Abstract
Although the vertebrate embryonic midline plays a critical role in determining the left/right asymmetric development of multiple organs, few genes expressed in the midline are known to function specifically in establishing laterality patterning. Here we show that a gene encoding protein disulfide isomerase P5 (PDI-P5) is expressed at high levels in the organizer and axial mesoderm and is required for establishing left/right asymmetries in the zebrafish embryo. pdi-p5 was discovered in a screen to detect genes down-regulated in the zebrafish midline mutant one-eyed pinhead and expressed predominantly in midline tissues of wild-type embryos. Depletion of the pdi-p5 product with morpholino antisense oligonucleotides results in loss of the asymmetric development of the heart, liver, pancreas, and gut. In addition, PDI-P5 depletion results in bilateral expression of all genes known to be expressed asymmetrically in the lateral plate mesoderm and the brain during embryogenesis. The laterality defects caused by pdi-p5 antisense treatment arise solely due to loss of the PDI-P5 protein, as they are reversed when treated embryos are supplied with an exogenous source of the PDI-P5 protein. Thus the spectrum of laterality defects resulting from depletion of the PDI-P5 protein fully recapitulates that resulting from loss of the midline. As loss of PDI-P5 does not appear to interfere with other aspects of midline development or function, we propose that PDI-P5 is specifically involved in the production of midline-derived signals required to establish left/right asymmetry.
Keywords: PDI, laterality patterning, midline, heart morphogenesis
Despite an external appearance of bilateral symmetry, the internal organs of vertebrate animals, including the viscera and the brain, exhibit asymmetries in morphological structure (chirality), location (situs), or both (Fujinaga 1997; Kosaki and Casey 1998; Concha et al. 2000; Liang et al. 2000). In many cases asymmetry in organ formation is preceded during somitogenesis by asymmetric expression of genes in the regions from which the organ anlage arise. Left-side expression in the lateral plate mesoderm (LPM) and diencephalon of genes that encode signaling molecules (e.g., nodal or lefty) and transcription factors (e.g., pitx2) correlates with and has been functionally linked to later morphogenetic events, such as heart looping, that result in overt organ asymmetries (for review, see Yost 1998; Burdine and Schier 2000; Capdevila et al. 2000).
Both asymmetric gene expression and subsequent asymmetric morphogenetic processes are regulated by the embryonic midline (Goldstein et al. 1998; Yost 1998; Roessler and Muenke 2001). Surgical removal of midline tissues in chick, Xenopus, or zebrafish (Danos and Yost 1996; Lohr et al. 1997; Concha et al. 2000) or deletion of midline tissues in embryonic zebrafish or mouse mutants (Danos and Yost 1996; Chen et al. 1997; Dufort et al. 1998; King et al. 1998; Melloy et al. 1998; Izraeli et al. 1999; Bisgrove et al. 2000; Chin et al. 2000) leads to bilateral expression of nodal, lefty, and pitx2 and produces laterality defects in the formation of the heart, gut, and brain. The bilateral gene expression resulting from loss of midline structures has been interpreted as arising from the loss of a midline-produced barrier to signals that promote left-side gene expression and/or the loss of signals that inhibit right-side gene expression. The finding that left-side gene expression is often absent in the right-side partner of conjoined twins lends support to the idea that the midline actively produces a signal that antagonizes gene expression on the right side (Levin et al. 1996; Nascone and Mercola 1997).
Although many mutations with pleiotropic effects on midline development affect the establishment of left-right asymmetries, few midline-expressed genes have been linked specifically to the midline signaling process. Loss-of-function of mouse lefty1, which is expressed in the left lateral portion of the floorplate, leads to bilateral gene expression of nodal and lefty2 in the LPM and to left lung isomerism, but does not significantly perturb other features of midline development (Meno et al. 1998). As Lefty factors function as antagonists of Nodal signaling (Bisgrove et al. 1999; Meno et al. 1999), the phenotype of the mutant is consistent with evidence indicating that Nodal signaling is an important determinant of left/right asymmetries (for review, see Burdine and Schier 2000; Roessler and Muenke 2001). In contrast, it is difficult to distinguish between the direct and indirect effects of other genes, such as sonic hedgehog, SIL, and no turning, that appear to be required for both midline development and laterality patterning (Chiang et al. 1996; Melloy et al. 1998; Izraeli et al. 1999; Tsukui et al. 1999; Hamada et al. 2002). It is likely that additional midline genes that function specifically to regulate the midline-derived signals essential for left/right patterning are yet to be discovered.
To discover novel genes expressed in the embryonic midline of the zebrafish that might function in establishing laterality patterning, we developed a screen to detect genes down-regulated in zygotic one-eyed pinhead (oep) mutant embryos, which exhibit multiple deficiencies in midline development yet form a morphologically normal notochord (Schier et al. 1997). Using subtraction hybridization methods to generate a cDNA population enriched for gene sequences expressed in wild-type but not mutant embryos at the onset of midline tissue differentiation, followed by hybridization screening of a cDNA array, we identified genes expressed in midline tissues of the zebrafish. Functional analysis of one of these genes, which encodes protein disulfide isomerase P5 (PDI-P5), demonstrated that it is specifically involved in laterality patterning. Depletion of the PDI-P5 protein with antisense morpholino oligonucleotides (Summerton and Weller 1997; Nasevicius and Ekker 2000) revealed that pdi-p5 is required for establishing asymmetric gene expression in the LPM and the brain and for regulating definitive morphological asymmetries in the development of the heart, liver, and pancreas. Interference with pdi-p5 function did not perturb other detectable aspects of midline development or function, indicating that pdi-p5 has a specific role in laterality patterning. Our results demonstrate that zebrafish developmental mutants can be harnessed effectively for gene discovery and the functional dissection of developmental pathways.
Results
pdi-p5 is expressed in midline tissues of gastrula-stage embryos
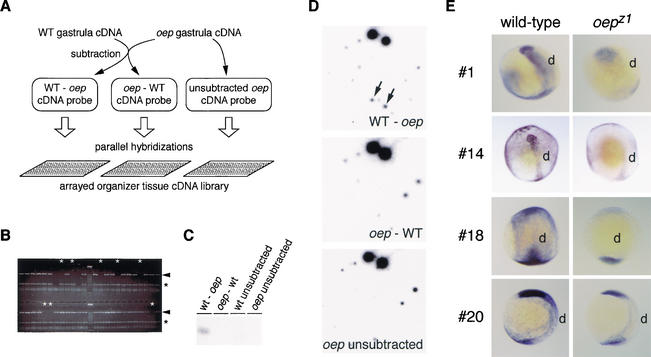
Zygotic one-eyed pinhead mutants have diminished Nodal signaling, resulting in deficiencies in the development of the gastrula organizer and the midline tissues derived from it, the axial mesoderm, and the floorplate of the nervous system (Schier et al. 1997; Schier and Shen 2000). We used the zebrafish oep mutant to identify novel midline-expressed genes that might function in the establishment of left/right asymmetry. An array of cDNA sequences expressed in the organizer was analyzed by differential hybridization screening for genes expressed in wild-type (WT) but not oep mutant embryos (Fig. 1A). mRNA was extracted from individually genotyped midgastrula embryos (Fig. 1B), and subtractive hybridization between WT and mutant cDNA populations was used to generate probes enriched for WT-specific or mutant-specific sequences (Fig. 1C). Replicate samples of the arrayed library were screened (Fig. 1D), resulting in identification of four candidate sequences whose expression was significantly reduced in mutant embryos harboring either the oepz1 or oepm134 mutant alleles (Fig. 1E; data not shown). Detailed analysis of candidate gene #14, which is expressed prominently in the axial mesoderm of midgastrula WT embryos and significantly down-regulated in oep mutant embryos [29% (n = 134) progeny of oepz1/+ inter se matings; 22% (n = 59) progeny of oepm134/+ inter se matings], is described below.
