP, and various amounts of FtsZ and ZapA. Polymerization was started by shifting samples to 37°C. At the desired times, 30-μL aliquots were removed, mixed with 300 μL of 5% charcoal in 20 mM phosphoric acid, and centrifuged briefly. Radioactivity in aliquotes of the supernatant fluid containing 32Pi were measured using a scintillation counter.
Acknowledgments
We thank P. Levin for strains and plasmids, M. Erickson and L. Romberg for help with electron microscopy, J. McCormick for drawing our attention to the presence of a zapA ortholog in E. coli, the Hastings laboratory for the use of their fluorimeter, and P. Levin, J. McCormick, S. Ben-Yehuda, D. Rudner, J. Dworkin, K. Carniol, and C. van Ooij for comments on the manuscript. F.J.G.-F. was a Helen Hay Whitney postdoctoral fellow. This work was supported by NIH grant GM18568 to R.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL losick@mcb.harvard.edu; FAX: (617) 496-4642.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1014102.
References
- Addinall SG, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall SG, Cao C, Lutkenhaus J. Temperature shift experiments with an ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J Bacteriol. 1997;179:4277–4284. doi: 10.1128/jb.179.13.4277-4284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigoni F, Pogliano K, Webb CD, Stragier P, Losick R. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science. 1995;270:637–640. doi: 10.1126/science.270.5236.637. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Losick R. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell. 2002;109:257–266. doi: 10.1016/s0092-8674(02)00698-0. [DOI] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Cha JH, Stewart GC. The divIVA minicell locus of Bacillus subtilis. J Bacteriol. 1997;179:1671–1683. doi: 10.1128/jb.179.5.1671-1683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Lutkenhaus J. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol. 1991;173:3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PA, Crossley RE, Rothfield LI. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- Edwards DH, Errington J. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol. 1997;24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- Erickson HP. FtsZ, a prokaryotic homolog of tubulin? Cell. 1995;80:367–370. doi: 10.1016/0092-8674(95)90486-7. [DOI] [PubMed] [Google Scholar]
- Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J, Daniel RA. Cell division during growth and sporulation. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and its closest relatives. Washington, D.C.: ASM Press; 2002. pp. 97–112. [Google Scholar]
- Garrido T, Sanchez M, Palacios P, Aldea M, Vicente M. Transcription of ftsZ oscillates during the cell cycle of Escherichia coli. EMBO J. 1993;12:3957–3965. doi: 10.1002/j.1460-2075.1993.tb06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PA. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, Rhee AC, de Boer PA. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J Bacteriol. 2000;182:5153–5166. doi: 10.1128/jb.182.18.5153-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry EJ. Bacterial cell division: Regulating Z-ring formation. Mol Microbiol. 2001;40:795–803. doi: 10.1046/j.1365-2958.2001.02370.x. [DOI] [PubMed] [Google Scholar]
- Harry EJ, Rodwell J, Wake RG. Co-ordinating DNA replication with cell division in bacteria: A link between the early stages of a round of replication and mid-cell Z ring assembly. Mol Microbiol. 1999;33:33–40. doi: 10.1046/j.1365-2958.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- Hiraga S, Niki H, Ogura T, Ichinose C, Mori H, Ezaki B, Jaffe A. Chromosome partitioning in Escherichia coli: Novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. Microtubule organization and dynamics dependent on microtubule-associated proteins. Curr Opin Cell Biol. 1994;6:74–81. doi: 10.1016/0955-0674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K, Gunther NW, IV, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice SS, Garcia-Lara J, Rothfield LI. Cell division inhibitors SulA and MinC/MinD block septum formation at different steps in the assembly of the Escherichia coli division machinery. Mol Microbiol. 2000;37:410–423. doi: 10.1046/j.1365-2958.2000.02007.x. [DOI] [PubMed] [Google Scholar]
- King N, Dreesen O, Stragier P, Pogliano K, Losick R. Septation, dephosphorylation, and the activation of sigmaF during sporulation in Bacillus subtilis. Genes & Dev. 1999;13:1156–1167. doi: 10.1101/gad.13.9.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Arnal I, Desai A, Drechsel DN, Hyman AA. Reconstitution of physiological microtubule dynamics using purified components. Science. 2001;294:1340–1343. doi: 10.1126/science.1064629. [DOI] [PubMed] [Google Scholar]
