Abstract
A variety of chromatin remodeling complexes are thought to assist sequence-specific transcription factors. The complexes described to date are expressed ubiquitously, suggesting that they have general transcriptional functions. We show that vertebrate neurons have a specialized chromatin remodeling complex, bBAF, specifically containing the actin-related protein, BAF53b, which is first expressed in postmitotic neurons at about murine embryonic day 12.5 (E12.5). BAF53b is combinatorially assembled into polymorphic complexes with ubiquitous subunits including the two ATPases BRG1 and BRM. We speculate that bBAF complexes create neuronal-specific patterns of chromatin accessibility, thereby imparting new regulatory characteristics to ubiquitous sequence-specific transcription factors in neurons.
Keywords: Chromatin remodeling, bBAF complexes, BAF53b, neuron development
The recent discovery that the mammalian genome encodes only about two- to threefold as many genes as invertebrates was surprising, particularly in light of the more complex nervous systems of mammals. In addition, many of the mammalian genes seem to have arisen by expansion of gene families, the members of which often have at least partially redundant functions. Thus the increase in gene number in mammals compared to invertebrates may have little additional informational content at the protein level. This conundrum may be partially addressed by the clear expansion of noncoding regulatory sequences in the mammalian genome and the growing notion that the function of a gene is diversified by its pattern of expression, that is, the context in which it functions.
Specialized chromatin remodeling complexes may change the overall pattern of expression of mammalian genes, allowing a common set of transcription factors and signaling pathways to produce different genomic transcriptional responses to common signals. This mechanism would diversify the function of each gene and might be particularly useful in the neuronal cell lineage, which gives rise to hundreds of cell types of diverse form and function (Stevens 1998). The expected characteristics of such a chromatin remodeling complex would be that it be expressed in all neuronal cell types, not be expressed outside the nervous system, and that it be activated at or near the time of neuronal subtype differentiation. We describe a complex, bBAF, for brain-specific BAF complex, that fits these criteria.
Results
Isolation of BAF53b
We previously cloned and characterized BAF53a (GenBank/EMBL accession no. AF041474), an actin-related protein (ARP), which is a subunit of mammalian BAF chromatin remodeling complexes (Zhao et al. 1998; Xue et al. 2000; Olave et al. 2002). BAF complexes (Khavari et al. 1993; Kwon et al. 1994; Wang et al. 1996a; Xue et al. 2000; Lemon et al. 2001) are related to the yeast SWI/SNF (Stern et al. 1984; Peterson and Herskowitz 1992; Cote et al. 1994), RSC (Cairns et al. 1996), INO80 (Shen et al. 2000), and the Drosophila BAP complex (Papoulas et al. 1998), but unlike invertebrate complexes are combinatorially assembled and functionally diverse (Wang et al. 1996a,b; Chi et al. 2002). Screening of a human brain cDNA library with the BAF53a probe resulted in the isolation of a 1492-bp cDNA encoding the entire open reading frame of a protein that is 84% identical (93% similar) to BAF53a over its 426 amino acids (GenBank/EMBL accession no. AF041475). A region of approximately 42 amino acids (amino acids 40–82 on BAF53b) at the N terminus is divergent with only 39% similarity (Fig. 1). This allowed us to raise specific antibodies to BAF53a and the related protein, which we termed BAF53b.
Figure 1.
Protein sequence alignment of human BAF53 proteins and homologs in Drosophila and C. elegans. Human (Hu) BAF53 proteins have a single homolog in Drosophila (Dm) and in C.Elegans (Ce). Protein sequence alignment was done using the CLUSTAL method. Conserved amino acid residues are boxed in black.
Sequence database searches revealed only one protein homologous to BAF53a and b in Drosophila (CG6546) and a hypothetical protein in C. elegans (ZK616.4) with 54% and 46% respective identity that extended through the entire sequence of hBAF53b (Fig. 1). Thus, while mammals have two distinct genes present at different genomic loci encoding highly homologous actin-related BAF53 proteins, Drosophila and C. elegans have only one. This observation is consistent with the notion that combinatorial assembly of BAF complexes is confined to vertebrates.
BAF53b is part of a brain-specific chromatin remodeling complex
The isolation of BAF53b cDNA from a human brain cDNA library and an analysis of EST databases suggested that BAF53b is expressed selectively in the brain. Indeed, RNase protection assays (Fig. 2A) revealed that while BAF53a is expressed in all adult mouse tissues examined, BAF53b transcripts appear to be restricted exclusively to the brain and peripheral nervous tissue (see below). Although BAF53a transcripts are detected at low levels in the adult brain, we have been unable to detect BAF53a protein in neurons with our affinity-purified antibodies, indicating that BAF53a transcripts in the brain probably arise from meninges and vascular components (see below).
Figure 2.
