Abstract
Translational control plays a key role in many biological processes including pattern formation during early Drosophila embryogenesis. In this process, the anterior determinant Bicoid (BCD) acts not only as a transcriptional activator of segmentation genes but also causes specific translational repression of ubiquitously distributed caudal (cad) mRNA in the anterior region of the embryo. We show that translational repression of cad mRNA is dependent on a functional eIF4E-binding motif. The results suggest a novel mode of translational repression, which combines the strategy of target-specific binding to 3′-untranslated sequences and interference with 5′-cap-dependent translation initiation in one protein.
Keywords: Bicoid, translational repression, caudal, eIF4E, cap-dependence
Pattern formation during early Drosophila embryogenesis is initiated by an asymmetric distribution of the maternal transcription factors Bicoid (BCD), Hunchback (HB), and Caudal (CAD) in a single cell, the egg (for review, see St Johnston and Nüsslein-Volhard 1992; Rivera-Pomar and Jäckle 1996). HB and CAD form concentration gradients along the longitudinal axis that are generated by the spatially restricted translational repression of evenly distributed maternal mRNA (Curtis et al. 1995; for review, see Wickens et al. 2000). In contrast, the concentration gradient of the anterior determinant BCD derives from prelocalized maternal mRNA in the anterior pole region of the embryo (for review, see St Johnston and Nüsslein-Volhard 1992; Hake and Richter 1997). BCD acts as a transcriptional activator of segmentation genes (for review, see Driever 1993) and causes specific translational repression of caudal (cad) mRNA in the anterior region of the embryo (Tautz 1988; Driever and Nüsslein-Volhard 1989; Struhl et al. 1989; Dubnau and Struhl 1996; Rivera-Pomar et al. 1996). Translational repression of cad mRNA involves the binding of BCD to a distinct cis-acting element within the 3′-untranslated region (UTR) of the mRNA (Dubnau and Struhl 1996; Rivera-Pomar et al. 1996) and functions in a 5′-cap-dependent manner in cell culture (Niessing et al. 1999).
The cap-dependent mode of translation depends on the assembly of an evolutionarily conserved protein complex that is initiated by the binding of the translation initiation factor 4E (eIF4E) to the m7GpppN-cap structure (Shatkin 1976; for review, see Merrick and Hershey 2000; Raught et al. 2000). Subsequently, the adapter protein eIF4G binds to eIF4E and allows additional factors (including eIF4A, eIF4B, eIF1, eIF1A, eIF2, eIF3, and the ribosomal subunits) to assemble into a complex that initiates translation (for review, see Merrick and Hershey 2000; Raught et al. 2000). The cap-dependent translation initiation process can be regulated by eIF4E-binding proteins such as BP1, BP2, and Maskin (for review, see Raught et al. 2000; Sachs and Varani 2000). They block the eIF4E ∷ eIF4G association through outcompeting binding to eIF4E, involving a small eIF4E-binding motif of the minimal consensus sequence YxxxxL (for review, see Raught et al. 2000; Richter 2000; Sachs and Varani 2000). Here we show that BCD contains a functional eIF4E-binding motif and that the translational repression of cad mRNA is dependent on this motif in vivo. The results suggest that 3′-UTR-bound BCD interferes with the assembly of the initiation complex and thereby causes repression of cad mRNA translation.
Results
BCD associates with a 5′-cap-bound protein in vitro
The cap-dependence of cad mRNA translational inhibition suggested that 3′-UTR-bound BCD interacts with one or several components of the translation initiation complex at the 5′-end (Niessing et al. 1999). To test whether BCD can associate with these cap-bound proteins, we produced cytoplasmic extracts of early Drosophila embryos and asked whether BCD can associate with m7GTP-sepharose, serving as a cap analog (Edery et al. 1988; Pyronnet et al. 2001). The m7GTP-sepharose-bound protein fraction of wild-type extracts contained a series of proteins including eIF4E (Edery et al. 1988; Pyronnet et al. 2001; data now shown) and a single protein that was absent from cytoplasmic extracts of embryos derived from homozygous bcd mutant females (Fig. 1a, lanes 1,3). Anti-BCD antibody staining of Western blots (Fig. 1a, lanes 2,4) showed that this protein is BCD. We also examined protein extracts from embryos that expressed a transgene-derived, cDNA-based GFP–BCD fusion protein (Hazelrigg et al. 1998). The GFP–BCD fusion protein was recovered from the m7GTP-sepharose-bound protein fraction as shown by SDS-PAGE analysis (Fig. 1b, lanes 1,2) and Western blots stained with anti-BCD (Fig. 1b, lanes 3,4) and anti-GFP (Fig. 1b, lanes 5,6) antibodies, respectively.
