Abstract
We have investigated whether the Ku complex is involved in regulating DNA replication in the yeast Saccharomyces cerevisiae. We find that Ku proteins control the replication time of telomeric regions; replication origins located close to telomeres or within subtelomeric repeat sequences normally initiate late, but are activated much earlier in mutants lacking Ku function. In contrast, origins distant from telomeres initiate replication at the normal time. Ku is one of the first components identified as important for replication timing, and specification of the replication time of chromosome ends by Ku is consistent with its role in maintaining telomere localization.
Keywords: Ku, replication, origin, telomere, Saccharomyces cerevisiae
Eukaryotic cells replicate their chromosomes during S phase of the cell cycle. Progress through S phase is controlled by regulating when origins of DNA replication are activated, with the various origins initiating replication in a defined and reproducible temporal sequence. Replication origin structure and function are best understood in Saccharomyces cerevisiae, for which the replicon organization of the entire genome has been mapped recently, all putative origins having been located by use of microarray analyses of replication time and initiation factor binding sites (Raghuraman et al. 2001; Wyrick et al. 2001).
Despite the wealth of information available concerning origin location and replication time in the yeast genome, our understanding of how the temporal program of origin firing is controlled is far from complete. It is known that chromosome context plays an important role in determining when an origin initiates (for review, see Gilbert 2001). Origin rearrangement studies indicated that an early activation time generally represents a default state, whereas certain chromosome contexts can impose late initiation. The chromosome end is an example of a late context; most origins located either within ∼35 kb of the telomere (telomere-proximal context) or actually in the subtelomeric repeat sequences (subtelomeric context) are activated late in S phase (Ferguson and Fangman 1992; Raghuraman et al. 1997). The yeast genome also contains late-activated origins that lie far from any telomere (internal context); in these cases as well, some feature of the long-range surrounding chromosome context imposes late initiation (Friedman et al. 1996). In molecular terms, however, our understanding of the mechanisms controlling origin firing time is limited. The late replication time of telomere-proximal origins appears to be preprogrammed during early G1 phase (Raghuraman et al. 1997), but molecules involved in programming telomere-proximal origins have not been identified previously. Likewise, nothing is known of the mechanism that establishes the internal late-origin context. Replication of a subtelomeric late origin, the chromosome V right Y‘ origin, is somewhat earlier than normal in a sir3 mutant, but the influence of Sir3 on origin firing time is limited to subtelomeric origins (Stevenson and Gottschling 1999). Cdc7 kinase and cyclin-dependent kinase are required for the normal origin firing sequence, but are probably involved in execution rather than determination of the origin temporal program (Bousset and Diffley 1998; Donaldson et al. 1998a,b).
Because Ku proteins function at eukaryotic telomeres, we became interested in the possibility that they are involved in controlling origin activation timing. Ku consists of two protein subunits that are related in sequence and form a heterodimeric complex involved in multiple cellular processes (for review, see Featherstone and Jackson 1999). Ku is important for the repair of double-stranded DNA breaks and has high affinity for DNA ends, and it has emerged that Ku also functions at telomeres. The yeast Ku proteins Yku70 and Yku80 (also called Hdf1 and Hdf2) are required for correct localization of telomeres to the nuclear periphery, and contribute to recruitment of telomerase and transcriptional repression by Sir proteins (Gravel et al. 1998; Laroche et al. 1998; Polotnianka et al. 1998; Peterson et al. 2001). Ku also binds to mammalian telomeres, and acts to maintain telomere length and prevent chromosome end-joining events (Bailey et al. 1999; Hsu et al. 2000; d’Adda di Fagagna et al. 2001). Mammalian telomeres associate with nuclear matrix components rather than with the nuclear periphery (Luderus et al. 1996), and it is not yet known whether Ku is involved in mediating this interaction.
A number of researchers have proposed that Ku functions in S-phase progression in yeast and mammalian cells (Shakibai et al. 1996; Barnes and Rio 1997; Novac et al. 2001). To establish whether S. cerevisiae Ku is required for a normal S phase, we have examined chromosome replication in strains lacking Ku function. We find that Ku is involved specifically in determining the activation time of subtelomeric and telomere-proximal replication origins.