- Kunst F, Rapoport G. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J Bacteriol. 1995;177:2403–2407. doi: 10.1128/jb.177.9.2403-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes & Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- Levin PA, Kurtser IG, Grossman AD. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci. 1999;96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Schwartz RL, Grossman AD. Polymer stability plays an important role in the positional regulation of FtsZ. J Bacteriol. 2001;183:5449–5452. doi: 10.1128/JB.183.18.5449-5452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Mukherjee A, Lutkenhaus J. Recruitment of ZipA to the division site by interaction with FtsZ. Mol Microbiol. 1999;31:1853–1861. doi: 10.1046/j.1365-2958.1999.01322.x. [DOI] [PubMed] [Google Scholar]
- Lowe J, Amos LA. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 1999;18:2364–2371. doi: 10.1093/emboj/18.9.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Stricker J, Erickson HP. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima–quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskeleton. 1998;40:71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Lu C, Reedy M, Erickson HP. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol. 2000;182:164–170. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. Spatial regulation of cytokinesis in bacteria. Curr Opin Microbiol. 2001;4:647–652. doi: 10.1016/s1369-5274(01)00264-8. [DOI] [PubMed] [Google Scholar]
- Marston AL, Errington J. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol Microbiol. 1999;33:84–96. doi: 10.1046/j.1365-2958.1999.01450.x. [DOI] [PubMed] [Google Scholar]
- Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes & Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J Bacteriol. 1999;181:823–832. doi: 10.1128/jb.181.3.823-832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol. 1999a;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci. 1999b;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 1999;18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg L, Simon M, Erickson HP. Polymerization of FtsZ, a bacterial homolog of tubulin. Is assembly cooperative? J Biol Chem. 2001;276:11743–11753. doi: 10.1074/jbc.M009033200. [DOI] [PubMed] [Google Scholar]
- Rowland SL, Fu X, Sayed MA, Zhang Y, Cook WR, Rothfield LI. Membrane redistribution of the Escherichia coli MinD protein induced by MinE. J Bacteriol. 2000;182:613–619. doi: 10.1128/jb.182.3.613-619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular cloning. A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Scheffers DJ, de Wit JG, den Blaauwen T, Driessen AJ. GTP hydrolysis of cell division protein FtsZ: Evidence that the active site is formed by the association of monomers. Biochemistry. 2002;41:521–529. doi: 10.1021/bi011370i. [DOI] [PubMed] [Google Scholar]
- Sossong TM, Jr, Brigham-Burke MR, Hensley P, Pearce KH., Jr Self-activation of guanosine triphosphatase activity by oligomerization of the bacterial cell division protein FtsZ. Biochemistry. 1999;38:14843–14850. doi: 10.1021/bi990917e. [DOI] [PubMed] [Google Scholar]
- Stricker J, Maddox P, Salmon ED, Erickson HP. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci. 2002;99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Margolin W. FtsZ dynamics during the division cycle of live Escherichia coli cells. J Bacteriol. 1998;180:2050–2056. doi: 10.1128/jb.180.8.2050-2056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Yu XC, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol Microbiol. 1998;29:491–503. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- Tournebize R, Popov A, Kinoshita K, Ashford AJ, Rybina S, Pozniakovsky A, Mayer TU, Walczak CE, Karsenti E, Hyman AA. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat Cell Biol. 2000;2:13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- Trueba FJ. On the precision and accuracy achieved by Escherichia coli cells at fission about their middle. Arch Microbiol. 1982;131:55–59. doi: 10.1007/BF00451499. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Amos L, Lowe J. Bacterial ancestry of actin and tubulin. Curr Opin Microbiol. 2001;4:634–638. doi: 10.1016/s1369-5274(01)00262-4. [DOI] [PubMed] [Google Scholar]
- Wang L, Lutkenhaus J. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Lutkenhaus J. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol Microbiol. 1993;9:435–442. doi: 10.1111/j.1365-2958.1993.tb01705.x. [DOI] [PubMed] [Google Scholar]
- Woldringh CL, Mulder E, Valkenburg JA, Wientjes FB, Zaritsky A, Nanninga N. Role of the nucleoid in the toporegulation of division. Res Microbiol. 1990;141:39–49. doi: 10.1016/0923-2508(90)90096-9. [DOI] [PubMed] [Google Scholar]
- Youngman PJ, Perkins JB, Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci. 1983;80:2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XC, Margolin W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 1997;16:5455–5463. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— FtsZ ring clusters in min and partition mutants: Role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol. 1999;32:315–326. doi: 10.1046/j.1365-2958.1999.01351.x. [DOI] [PubMed] [Google Scholar]