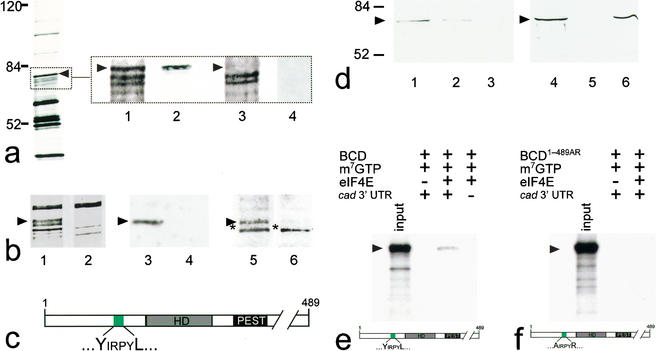
Figure 1.
BCD copurifies with 5′-cap-bound proteins. Cytoplasmic protein extracts of young embryos were affinity-purified using a cap-analog m7GTP-sepharose resin (Edery et al. 1988). (a) Silver-stained SDS-PAGE of affinity-purified proteins contained within cytoplasmic extracts of wild-type and bcd mutant Drosophila embryos. Arrowhead marks the protein band that was subsequently identified as BCD (left, see also lanes 1–4 in b). Note that eIF4E, the most abundant 30-kD component among the purified proteins, is run off the gel (left) to obtain maximum resolution of the relevant range of protein bands between 50 and 100 kD. (Lanes 1–4) Relevant portions of silver-stained SDS-PAGE (dotted box) including a ∼70-kD protein (lane 1, arrowhead) that is recognized by monospecific anti-BCD antibodies on Western blots (lane 2). This protein is not present in silver-stained SDS-PAGE (lane 3), and anti-BCD antibodies stained Western blots (lane 4) of corresponding extracts from embryos that derived from homozygous bcd mutant females. (b) Relevant portions of silver-stained SDS-PAGE showing that the GFP–BCD fusion protein (lane 1, 95 kD) is absent in wild-type embryos (lane 2). Corresponding Western blots show that the 95-kD fusion protein reacts with both anti-BCD (lane 3; lane 4 is a wild-type control lacking the GFP–BCD transgene) and anti-GFP antibodies (lane 5; lane 6 is a wild-type control lacking the GFP–BCD transgene; asterisk marks cross-reacting protein of unknown identity). (c) Schematic diagram of BCD (positions refer to sequence according to Berleth et al. 1988) showing the homeodomain (HD, gray box; position 91–154), the PEST domain (PEST, black box; position 170–203), and a YIRPYL motif (green box; position 68–73). The eIF4E-binding properties of this motif have been recently analyzed in great detail in the context of human BP1 (Marcotrigiano et al. 1999; for review, see Raught et al. 2000; Sachs and Varani 2000; Miron et al. 2001). (d) Western blots showing that BCD bound to m7GTP-sepharose coupled recombinant eIF4E (lanes 1,4) can be competed for with 100 nM (lane 2) and 1 mM (lanes 3,5) of a peptide containing the YDRKFL motif of human BP1, whereas 1 mM of a mutated human BP1 peptide (mutated motif is ADRKFR) did not interfere with the binding of BCD to eIF4E (lane 6). (e,f) Autoradiogram showing that in vitro translated 35S-labeled BCD (input; a schematic representation of the protein is shown at the bottom of e) is capable of interacting with m7GTP-sepharose-bound recombinant eIF4E in the presence of in vitro transcribed cad 3′-UTR (e), whereas in vitro translated 35S-labeled BCD1–489AR (input; a schematic representation of the protein is shown at the bottom of f) showed no significant binding to eIF4E (f). Note that there is no unspecific binding of in vitro translated protein to m7GTP-beads without precoupled eIF4E. For details, see Materials and Methods.
BCD contains a functional eIF4E-binding motif
In searching for the cap-bound protein with which BCD associates, we noted a potential eIF4E-binding motif (YIRPYL) N-terminal to the BCD homeodomain (Fig. 1c). To test whether cap association of BCD could be mediated by eIF4E, we precoupled recombinant eIF4E to m7GTP-sepharose, incubated it with protein extracts from preblastoderm stage Drosophila embryos, and examined the eIF4E-associated proteins. SDS-PAGE followed by Western blot analysis identified BCD among the retained proteins (Fig. 1d, lanes 1,4). We next asked whether BCD binding could be competed for by adding increasing amounts of the YxxxxL-containing peptide of human BP1, which had been shown to compete efficiently for binding at the eIF4G-binding site of eIF4E (Marcotrigiano et al. 1999; Ptushkina et al. 1999; for review, see Raught et al. 2000; Richter 2000; Sachs and Varani 2000). Figure 1d indicates that the YxxxxL-containing peptide (Fig. 1d, lanes 2,3,5) competes for the binding of BCD to m7GTP-sepharose-associated proteins, whereas the corresponding peptide in which the conserved Y and L residues of the eIF4E-binding motif were replaced by A and R (Fig. 1d, cf. lanes 4–6) does not. This finding is consistent with earlier results showing that mutations in corresponding positions of the motif were able to abolish eIF4E binding (Miron et al. 2001). The data therefore suggest that BCD interacts with the eIF4E-containing 5′-cap complex via the eIF4E-binding site in a manner similar to BP1.