Results and Discussion
Ku complex mediates late replication of the chromosome V right region
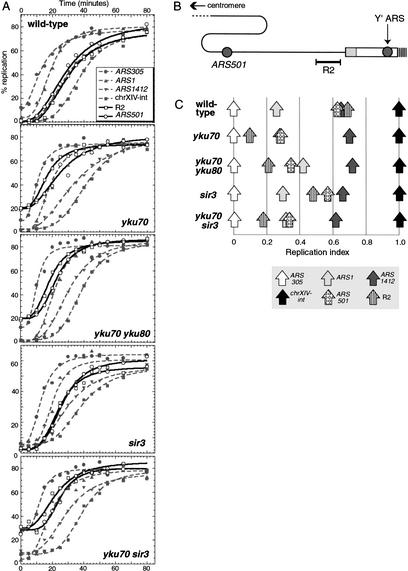
We used the dense isotope transfer method to monitor replication as cells progress through S phase (McCarroll and Fangman 1988). In wild-type yeast cells, chromosomal sequences replicate in a defined order due to the temporal program of origin activation (Fig. 1A, top). The replication origins ARS305 and ARS1 are activated early in S phase, leading to early replication of these two sequences, whereas ARS1412 is a late-replicating origin. A non-origin internal sequence on chromosome XIV (chr XIV-int) was used as a marker sequence for very late replication (Friedman et al. 1996).
Figure 1.
Sequences close to the chromosome V right telomere replicate early in yku mutants. (A) Timing experiments on wild-type, yku70, yku70 yku80, sir3, and yku70 sir3 strains released from a cdc7ts block. Replication kinetics are plotted for four internal sequences (ARS305, ARS1, ARS1412, and chr XIV-int) and two telomere-associated sequences (ARS501 and R2); see text for details. For unknown reasons, a lower proportion of sir3 cells released from the cdc7ts block; the percentage replication has been plotted on a different scale for ease of comparison. (B) Structure of the chromosome V right telomeric region. The two origins that are active in a wild-type strain (ARS501 and the Y‘ ARS) are drawn as gray circles. The Y‘ subtelomeric element is shown as an unfilled rectangle, the X element as a gray rectangle, and the TG1–3 terminal repeats as vertical hatching. ARS501 lies 27.3 kb and the Y‘ ARS 159 bp from the chromosome end (not including the TG1–3 tract). The R2 fragment is 4.4 kb long and is centered 11.9 kb from the chromosome end. (C) Replication times of internal and telomere-associated sequences plotted according to replication index (RI) values; see text for explanation.
The chromosome V right region, which contains two active replication origins, served as our model for telomere replication (Fig. 1B). The telomere-proximal origin ARS501 replicates late. Replication of the chromosome V right Y‘ ARS cannot be directly measured independent of other telomeres, because it lies within the repeated Y‘ subtelomeric element; instead, we followed replication of the last unique sequence on chromosome V (called R2) to monitor the late-activated subtelomeric Y‘ ARS (Stevenson and Gottschling 1999).
We found that in a yku70 deletion mutant strain, the replication kinetics of the four internally located chromosome loci (early origins ARS305 and ARS1, late origin ARS1412, and late sequence chrXIV-int) were very similar to wild type (Fig. 1A). Replication timing of the telomere-associated loci was, however, dramatically altered in the yku70 strain, with both ARS501 and R2 replicating much earlier than normal. In yku70 cells, ARS501 replicated at about the same time as the early origin ARS1, and the R2 sequence even earlier. About 20% of yku70 cells replicated the ARS501 and R2 sequences before release from the cdc7ts block used for synchronization. This phenomenon, termed escape replication, has been reported previously for very early-replicating sequences such as ARS305 (Reynolds et al. 1989). Escape replication has been difficult to study because its levels tend to vary in different experiments, but the observation of apparent escape replication of ARS501 and R2 in the yku70 strain suggests that these telomere-associated sequences have acquired properties normally associated with early-replicating regions. An alternative possibility is that the replication of ARS501 and R2 at the cdc7 block in the yku70 strain results from a mechanism unrelated to previously described escape replication, for example, a recombination-induced replication mechanism (Kraus et al. 2001).
The replication program of a yku70 yku80 double deletion mutant was very similar to that of yku70, with internal sequences replicating close to their normal times, but both the ARS501 and R2 sequences replicating abnormally early (Fig. 1A, third panel).
