Abstract
Aims
The aim was to develop a fast detection method for the polymorphic alleles related to impaired CYP2C9-mediated metabolism.
Methods
CYP2C9 genotypes were identified in 118 DNA samples using real-time PCR (LightCycler) followed by melting curve analysis. All samples were re-tested by validated PCR-RFLP methodology.
Results
The concordance between the two methods was 100% for two variant alleles. The frequencies of CYP2C9*1 (wild type), CYP2C9*2 and CYP2C9*3 (with 95% confidence intervals) were 0.81(0.05), 0.14(0.04) and 0.05(0.03), respectively, and are similar to those observed in other Caucasian populations.
Conclusions
This assay is simple and rapid and may be used for CYP2C9-genotyping in a clinical setting.
Keywords: CYP2C9, ethnic differences, genetic polymorphism, LightCycler
Introduction
CYP2C9, a major isoform of the cytochrome P450 in humans, is responsible for the oxidation of a wide array of drugs, including S-warfarin, phenytoin, tolbutamide, losartan and torsemide. Several nonsteroidal anti-inflammatory drugs (NSAIDs), including diclofenac, naproxen, piroxicam and ibuprofen, as well as the selective COX-2 inhibitor celecoxib are also metabolized mainly by the CYP2C9.
CYP2C9 displays a functional genetic polymorphism, where two alleles, CYP2C9*2 and CYP2C9*3, have been related unambiguously to impaired CYP2C9-mediated metabolism. The CYP2C9*2 allele, which is derived from a C430→T transversion in exon 3, encodes the Arg144 Cys substitution [1], whereas CYP2C9*3 with a A1075→C mutation in exon 7 results in the Ile359 Leu change [2].
Since there is evidence that the genetic polymorphism of CYP2C9 is associated with the increased toxicity of CYP2C9 substrates (e.g. warfarin [3] and phenytoin [4], a clinically applicable test for the functional CYP2C9 variants is needed. Although numerous detection methods for CYP2C9 polymorphisms have been developed recently, their ‘high-throughput’ application in a clinical setting is limited because of the labour-intensive nature of the methodology. Here, we describe a rapid single-step automated assay for detection of CYP2C9*1, CYP2C9*2 and CYP2C9*3 alleles using a real-time fluorescence polymerase chain reaction (PCR), which may be easily implemented as a high-throughput genotyping method into clinical practice.
Methods
DNA isolation
DNA was isolated from blood (200 µl) of 118 unrelated Caucasian subjects using a High Pure PCR Template Preparation Kit (Roche, Mannheim, Germany). All participants gave informed consent and the study protocol was approved by the ethics committee of the Medical Faculty of University of Frankfurt.
Real-time PCR
Real-time fluorescence PCR was performed on a LightCycler (Roche, Mannheim, Germany) using fluorogenic hybridization probes (TIB MOLBIOL, Berlin, Germany) and a LightCycler–FastStartDNA Master Hybridization Kit (Roche). Hybridization of the probes to the target DNA results in fluorescence resonance energy transfer (FRET) between two fluorophores. During melting of the final product, the sequence alteration is detected as a change in the melting temperature of the sensor probe. The sequences of primers and hybridization probes together with the PCR conditions are summarized in Table 1.
Table 1.
Real-time PCR primers, hybridization probes and LightCycler conditions for the genotyping of the CYP2C9*2 and CYP2C9*3 variant alleles.