To show that BCD and eIF4E can interact directly, we coupled recombinant eIF4E to m7GTP-sepharose and examined its association with in vitro translated 35S-labeled full-length BCD (Fig. 1e) and BCD1–489AR mutant protein (Fig. 1f). We observed specific binding of in vitro translated BCD to recombinant eIF4E (Fig. 1e). This interaction was absent in the case of BCD1–489AR mutant protein (Fig. 1, cf. e and f) and depended on the presence of cad 3′-UTR mRNA in the reaction mixture (Fig. 1e), implying that the association of BCD with target mRNA is a prerequisite for the binding. Collectively, these results suggest that BCD binds eIF4E directly and that the binding requires the intact YxxxxL motif. We would like to emphasize that the in vitro interaction of BCD and eIF4E may require cofactors present in the reticulocyte lysate used for the in vitro translation of BCD.
Translational repression of cad mRNA depends on the eIF4E-binding motif of BCD
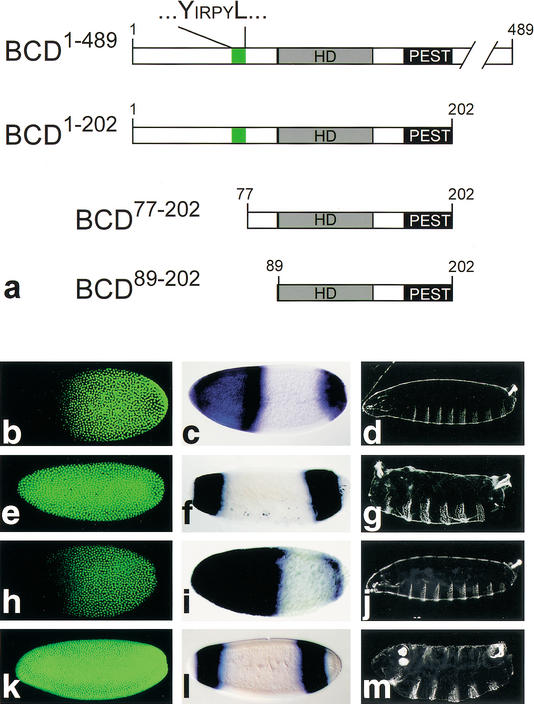
We next asked whether the eIF4E-binding motif of BCD is necessary for mediating translational repression of cad mRNA in the embryo. We generated BCD deletion mutants (Fig. 2a) and examined their transgene-derived activities in embryos from homozygous bcd mutant females (Frohnhöfer and Nüsslein-Volhard 1986). Embryos without BCD activity fail to repress translation of cad mRNA in the anterior region (Fig. 2, cf. b and e), lack anterior hb activation (Fig. 2, cf. c and f) and head and thorax development (Fig. 2, cf. d and g; Frohnhöfer and Nüsslein-Volhard 1986; Berleth et al. 1988; Driever et al. 1989; for review, see Driever 1993). Transgene-derived expression of full-size BCD1–489 (Fig. 2a) rescued all aspects of BCD requirement during Drosophila embryogenesis (Frohnhöfer and Nüsslein-Volhard 1986; Berleth et al. 1988; Driever et al. 1989; for review, see Driever 1993) including repression of cad mRNA translation (Fig. 2h–j). Transgene-dependent expression of the deletion mutant BCD1–202, containing the N-terminal half of BCD that includes the eIF4E-binding motif, the RNA-binding homeodomain, and the PEST domain (Fig. 2a), restores translational repression of cad mRNA in the anterior pole region of the embryo (Niessing et al. 1999). In contrast, BCD deletion mutants lacking the eIF4E-binding motif, such as BCD77–202 or BCD89–202 (Fig. 2a), did not repress translation (Fig. 2k). Furthermore, earlier studies had shown that a deletion mutant that lacks the N-terminal 29 amino acids is able to repress cad mRNA translation (Dubnau and Struhl 1996). The 47-amino-acid sequence interval between amino residues 29 and 77 of BCD, which includes the eIF4E-binding motif in position 68–73 (Fig. 2a), is therefore necessary for the BCD-dependent translational control in vivo.
Figure 2.