Replication time of a sequence in these experiments can be defined as the time at which half of the final level of replication has occurred. Replication times were calculated for all six sequences (see Supplementary Table 1, see Supplementary Material at http://www.genesdev.org) and standardized to ARS305 and chr XIV-int, whose values were set to 0 and 1, respectively, for each experiment (Fig. 1C). This method of calculating replication index (RI) adjusts for variations in the speed with which cultures release from synchrony and progress through S phase (Friedman et al. 1996). This analysis confirmed that the chromosome V right telomere-associated sequences (ARS501 and R2) replicate much earlier than normal in the yku70 and yku70 yku80 mutant strains (RI values for ARS501 and R2 shifted by 0.34 and 0.59, respectively, in the yku70 strain when compared with the wild type). The replication indices of internal sequences (ARS1 and ARS1412) were approximately normal in the yku mutant strains. ARS1 replicated slightly later than normal in the yku70 yku80 mutant, but the significance of this small shift in ARS1 replication time is unclear.
A previous study identified Sir3 as influencing the replication time of chromosome V right telomeric sequences. Stevenson and Gottschling (1999) found that deletion of SIR3 caused somewhat earlier activation of the Y‘ ARS, leading to slightly advanced replication of the R2 sequence. We analyzed the replication program of the sir3 mutant (Fig. 1A,C) for comparison with the yku phenotype. Our results were very similar to those described previously for sir3, with R2 somewhat advanced in replication time, but ARS501 only slightly affected. Comparing the data from the yku70 and sir3 strains therefore suggests that the effects of deleting Ku on ARS501 and the Y‘ ARS do not result simply from disrupted telomeric silencing. Replication kinetics of a yku70 sir3 double mutant were almost identical to those in the yku70 strain (Fig. 1A,C), both R2 and ARS501 replicating substantially earlier than in the sir3 single mutant.
ARS501 initiates replication in the yku70 mutant
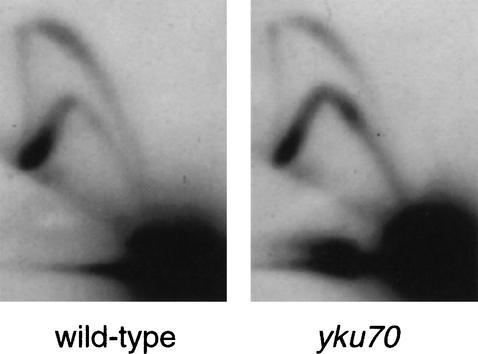
One interpretation of these results is that both ARS501 and the chromosome V right Y‘ ARS (which replicates R2) initiate abnormally early in yku mutants. However, because in the yku strains ARS501 replicated after R2, an alternative possibility could be that activation of only the Y‘ ARS is affected by the absence of Yku70. Precocious activation of the Y‘ ARS could result in abnormally early but passive replication of ARS501 by a leftward-moving replication fork. In this case, ARS501 would be inactive in the yku70 strain. We analyzed ARS501 activity in the yku70 strain using two-dimensional gel electrophoresis (Brewer and Fangman 1987), in which the presence of a bubble arc indicates that an origin initiates replication. A clear bubble arc showed that ARS501 initiates replication in a significant proportion of yku70 cells (Fig. 2). Measurement of the activation efficiency using fork direction analysis showed that ARS501 initiates replication in 65% of wild-type and 35% of yku70 cells (Supplementary Fig. 1, see Supplementary Material at http://www.genesdev.org). In those yku70 cells in which ARS501 is not activated, it was instead replicated by a leftward-moving fork coming from the telomere region. These studies therefore suggested that the early ARS501 replication time in the yku70 mutant results from a combination of precocious ARS501 initiation and passive replication from the prematurely activated Y‘ ARS.
Figure 2.
ARS501 is active in the yku70 mutant. Two-dimensional gel analysis of replicating structures at ARS501 in wild-type and yku70 mutant strains.
ARS501 activation time is affected by YKU70
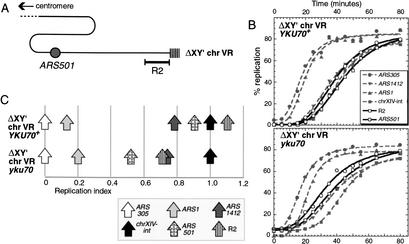
Because ARS501 initiated replication and its replication time was early in the yku70 strain, our two-dimensional gel analysis was consistent with the notion that, unlike Sir3, Ku influences ARS501 activation time. We tested this possibility more directly by examining the effect of a yku mutation in a strain in which any effects of the chromosome V right Y‘ ARS on ARS501 replication are eliminated. For this experiment, we used a truncated chromosome V construct from which both the X and Y‘ elements, including the Y‘ ARS, have been deleted, so that the R2 sequence abuts the terminal TG1–3 repeats (Fig. 3A). In this truncated chromosome construct, ARS501 is the rightmost replication origin on chromosome V, so that ARS501 replication time is not affected by replication forks from origins located closer to the telomere (Stevenson and Gottschling 1999).