Figure 1.
Identification of genes dependent on oep gene function. (A) Summary of screening strategy. cDNA populations from WT or oep mutant embryos were used to generate three cDNA probes: (1) WT-oep cDNA, enriched for sequences specific to the WT pool, was generated by subtractive hybridization using oep sequences as the subtractive driver; (2) oep-WT cDNA; and (3) unsubtracted oep cDNA served as control probes that detected sequences present in mutant embryos. Replicate filter sets of an arrayed organizer tissue cDNA library were hybridized with each probe, and candidate sequences were identified as those that hybridized only with the WT-oep cDNA probe. (B) To isolate RNA from oep mutant embryos prior to the expression of a distinguishing morphological phenotype, individual embryos from an oepz1/+ intercross were genotyped by PCR analysis to detect sequences removed by the oepz1 deletion mutation (arrowhead). White asterisks indicate oepz1/oepz1 DNA samples in which unlinked no tail sequences (black asterisks) but not oep-linked sequences could be detected. Following genotyping, RNA samples from mutant embryos were pooled. (C) Equal amounts of starting cDNA pools (WT unsubtracted and oep unsubtracted) or subtracted cDNA pools (WT-oep and oep-WT) were analyzed by Southern hybridization for presence of the organizer-specific gene, goosecoid (gsc). Only the WT-oep cDNA pool contained a detectable abundance of gsc, which is down-regulated in midgastrula oep mutant embryos. (D) An example of the results of differential hybridization screening. Hybridization of the three cDNA probes to one region of the arrayed library, containing ∼128 independent organizer gene sequences. Arrows indicate one gene sequence, spotted in duplicate, that hybridized solely with the WT-oep cDNA probe. In all, 23 unique sequences out of 5376 in the array hybridized with the WT-oep probe specifically. (E) Expression of four candidate oep-regulated genes in late gastrula embryos from oepz1/+ intercrosses. Candidates #1, #14, #18, and #20 are expressed at high levels in dorsal tissue (d) of late-gastrula stage WT embryos, but at markedly reduced levels in one-fourth of the embryos, presumed to be oepz1 mutants. Dorsolateral (#1, #14), dorsal (#18), or lateral (#20) views of embryos are shown. Candidate #1 encodes ribophorin I, a subunit of oligosaccharyltransferase (Kelleher et al. 1992), candidate #14 encodes protein disulfide isomerase-related protein p5 (see text), candidate #18 encodes chorein, a gene responsible for choera-acanthocytosis (Rampoldi et al. 2001; Ueno et al. 2001), and candidate #20 has weak homology to yeast tf2-transposon.
Candidate #14 encodes a member of the protein disulfide isomerase (PDI) superfamily (Freedman et al. 1994; Ferrari and Soling 1999), being most closely related to the P5 family of proteins (Fig. 2A). PDIs are known to facilitate posttranslational processing, and at least one member of this family, the Drosophila Windbeutel protein, has a specific developmental role in embryonic patterning (Konsolaki and Schupbach 1998). The maternally supplied pdi-p5 transcript (Fig. 2B, 1-cell stage) is initially distributed throughout the blastoderm of blastula-stage embryos. However, during the course of gastrulation the widespread presence of pdi-p5 transcripts fades whereas high levels of expression are maintained predominantly in the nascent embryonic shield and axial mesoderm (Fig. 2B, 70% epiboly and 90% epiboly). By the 6-somite stage, the gene is expressed specifically in the anterior prechordal plate and in the presumptive notochord tissue (Fig. 2B, 6-som). As the notochord differentiates in a rostral to caudal progression, expression in the notochord is extinguished anteriorly so that at the 18-somite stage, high levels of transcripts in the axial mesoderm are found only in the developing tail, where tissue differentiation is still not complete (Fig. 2B, 18-som). At this stage, pdi-p5 is also expressed in two midline tissues that lie directly above and below the notochord, the floorplate of the nervous system and the hypochord, respectively. As development proceeds over the next two days of embryogenesis, high levels of pdi-p5 expression can be detected in selected tissues, including the developing otic vesicles, the lateral line, the developing caudal and pectoral fins, the liver, and the pharyngeal cartilage (Fig. 2B, 18-som, 48 h, 60 h; data not shown).
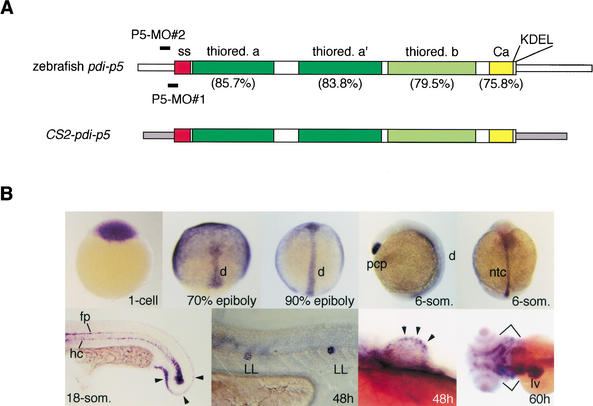
Figure 2.
Candidate #14 encodes zebrafish protein disulfide isomerase-p5. (A) Schematic representation of the zebrafish pdi-p5 mRNA. Narrow open boxes indicate 5′ and 3′UTR sequences. Wider internal boxes indicate coding sequences. Individual functional domains of the PDI (Ferrari and Soling 1999) are indicated: SS (red), signal sequence; thiored a and a‘ (dark green), potentially active thioredoxin-like domains; thiored. b (light green), likely inactive thioredoxin domain; Ca (yellow), potential Ca++-binding domain; KDEL, endoplasmic reticulum retention signal. Sequence identities between each of the zebrafish protein domains and the corresponding domains of the human PDI-P5 protein are indicated in parentheses. The overall identity between the zebrafish and human proteins is 76%. Two antisense morpholino oligonucleotides, P5-MO#1 and P5-MO#2, were used to analyze pdi-p5 function. P5-MO#1 is complementary to nucleotides −21nt to +5nt and P5-MO#2 is complementary to nucleotides –33 to −9nt with respect to the translation start site of pdi-p5 mRNA. The CS2-pdi-p5 chimeric mRNA, in which pdi-p5 coding sequences are flanked by 5′ and 3′ UTR sequences (narrow gray boxes) derived from the CS2+ vector, was designed to prevent antisense morpholinos from annealing the transcript. (B) Expression of pdi-p5 in WT zebrafish embryos. pdi-p5 transcripts are maternally supplied and widely distributed initially. During gastrulation, high levels of transcripts are present only in the axial mesoderm, including the prechordal plate and the presumptive notochord. At later developmental stages, pdi-p5 is expressed in the floorplate, hypochord, fin buds, lateral line, liver, and pharyngeal cartilage. Arrowheads indicate expression in the developing caudal fin (18-som) and pectoral fin bud (48 h). Brackets (60 h) indicate expression in the pharyngeal cartilage. d, dorsal; h, hours postfertilizaton; hc, hypochord; fp, floorplate; LL, lateral line; lv, liver; ntc, notochord; pcp, prechordal plate; som, somites.