Functional analysis of mutant BCD by transgene-dependent expression in bcd mutant embryos. (a) Schematic representation of the BCD-deletion mutants (cf. Fig. 1c, wild-type BCD). (b–d) Wild-type embryos are characterized by translational repression of cad mRNA (absence of anti-CAD antibody staining (green) in the anterior region of preblastoderm embryos (b; Dubnau and Struhl 1996; Rivera-Pomar et al. 1996), by activation of zygotic hb transcription as revealed by whole mount in situ hybridization with an hb cDNA probe (c; Klingler and Gergen 1993; reviewed in Martinez Arias 1993), and by the wild-type cuticle pattern (d). (e–g) Embryos derived from homozygous bcdE1 mutant females fail to repress cad mRNA translation (e), lack the anterior hb expression domain (which is replaced by a duplication of the posterior, BCD-independent hb expression domain; f), and show a bcd mutant cuticle phenotype (g; Frohnhöfer and Nüsslein-Volhard 1986). (h–j) Transgene-derived BCD1–489 expression restores all aspects of the bcd mutant phenotype including translational repression of cad mRNA (h), anterior hb expression (i), and the larval cuticle phenotype (j). (k–m) cad mRNA translation is not repressed in response to transgene-derived BCD77–202 (k) or BCD89–202 (data not shown). In both cases, the transgene-expressed BCD deletion mutants also fail to restore anterior hb expression (l) and develop a bcd mutant cuticle phenotype (m) because of the absence of the C-terminal transactivation domains (Sauer et al. 1995; Schaeffer et al. 1999). Orientation of embryos is anterior left and dorsal up.
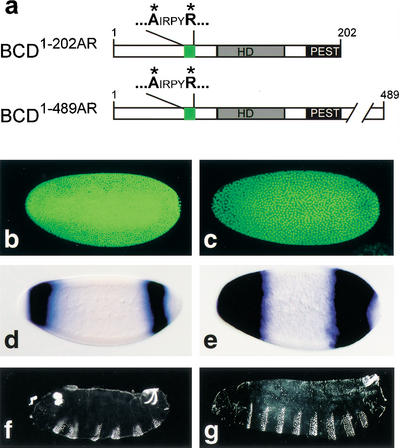
To show that the eIF4E-binding motif of BCD itself is needed to exert translational repression on cad mRNA, we performed transgene-dependent rescue experiments with bcd mutant embryos. We expressed mutant BCD proteins in which Y and L of the eIF4E-binding motif had been replaced by A and R residues. Transgene-dependent expression of mutant BCD1–202AR protein (Fig. 3a) did not repress translation of cad mRNA in the anterior region of the embryo (Fig. 3b) and was unable to activate hb transcription (Fig. 3d) or rescue the segmentation defects of the mutants (Fig. 3f). Expression of BCD1–489AR, which in contrast to BCD1–202AR contains the C-terminal transcriptional activation domains (Sauer et al. 1995; Schaeffer et al. 1999), also failed to restore the translational repression of cad mRNA (Fig. 3c). However, it supported transcriptional activation of BCD-dependent hb expression (Fig. 3e) and head and thorax development (Fig. 3g). These results indicate that the eIF4E-binding motif of BCD is not essential for transcriptional activity of BCD but is specifically required for translational repression of cad mRNA.
Figure 3.
Translational repression of cad mRNA, transcriptional activation of anterior hb expression, and rescue of segmentation defects in response to transgene-expressed BCD replacement mutants. (a) Schematic representation of the BCD replacement mutants BCD1–202AR and BCD1–489AR in which the YIRPYL motif is changed into AIRPYR (green box; for other details, see Fig. 1c). (b,d,f) Embryos from homozygous bcdE1 females expressing transgene-derived BCD1–202AR fail to repress cad mRNA translation (b) or to activate anterior hb expression (d), and develop a bcd mutant cuticle phenotype (f). (c,e,g) Embryos from homozygous bcdE1 females expressing transgene-derived BCD1–489AR fail to repress cad mRNA translation (c) but activate anterior hb expression (e) and develop the normal head and thoracic pattern elements (g). For further details, see text.
Discussion
The results show that BCD can associate with cap-associated eIF4E in vitro and that the eIF4E-binding motif of BCD is necessary for BCD-dependent translational repression of cad mRNA in the embryo. These findings suggest a repression mechanism in which BCD blocks the eIF4G ∷ eIF4E interaction necessary for the initiation of cap-dependent cad mRNA translation. Because no interaction between recombinant eIF4E and BCD could be detected in the absence of cad mRNA, we conclude that the binding of BCD to the cad 3′-UTR is most likely a prerequisite for their interaction. This interpretation is consistent with previous findings where a mutant BCD, which specifically lacks its ability to bind cad mRNA, was also unable to repress translation (Niessing et al. 2000).