Figure 3.
The time of replication initiation by ARS501 is affected by deletion of YKU70. (A) The truncated chromosome construct ΔXY‘ chr VR, in which sequences to the right of R2 are replaced with telomeric TG1–3 repeats. This cartoon should be compared with that of the wild-type chromosome V right region shown in Figure 1B. (B) Timing experiments on ΔXY‘ chr VR YKU70+ and ΔXY‘ chr VR yku70 strains released from a cdc7ts block. (C) Replication times in the same experiments plotted according to replication index (RI) values.
We compared the replication programs of YKU70+ and yku70 strains carrying this truncated chromosome, referred to as ΔXY‘ chrVR (Fig. 3B,C). The ARS501 replication time was substantially earlier in the yku70 ΔXY‘ chrVR strain (RI value of 0.52) than in YKU70+ ΔXY‘ chrVR (RI value of 0.90). The magnitude of the shift in ARS501 replication time on deletion of YKU70 was therefore similar in the truncated and unmodified chromosome contexts (RI shifts of 0.38 and 0.34, respectively; see Fig. 1C). Because deleting YKU70 has a substantial effect on ARS501 even when the X and Y‘ telomeric elements are absent, we conclude that Ku affects ARS501 activation time independent of changes in the time of Y‘ ARS activation.
ARS501 replicated later in a YKU70+ strain carrying the truncated chromosome than in the full-length chromosome context (cf. Fig. 1C, wild-type), perhaps, because in the truncated chromosome, the normal contribution to ARS501 replication by leftward-moving forks from the Y‘ origin is removed. R2 also replicated later than usual in the truncated chromosome context, consistent with its being replicated from ARS501 (Stevenson and Gottschling 1999).
The Ku complex mediates late replication of other telomeric regions
Our results so far have shown that the Ku complex influences the activation time of origins associated with the chromosome V right telomere. To establish whether Ku affects the activation time of origins close to yeast telomeres generally, we determined the replication time of other telomeric regions in the experiments shown in Figure 1.
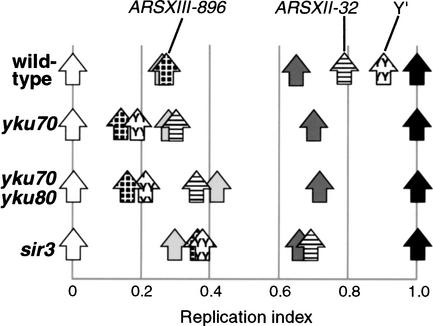
Telomeric Y‘ elements replicate on average very late in wild-type cells (Fig. 4), but much earlier in the yku70 strain (an RI shift of 0.71) and in the yku70 yku80 double mutant. The Y‘ replication values do not provide information about specific telomeres, but do indicate that the telomeric sequences of most yeast chromosomes replicate early in the absence of Ku.
Figure 4.
Replication timing of telomere regions on other chromosomes depends on the Ku complex. RI values of other telomeric sequences in wild-type, yku70, yku70 yku80, and sir3 strains. Replication times of two telomere-proximal origins are shown; late origin ARSXII-32 (horizontally hatched arrow) and early origin ARSXIII-896 (checkered arrow). The average RI for all Y‘ elements in the cell is indicated by a Y-hatched arrow. Also shown are the internal sequences ARS305 (white), ARS1 (light gray), ARS1412 (dark gray), and chr XIV-int (black).
We examined the replication of a number of telomere-proximal origins identified in the whole-genome analysis performed by Raghuraman et al. (2001). We have called one such origin ARSXII-32 according to its location ∼32 kb from the chromosome XII left telomere. ARSXII-32 replicates late in the wild-type S phase (Fig. 4). Like ARS501, the ARSXII-32 replication time shifts to early in yku mutant strains, but remains late in the sir3 mutant (Fig. 4). Therefore, Ku complex appears to be more generally responsible for the replication time of late telomere-proximal origins on yeast chromosomes, and ARS501 is not an isolated case.