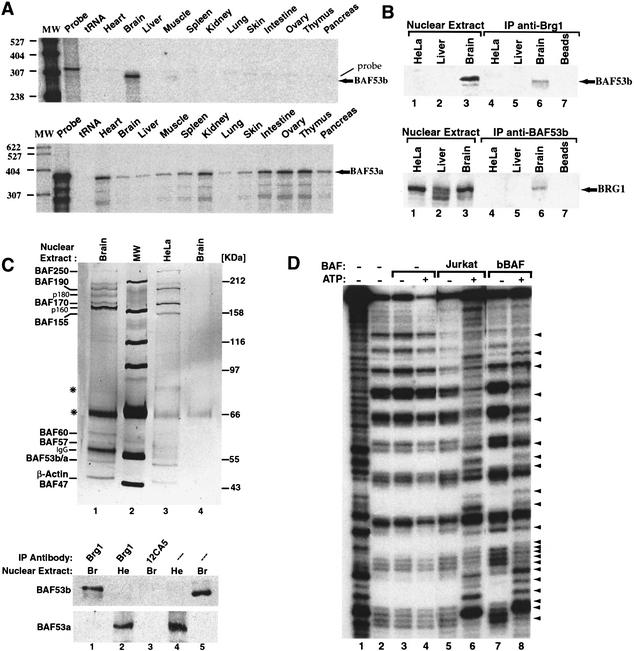
BAF53b is a subunit of a brain-specific chromatin remodeling complex. (A) BAF53b mRNA expression is restricted to the nervous system. Expression patterns for BAF53b (top) and BAF53a (bottom) in different adult mice tissues using ribonuclease protection assay. Arrows, protected fragments for both RNAs. Note that the faint upper band in all lanes of the BAF53b gel is the undigested probe. (B) BAF53b protein is expressed exclusively in mouse brain nuclear extract and reciprocally immunoprecipitates with the BAF ATPase subunit BRG1. Western blots for BAF53b (top) and BRG1 (bottom) using anti-BAF53b or anti-BRG1 antibodies. (Lanes 1–3) Nuclear extracts (10 μg) prior to purification. Immunoprecipitations using the antibodies shown on top of each panel are in lanes 4–7. (C) The bBAF complex is unique. (Top) Silver-stained SDS-PAGE gel (7.5%) showing immunopurified chromatin remodeling complexes from mouse brain nuclear extracts (Brain, lane 1) or HeLa cells (lane 3) using anti-BRG1 antibodies. (Lane 4) Immunopurification control of brain extracts with 12CA5 antibodies. Molecular weight markers (MW, lane 2) are indicated at the right. BAF subunits are shown at the left. Asterisks indicate nonspecific proteins. IgG is immunoglobulin heavy chain. (Bottom) BAF53b and BAF53a are mutually exclusive subunits of BAF chromatin remodeling complexes. After immunopurification of BAF complexes with the indicated antibodies, blots were analyzed with BAF53b- and BAF53a-specific antibodies. (Lane 3) Control immunoprecipitation using 12CA5 antibody. (Lanes 4,5) Input nuclear extracts [HeLa (He); Brain (Br); 10 μg]. (D) bBAF has ATP-dependent mononucleosome disruption activity. bBAF complexes attached to beads were immunopurified as described above and assayed on mononucleosomes in the absence (lane 7) or presence (lane 8) of 1 mM ATP. As a control, BAF complexes from Jurkat cells were used (lanes 5,6). No disruption is observed in the absence of chromatin remodeling complexes (lanes 3,4). Arrows show the ATP-dependent disruption of mononucleosomes (cf. lanes 5–8 and 3,4). Lanes 1 and 2 show controls for naked DNA and mononucleosomes alone, respectively.
The knowledge that BAF53a is a subunit of human SWI/SNF complexes (Zhao et al. 1998) and the extensive amino acid sequence identity between BAF53a and BAF53b prompted us to test whether BAF53b is a member of a brain-specific chromatin remodeling complex. By Western analysis we found that, in contrast to BAF53a, BAF53b protein is expressed in murine brain but not in liver or HeLa cell nuclear extracts (Fig. 2B, top, lanes 1–3) as expected from the pattern of expression of its mRNA (Fig. 2A). We used antibodies against BRG1, a DNA-dependent ATPase subunit, to immunoprecipitate BAF complexes. The J1 BRG1 antibody coimmunoprecipitated BAF53b from brain but not from HeLa or liver nuclear extracts (Fig. 2B, top, lanes 4–7). As expected, anti-BRG1 antibodies immunoprecipitated BRG from all extracts examined (Fig. 2B, bottom, lanes 1–3). Antibodies to BAF53b coimmunoprecipitated BRG1 protein from mouse brain but not from HeLa cell or liver nuclear extracts (Fig. 2B, bottom, lanes 4–7), confirming the association of these two proteins specifically in brain nuclear extracts. The conditions used for immunoprecipitation were highly stringent, and complexes remained intact even in the presence of 2 M salt and extensive washing over a 3-h period.