BCD-dependent control of translation of cad mRNA is likely to function in a manner similar to BP1, BP2, and Maskin (for review, see Raught et al. 2000; Richter 2000). However, despite the intriguing similarities among BP1/BP2, Maskin, and BCD, the modes of how they exert translational repression are distinct (Fig. 4). BP1 and BP2 are part of a general mRNA repression system, which blocks eIF4E ∷ eIF4G interaction in a reversible, cell-growth-dependent manner in response to insulin receptor signaling (Fig. 4a; for review, see Raught et al. 2000; Sachs and Varani 2000). In contrast, Maskin represses translation in an mRNA-specific manner. It binds to the cytoplasmic polyadenylation element-binding protein (CPEB), a factor that interacts with a short uridine-rich cytoplasmic polyadenylation element (CPE) of cyclin B mRNA. CPEB-tethered Maskin acts from the 3′-end of specific mRNAs by binding to eIF4E and blocking an association of eIF4E and eIF4G (Fig. 4b; for review, see Richter 2000; Richter and Theurkauf 2001). In this mode of repression, target specificity of repression is provided by the interaction of CPEB with the CPE, whereas the repression of translation at the 5′-end is executed by Maskin. BCD uses a strategy that combines these two features of CPEB and Maskin. Its homeodomain directly binds to the BCD response element (BRE) in the 3′-UTR of cad mRNA (Dubnau and Struhl 1996; Rivera-Pomar et al. 1996) and provides also a direct link to the 5′-cap-bound complex involving the eIF4E-interaction motif.
Figure 4.
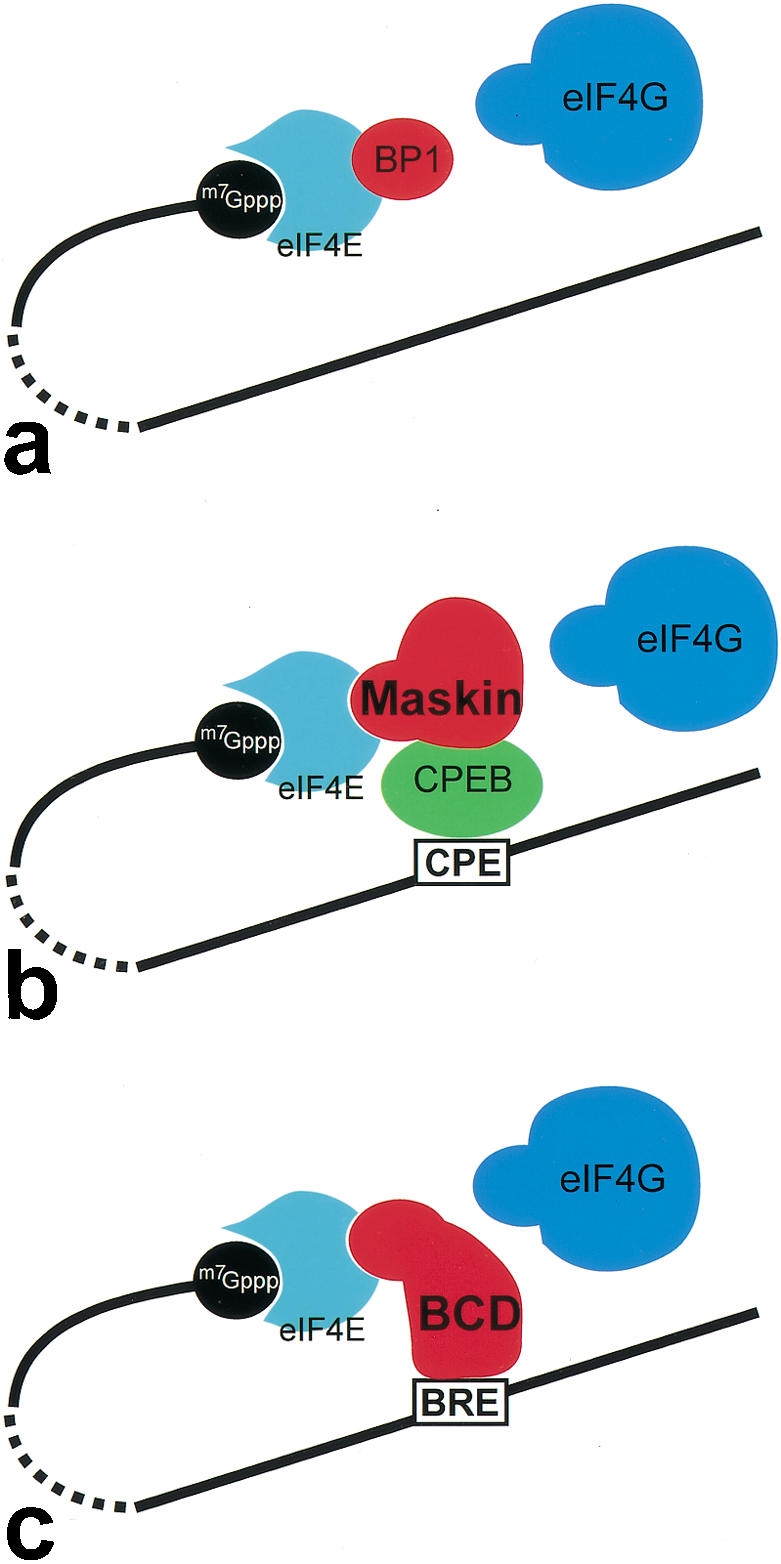
Different modes of cap-dependent translational repression by interference with the assembly of the eIF4E ∷ eIF4G interaction complex. (a) Binding of BP to eIF4E blocks eIF4G-binding and modulates translation efficiency of 5′-capped mRNAs in an insulin signaling-dependent manner (for review, see Raught et al. 2000; Sachs and Varani 2000). (b) Translational repression of mRNAs, which anchor CPEB through a CP element in their 3′-UTR. CPEB is able to associate with Maskin, which successively blocks the eIF4E ∷ eIF4G interaction by binding to eIF4E (for review, see Richter 2000; Richter and Theurkauf 2001). (c) BCD uses a similar strategy of repression by combining the binding properties of both CPEB and Maskin. BCD binds directly to the BRE in the 3′-UTR and blocks the eIF4E ∷ eIF4G interaction at the 5′-end. In each case (a–c), the eIF4E interaction involves the YxxxxL motif of the translational repressors.