Asymmetric morphogenesis and laterality patterning requires pdi-p5 function
To test the role of pdi-p5 in the early embryo, we analyzed the consequences of depleting the pdi-p5 gene product in vivo with antisense morpholino oligonucleotides, which bind stably to target mRNAs and can thereby inhibit translation (Summerton and Weller 1997; Summerton 1999). Two antisense oligonucleotides, P5-MO#1 and P5-MO#2, complementary to 5′UTR sequences near the initiation codon (Fig. 2A; Materials and Methods) and capable of inhibiting translation of synthetic pdi-p5 mRNA in vitro (Fig. 3A), were used in these experiments. The syndromes caused by each morpholino were identical.
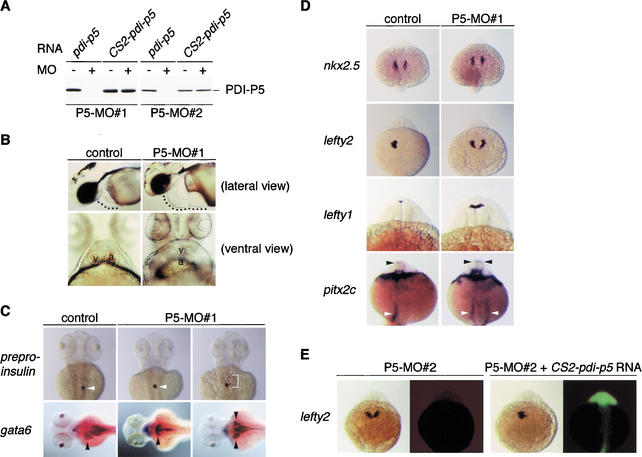
Figure 3.
Depletion of the PDI-P5 product causes laterality defects in organ morphogenesis and gene expression patterning. (A) The P5-MO#1 and P5-MO#2 morpholinos inhibit translation from pdi-p5 mRNA but not from CS2-pdi-p5 mRNA. pdi-p5 or CS2-pdi-p5 mRNA was used as a template for in vitro translation in the presence (+) or absence (−) of antisense morpholinos. (B) Injection of P5-MO#1 antisense morpholinos causes laterality defects affecting heart development. Following injection of 2 ng P5-MO#1 at the 1-cell stage, 2-d embryos exhibited pericardiac edema, an indicator of defective heart development (pericardial cavity is indicated). In a ventral view of a normal 2-d embryo, the ventricle (v) appears on the left side of the atrium (a). A similar view of a 2-d embryo that had been injected with P5-MO#1 shows that both chambers lie directly on the midline, with the ventricle anterior to the atrium. (C) P5-MO#1 antisense morpholinos produce laterality defects in pancreas and liver development. Expression of preproinsulin in pancreatic tissue (white arrowheads) and gata6 in liver (black arrowheads) visualized in dorsal views of 60-h embryos reveals that the pancreas normally develops on the right side and the liver develops on the left side of the midline in control embryos. In embryos injected at the 1-cell stage with the P5-MO#1 antisense morpholino, these organs usually developed directly on the midline, although isomerized development of the liver was also observed (gata6, right panel). In some antisense-treated embryos, pancreatic development was disrupted, producing fragments of preproinsulin-expressing tissue that developed near the midline (preproinsulin, right panel). (D) P5-MO#1 antisense morpholinos disrupt gene expression indicators of laterality patterning of the heart, brain, and gut. The initial development of the heart field, marked by nkx2.5 expression at the 18-somite stage, is unaffected in PDI-P5-depleted embryos. However, by the 22–24-somite stage, genes normally expressed only on the left side of tissue primordia are expressed bilaterally in most antisense-treated embryos. Left-side only expression of lefty2 in the heart primordium, lefty1 in the diencephalon, and pitx2c in the diencephalon (black arrowhead) and gut (white arrowhead) was observed in 22–24-somite-stage control embryos. Injection of the P5-MO#1 antisense morpholino resulted in bilateral expression of each of these genes, but did not alter their appropriate tissue-restricted patterns. All embryos shown in dorsal views. (E) The heart laterality defects caused by injection of pdi-p5 antisense morpholinos are due specifically to depletion of PDI-P5 product. Embryos were first injected with the P5-MO#2 morpholino, and a fraction of the antisense-treated embryos were subsequently injected with CS2-pdi-p5 mRNA together with fluorescent lineage tracer dye. All embryos were processed to reveal expression of lefty2 mRNA (left panel of each pair) and fluorescent lineage tracer dye (right panel of each pair). Embryos that received only the P5-MO#2 morpholino usually expressed lefty2 bilaterally in the LPM. Coinjection of the CS2-pdi-p5 mRNA resulted in a significant increase in the proportion of embryos that expressed lefty2 only on the left side of the LPM.
Injection of morpholino antisense oligonucleotides into WT embryos at the 1-cell stage consistently produced embryos with overall normal morphology but with pericardiac edema, a syndrome indicative of defective heart development (Fig. 3B, Table 1). The most overt defect in heart development observed following antisense treatment was the incorrect positioning of the heart chambers, which can be readily seen in ventral views of 48-h embryos (Fig. 3B). During heart morphogenesis, the posterior ventricular segment of the heart tube loops rightward (D-looping) so that the ventricle eventually comes to lie to the right of and slightly anterior to the atrium (Chen et al. 1997; Chin et al. 2000; Stainier 2001). Approximately 96% of the WT embryos used in the present study displayed clear evidence of D-looping, and the remaining 4% of embryos had hearts lying directly on the midline (O-looping; Table 1). In contrast, 67%–73% of the embryos injected with either the P5-MO#1 or P5-MO#2 oligonucleotide failed to exhibit any heart looping (Fig. 3B, Table 1). Among the remaining embryos, many displayed normal D-looping, but others exhibited L-looping (leftward) or failed to form a normal heart tube.
Table 1.
Heart development in pdi-p5 antisense morpholino-injected embryos
|
|
No.analyzed
|
No. with pericardiac edema
|
Cardiac looping
|
Defective heart formationa
|
|||
|---|---|---|---|---|---|---|---|
| D-loop
|
O-loop
|
L-loop
|
rudiments
|
cardia bifida
|
|||
| Morpholino injected
| |||||||
| None | 144 | 1 | 96% | 4% | 0 | 0 | 0 |
| P5-MO#1 | 377 | 359 | 19% | 67% | 5% | 6% | 3% |
| P5-MO#2 | 129 | 113 | 14% | 73% | 3% | 9% | 1% |
Morpholino oligonucleotides (2 ng P5-MO#1 or 10 ng P5-MO#2) were injected into the yolk of 1-cell embryos. Two-day embryos were visually inspected for formation of heart and direction of looping.
Some embryos exhibited heart defects that precluded ability to score looping. Defects included rudimentary heart formation and cardia bifida.