| Variant allele | Reagents | Per sample |
|---|---|---|
| CYP2C9*2/CYP2C9*1 | PCR primer | |
| 5′- CACTGGCTGAAAGAGCTAACAGAG −3′a | 0.5 µm | |
| 5′- GTGATATGGAGTAGGGTCACCCAC −3′a | 0.5 µm | |
| Hybridization probes | ||
| 5′- CATTGAGGACCGTGTTCAAGAGGAA- FL−3′(sensor) | 0.2 µm | |
| 5′- LCR640– CCCGCTGCCTTGTGGAGGAGTTGAG 3′(anchor) | 0.2 µm | |
| MgCl2 | 3 mm | |
| FastStartDNA | 2 µl | |
| Genomic DNA | 60 ng | |
| Initial denaturation 95 °C for 10 min | ||
| Amplification (45 cyles) | ||
| Denaturation 95 °C for 5 s | ||
| Annealing at 67 °C for 10 s | ||
| Extension at 72 °C for 15 s | ||
| Melting curve analysis by heating 0.2 °C s−1 from 40 °C to 85 °C. | ||
| CYP2C9*3/CYP2C9*1 | PCR primer | |
| 5′- AGGAAGAGATTGAACGTGTGA- 3′a | 0.5 µm | |
| 5′- CCTTGGGAATGAGATAGTTTCTGAA- 3′ | 0.5 µm | |
| Hybridization probes | ||
| 5′- CCAGAGATACCTTGACCTTC- FL−3′ (sensor) | 0.2 µm | |
| 5′- LCR640– CCCACCAGCCTGCCCCATGC −3′ (anchor) | 0.2 µm | |
| MgCl2 | 4 mm | |
| FastStartDNA | 2 µl | |
| Genomic DNA | 40 ng | |
| Initial denaturation 95 °C for 10 min | ||
| Amplification (50 cyles) | ||
| Denaturation 95 °C for 5 s | ||
| Annealing at 58 °C for 10 s | ||
| Extension at 72 °C for 5 s | ||
| Melting curve analysis heating 0.2 °C s−1 from 40 °C to 85 °C. |
The fluorescein-labelled (FL) oligonucleotide probes (sensor) laying over the polymorphism sites were designed to recognize the wild-type sequence in CYP2C9*2 assay and the mutated sequence in CYP2C9*3 assay.The acceptor hybridization probe (anchor) is labelled at the 5’-end with a LightCycler-Red fluorophore (LCR640).
The primers were adapted from PCR-RFLP method as described by Aynacioglu et al. [7].
PCR-RFLP
All samples were re-assayed using a validated polymerase chain reaction-based restriction length polymorphism (PCR-RFLP) method.
CYP2C9*2. PCR was performed according to the protocol described above. 8 µl of the PCR product (375 bp) was digested with 5 U of the endonuclease AvaII (New England Biolabs, Herts UK). After overnight incubation at 37 °C, the digested products were separated by electrophoresis using a 3% agarose gel. The presence of the wild-type allele (CYP2C9*1) created an AvaII restriction site, producing 296 bp and 79 bp fragments, whereas the variant allele CYPC9*2 was not digested.
CYP2C9*3. The genotyping for CYP2C9*3 was performed according to the RFLP-PCR method previously described by Sullivan-Klose et al. [2] Briefly, two separate mismatched forward PCR primers (AATAATAATATGCACGAGGTCCAGAGATG(GTA)C) and an intron 7 reverse primer (GATACTATGAATTTGGGACTTC) were required to introduce a NsiI site into the CYP2C9*1 allele and a KpnI site into the CYP2C9*3 allele, respectively. The amplified products (141 bp) were digested with 5 U of either NsiI or KpnI (New England Biolabs). After overnight incubation at 37 °C, the cleavage products (112 + 29 bp by CYP2C9*1 and 111 + 30 bp by CYP2C9*3) were separated on a 3% agarose gel.
Results
Figure 1 shows the results of the melting curve analysis performed for the different CYP2C9*2 genotypes. In the presence of the wild-type DNA, hybridization probes form an exact match with the target, resulting in a higher stability of the complex and the highest melting peak temperature Tm (65.6 °C). A lower Tm was observed in case of the mismatch C430→T (58.5 °C), denoting the CYP2C9*2 variant. The heterozygous samples showed two distinct melting peaks at Tm of 58.5 °C and 65.6 °C. The temperature shift of about 7 °C was sufficient to allow unambiguous assignment of genotype.
Figure 1.