The simplest model to account for BCD-dependent repression of translation therefore involves three essential steps, which are (1) target recognition by binding to the specific target site within the 3′-UTR, a process mediated by BCD's arginine-rich RNA-binding motif in the homeodomain (Niessing et al. 2000), (2) looping of cad mRNA to allow for interaction of the 3′-UTR-bound BCD with 5′-cap-bound eIF4E, which (3) causes a BP1/BP2-like blocking of the eIF4G-binding site on eIF4E (Fig. 4c) to prevent the assembly of a functional translation initiation complex. The mode of BCD-dependent repression of translation, therefore, combines the strategy of target-specific binding to 3′-UTRs as shown for a number of other translational repressors (Curtis et al. 1995; Hake and Richter 1997; for review, see Wickens et al. 2000) with a repression mechanism known from growth regulation (for review, see Raught et al. 2000; Sachs and Varani 2000) and cyclin B-dependent cell cycle regulation (for review, see Richter 2000; Richter and Theurkauf 2001).
Materials and methods
Binding assays
About 300 μL of embryos (0–3 h) was homogenized with three strokes in 1.5 mL of buffer A (0.1 M KCl, 20 mM HEPES at pH 7.5, 0.2 mM EDTA, protease inhibitor cocktail; Boehringer Mannheim; 2-ml douncer, 4°C; Edery et al. 1988). The homogenate was centrifuged (microcentrifuge, full speed; 4°C), and the supernatant was transferred into a new vial. Centrifugation was repeated twice, and the aqueous phase was then transferred into a 15-mL falcon tube containing 5 mL of buffer A and 0.5 mL of m7GTP-sepharose (Pharmacia). After incubation (1 h with slight agitation at 4°C), resins were washed 6× with 5 mL of buffer A + 100 μM GDP and centrifugation steps in between following a standard protocol (Edery et al. 1988). Proteins bound to m7GTP-sepharose were eluted (two incubations in 1 mL of buffer A containing 100 μM m7GTP; 5 min each), and samples were concentrated and subsequently separated by SDS-PAGE (10% gel), followed by silver-staining or Western-blotting (monoclonal anti-BCD antibody, dilution 1:50, and secondary goat anti-mouse antibodies, dilution 1:2000 or rabbit anti-GFP, Santa Cruz, dilution 1:500, and secondary goat anti-rabbit antibodies, dilution 1:2000). Secondary antibodies were peroxidase-coupled. Protein was visualized with SuperSignal Chemiluminescent Substrate (Pierce).
For the peptide competition assay, 600 μL of m7GTP-beads was incubated with 750 μL of recombinant Drosophila eIF4E-ΔN-term (Marcotrigiano et al. 1999; 0.29 μg/μL) at 4°C for 2 h. Subsequently, the resin with bound 4E was washed 3× with 5 mL of buffer and incubated with 4 mL of lysate derived from 0–3-h embryos in buffer A. After incubation at 4°C for 2 h with slight agitation, the resin was washed 3× with buffer A and afterward split into three parts, which were incubated 3× at 4°C for 10 min under slight agitation either with buffer A containing 100 μM GDP, or with buffer A containing 100 μM GDP and 1 mM human BP1 peptide (STTPGGTRIIYDRKFLMECRNSPVTKT) or the mutant version (STTPGGTRIIADRKFRMECRNSPVTKT) of it. Proteins bound to the resin were subsequently eluted (two incubations in 200 μL of buffer A containing 500 μM m7GTP; 5 min under slight agitation), concentrated, and separated by SDS-PAGE (12% gel), followed by Western-blotting (monoclonal anti-BCD, dilution 1:50, and secondary biotinylated rat anti-mouse antibodies, dilution 1:1000). Proteins were visualized using the AP-conjugated Vectastain ABC-kit (Vector Laboratories).