As defects in heart looping morphogenesis are commonly associated with heterotaxy (Goldstein et al. 1998; Kosaki and Casey 1998; Bisgrove and Yost 2001), we analyzed the development of additional organ systems in PDI-P5-depleted embryos. The pancreas and liver, which can be detected by virtue of expression of the preproinsulin and gata-6 genes, respectively, are both situated asymmetrically in most WT embryos—the pancreas to the right of the midline, and the liver to the left of the midline (Fig. 3C). PDI-P5 depletion caused a significant reduction in the fraction of embryos that displayed normal situs of these organs (Table 2). In most of the treated embryos, the pancreas and liver developed directly on the embryonic midline, although embryos exhibiting situs inversus or isomerization of the liver, in which the paired organs often appeared joined at the midline, were also observed (Fig. 3C). As was true for the heart, depletion of PDI-P5 protein sometimes interfered with development of the pancreas, resulting in the production of fragments of pancreatic tissue that were not scored for situs (Fig. 3C). In addition to the loss of asymmetric morphogenesis of the heart, pancreas, and liver, embryos depleted for PDI-P5 protein also exhibited laterality defects affecting development of the gut. Normal morphogenesis, or coiling, of the gut is associated with left side-restricted expression of pitx2c in the posterior LPM (Fig. 3D, Table 3; Essner et al. 2000). In PDI-P5-depleted embryos, pitx2c was expressed bilaterally in the LPM (Fig. 3D, Table 3). In sum, treatment of embryos with pdi-p5 antisense morpholino oligonucleotides blocks the asymmetric development of many, if not all, organ systems.
Table 2.
Depletion of PDI-P5 disrupts asymmetric localization of the visceral organs
|
|
No.analyzed
|
Position of organ
|
||||
|---|---|---|---|---|---|---|
| Left
|
Right
|
Middle
|
Bilateral
|
Unscoreda
|
||
| Liver
| ||||||
| No injection | 71 | 80% | 3% | 0 | 17% | 0 |
| P5-MO#1 | 40 | 5% | 10% | 62% | 23% | 0 |
| PS-MO#2 | 35 | 11% | 14% | 46% | 29% | 0 |
| Pancreas | ||||||
| No injection | 49 | 8% | 88% | 0 | 0 | 4% |
| P5-MO#1 | 38 | 0 | 0 | 58% | 0 | 42% |
| P5-MO#2 | 37 | 19% | 24% | 54% | 0 | 3% |
Morpholino oligonucleotides (2 ng P5-MO#1 or 10 ng P5-MO#2) were injected into the yolk of 1-cell embryos. embryos (60 hpf) were analyzed by whole-mount in situ hybridization with gataó and preproinsulin to identify the liver and the pancreas, respectively.
Expression was observed in tissue fragments.
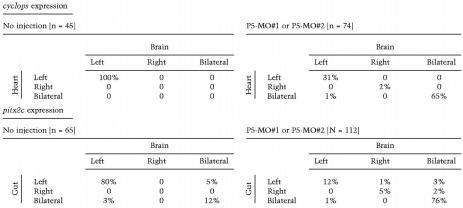
Table 3.
Coincident disruption of laterality patterning at multiple sites in PDI-P5 depleted embryos
Morpholino oligonucleotides (2 ng P5-MO#1 or 10 ng P5-MO#2) were injected into the yolk of 1-cell embryos. Results from experiments with each antisense morpholino were pooled. Embryos (20- to 24-somite stage) were analyzed whole-mount in situ hybridization for expression of cyclops or pitx2c in the indicated tissues. Embryos without staining were not scored in these analyses.
The disruption of the asymmetric development of multiple organs caused by depletion of the PDI-P5 protein is reminiscent of the panoply of laterality defects associated with loss of midline tissues or function. To determine whether PDI-P5 might be a required component of the midline's function in laterality patterning preceding overt morphogenesis, we analyzed antisense-treated embryos for expression of the earliest asymmetrically expressed markers of left/right patterning. During normal development, progenitors of the heart arise within the anterior portion of the LPM as bilaterally symmetric fields of cells that express nkx2.5 (Fig. 3D; Chen and Fishman 1996). Asymmetric development is first evidenced at the 22–24-somite stage, when only the left-side heart primordium expresses lefty2 and cyclops (Fig. 3D, Tables 3, 4; Rebagliati et al. 1998; Sampath et al. 1998; Bisgrove et al. 1999; Thisse and Thisse 1999). Although the initial development of the heart fields appeared normal in antisense-treated 18-somite embryos, as evidenced by the bilateral expression of nkx2.5 in the LPM, by the 24-somite stage the vast majority of antisense-treated embryos displayed laterality patterning defects as judged by expression of lefty2 or cyclops in the LPM (Fig. 3D, Tables 3, 4). Similarly, whereas lefty1, pitx2c, and cyclops are normally expressed only in the left side of the diencephalon (Rebagliati et al. 1998; Sampath et al. 1998; Bisgrove et al. 1999; Thisse and Thisse 1999; Essner et al. 2000), these genes were expressed bilaterally in the brains of antisense-treated embryos (Fig. 3D, Tables 3, 4).
Table 4.
Depletion of PDI-P5 disrupts asymmetric lefty expression
|
|
No.analyzed
|
Expression pattern
|
||
|---|---|---|---|---|
| Left
|
Right
|
Bilateral
|
||
|
lefty2 in the heart
| ||||
| No injection | 111 | 91% | 0 | 9% |
| P5-MO#1 | 93 | 13% | 5% | 82%a |
| P5-MO#2 | 29 | 14% | 7% | 79%a |
| lefty1 in the brain | ||||
| No injection | 36 | 81% | 5% | 14% |
| P5-MO#1 | 42 | 26% | 7% | 67%a |
| P5-MO#2 | 14 | 14% | 14% | 71%a |
Morpholino oligonucleotides (2 ng P5-MO#1 or 10 ng P5-MO#2) were injected into the yolk of 1-cell embryos. Embryos (20- to 24-somite stage) were analyzed by whole-mount in situ hybridization for expression of lefty2 or lefty1 in the indicated tissues. Embryos without staining were not scored in these analyses.
Significantly different from controls, P < 0.001.
To determine whether the laterality defects observed in antisense-treated embryos resulted solely from depletion of the PDI-P5 product, we investigated whether supplying treated embryos with an exogenous source of PDI-P5 protein could rescue the embryos to a WT phenotype. In these experiments, embryos were injected with the P5-MO#1 or P5-MO#2 morpholino oligonucleotides, with or without CS2-pdi-p5 mRNA, which is not complementary to either antisense oligonucleotide (Fig. 2A), is capable of programming synthesis of PDI-P5 in their presence (Fig. 3A) and has no discernible effect alone on laterality patterning (Table 5). As a simple uniform measurement of the laterality defects associated with each morpholino with or without CS2-pdi-p5 mRNA, the fractions of embryos with bilateral or unilateral gene expression in the heart field (lefty2), brain (pitx2c), or gut (pitx2c), were compared. Supplying antisense-treated embryos with exogenous PDI-P5 protein restored normal regulation of laterality patterning, measured in multiple tissues, indicating that depletion of the PDI-P5 protein was the specific cause of the laterality defects induced by the antisense treatment (Fig. 3E, Table 5).
Table 5.