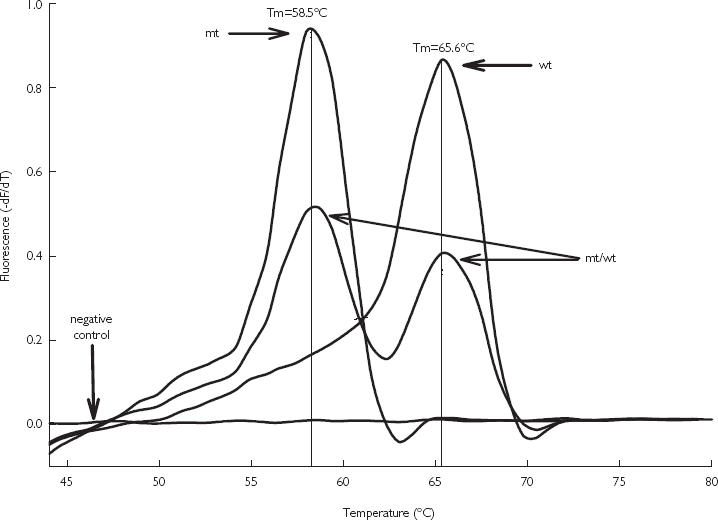
Melting curve analysis of the CYP2C9*2 variant in the different CYP2C9 genotypes. For easier analysis of the melting peaks, the melting temperature curve is plotted as the first negative derivative of the fluorescence vs temperature (– dF/dT). In the presence of wild-type DNA (wt), hybridization probes form an exact match with the target, resulting in higher stability of the complex and the highest Tm (65.6 °C). A lower Tm was observed in case of the mismatch (58.5 °C), denoting the CYP2C9*2 variant (mt). The heterozygous samples (mt/wt) showed two distinct melting peaks at Tm of 58.5 °C and 65.6 °C. The negative control without template shows no fluorescent signal.
Similarly, a single base mismatch A1075→C in the CYP2C9*3 allele resulted in a melting temperature shift from 52 °C to 62 °C. Since the sensor probe for CYP2C9*3 genotyping was designed to hybridize the mutated sequence, the hybridized variant allele was characterized by higher Tm than the wild-type allele.
The results of the real-time PCR analysis were compared with the validated PCR-RFLP method. In 118 samples studied, the concordance rate was 100% for both polymorphic sites. The frequencies (± 95% confidence limits) of the alleles CYP2C9*1, CYP2C9*2 and CYP2C9*3 were 0.81 ± 0.05, 0.14 ± 0.04 and 0.05 ± 0.03, respectively. Only one subject was homozygous for the CYP2C9*2 allele, and no homozygotes for the CYP2C9*3 allele were detected. Genotyping has revealed that one subject was heterozygous for both mutations, which was interpreted as the CYP2C9*2/*3 genotype, since no evidence for a linkage between CYP2C9*2 and CYP2C9*3 on the same chromosome exists [5]. The distribution of the CYP2C9 genotypes was in agreement with Hardy–Weinberg equation, where the expected genotype frequencies lay within the 95% confidence interval of those found. Since no individuals homozygous for CYP2C9*3 were detected amongst the subjects studied, the ability of the method to identify this genotype was confirmed by analysing the DNA from a CYP2C9*3 homozygote obtained from another laboratory (courtesy of Professor J. Brockmoeller).
Discussion
Polymorphisms in CYP2C9 have the potential to increase the toxicity of CYP2C9-metabolized drugs, especially those with low therapeutic indices [3, 6]. However, data on functional relevance of the CYP2C9 polymorphism are incomplete, partly because of the limitations of the genetic tests available. Numerous detection methods for CYP2C9 polymorphisms have been recently developed, including PCR-RFLP protocols [2, 7], allele-specific PCR [8] and single-strand conformation polymorphism (SSCP) [5]. However, in all of these methods, processing of samples and analysis of the separated gene products require several labour-intensive manual steps.
In this paper, we describe a rapid single-step method for the genotyping of CYP2C9 using real-time fluorescence PCR. One of the specific features of the method is the rapid processing of samples (30 min) due to the combined amplification and detection steps. Furthermore, the assay does not require post-PCR electrophoresis and is therefore specific because it is unaffected by the formation of spurious PCR amplification products. In contrast to previous assays, which depend on the analysis of ethidium bromide-stained gels, the amplified products are identified by the melting peak allowing more reliable distinction between genotypes. These characteristics may be particularly suitable in the clinical setting where high-throughput diagnostic procedures are required.