Although most replication origins located close to telomeres (i.e., within ∼35 kb) are activated late, there are a few cases in which early origins lie close to telomeres. ARSXIII-896 is one such origin, lying about 28.6 kb from the chromosome XIII right telomere, and replicated at about the same time as ARS1 (Fig. 4). In the Ku mutants, ARSXIII-896 replicated slightly earlier than in the wild-type strain, although the shift in replication time was not nearly as pronounced as for normally late telomere-proximal origins. The replication time of early telomere-proximal origins may therefore also be slightly influenced by the Ku complex.
Effect of the Ku complex on subtelomeric and telomere-proximal replication origins
In this study, we have shown that the Ku complex is crucial for replication of yeast telomeres at the correct time in S phase, and that cells lacking Ku activate both telomeric and telomere-proximal replication origins abnormally early. This effect of Ku is specific to origins in telomeric regions and does not derail the replication program generally, because internally located sequences replicate at the normal time in yku mutants (Fig. 1). Previous data suggesting that in yku70 mutants, the time of S phase entry is advanced (Barnes and Rio 1997) might conceivably result from the advancement in telomere replication time that we have found in yku strains. It has been suggested that yku70 mutants over-replicate their DNA (Feldmann and Winnacker 1993), but we found no evidence for re-replication of DNA in our isotopic labeling experiments.
The effect of Ku at telomeres differs from that of Sir3, the only gene product described previously as being involved in specifying the chromosome replication program. Deletion of SIR3 leads to premature activation of Y‘ origins (Stevenson and Gottschling 1999). The Ku complex at telomeres cooperates with Rap1 in recruiting Sir3 and Sir4 (Mishra and Shore 1999; Dubrana et al. 2001), and the replication program of a yku70 sir3 mutant was very similar to that of the yku70 single mutant, consistent with the idea that Ku is required for the Sir3 effect on Y‘ origins. However, yku mutations have a greater effect than the sir3 mutation on the replication time of R2 and Y‘ sequences, suggesting that Ku mediates late activation of subtelomeric origins by additional mechanisms as well as establishment of Sir-repressed chromatin. Stevenson and Gottschling (1999) showed that the sir3 mutation also leads to slight activation of a normally inactive ARS within the chromosome V right X subtelomeric element; as expected, we found that the chromosome V right X ARS is also activated inefficiently in the yku70 mutant (data not shown).
The telomere-proximal origin ARS501 replicates slightly earlier than normal if SIR3 is deleted (Fig. 1; Stevenson and Gottschling 1999). This slight effect is probably caused by passive replication from the precociously activated Y‘ ARS, because in the ΔXY‘ chromosome V right truncation context deleting SIR3 does not advance, but in fact slightly delays, ARS501 activation (Stevenson and Gottschling 1999). yku mutations, in contrast, lead to substantially earlier replication of ARS501 and other origins in telomere-proximal locations, and our analysis of the ΔXY‘ chromosome V right truncation strain shows that Ku influences ARS501 activation time independent of the Y‘ ARS.
By what mechanism does Ku affect the activation time of ARS501? One possibility is that Ku complex binds telomere-proximal and subtelomeric replication origins and affects their activation time. Ku binds a linear ARS121 yeast origin fragment in vitro (Shakibai et al. 1996) and associates with putative origin sequences in a monkey cell line (Novac et al. 2001). However, there is currently no direct evidence that Ku proteins are required for pre-replication complex assembly, or associate with yeast replication origins in vivo, and our results do not suggest any general requirement for Ku to initiate replication at origins. Ku has, on the other hand, clearly been shown to bind to telomeres (Gravel et al. 1998; Martin et al. 1999). We therefore favor a model in which Ku complex bound to telomeres is involved in setting up a specialized chromatin context that affects the activation time of telomere-proximal origins. Ku is already known to be involved in Sir-mediated transcriptional silencing of telomeric regions, but transcriptional silencing by Sir proteins extends at most a few kilobases from the telomere (Pryde and Louis 1999), not nearly far enough to affect origins such as ARS501 and ARSXII-32. A specialized chromatin domain established by Ku and affecting these origins would have to act over a much longer range than the Sir-mediated telomeric silencing effect. Because the sir3 mutation does not directly affect ARS501, any such chromatin domain presumably does not depend on Sir proteins. We are testing genes known to play a role in chromatin structure for involvement in specifying the replication time of telomere-proximal origins.