Antisense pdi-p5 morpholino effects on laterality result from depletion of the PDI-P5 protein
|
|
No. analyzed
|
Gene expression pattern
|
P-value
|
|
|---|---|---|---|---|
| Unilateral
|
Bilateral
|
|||
|
lefty2 in the heart
| ||||
| P5-MO#2 | 19 | 26% | 74% | |
| P5-MO#2 + CS2-pdi-p5 RNA | 43 | 56% | 44% | 0.0523 |
| CS2-pdi-p5 RNA | 24 | 96% | 4% | |
| pitx2c in the brain | ||||
| P5-MO#1 | 126 | 19% | 81% | |
| P5-MO#1 + CS2-pd-p5 RNA | 196 | 42% | 58% | <0.0001 |
| P5-MO#2 | 50 | 34% | 66% | |
| P5-MO#2 + CS2-pdi-p5 RNA | 123 | 68% | 32% | <0.0001 |
| pitx2c in the gut | ||||
| P5-MO#1 | 125 | 18% | 82% | |
| P5-MO#1 + CS2-pdi-p5 RNA | 195 | 33% | 67% | <0.0001 |
| P5-MO#2 | 58 | 48% | 52% | |
| P5-MO#2 + CS2 pdi-p5 RNA | 151 | 67% | 33% | <0.0001 |
P5-MO#1 (1.5ng or 2ng) or P5-MO#2 (10ng) morpholino oligonucleotides were injected with or without CS2-pdi-p5 RNA (50pg or 400pg) into 1-cell embryos. Some embryos analyzed for lefty2 expression were injected only with RNA. Embryos (22- to 24-somite stage) were analyzed by whole-mount in situ hybridization for expression of lefty2 or pitx2c in the indicated tissues. P values (Fisher's exact test) indicate that rescue of lefty2 expression in the heart was marginally significant whereas rescue of pitx2c expression in the brain or gut was extremely significant.
Inhibition of pdi-p5 function disrupted laterality patterning at several sites. To determine whether our experiments indicated that patterning at one site could be disrupted independently of patterning at another site, we analyzed individual embryos for expression of cyclops, which normally exhibits left-side only expression in the heart and brain, or pitx2c, which usually exhibits left-side only expression in the developing gut and brain. With rare exceptions, when laterality patterning was perturbed at one site as a consequence of antisense treatment, it was perturbed at the second site (Table 3). Thus our data fail to indicate separable region-specific requirements for pdi-p5 function.
Midline tissue develops normally in PDI-P5-depleted embryos
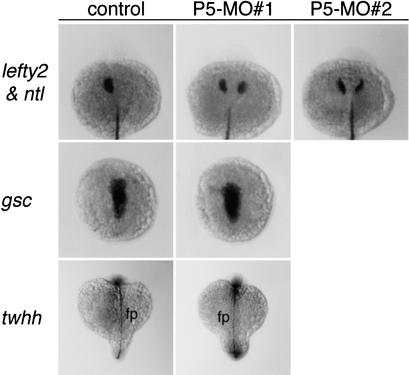
The laterality defects resulting from depletion of the PDI-P5 product are similar to those observed in zebrafish mutants, such as no tail and floating head, in which embryonic midline development is severely disrupted (Bisgrove et al. 2000; Chin et al. 2000). Therefore it is possible that pdi-p5 contributes indirectly to laterality by providing a function needed for normal midline development. However, several lines of evidence indicate that depletion of PDI-P5 does not compromise midline development. Whereas embryos injected with either P5-MO#1 or P5-MO#2 antisense oligonucleotides exhibited bilateral expression of lefty2 in the LPM, they had normal expression of no tail, a marker of posterior axial mesoderm (Fig. 4). Similarly, goosecoid expression, a marker of anterior axial development at late gastrula stages, was normal in antisense-treated embryos (Fig. 4). In addition, development of the floorplate of the nervous system appeared normal in PDI-P5-depleted embryos as judged by expression of tiggy-winkle hedgehog (twhh) at the 14-somite stage (Fig. 4) and by the finding that antisense-treated embryos with heart laterality defects at 2 d had well separated eyes (Fig. 3B). These findings indicate that depletion of PDI-P5 causes laterality defects without significantly disrupting midline development.
Figure 4.
Depletion of the PDI-P5 product does not disrupt normal midline development. Whereas embryos injected with either P5-MO#1 or P5-MO#2 antisense oligonucleotides have bilateral expression of lefty2 in the LPM at the 22–24-somite stage, these embryos exhibit a WT pattern of no tail (ntl) expression in the posterior axial mesoderm. Other markers of axial development also appear normal, including goosecoid (gsc), which marks the anterior axial mesoderm, and tiggy-winkle hedgehog (twhh), which marks the floorplate of the nervous system (fp).
Discussion
Use of zebrafish mutants to discover new tissue-restricted genes or pathway targets
Identification of genes that fail to be expressed in developmental mutants is a growing tool for the discovery of genes involved in tissue development (Shimono and Behringer 1999; Rubinstein et al. 2000). Our results demonstrate that the zebrafish system is exceedingly well suited for both discovering and probing the function of novel genes that are misregulated in developmental mutants. As zebrafish embryos can be staged with very high precision, comparative analysis of gene expression can be used to identify genes that are among the earliest to be misregulated as a consequence of loss of function of a well defined regulatory gene. Here we used the approach to discover new genes expressed in the embryonic midline and linked one of them to a known activity of the midline. These methods can also be applied to identify candidate targets of transcription factors or signaling pathways. The combination of gene discovery and functional perturbation methods can be used to dissect the highly pleiotropic effects often associated with mutations in regulatory genes that govern early development.
Although the approach taken here emphasizes the use of zebrafish mutants, we recognize that comparative analyses of gene expression patterns in closely related tissues has a long and successful history. In the past, such studies have been used to identify genes expressed specifically in one cell type (Hedrick et al. 1984), in one region of an egg (Weeks and Melton 1987), at one developmental stage (Zimmermann et al. 1980), in response to hormone stimulation (Brown et al. 1996), or in cancerous derivatives of a given tissue (Perou et al. 2000). With the advent of array technologies and procedures for sampling gene expression in small numbers of cells (Dulac and Axel 1995; Shimono and Behringer 1999), comparative analysis of gene expression profiles will be used increasingly to discover genes that are associated with cell-specific properties. The work described here illustrates that antisense morpholino oligonucleotide technology can be used as an insightful complement to gene discovery studies in the zebrafish.
Function of pdi-p5 in laterality patterning
The midline plays a critical role in left/right development. Ablation of embryonic midline tissues, whether by surgical extirpation or as a result of mutations in genes that regulate midline development, results in bilateral expression of nodal, lefty, and pitx2 genes and consequently produces heterotaxy (for review, see Yost 1998; Burdine and Schier 2000; Hamada et al. 2002). Although the exact role of the midline in laterality patterning is unknown, it is believed to exert its influence following the initial breaking of symmetry in the embryo, affecting the side-specific restriction of signals that regulate asymmetric expression of nodal genes in tissue primordia. Two models, which are not mutually exclusive, are generally set forth to explain the role of the midline (Burdine and Schier 2000; Hamada et al. 2002). First, the midline might provide a physicochemical barrier to the rightward propagation of signals that emanate from the left side and promote left side-specific gene expression. This model is supported by experiments indicating that an intact extracellular matrix is required for maintenance of asymmetries (Yost 1992). Second, the midline might serve as a source of a right-side antagonist of left-side gene expression. A candidate for such an antagonist is mouse Lefty1, which is expressed specifically in the floorplate of the nervous system in the mouse and is capable of antagonizing Nodal signaling. Mutants that lack lefty1 exhibit bilateral expression of left side-specific genes and heterotaxy (Meno et al. 1998). Among mutations that reveal the barrier or antagonistic factor function of the midline, lefty1 is the only one that appears not to interfere with other aspects of midline development (Hamada et al. 2002).