To investigate the subunit composition of complexes containing BAF53b, we purified the complexes from nuclear extracts derived from either mouse brain or HeLa cells. Specific and distinctive complexes were affinity-purified using the J1 anti-BRG1 antibody (Fig. 2C) compared to the control 12CA5 antibody (Fig. 2C, lane 4) or normal rabbit serum (data not shown). The most significant difference between these two BAF complexes is with the subunits ranging from 155 to 180 kD. The differences are not due to degradation, since Western analysis of the extracts revealed no protein fragmentation. Two new specific bands, p180 and p160, remained associated with the brain BAF complex even after purification under highly stringent conditions and are either modifications of BAF170 and BAF155, or new subunits. Importantly, while BAF53b was present only in the complex purified from brain nuclear extracts, BAF53a was present in complexes from HeLa cells and all mouse tissues examined except the brain (Fig. 2C, bottom; data not shown). This suggests that assembly of the two subunits is mutually exclusive, with BAF53b replacing BAF53a in BAF complexes present in the murine nervous system. We call the brain-specific complex “bBAF.”
If the bBAF complex is involved in controlling accessibility of regulatory regions to DNA binding proteins, one would expect that it would remodel nucleosomes in vitro similarly to other mammalian chromatin remodeling complexes (Wang et al. 1996a; Nie et al. 2000; Xue et al. 2000; Lemon et al. 2001). Indeed, highly purified bBAF complexes (Fig. 2C, lane 1) were able to remodel mononucleosomes in an ATP-dependent fashion (Fig. 2D). To our knowledge, the bBAF complex represents the first tissue-specific chromatin remodeling complex described to date.
bBAF is polymorphic and combinatorially assembled
Our previous studies showed that the subunits of BAF complexes are combinatorially assembled from the products of gene families encoding the different subunits (Wang et al. 1996a,b). For example, the ATPase subunit can either be BRG1 or hBRM, either of which can be paired with BAF60a, BAF60b, or BAF60c. Despite their coexpression in all tissues studied to date, this biochemical polymorphism produces complexes with apparently unique functions, because BRG1 and BRM deletions in mice give different phenotypes (Reyes et al. 1998; Bultman et al. 2000; Chi et al. 2002). To determine whether the brain-specific bBAF complex is also combinatorially assembled, we investigated the association of these complexes with the homologous BRG1 and BRM ATPases. Immunoprecipitation of BAF53b from murine brain nuclear extracts revealed that it was tightly associated with the BRG1 ATPase (Fig. 2B) and also the BRM ATPase (Fig. 3B). Since these subunits occupy mutually exclusive positions in BAF complexes (Wang et al. 1996b), we concluded that bBAF complexes are polymorphic, at least with respect to the ATPase involved. In addition, since the brain expresses BAF60a, b, and c (the products of three distinct genetic loci in mammals) as well as BAF250a and b (W. Wang and G.R. Crabtree, unpubl.), it is likely that bBAF complexes are combinatorially assembled with respect to other subunit family members.
Figure 3.
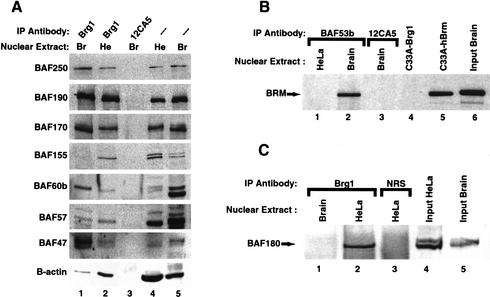
bBAF is a combinatorially assembled chromatin remodeling complex distinct from BAF and PBAF. (A) BAF complexes from Brain (Br) and Hela cells (He) were immunoprecipitated and blotted with antibodies to different BAF subunits. Brain (Br, lane 1) and HeLa-derived BAF complexes (He, lane 2). (Lane 3) Control immunopurification using 12CA5 antibodies. (Lanes 4,5) Input nuclear extracts (10 μg). (B) BAF53b containing bBAF complexes also associates with BRM ATPase subunit. Anti-BAF53b antibodies were used to immunoprecipitate BAF complexes from either HeLa cells or mouse brain nuclear extracts. Western blots with anti-hBRM antibodies (dil 1:1000) show that BRM ATPase coimmunoprecipitates with BAF53b in brain (lane 2) but not from HeLa nuclear extracts (lane 1). Lane 3 is a control immunoprecipitation using 12CA5 antibodies. Whole RIPA cell extracts (15 μg) of the BRG1/BRM-deficient cell line C33A transiently transfected (36 h) with Brg1 (lane 4) or hBrm (lane 5) expression vectors show the specificity of the anti-BRM antibody. (Lane 6) Input mouse brain nuclear extract (10 μg) used in the immunoprecipitation. (C) BAF180 is not a subunit of bBAF. Anti-BRG1 antibodies were used to immunoprecipitate BAF complexes from mouse brain or HeLa nuclear extracts and analyzed for the presence of BAF180 using specific antibodies. BAF180 is present in the HeLa (BAF)- but not brain (bBAF)-derived complex (lanes 1,2), although both input nuclear extracts (10 μg) contain 180 kD proteins (lanes 4,5). Note that the brain p180 runs at a different mobility, and at this time it is not certain that the band seen from the brain represents authentic p180 or a cross-reacting protein. (Lane 3) Control immunoprecipitation using normal rabbit serum (NRS).