Since the occurrence of the CYP2C9 polymorphism seems to be strongly dependent on ethnic origin, we compared the frequencies of the CYP2C9 variants in a German population with these other ethnic groups. As expected, the allele frequencies in German subjects corresponded to those previously estimated for other Northern Europeans [9–11]. In contrast to Caucasians, who are characterized by the highest degree of genetic variability in CYP2C9, CYP2C9 polymorphic variants are less common in Asians, suggesting that the mutations might have evolved quite recently, after the separation of the Caucasian and Asian racial groups.
In conclusion, we have developed a rapid single-step genotyping assay for the identification of two functionally significant CYP2C9 allelic variants using a real-time fluorescence polymerase chain reaction which is applicable as a high-throughput assay in the clinical setting.
Acknowledgments
We thank Professor Juergen Brockmoeller and Dr Julia Kirchheiner for providing us with a reference DNA sample (CYP2C9*3-homozygote). We also thank TIB MOLBIOL, Berlin, Germany for construction of the hybridization probes and excellent technical support. The work was supported by the Deutsche Forschungsgemeinschaft DFG GE 695/1–2.
References
- 1.Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics. 1994;4:39–42. doi: 10.1097/00008571-199402000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan-Klose TH, Ghanayem BI, Bell DA, et al. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6:341–349. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Steward DJ, Haining RL, Henne KR, et al. Genetic association between sensitivity to warfarin and expression of CYP2C9*3. Pharmacogenetics. 1997;7:361–367. doi: 10.1097/00008571-199710000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Ninomiya H, Mamiya K, Matsuo S, Ieiri I, Higuchi S, Tashiro N. Genetic polymorphism of the CYP2C subfamily and excessive serum phenytoin concentration with central nervous system intoxication. Ther Drug Monit. 2000;22:230–232. doi: 10.1097/00007691-200004000-00016. [DOI] [PubMed] [Google Scholar]
- 5.Stubbins MJ, Harries LW, Smith G, Tarbit MH, Wolf CR. Genetic analysis of the human cytochrome P450 CYP2C9 locus. Pharmacogenetics. 1996;6:429–439. doi: 10.1097/00008571-199610000-00007. [DOI] [PubMed] [Google Scholar]
- 6.van der Weide J, Steijns LS, van Weelden MJ, de Haan K. The effect of genetic polymorphism of cytochrome P450 CYP2C9 on phenytoin dose requirement. Pharmacogenetics. 2001;11:287–291. doi: 10.1097/00008571-200106000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Aynacioglu AS, Brockmoller J, Bauer S, et al. Frequency of cytochrome P450 CYP2C9 variants in a Turkish population and functional relevance for phenytoin. Br J Clin Pharmacol. 1999;48:409–415. doi: 10.1046/j.1365-2125.1999.00012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhasker CR, Miners JO, Coulter S, Birkett DJ. Allelic and functional variability of cytochrome P4502C9. Pharmacogenetics. 1997;7:51–58. doi: 10.1097/00008571-199702000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Scordo MG, Aklillu E, Yasar U, Dahl ML, Spina E, Ingelman-Sundberg M. Genetic polymorphism of cytochrome P460 2C9 in a Caucasian and a black African population. Br J Clin Pharmacol. 2001;52:447–50. doi: 10.1046/j.0306-5251.2001.01460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Martin E, Martinez C, Ladero JM, Gamito FJ, Agundez JA. High frequency of mutations related to impaired CYP2C9 metabolism in a Caucasian population. Eur J Clin Pharmacol. 2001;57:47–9. doi: 10.1007/s002280100264. [DOI] [PubMed] [Google Scholar]
- 11.Yoon YR, Shon JH, Kim MK, Lim YC, Lee HR, Park JY, et al. Frequency of cytochrome P450 2C9 mutant alleles in a Korean population. Br J Clin Pharmacol. 2001;51:277–80. doi: 10.1046/j.1365-2125.2001.00340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