Depletion of PDI-P5 also causes disruption of left/right asymmetries without any pleiotropic midline defects. The findings that pdi-p5 function is required for laterality patterning of the brain, heart, and gut primordia and for asymmetric morphogenesis of the heart, liver, and pancreas indicate that this gene is an essential component of the regulation of most if not all aspects of left/right asymmetric development. Although the early widespread expression of pdi-p5 may contribute to laterality patterning, pdi-p5 expression is restricted to the midline at the time that left-side gene expression is initiated in the LPM and brain. Therefore, PDI-P5 appears to be a novel essential component of the midline function that is responsible for establishing left/right asymmetries. Moreover, our findings indicate the existence of common elements, such as PDI-P5, shared by the midline signaling pathways that govern laterality development in both the brain and the visceral organs.
The function provided by PDI-P5 in the midline is not known. PDIs are best known for catalyzing reducing as well oxidizing reactions affecting disulfide bond formation, for facilitating disulfide bond isomerization of synthesized peptides in endoplasmic reticulum, and for chaperone activity (Freedman et al. 1994; Ferrari and Soling 1999). However, with respect to the potential function in laterality patterning of the midline-expressed PDI-P5, it is intriguing that PDIs are associated with multiple aspects of the secretory pathway and can act at the cell surface to process receptor-bound ligands for cell internalization (for review, see Noiva 1999). For example, the PDI encoded by windbeutel has been shown to be an essential element in the maternally encoded dorsoventral patterning pathway in Drosophila that ultimately generates ventral-specific activated Toll ligand (Konsolaki and Schupbach 1998). In this pathway, Windbeutel functions to transport Pipe, a glycosaminoglycan-modifying enzyme, from the endoplasmic reticulum to the Golgi where Pipe is needed to modify GAGs destined to be secreted (Sen et al. 2000). As extracellular matrix substrates have been implicated in the signaling leading to laterality patterning (Yost 1992; Metzler et al. 1994), zebrafish PDI-P5 might play a role in vertebrate left/right patterning similar to that which Windbeutel plays in dorsoventral patterning. Alternatively, PDI-P5 might have a more direct role in the posttranslational processing of a TGFβ signaling factor produced in the midline, such as a Lefty antagonist of Nodal signaling (Meno et al. 1998) or a putative novel Nodal factor (Concha et al. 2000).
Materials and methods
Animals
Zebrafish embryos were generated by natural spawnings and cultured at 28°C under standard conditions (Westerfield 1995). WT animals were of the *AB strain. Two alleles of one-eyed pinhead were used: oepz1, a simple deletion of the oep locus, and oepm134, a point mutation (Schier et al. 1997; Zhang et al. 1998).
Preparation of an arrayed shield region cDNA library
Embryonic shields containing organizer as well as minimal neighboring tissue were manually dissected from ∼300 60%–70% epiboly WT embryos in MBS (88 mM NaCl, 1 mM KCl, 0.7 mM CaCl2, 1 mM MgSO4, 2.5 mM NaHCO3, 5 mM HEPES at pH 7.8). Following extraction of RNA with TRIZOL Reagent (GIBCO-BRL), cDNA was prepared from 15 μg total RNA using oligo dT-priming and cloned directionally into the λZapII vector (Stratagene). Individual plaques from primary plating of the library were picked into wells of 56 96-well plates containing 22 μL λTaqDil (2.5 mM Tris-HCl at pH 7.5, 2.5 mM MgCl2, 0.01% gelatin) supplemented with 7% DMSO. Aliquots of plaque suspensions were transferred to new wells, and cDNA inserts were PCR-amplified using m13 Forward and Reverse primers (M13-UL, 5′-CGACGTTGTAAAACGACGGCCAGTG-3′; M13-RL, 5′-GGAAACAGCTATGACCATGATTACG-3′). Greater than 90% of the PCR reactions generated products. A BioMek 2000 (Beckman) pipetting workstation was used to transfer PCR products onto nylon membranes (Stratagene) in a high density grid such that: (1) each DNA sample was spotted twice in positions that together denoted a unique signature, and (2) DNA samples from eight 96-well plates were spotted in a 4 × 4 matrix onto a membrane the size of single 96-well plate (http://Metherall.genetics.utah.edu/HTR.html). In total, 5376 DNA samples were arrayed onto a set of seven filters, and replicate sets of filters were prepared. Analysis of the arrayed library indicated that ∼1% of the total sequences hybridized with a probe for the abundantly expressed gene, ef1α, and 0–2 sequences (∼0.02% of the total) hybridized with probes for tissue-specific genes, such as goosecoid, no tail, floating head, or lim1.
Differential hybridization screening
To enhance isolation of sequences expressed in the organizer tissue of WT or oep mutant embryos, RNA was prepared from dorsalized midgastrula stage (70% epiboly) embryos that had been incubated in 0.3 M LiCl for 10 min at the 128–256-cell stage. This treatment leads to overexpression of organizer genes (Stachel et al. 1993). WT RNA from midgastrula embryos was extracted with TRIZOL Reagent. To prepare RNA from oepz1 homozygous dorsalized midgastrula embryos, embryos generated from oepz1/+ intercrosses were individually genotyped. A small piece of animal cap tissue was removed from each embryo and assayed by PCR for presence of sequences tightly linked to the oep gene and removed by the oepz1 deletion mutation (forward primer, 5′-CCAGTCCCGAGTGTGGACACC-3′; reverse primer, 5′-CCAGACTTGTCTACAGAAGAC-3′) and for presence of the no tail gene [as an internal control for the PCR reaction (see Fig. 1B); forward primer, 5′-AATGTACTCG GTCCTGCTGG-3′; reverse primer, 5′-GTGAGTCCGGGTG GATGTAG-3′]. RNA was extracted from the remaining portion of each embryo with TRIZOL Reagent. Samples from 55 individual oepz1 homozygous embryos were pooled, and RNA was prepared according to standard procedures.
WT or oep cDNA pools were generated as described previously (Dulac and Axel 1995). WT-oep and oep-WT subtracted cDNA probes were prepared using the PCR-Select cDNA Subtraction Kit (CLONTECH). The oep-unsubtracted probe was prepared directly from the cDNA pool generated from oepz1 homozygous embryos. For differential hybridization analysis, each cDNA pool was radiolabeled and hybridized to a replicate set of the seven filters containing the arrayed shield region library. Filters were washed, exposed to X-ray film, and visually inspected to identify sequences that hybridized only with WT-oep probe.
RNA and morpholino oligonucleotide injections
Two antisense morpholino oligonucleotides were synthesized (Gene Tools): P5-MO#1 (5′-CGCATGGTTGTTCTTTGTGCT TATT-3′) and P5-MO#2 (5′-TTTGTGCTTATTCAATGTATG GTTC-3′). Approximately 1 nL morpholino oligonucleotides in 1× Danieau buffer [58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, 5 mM HEPES at pH 7.6] was injected into the yolk of 1-cell embryos. To assess the ability of exogenously supplied PDI-P5 to rescue antisense-injected embryos, a chimeric gene, CS2-pdi-p5, was constructed by placing pdi-p5 coding sequences into the CS2+ plasmid (Turner and Weintraub 1994). In the chimeric gene, 5′UTR sequences are derived from the Xenopus β-globin gene. For experiments aimed at rescuing defects resulting from the P5-MO#2 morpholino, 1-cell embryos were injected with 10 ng P5-MO#2 oligonucleotides, and a subset of the embryos was subsequently injected with CS2-pdi-p5 mRNA (50 or 400 pg). The two simultaneously produced sets of embryos were analyzed at the 24-somite stage for expression of lefty2 or pitx2c. For rescue experiments involving P5-MO#1, parallel sets of 1-cell embryos were injected with morpholino oligonucleotides alone, or with a mixture of the morpholino and CS2-pdi-p5 mRNA (50 or 400 pg).