BAF complexes in brain extracts also appear to have different subunit composition than the well studied HeLa SWI/SNF-like BAF complexes. Western analysis of purified complexes revealed that BAF250, BRG1 (BAF190), BAF170, BAF155, BAF60b, BAF57, BAF47, and β-actin subunits were all present in affinity-purified complexes from both the brain and HeLa cells. However, BAF155, which shares several domains with yeast Swi3p (Wang et al. 1996b), is present in sub-stoichiometric amounts in the affinity-purified bBAF complex compared to the complex derived from HeLa cells (Fig. 3A, lanes 1,2 on BAF155 blot). A clear difference in BAF155 expression between the brain and the HeLa complex is also observed on a silver-stained gel of a separate purification (Fig. 2C). Importantly, degradation products were not present in either extract. Thus, the sub-stoichiometric amounts of BAF155, the presence of p180 and p160 polypeptides described above, and the exclusive presence of BAF53b but not BAF53a distinguishes bBAF from BAF complexes in nonneuronal tissues. Examples of specialized BAF complexes include E-RC1, which is involved in the efficient in vitro expression of the β-globin gene by EKLF (Armstrong et al. 1998; Lee et al. 1999; Kadam et al. 2000), and more recently PBAF, which has been shown to be necessary for ligand-dependent transactivation by nuclear hormone receptors (Xue et al. 2000; Lemon et al. 2001). One of the biochemical hallmarks of the PBAF complex is the absence of BAF250 and the presence of the polybromo subunit BAF180 (Xue et al. 2000). The bBAF complex contains BAF250 and a component of 180 kD (p180) which could be the previously identified BAF180 (Xue et al. 2000). We determined whether p180 was BAF180 by using immunopurification of bBAF and Western blotting for BAF180 (Fig. 3C). We found that BAF180 is present in BAF but not bBAF complexes, even though it is present in the input brain nuclear extract used in the immunopurification. Together, these results suggest that bBAF complexes are unique in their subunit composition.
Developmental expression of BAF53b
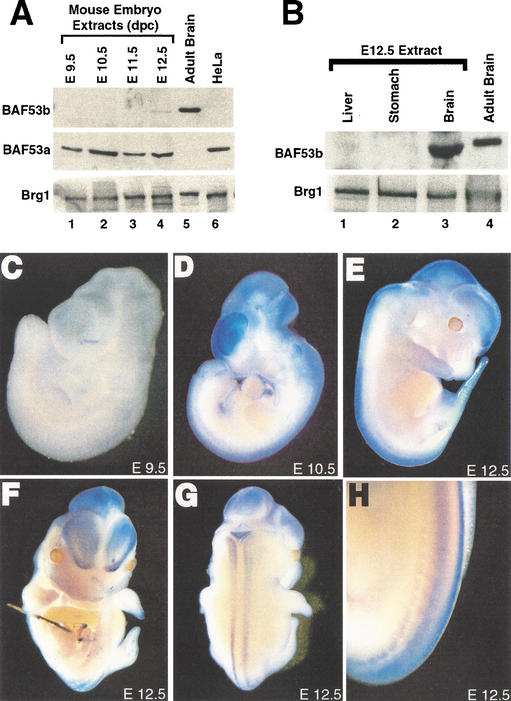
Cell-type specification in the murine nervous system depends on both extrinsic and intrinsic signaling mechanisms, and in mice begins between embryonic day 9.5 (E9.5) and E10.5 and continues until after birth in the case of glial cells (for review, see Edlund and Jessell 1999; Anderson 2001; Livesey and Cepko 2001; Zhang 2001). In agreement with previous reports (Zhao et al. 1998; Bultman et al. 2000; Machida et al. 2001), BAF53a and BRG1 were ubiquitously expressed at all stages of murine development examined. In contrast, BAF53b protein was not observed in whole mouse embryo extracts until E12.5 (Fig. 4A) and is specifically expressed in nervous tissue such as brain (Figs. 2A, 4A,B).
Figure 4.
BAF53b expression during murine development. (A) Western blots for BAF53b (top), BAF53a (middle), and BRG1 (bottom) expression in whole cell RIPA extracts (15 μg) from murine embryos of different developmental stages (lanes 1–4). Lanes 5 and 6 show control nuclear extracts (10 μg) from adult mouse brain and HeLa cells, respectively. (B) BAF53b is not expressed in endoderm-derived tissues of E12.5 mouse embryo. Western blots for BAF53b (top) and BRG1 (bottom) expression in liver, stomach, and brain RIPA extracts (30 μg; lanes 1–3). (Lane 4) Control nuclear extract (10 μg) from adult mice brains. (C–H) BAF53b is expressed in the nervous system. Whole-mount in situ hybridization using a specific antisense probe for BAF53b. Sense probe (data not shown) showed minimal staining. (C,D) Lateral view of E9.5 and E10.5 embryos, respectively. (E–H) E12.5 embryos showing lateral (E), frontal (F), dorsal (G), and dorsolateral (H) views. Dorsal root ganglia derived from the neural crest are clearly staining in H.