In vitro binding assays were carried out with 100 μL of m7GTP-beads. They were precoupled with 50 μL of recombinant Drosophila eIF4E-ΔN-term (0.29 μg/μL) and blocked with 5 μL of purified BSA (100 μg/μL) at 4°C for 2 h. Subsequently, the resin with bound eIF4E was washed 3 times with 1 mL of buffer A. Full-length BCD and BCD1–489AR were in vitro translated and 35S-labeled using the TNT T7 Quick Coupled Transcription/Translation System (PROMEGA) and the Pro Mix 35S-methionine/cysteine in vitro cell-labeling-mix (Amersham Biosciences) according to the manufacturers' instructions. Of the 50 μL of in vitro translation mix, 10 μL was preincubated with ∼20 μg of in vitro transcribed cad 3′-UTR mRNA including the BCD response element (Dubnau and Struhl 1996; Rivera-Pomera et al. 1996) at 4°C for 1 h. This preincubation mix was incubated with the washed eIF4E-coupled m7GTP-beads, incubated at 4°C for 2 h, and afterward washed 5× for 10 min with buffer A. Subsequently, eIF4E and associated proteins were specifically eluted in 50 μL of buffer A containing 500 μM m7GTP (20 min on ice under slight agitation). Eluted proteins as well as 3 μL of the in vitro translation input were analyzed by SDS-PAGE followed by autoradiography.
DNA encoding BCD1–489AR was generated by PCR-based mutagenesis involving a full-size BCD cDNA clone as described in Niessing et al. (1999), and the sequence was verified by DNA sequencing.
Transgenes and mutant analysis
In vitro mutagenized bcd cDNAs (QuikChange kit, Stratagene) were cloned into the P-element-based pCaSpeR vector DNA bearing the nanos 5′ sequences and the bcd 3′-UTR (Niessing et al. 1999). Several transgenic lines were established by P-element-mediated germ-line transformation as described (Niessing et al. 1999, 2000) and crossed to bcdE1 mutants. Transgene-dependent zygotic hb expression and Cad gradient formation were monitored by whole mount in situ hybridization (Tautz and Pfeifle 1989; Klingler and Gergen 1993) and antibody staining (Niessing et al. 1999, 2000) using guinea-pig anti-Cad antibodies (dilution 1:300) and goat anti-guinea-pig Cy3-labeled antibodies (dilution 1:500; after preabsorption). Larval cuticles were prepared and analyzed as described (Martinez Arias 1993).
Acknowledgments
We thank H. Taubert, G. Dowe, W. Gerber, and E.A. Wimmer for various important contributions; J. Reinitz and D. Kosman for their generous gift of antibodies; and J. Marcotrigiano for providing eIF4E cDNA and protein. We thank S.K. Burley for helpful discussions and for his support, T. Hazelrigg for providing the GFP–BCD-expressing fly strain, and J. Wehland for the synthesis of peptides. The work was supported by the Max-Planck-Society and the Fonds der Chemischen Industrie (H.J.). D.N. was a fellow of the Boehringer Ingelheim Fonds and holds an HFSP fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hjaeckl@gwdg.de; FAX 49-551-201-1755.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.240002.
References
- Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nüsslein-Vollhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Lehmann R, Zamore PD. Translational regulation in development. Cell. 1995;81:171–178. doi: 10.1016/0092-8674(95)90325-9. [DOI] [PubMed] [Google Scholar]
- Driever W. Maternal control of anterior development in the Drosophila embryo. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 301–324. [Google Scholar]
- Driever W, Nüsslein-Volhard C. The Bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- Driever W, Ma J, Nüsslein-Volhard C, Ptashne M. Rescue of bicoid mutant Drosophila embryos by bicoid fusion proteins containing heterologous activating sequences. Nature. 1989;342:149–154. doi: 10.1038/342149a0. [DOI] [PubMed] [Google Scholar]
- Dubnau J, Struhl G. RNA recognition and translational regulation by a homeodomain protein. Nature. 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- Edery I, Altmann M, Sonenberg N. High level synthesis in Escherichia coli of functional cap-binding eukaryotic initiation factor eIF4E and affinity purification using a simplified cap-analog resin. Gene. 1988;74:517–525. doi: 10.1016/0378-1119(88)90184-9. [DOI] [PubMed] [Google Scholar]
- Frohnhöfer HG, Nüsslein-Volhard C. Organization of the pattern in the Drosophila embryo by the maternal gene bicoid. Nature. 1986;324:120–125. [Google Scholar]
- Hake LE, Richter JD. Translational regulation of maternal mRNA. Biochim Biophys Acta. 1997;1332:M31–M38. doi: 10.1016/s0304-419x(96)00039-x. [DOI] [PubMed] [Google Scholar]
- Hazelrigg T, Liu N, Hong Y, Wang S. GFP expression in Drosophila tissues: Time requirement for formation of a fluorescent product. Dev Biol. 1998;199:245–249. doi: 10.1006/dbio.1998.8922. [DOI] [PubMed] [Google Scholar]