In vitro translation assay
In vitro transcribed pdi-p5 or CS2-pdi-p5 mRNA (1 μg) was incubated with or without 20 μg P5-MO#1 or P5-MO#2 in 25 μL in vitro translation reaction mixture containing rabbit reticulocyte lysate (Promega). To compare translation efficiencies, the reaction was performed in the presence of 35S-labeled methionine, and in vitro translated fractions were resolved in a SDS-10% polyacrylamide gel, dried and placed on X-ray film.
Accession numbers
GenBank accession nos.: pdi-p5, AF387900; ribophorin I (candidate #1), AF533661; chorein (candidate #18), AF533660; and candidate #20, AF533659.
Acknowledgments
We thank our colleagues Lisa Goering, Anthea Letsou, and Joe Yost for their critical advice and Jim Raimey and Sharon Johnson for their wonderful technical expertise. K.H. was supported by a postdoctoral fellowship from the Japan Society for the Promotion of Science. This work was supported by grants from the University of Utah Research Foundation and the NIH (HD37572).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL grunwald@genetics.utah.edu; FAX (801) 581-5374.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1001302.
References
- Bisgrove BW, Yost HJ. Classification of left-right patterning defects in zebrafish, mice, and humans. Am J Med Genet. 2001;101:315–323. doi: 10.1002/ajmg.1180. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Essner JJ, Yost HJ. Regulation of midline development by antagonism of lefty and nodal signaling. Development. 1999;126:3253–3262. doi: 10.1242/dev.126.14.3253. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Essner JJ, Yost HJ. Multiple pathways in the midline regulate concordant brain, heart and gut left-right asymmetry. Development. 2000;127:3567–3579. doi: 10.1242/dev.127.16.3567. [DOI] [PubMed] [Google Scholar]
- Brown DD, Wang Z, Furlow JD, Kanamori A, Schwartzman RA, Remo BF, Pinder A. The thyroid hormone-induced tail resorption program during Xenopus laevis metamorphosis. Proc Natl Acad Sci. 1996;93:1924–1929. doi: 10.1073/pnas.93.5.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine RD, Schier AF. Conserved and divergent mechanisms in left-right axis formation. Genes & Dev. 2000;14:763–776. [PubMed] [Google Scholar]
- Capdevila J, Vogan KJ, Tabin CJ, Izpisua Belmonte JC. Mechanisms of left-right determination in vertebrates. Cell. 2000;101:9–21. doi: 10.1016/S0092-8674(00)80619-4. [DOI] [PubMed] [Google Scholar]
- Chen JN, Fishman MC. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- Chen JN, van Eeden FJ, Warren KS, Chin A, Nusslein-Volhard C, Haffter P, Fishman MC. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development. 1997;124:4373–4382. doi: 10.1242/dev.124.21.4373. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Chin AJ, Tsang M, Weinberg ES. Heart and gut chiralities are controlled independently from initial heart position in the developing zebrafish. Dev Biol. 2000;227:403–421. doi: 10.1006/dbio.2000.9924. [DOI] [PubMed] [Google Scholar]
- Concha ML, Burdine RD, Russell C, Schier AF, Wilson SW. A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron. 2000;28:399–409. doi: 10.1016/s0896-6273(00)00120-3. [DOI] [PubMed] [Google Scholar]
- Danos MC, Yost HJ. Role of notochord in specification of cardiac left-right orientation in zebrafish and Xenopus. Dev Biol. 1996;177:96–103. doi: 10.1006/dbio.1996.0148. [DOI] [PubMed] [Google Scholar]
- Dufort D, Schwartz L, Harpal K, Rossant J. The transcription factor HNF3β is required in visceral endoderm for normal primitive streak morphogenesis. Development. 1998;125:3015–3025. doi: 10.1242/dev.125.16.3015. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Branford WW, Zhang J, Yost HJ. Mesendoderm and left-right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development. 2000;127:1081–1093. doi: 10.1242/dev.127.5.1081. [DOI] [PubMed] [Google Scholar]
- Ferrari DM, Soling HD. The protein disulphide-isomerase family: Unraveling a string of folds. Biochem J. 1999;339:1–10. [PMC free article] [PubMed] [Google Scholar]
- Freedman RB, Hirst TR, Tuite MF. Protein disulphide isomerase: Building bridges in protein folding. Trends Biochem Sci. 1994;19:331–336. doi: 10.1016/0968-0004(94)90072-8. [DOI] [PubMed] [Google Scholar]
- Fujinaga M. Development of sidedness of asymmetric body structures in vertebrates. Int J Dev Biol. 1997;41:153–186. [PubMed] [Google Scholar]
- Goldstein AM, Ticho BS, Fishman MC. Patterning the heart's left-right axis: From zebrafish to man. Dev Genet. 1998;22:278–287. doi: 10.1002/(SICI)1520-6408(1998)22:3<278::AID-DVG9>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Hamada H, Meno C, Watanabe D, Saijoh Y. Establishment of vertebrate left-right asymmetry. Nat Rev Genet. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- Hedrick SM, Cohen DI, Nielsen EA, Davis MM. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984;308:149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- Izraeli S, Lowe LA, Bertness VL, Good DJ, Dorward DW, Kirsch IR, Kuehn MR. The SIL gene is required for mouse embryonic axial development and left-right specification. Nature. 1999;399:691–694. doi: 10.1038/21429. [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Kreibich G, Gilmore R. Oligosaccharyltransferase activity is associated with a protein complex composed of ribophorins I and II and a 48 kd protein. Cell. 1992;69:55–65. doi: 10.1016/0092-8674(92)90118-v. [DOI] [PubMed] [Google Scholar]
- King T, Beddington RS, Brown NA. The role of the brachyury gene in heart development and left-right specification in the mouse. Mech Dev. 1998;79:29–37. doi: 10.1016/s0925-4773(98)00166-x. [DOI] [PubMed] [Google Scholar]
- Konsolaki M, Schupbach T. windbeutel, a gene required for dorsoventral patterning in Drosophila, encodes a protein that has homologies to vertebrate proteins of the endoplasmic reticulum. Genes & Dev. 1998;12:120–131. doi: 10.1101/gad.12.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaki K, Casey B. Genetics of human left-right axis malformations. Semin Cell Dev Biol. 1998;9:89–99. doi: 10.1006/scdb.1997.0187. [DOI] [PubMed] [Google Scholar]
- Levin M, Roberts DJ, Holmes LB, Tabin C. Laterality defects in conjoined twins. Nature. 1996;384:321. doi: 10.1038/384321a0. [DOI] [PubMed] [Google Scholar]
- Liang JO, Etheridge A, Hantsoo L, Rubinstein AL, Nowak SJ, Izpisua Belmonte JC, Halpern ME. Asymmetric nodal signaling in the zebrafish diencephalon positions the pineal organ. Development. 2000;127:5101–5112. doi: 10.1242/dev.127.23.5101. [DOI] [PubMed] [Google Scholar]