Whole-mount in situ hybridization experiments revealed that BAF53b mRNA was undetectable in E9.5 embryos (Fig. 4C). However, by E10.5, BAF53b expression was apparent in the nervous system, with high levels observed in the telencephalon, and less in the diencephalon, mesencephalon, and spinal cord (Fig. 4D). This pattern of expression of BAF53b persisted and became more defined in embryos at E12.5 (Fig. 4E–H) and at later stages of development (Fig. 2A; data not shown). Specific neural crest derivatives that give rise to neurons, such as the dorsal root ganglia, were also BAF53b-positive (Fig. 4H). However, neural crest derivatives that have nonneuronal characteristics, such as the great vessels, were not BAF53b-positive. These results demonstrate that BAF53b expression begins near the time of neuronal cell type specification. The disparity between the appearance of BAF53b protein (E12.5) and mRNA (E10.5) is likely related to the relative insensitivity of the whole embryo protein analysis or to posttranscriptional regulation of BAF53b mRNA.
BAF53b is expressed exclusively in postmitotic neurons
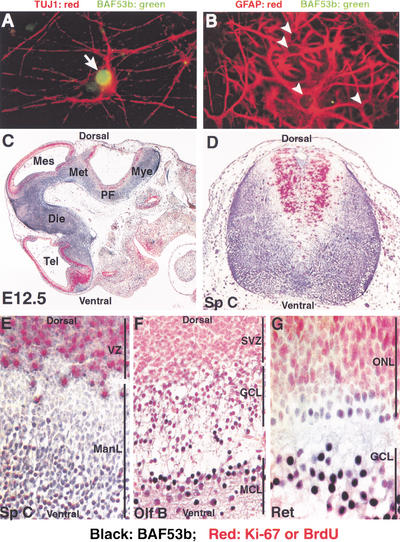
To determine whether BAF53b and its chromatin remodeling complexes were restricted to specific cell types within the nervous system, we analyzed its expression in a subpopulation of cells derived from post-natal day 0 (P0) rat hippocampus. Double immunofluorescence experiments showed that BAF53b was a nuclear protein (green label) expressed exclusively in neurons, which express the neuron-specific type III β-tubulin (TUJ-1; Fig. 5A, white arrow). BAF53b was not expressed in glial cells, which express glial acidic fibrillary protein (GFAP; Fig. 5B, arrowheads). We found that BAF53b, but not BAF53a, was present in virtually all neurons in the cerebral neocortex (layers II–VI), hippocampus (CA1–CA3 region and dentate gyrus), cerebellum (molecular, granular, and Purkinje cell layers), spinal cord (dorsally and ventrally), dorsal root ganglion, retina, and the olfactory bulb (mitral and granule cell layers) of adult mice (I. Olave and G.R. Crabtree, unpubl.; data not shown). The above observations indicate that BAF53b-containing bBAF chromatin remodeling complexes are likely to be present in all types of neurons, raising the issue of when BAF53b expression begins relative to the time that multipotential neural precursor cells begin to differentiate.
Figure 5.
BAF53b is expressed in postmitotic neurons but not glia. (A,B) BAF53b is expressed in the nucleus of neurons but not glia cells. A mixed population of neurons and glia cells derived from P0 rat hippocampus were cultured for 10 d, and BAF53b detected by immunofluorescence. (A) Costaining of neurons with the neuronal-specific marker TUJ1 (red) and BAF53b (green) antibodies as indicated. (B) Costaining with the astrocyte-specific marker GFAP (red) and BAF53b (green) antibodies as indicated. Note the empty nuclei denoting no detectable BAF53b staining. (C–G) BAF53b is expressed in postmitotic neurons. Double immunohistochemistry on paraffin-embedded embryo sections using antibodies against BAF53b (black-labeled cells) and the proliferation markers Ki-67 (C, E–G) or BrdU (D, red-labeled cells). (C) Sagittal section of E12.5 embryo head. BAF53b is expressed in the postmitotic cells of the nervous system. (D) Transverse section of cervical spinal cord from E12.5 embryo pulse-labeled in utero with BrdU. (E) Sagittal section of spinal cord from E12.5 embryo. (F) Sagittal section of olfactory bulb from E15.5 embryo. (G) Sagittal section of retina from P0 mouse. Tel, telencephalon; Die, diencephalon; Met, metencephalon; Mye, myelencephalon; PF, pontine flexure; Sp C, spinal cord; VZ, ventricular zone; ManL, mantle layer; Olf B, olfactory bulb; SVZ, subventricular zone; GCL, granule cell layer; MCL, mitral cell layer; Ret, retina; ONL, outer neuroblastic layer; GCL, ganglia cell layer.