There have been several reports of a relationship between intranuclear position of sequences and replication time. In general, peripheral nuclear position appears to correlate with late replication in mammalian cells. Heun et al. (2001) showed that in yeast cells, late origins tend to be close to the nuclear periphery during the cell cycle interval when replication time is established, whereas Dimitrova and Gilbert (1999) found that in mammalian nuclei, programming of origin activation time coincides with repositioning of sequences following mitosis. These reports have led to the suggestion that peripheral positioning leads to programming of origins for late replication, perhaps by locating them in a nuclear compartment containing chromatin-modifying factors that can establish a late-activation domain (Gilbert 2001). This idea leads to a possible model for the effect of the Ku on telomere-proximal origins, in which Ku influences the origin activation time via its role in intranuclear positioning of telomeres. Ku is responsible for tethering yeast telomeres at the nuclear envelope, and in yku mutants, the tethering mechanism fails and telomeres are instead distributed throughout the nuclear space (Laroche et al. 1998); a caveat is that truncated telomeres may not be localized by the same mechanism (Tham et al. 2001). Nevertheless, one attractive model would suggest that Ku influences the replication time of telomere-proximal origins by localizing them to a peripheral nuclear compartment that is conducive to establishment of a late activation context, the molecular nature of which is not yet clear.
Materials and methods
Yeast strains
All experiments were carried out in the A364A strain background (Hartwell 1967), its cdc7-1 derivative RM14-3a, or its cdc7-1 HMLa MATa HMRa derivative AR120 (Stevenson and Gottschling 1999). yku70 ∷ URA3 and yku80 ∷ TRP1 versions of these strains were made by standard one-step gene disruption and confirmed by PCR; details of primers used are available on request. Strains were AW104 (cdc7-1), AW101 (cdc7-1 yku70), AW116 (cdc7-1 yku70 yku80), UCC4403 (cdc7-1 sir3), and AC10 (cdc7-1 yku70 sir3). Strain AW99 (CDC7 yku70) was used for dense isotope analysis using α-factor-only synchronization. Construction of strain UCC4431(ΔXY‘ chr VR) is described in Stevenson and Gottschling (1999). The yku70 derivative of UCC4431 used in this study is AW123 (ΔXY‘ chr VR yku70).
Dense isotope transfer determination of replication times
Dense isotope transfer analysis was carried out as described previously (Friedman et al. 1996; Donaldson et al. 1998b). Probe fragments prepared by PCR amplification of appropriate sequences were used to detect the following genomic EcoRI fragments (numbers referring to the sequences given in the Saccharomyces Genome Database): ARS305, chromosome III 36,591-42,445; ARS1, chr IV 463,189-465,909; ARS1412, chr XIV 194,656-198,154; chrXIV-int, chr XIV 221,459-226,536; ARS501, chr V 544,359-552,288; R2, chr V 562,741-567,161; ARSXII-32, chr XII 26,154-31,578; and ARSXIII-896, chr XIII 894,552-900,786. The Y‘ probe was a 738 bp Hpa2 probe containing the Y‘ ARS. Timing analysis carried out on a yku70 strain released from an α-factor block gave results consistent with those shown here, with telomere-associated (but not internal) sequences replicating prematurely; S phase occurred at the normal time relative to bud emergence (data not shown).
Two-dimensional gel analysis
Two-dimensional gel analysis was carried out as described in Friedman and Brewer (1995). A 3.195-kb XbaI fragment was probed to analyze ARS305, and a 4.137-kb BamHI/SalI fragment was probed to analyze the chromosome V right X ARS. For analysis of fork direction to the right of ARS501, a 6013-bp ClaI fragment was cleaved in-gel with EcoRV; to the left of ARS501, a 4955-bp SnaBI fragment was cleaved in-gel with NcoI. Probe fragments were prepared by PCR amplification of appropriate genomic sequences.
Acknowledgments
We thank Julian Blow, Mike Stark, and Doug Stirling for discussion, and Walt Fangman and Julian Blow for comments on the manuscript. We are grateful to Dan Gottschling and Christine Smith for the gift of strains. A.J.C is supported by an MRC studentship. A.D.D. is a Royal Society University Research Fellow and an EMBO Young Investigator.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL a.d.donaldson@dundee.ac.uk; FAX 44-1382-345893.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.231602.