- Lohr JL, Danos MC, Yost HJ. Left-right asymmetry of a nodal-related gene is regulated by dorsoanterior midline structures during Xenopus development. Development. 1997;124:1465–1472. doi: 10.1242/dev.124.8.1465. [DOI] [PubMed] [Google Scholar]
- Melloy PG, Ewart JL, Cohen MF, Desmond ME, Kuehn MR, Lo CW. No turning, a mouse mutation causing left-right and axial patterning defects. Dev Biol. 1998;193:77–89. doi: 10.1006/dbio.1997.8787. [DOI] [PubMed] [Google Scholar]
- Meno C, Shimono A, Saijoh Y, Yashiro K, Mochida K, Ohishi S, Noji S, Kondoh H, Hamada H. lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell. 1998;94:287–297. doi: 10.1016/s0092-8674(00)81472-5. [DOI] [PubMed] [Google Scholar]
- Meno C, Gritsman K, Ohishi S, Ohfuji Y, Heckscher E, Mochida K, Shimono A, Kondoh H, Talbot WS, Robertson EJ, et al. Mouse Lefty2 and zebrafish antivin are feedback inhibitors of nodal signaling during vertebrate gastrulation. Mol Cell. 1999;4:287–298. doi: 10.1016/s1097-2765(00)80331-7. [DOI] [PubMed] [Google Scholar]
- Metzler M, Gertz A, Sarkar M, Schachter H, Schrader JW, Marth JD. Complex asparagine-linked oligosaccharides are required for morphogenic events during post-implantation development. EMBO J. 1994;13:2056–2065. doi: 10.1002/j.1460-2075.1994.tb06480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascone N, Mercola M. Organizer induction determines left-right asymmetry in Xenopus. Dev Biol. 1997;189:68–78. doi: 10.1006/dbio.1997.8635. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Noiva R. Protein disulfide isomerase: The multifunctional redox chaperone of the endoplasmic reticulum. Semin Cell Dev Biol. 1999;10:481–493. doi: 10.1006/scdb.1999.0319. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Rampoldi L, Dobson-Stone C, Rubio JP, Danek A, Chalmers RM, Wood NW, Verellen C, Ferrer X, Malandrini A, Fabrizi GM, et al. A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat Genet. 2001;28:119–120. doi: 10.1038/88821. [DOI] [PubMed] [Google Scholar]
- Rebagliati MR, Toyama R, Fricke C, Haffter P, Dawid IB. Zebrafish nodal-related genes are implicated in axial patterning and establishing left-right asymmetry. Dev Biol. 1998;199:261–272. doi: 10.1006/dbio.1998.8935. [DOI] [PubMed] [Google Scholar]
- Roessler E, Muenke M. Midline and laterality defects: Left and right meet in the middle. Bioessays. 2001;23:888–900. doi: 10.1002/bies.1130. [DOI] [PubMed] [Google Scholar]
- Rubinstein AL, Lee D, Luo R, Henion PD, Halpern ME. Genes dependent on zebrafish cyclops function identified by AFLP differential gene expression screen. Genesis. 2000;26:86–97. doi: 10.1002/(sici)1526-968x(200001)26:1<86::aid-gene11>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Sampath K, Rubinstein AL, Cheng AM, Liang JO, Fekany K, Solnica-Krezel L, Korzh V, Halpern ME, Wright CV. Induction of the zebrafish ventral brain and floorplate requires cyclops/nodal signalling. Nature. 1998;395:185–189. doi: 10.1038/26020. [DOI] [PubMed] [Google Scholar]
- Schier AF, Shen MM. Nodal signalling in vertebrate development. Nature. 2000;403:385–389. doi: 10.1038/35000126. [DOI] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SC, Helde KA, Talbot WS, Driever W. The one-eyed pinhead gene functions in mesoderm and endoderm formation in zebrafish and interacts with no tail. Development. 1997;124:327–342. doi: 10.1242/dev.124.2.327. [DOI] [PubMed] [Google Scholar]
- Sen J, Goltz JS, Konsolaki M, Schupbach T, Stein D. Windbeutel is required for function and correct subcellular localization of the Drosophila patterning protein Pipe. Development. 2000;127:5541–5550. doi: 10.1242/dev.127.24.5541. [DOI] [PubMed] [Google Scholar]
- Shimono A, Behringer RR. Isolation of novel cDNAs by subtractions between the anterior mesendoderm of single mouse gastrula stage embryos. Dev Biol. 1999;209:369–380. doi: 10.1006/dbio.1999.9256. [DOI] [PubMed] [Google Scholar]
- Stachel SE, Grunwald DJ, Myers PZ. Lithium perturbation and goosecoid expression identify a dorsal specification pathway in the pregastrula zebrafish. Development. 1993;117:1261–1274. doi: 10.1242/dev.117.4.1261. [DOI] [PubMed] [Google Scholar]
- Stainier DY. Zebrafish genetics and vertebrate heart formation. Nat Rev Genet. 2001;2:39–48. doi: 10.1038/35047564. [DOI] [PubMed] [Google Scholar]
- Summerton J. Morpholino antisense oligomers: The case for an RNase H-independent structural type. Biochim Biophys Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- Summerton J, Weller D. Morpholino antisense oligomers: Design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. Antivin, a novel and divergent member of the TGFβ superfamily, negatively regulates mesoderm induction. Development. 1999;126:229–240. doi: 10.1242/dev.126.2.229. [DOI] [PubMed] [Google Scholar]
- Tsukui T, Capdevila J, Tamura K, Ruiz-Lozano P, Rodriguez-Esteban C, Yonei-Tamura S, Magallon J, Chandraratna RA, Chien K, Blumberg B, et al. Multiple left-right asymmetry defects in Shh(−/−) mutant mice unveil a convergence of the shh and retinoic acid pathways in the control of Lefty-1. Proc Natl Acad Sci. 1999;96:11376–11381. doi: 10.1073/pnas.96.20.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes & Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Ueno S, Maruki Y, Nakamura M, Tomemori Y, Kamae K, Tanabe H, Yamashita Y, Matsuda S, Kaneko S, Sano A. The gene encoding a newly discovered protein, chorein, is mutated in chorea-acanthocytosis. Nat Genet. 2001;28:121–122. doi: 10.1038/88825. [DOI] [PubMed] [Google Scholar]
- Weeks DL, Melton DA. A maternal mRNA localized to the vegetal hemisphere in Xenopus eggs codes for a growth factor related to TGF-beta. Cell. 1987;51:861–867. doi: 10.1016/0092-8674(87)90109-7. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. Eugene, OR: University of Oregon Press; 1995. [Google Scholar]
- Yost HJ. Regulation of vertebrate left-right asymmetries by extracellular matrix. Nature. 1992;357:158–161. doi: 10.1038/357158a0. [DOI] [PubMed] [Google Scholar]
- Yost HJ. Left-right development in Xenopus and zebrafish. Semin Cell Dev Biol. 1998;9:61–66. doi: 10.1006/scdb.1997.0191. [DOI] [PubMed] [Google Scholar]
- Zhang J, Talbot WS, Schier AF. Positional cloning identifies zebrafish one-eyed pinhead as a permissive EGF-related ligand required during gastrulation. Cell. 1998;92:241–251. doi: 10.1016/s0092-8674(00)80918-6. [DOI] [PubMed] [Google Scholar]
- Zimmermann CR, Orr WC, Leclerc RF, Barnard EC, Timberlake WE. Molecular cloning and selection of genes regulated in Aspergillus development. Cell. 1980;21:709–715. doi: 10.1016/0092-8674(80)90434-1. [DOI] [PubMed] [Google Scholar]