Neuronal differentiation occurs by an anatomically orderly process in which precursors to both glia and neurons originate in a germinal or proliferative zone. Proliferation of multipotential precursor cells is followed by migration of postmitotic neurons to specific locations or layers (Edlund and Jessell 1999; Lee and Jessell 1999; Briscoe et al. 2000; Desai and McConnell 2000) where they establish a rich variety of different forms and functions. To determine whether BAF53b is expressed in neuronal precursor cells located in the ventricular zones, we studied mouse embryos that had been either pulsed-labeled with BrdU (Fig. 5D) or stained with the cell proliferation marker Ki-67 (Fig. 5C, E–G). Double immunohistochemical analysis using anti-BAF53b (black staining) and anti-BrdU or Ki-67 antibodies (red staining) showed that BAF53b is expressed in postmitotic cells in the murine CNS (Fig. 5C). Importantly, BAF53b was not detected in actively proliferating precursor cells (Fig. 5C–G). In the spinal cord of E12.5 embryos, BAF53b expression was restricted to postmitotic neurons present in the mantle layer (ManL) but clearly absent from cells present in the ventricular zone (VZ, Fig. 5E). Ventrally located neurons, such as motor neurons, and the more dorsally located interneurons of the spinal cord of E12.5 embryos, were equally stained for BAF53b (Fig. 5D). In the olfactory bulb of E15.5 embryos (Fig. 5F), all neurons that were actively dividing in the subventricular zone (SVZ) did not express BAF53b. However, granule cells (GCL) and mitral cells (MCL) that were in the process of or terminated their differentiation showed light or strong staining for BAF53b, respectively. A clear example of BAF53b expression in postmitotic neurons was seen in the retina of neonate embryos (Fig. 5G). BAF53b expression was found only in retina ganglion neurons in the ganglia cell layer, which are the first type of neuron to differentiate during retinal development. In contrast, mitotically active cells present in the outer neuroblastic layer (ONL) lack BAF53b expression.
Discussion
We have shown that murine neurons have a specific chromatin remodeling complex (bBAF) based on the neuron-specific expression of BAF53b. bBAF is assembled with either BRG1 or BRM (which are expressed in all cell types) as the ATPase (Fig. 3B). Since BAF60a, b, and c as well as BAF250a and b are present in neurons, we assume that the complex is combinatorially assembled with respect to the other subunits, as well. However, definitive demonstration of polymorphism for these other subunits may be very difficult because of the cellular heterogeneity of neurons. Combinatorial assembly appears to be unique to vertebrates, because Drosophila and C. elegans have only one gene encoding each subunit including BAF53 (Fig. 1), and Tamkun et al. found no evidence of combinatorial assembly of the Drosophila complex (Papoulas et al. 1998). Analyses of the subunits present in bBAF suggest that these complexes are unique, distinguishing themselves from other mammalian SWI/SNF-related chromatin remodeling complexes studied to date. The exclusive presence of BAF53b but not BAF53a (Fig. 2B,C), substoichiometric levels of BAF155 (Figs. 2C, 3A), and the presence of two new putative subunits, p160 and p180, that are different from the polybromo BAF180 present in PBAF (Figs. 2C, 3C) support this notion.
Our studies indicate that BAF53b protein is first expressed in neuronal precursor cells that have become postmitotic (Fig. 5). Thus, BAF53b expression appears after the determination of the neuronal lineage, but close to the time when neurons become functionally distinct. Studies of neuron cell lineage specification in the vertebrate spinal cord have indicated that homeodomain and bHLH transcription factors play critical roles (Tsuchida et al. 1994; Pfaff et al. 1996; Briscoe et al. 2000; for review, see Marquardt and Pfaff 2001). Although some of these transcription factors are expressed before the last cell cycle division of neurons (hence before BAF53b), others, such as the motor neuron specification transcription factor Islet-1, are expressed postmitotically (Pfaff et al. 1996). The polymorphic bBAF complexes present in postmitotic vertebrate neurons (Fig. 5C–G) have the capacity to remodel chromatin in vitro (Fig. 2D) and may functionally cooperate in vivo with such postmitotic lineage specification transcription factors in accessing their regulatory targets.
The remarkable neuron-specific expression of BAF53b (Figs. 4,5), and therefore bBAF complexes, contrast with the expression patterns found for other proteins/complexes involved in chromatin regulation to date. The murine acetyltransferase Querkopf and its human homolog, MORF (Champagne et al. 1999) have a wide pattern of expression during development. Consistent with this, Querkopf mutant mice show defects not only in neural but also in skeletal development (Thomas et al. 2000). The histone deacetylase RPD3 is highly expressed in the Drosophila nervous system but widespread defects are seen in rpd3 mutant flies, consistent with its expression in other tissues (De Rubertis et al. 1996; Chen et al. 1999; Mannervik and Levine 1999).
Certain subunits of the general transcription factor TFIID seem to have specialized functions, apparently arising from their restricted patterns of expression. For example, specific TBP-associated factors [TAF(II)s] have been found in B cells (Dikstein et al. 1996), granulosa cells of the murine ovarian follicle (Freiman et al. 2001), and in the male primary spermatocytes of Drosophila (Hiller et al. 2001).