- Klingler M, Gergen P. Regulation of runt transcription by Drosophila segmentation genes. Mech Dev. 1993;43:3–19. doi: 10.1016/0925-4773(93)90019-t. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, Gringras A-C, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- Martinez Arias A. Development and patterning of the larval epidermis of Drosophila. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 517–608. [Google Scholar]
- Merrick WC, Hershey JWB. Pathway and mechanism of initiation of protein synthesis. In: Hershey JWB, et al., editors. Translational control of gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 33–88. [Google Scholar]
- Miron M, Verdu J, Lachance PED, Birnbaum MJ, Lasko PF, Sonenberg N. Translational inhibitor 4E-BP is an effector of PI(3)K/Act signalling and cell growth in Drosophila. Nat Cell Biol. 2001;3:596–601. doi: 10.1038/35078571. [DOI] [PubMed] [Google Scholar]
- Niessing D, Dostatni N, Jäckle H, Rivera-Pomar R. Sequence interval within the PEST motif of Bicoid is important for translational repression of caudal mRNA in the anterior region of the Drosophila embryo. EMBO J. 1999;18:1966–1973. doi: 10.1093/emboj/18.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessing D, Driever W, Sprenger F, Taubert H, Jäckle H, Rivera-Pomar R. Homeodomain position 54 specifies transcriptional versus translational control by Bicoid. Mol Cell. 2000;5:395–401. doi: 10.1016/s1097-2765(00)80434-7. [DOI] [PubMed] [Google Scholar]
- Ptushkina M, von der Haar T, Karim MM, Hughes JM, McCarthy JE. Repressor binding to a dorsal regulatory site trap human eIF4E in a high cap-affinity state. EMBO J. 1999;18:4068–4075. doi: 10.1093/emboj/18.14.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Dostie J, Sonenberg N. Suppression of cap-dependent translation in mitosis. Genes & Dev. 2001;15:2083–2093. doi: 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Gingras A-C, Sonenberg N. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In: Hershey JWB, et al., editors. Translational control of gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 245–294. [Google Scholar]
- Richter JD. Influence of polyadenylation-induced translation on metazoan development and neuronal synaptic function. In: Hershey JWB, et al., editors. Translational control of gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 785–806. [Google Scholar]
- Richter JD, Theurkauf WE. Development. The message is in the translation. Science. 2001;293:60–62. doi: 10.1126/science.1062237. [DOI] [PubMed] [Google Scholar]
- Rivera-Pomar R, Jäckle H. From gradients to stripes in the Drosophila embryogenesis. Trends Genet. 1996;12:478–483. doi: 10.1016/0168-9525(96)10044-5. [DOI] [PubMed] [Google Scholar]
- Rivera-Pomar R, Niessing D, Schmidt Ott U, Gehring WJ, Jäckle H. RNA binding and translational suppression by bicoid. Nature. 1996;379:746–749. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- Sachs AB, Varani G. Eukaryotic translation initiation: There are (at least) two sides of every story. Nat Struct Biol. 2000;7:356–361. doi: 10.1038/75120. [DOI] [PubMed] [Google Scholar]
- Sauer F, Hansen SK, Tjian R. DNA-template and activator–coactivator requirements for transcriptional synergism by Drosophila bicoid. Science. 1995;270:1825–1828. doi: 10.1126/science.270.5243.1825. [DOI] [PubMed] [Google Scholar]
- Schaeffer V, Janody F, Loss C, Desplan C, Wimmer EA. Bicoid functions without its TATA-binding protein associated factor interaction domains. Proc Natl Acad Sci. 1999;96:4461–4466. doi: 10.1073/pnas.96.8.4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin AJ. Capping of eucaryotic mRNAs. Cell. 1976;9:645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- St. Johnston D, Nüsslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992;68:201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- Struhl G, Struhl K, Macdonald PM. The gradient morphogen bicoid is a concentration dependent transcriptional activator. Cell. 1989;57:1259–1273. doi: 10.1016/0092-8674(89)90062-7. [DOI] [PubMed] [Google Scholar]
- Tautz D. Regulation of the Drosophila segmentation gene hunchback by two maternal morphogenetic centres. Nature. 1988;332:281–284. doi: 10.1038/332281a0. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the location of specific RNAs in Drosophila embryos reveals a translational control of segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Wickens M, Goodwin EB, Kimble J, Strickland S, Hentze M. Translational control of developmental decisions. In: Hershey JWB, et al., editors. Translational control of gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 295–370. [Google Scholar]