References
- Bailey SM, Meyne J, Chen DJ, Kurimasa A, Li GC, Lehnert BE, Goodwin EH. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc Natl Acad Sci. 1999;96:14899–14904. doi: 10.1073/pnas.96.26.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes G, Rio D. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc Natl Acad Sci. 1997;94:867–872. doi: 10.1073/pnas.94.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousset K, Diffley JF. The Cdc7 protein kinase is required for origin firing during S phase. Genes & Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Hande MP, Tong WM, Roth D, Lansdorp PM, Wang ZQ, Jackson SP. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- Donaldson AD, Fangman WL, Brewer BJ. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes & Dev. 1998a;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, Fangman WL. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell. 1998b;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- Dubrana K, Perrod S, Gasser SM. Turning telomeres off and on. Curr Opin Cell Biol. 2001;13:281–289. doi: 10.1016/s0955-0674(00)00210-6. [DOI] [PubMed] [Google Scholar]
- Featherstone C, Jackson SP. Ku, a DNA repair protein with multiple cellular functions? Mutat Res. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- Feldmann H, Winnacker EL. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J Biol Chem. 1993;268:12895–12900. [PubMed] [Google Scholar]
- Ferguson BM, Fangman WL. A position effect on the time of replication origin activation in yeast. Cell. 1992;68:333–339. doi: 10.1016/0092-8674(92)90474-q. [DOI] [PubMed] [Google Scholar]
- Friedman KL, Brewer BJ. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enz. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- Friedman KL, Diller JD, Ferguson BM, Nyland SVM, Brewer BJ, Fangman WL. Multiple determinants controlling activation of yeast replication origins late in S phase. Genes & Dev. 1996;10:1595–1607. doi: 10.1101/gad.10.13.1595. [DOI] [PubMed] [Google Scholar]
- Gilbert DM. Nuclear position leaves its mark on replication timing. J Cell Biol. 2001;152:F11–F15. doi: 10.1083/jcb.152.2.f11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967;93:1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Laroche T, Raghuraman MK, Gasser SM. The positioning and dynamics of origins of replication in the budding yeast nucleus. J Cell Biol. 2001;152:385–400. doi: 10.1083/jcb.152.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Gilley D, Galande SA, Hande MP, Allen B, Kim SH, Li GC, Campisi J, Kohwi-Shigematsu T, Chen DJ. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes & Dev. 2000;14:2807–2812. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus E, Leung W, Haber JE. Break-induced replication: A review and an example in budding yeast. Proc Natl Acad Sci. 2001;98:8255–8262. doi: 10.1073/pnas.151008198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr Biol. 1998;8:653–656. doi: 10.1016/s0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- Luderus ME, van Steensel B, Chong L, Sibon OC, Cremers FF, de Lange T. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J Cell Biol. 1996;135:867–881. doi: 10.1083/jcb.135.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- McCarroll RM, Fangman WL. Time of replication of yeast centromeres and telomeres. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- Mishra K, Shore D. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr Biol. 1999;9:1123–1126. doi: 10.1016/s0960-9822(99)80483-7. [DOI] [PubMed] [Google Scholar]
- Novac O, Matheos D, Araujo FD, Price GB, Zannis-Hadjopoulos M. In vivo association of Ku with mammalian origins of DNA replication. Mol Biol Cell. 2001;12:3386–3401. doi: 10.1091/mbc.12.11.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- Polotnianka RM, Li J, Lustig AJ. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol. 1998;8:831–834. doi: 10.1016/s0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. EMBO J. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Brewer BJ, Fangman WL. Cell cycle-dependent establishment of a late replication program. Science. 1997;276:806–809. doi: 10.1126/science.276.5313.806. [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- Reynolds AE, McCarroll RM, Newlon CS, Fangman WL. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989;9:4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakibai N, Kumar V, Eisenberg S. The Ku-like protein from Saccharomyces cerevisiae is required in vitro for the assembly of a stable multiprotein complex at a eukaryotic origin of replication. Proc Natl Acad Sci. 1996;93:11569–11574. doi: 10.1073/pnas.93.21.11569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JB, Gottschling DE. Telomeric chromatin modulates replication timing near chromosome ends. Genes & Dev. 1999;13:146–151. doi: 10.1101/gad.13.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham WH, Wyithe JS, Ferrigno PK, Silver PA, Zakian VA. Localization of yeast telomeres to the nuclear periphery is separable from transcriptional repression and telomere stability functions. Mol Cell. 2001;8:189–199. doi: 10.1016/s1097-2765(01)00287-8. [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: High-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]