In the course of purifying and cloning the subunits of ATP-dependent chromatin remodeling complexes (Khavari et al. 1993; Wang et al. 1996a,b; Zhao et al. 1998), we searched extensively for tissue-specific subunits, and to date BAF53b is the only one that we and others have identified, raising the question of why neurons should have a dedicated chromatin remodeling complex. At least two possible answers can be suggested. The first is simply that bBAF is dedicated to the transcription of genes that distinguish neurons from other cell types, that is, those involved in processes such as axonal guidance and electrical excitability. A second possibility has to do with the very large number of cell types in the vertebrate nervous system, which may be equivalent to or exceed the number of all the other nonneuronal cell types (Stevens 1998). Production of neural-specific chromatin remodeling complexes based on BAF53b rather than BAF53a could aid in establishing neural-specific chromatin structures, thereby broadly changing the genomic regulatory regions accessible to ubiquitous transcriptional factors in neurons. Such a restructuring of chromatin could impart neural patterns of expression to genes used both in neurons and nonneurons. Further diversification of bBAF complexes by combinatorial assembly could allow any single gene to gain new patterns of expression, thereby increasing the potential to generate new cell types within the neuronal lineage. Analysis of neuronal development in mice with disruptions of the BAF53b gene will be necessary to test these speculative models of bBAF chromatin remodeling complexes.
Materials and methods
Immunoblots, immunoprecipitation, and affinity purification of bBAF
Rabbit anti-BAF53b antibodies were raised against an MBP fusion protein containing amino acids 30–119 of the BAF53b protein sequence. BAF53a cross-reacting antibodies were immunodepleted by successive passages of the BAF53b serum over an MBP-BAF53a affinity column. Finally, affinity purification of BAF53b-specific antibodies was performed by successive passages of the BAF53a-free cross-reactive serum over an affinity column containing GST-BAF53b fusion protein. Affinity-purified anti-BAF53a antibodies were produced in a similar way using the respective epitopes. Anti-BRG1 antibodies were described (Wang et al. 1996a). Anti-BRM serum was a gift from Dr. Moshe Yaniv.
Proteins were run on 8% SDS-PAGE mini-gels, transferred to PVDF membrane, blocked with 5% milk on PBST-0.3 M (0.1% Tween-20; PBS; 0.3 M NaCl) prior to incubation with primary antibodies. After washing with PBST-0.3M (PBST-0.6M for anti-BAF180 antibodies) and incubating with secondary antibody, blots were developed with enhanced chemiluminescent reagents (Amersham) and exposed to film.
Immunoprecipitations and affinity purification of bBAF were performed overnight at 4°C with 100 μg of nuclear extract and antibodies crosslinked to protein-A beads in a total volume of 200 μL using RIPA buffer (for anti-BRG1). Because the interaction with BAF53b antibodies is disrupted in RIPA buffer, 0.4 M Buffer D (Wang et al. 1996a) was used instead. Beads were washed, and interacting proteins were eluted with 0.1M glycine pH2.5 and neutralized with 1M Tris at pH 8.0 before SDS-PAGE and Western analysis.
BrdU pulse labeling, immunohistochemistry, immunofluorescence, and whole-mount in situ hybridization
For BrdU pulse labeling, staged pregnant CD1 female mice were injected intraperitoneally with bromodeoxyuridine (BrdU; Sigma) at 50 μg/g of body weight 3 h prior to sacrifice. Embryos were dissected, processed, paraffin-embedded, and sectioned (5 μm) using standard procedures. Immunocytochemistry was done using rabbit affinity-purified anti-BAF53b (dil 1:300) and anti-BrdU (dil 1:1000; Neomarkers clone BRD.2) or anti Ki-67 (dil 1:30; DAKO, clone TEC-3) monoclonal antibodies as indicated. Development was done with a Vectastain Elite ABC Kit (Vector Labs) using DAB staining (for BAF53b) or alkaline phosphatase (for BrdU or Ki-67) following kit instructions.
Immunofluorescence on rat (P0) hippocampus primary culture was performed using standard procedures. Primary antibodies used were TUJ1 (dil: 1:1000, Covance), a cocktail anti-GFAP (dil 1:100, PharMingen), and rabbit affinity-purified anti-BAF53b. Detection was performed with goat anti-rabbit Alexa 488 and goat anti-mouse Alexa 594 secondary antibodies (Molecular Probes). Imaging was performed using a Zeiss Axiophot fluorescence microscope.
Whole-mount in situ hybridizations were performed using standard procedures with digoxigenin-labeled probes specific for BAF53b. A 400-bp fragment that extended until 171 bp of the 3′-UTR of the Celera genomic mouse BAF53b sequence was cloned by PCR using genomic mouse DNA into pGEM3fz(+) vector. The construct was sequenced and used for in vitro transcription of antisense and sense probes.
Mononucleosome disruption assay and RNase protection
The mononucleosome disruption assay was done as described previously (Nie et al. 2000). For RNase protection, the BAF53a-specific riboprobe was done by cloning an EcoR1-KpnI (nt 992–1344) cDNA fragment into xhomid vector and transcribed with T7 RNA polymerase. The BAF53b riboprobe was obtained by transcribing an EST clone (AA003209) that corresponds to the region between nt 930 and nt 1220 of the published BAF53b cDNA sequence.
Acknowledgments
We thank Susan McConnell, Kryn Stankunas, Fan Wang, Isabella Graef, Joel Nielson, Joseph Bayle, and Elena Battaglioli for helpful comments on the manuscript, and the members of the Crabtree lab for invaluable discussions and suggestions. G.R.C. is an investigator with the Howard Hughes Medical Institute (HHMI). I.O. is supported by a postdoctoral fellowship (no. PF-99-255-01-DDC) from the American Cancer Society. This work was supported by the HHMI and grants from the NIH (W.W. and G.R.C., CA39612)
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL crabtree@cmgm.stanford.edu; FAX (650) 723-5158.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.992102.
References
- Anderson DJ. Stem cells and pattern formation in the nervous system: The possible versus the actual. Neuron. 2001;30:19–35. doi: 10.1016/s0896-6273(01)00260-4. [DOI] [PubMed] [Google Scholar]
- Armstrong JA, Bieker JJ, Emerson BM. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–103. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Champagne N, Bertos NR, Pelletier N, Wang AH, Vezmar M, Yang Y, Heng HH, Yang XJ. Identification of a human histone acetyltransferase related to monocytic leukemia zinc finger protein. J Biol Chem. 1999;274:28528–28536. doi: 10.1074/jbc.274.40.28528. [DOI] [PubMed] [Google Scholar]
- Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes & Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi TH, Wan M, Zhao K, Taniuchi I, Chen L, Littman DR, Crabtree GR. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418:195–199. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- De Rubertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature. 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- Desai AR, McConnell SK. Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development. 2000;127:2863–2872. doi: 10.1242/dev.127.13.2863. [DOI] [PubMed] [Google Scholar]
- Dikstein R, Zhou S, Tjian R. Human TAFII105 is a cell type-specific TFIID subunit related to hTAFII130. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- Edlund T, Jessell TM. Progression from extrinsic to intrinsic signaling in cell fate specification: A view from the nervous system. Cell. 1999;96:211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- Freiman RN, Albright SR, Zheng S, Sha WC, Hammer RE, Tjian R. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- Hiller MA, Lin TY, Wood C, Fuller MT. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes & Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes & Dev. 2000;14:2441–2451. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Lee C-H, Murphy MR, Lee J-S, Chung JH. Targeting a SWI/SNF-related chromatin remodeling complex to the β-globin promoter in erythroid cells. Proc Natl Acad Sci. 1999;96:12311–12315. doi: 10.1073/pnas.96.22.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KF, Jessell TM. The specification of dorsal cell fates in the vertebrate central nervous system. Annu Rev Neurosci. 1999;22:261–294. doi: 10.1146/annurev.neuro.22.1.261. [DOI] [PubMed] [Google Scholar]
- Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodeling cofactors for ligand-activated transcription. Nature. 2001;414:924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: Lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Machida Y, Murai K, Miyake K, Iijima S. Expression of chromatin remodeling factors during neural differentiation. J Biochem. 2001;129:43–49. doi: 10.1093/oxfordjournals.jbchem.a002834. [DOI] [PubMed] [Google Scholar]
- Mannervik M, Levine M. The Rpd3 histone deacetylase is required for segmentation of the Drosophila embryo. Proc Natl Acad Sci. 1999;96:6797–6801. doi: 10.1073/pnas.96.12.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T, Pfaff SL. Cracking the transcriptional code for cell specification in the neural tube. Cell. 2001;106:651–654. doi: 10.1016/s0092-8674(01)00499-8. [DOI] [PubMed] [Google Scholar]
- Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ, Archer TK, Wang W. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879–8888. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002;71:755–781. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- Papoulas O, Beek SJ, Moseley SL, McCallum CM, Sarte M, Shearn A, Tamkun JW. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodeling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- Stevens CF. Neuronal diversity: Too many cell types for comfort? Curr Biol. 1998;8:708–710. doi: 10.1016/s0960-9822(98)70454-3. [DOI] [PubMed] [Google Scholar]
- Thomas T, Voss AK, Chowdhury K, Gruss P. Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development. 2000;127:2537–2548. doi: 10.1242/dev.127.12.2537. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996a;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes & Dev. 1996b;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Xue Y, Canman JC, Lee CS, Nie Z, Yang D, Moreno GT, Young MK, Salmon ED, Wang W. The human SWI/SNF-B chromatin remodeling complex is related to yeast Rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HS. Defining glia cells during CNS development. Nat Rev Neurosci. 2001;2:840–843. doi: 10.1038/35097593. [DOI] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]