Abstract
In plant meristems, dividing cells interpret positional information and translate it into patterned cell differentiation. Here we report the molecular identification of the Arabidopsis HOBBIT gene that is required for cell division and cell differentiation in meristems. We show that it encodes a homolog of the CDC27 subunit of the anaphase-promoting complex (APC). HOBBIT partially complements a yeast nuc2/cdc27 mutant. Unlike other CDC27 homologs in Arabidopsis, its transcription is cell cycle regulated. Furthermore, hobbit mutants show a reduction in DR5 :: GUS auxin reporter gene expression and accumulate the AXR3/IAA17 repressor of auxin responses. HOBBIT activity may thus couple cell division to cell differentiation by regulating cell cycle progression in the meristem or by restricting the response to differentiation cues, such as auxin, to dividing cells.
Keywords: APC3, auxin, BimA, meristem, Nuc2, pattern formation
Meristems of higher plants are postembryonic regions of mitotic activity that continuously produce new cells (Steeves and Sussex 1989). These cells interpret positional cues for fate decisions, and already in the mitotic zone they specifically orient planes of cell division and acquire different shapes. Once departed from the meristem, the cells expand and fully differentiate. Thus, cell differentiation commences while cells are actively dividing and continues when mitotic activity ceases.
Cell cycle regulation in yeast and animals involves alternating phases of high and low activity of cyclin-dependent kinases. The anaphase-promoting complex (APC), a multisubunit ubiquitin ligase triggering proteolytic destruction of mitotic cyclins, is an important regulator of the low-activity phase (Zachariae and Nasmyth 1999). Plants contain cyclin-dependent kinases and associated factors, whose roles have been studied by dominant-negative and ectopic expression approaches (Hemerly et al. 1995; Doerner et al. 1996; Cockcroft et al. 2000). Furthermore, a proteolytic pathway acts in cyclin degradation, and a potential regulatory factor of the APC has been described (Cebolla et al. 1999; Criqui et al. 2000). Thus, plants appear to contain conserved regulatory components of the cell cycle, but there is limited knowledge on how cell cycle controls are integrated with the cell differentiation program in meristems.
Mutations in the Arabidopsis thaliana HOBBIT (HBT) gene interfere with postembryonic cell division and with the differentiation of the distally located quiescent center (QC), columella root cap, and lateral root cap cells (Willemsen et al. 1998). The differentiation of these three cell types, furthermore, depends on distal accumulation of the plant hormone auxin, and high auxin levels can promote their differentiation in more proximal regions; this response, however, is restricted to actively dividing cells (Sabatini et al. 1999). The earliest detected defect in hbt mutants is misorientation of the plane of cell division in a progenitor cell of distal root cell types. Similar early defects occur in the auxin-resistant mutants bodenlos (bdl) and auxin resistant6 (axr6; Hamann et al. 1999; Hobbie et al. 2000) and in plants mutant for the MONOPTEROS (MP) gene, encoding a transcription factor that binds to auxin-responsive promoter elements (Hardtke and Berleth 1998).
We report here that the HBT gene is required for progression of cell differentiation rather than for pattern formation and that it encodes a protein homologous to CDC27/Nuc2, a component of the anaphase-promoting complex (APC). HBT can partially complement cell division defects in a Schizosaccharomyces pombe nuc2 mutant, and the HBT transcript accumulates in a punctuate cell cycle-dependent pattern, linking HBT gene activity to the cell cycle. We further show accumulation of an Aux/IAA auxin-response transcriptional repressor in hbt mutants, suggesting that HBT gene activity can control, directly or indirectly, auxin-mediated cell division and differentiation responses.
Results
hobbit mutants are defective in progression of cell differentiation in root and shoot meristems
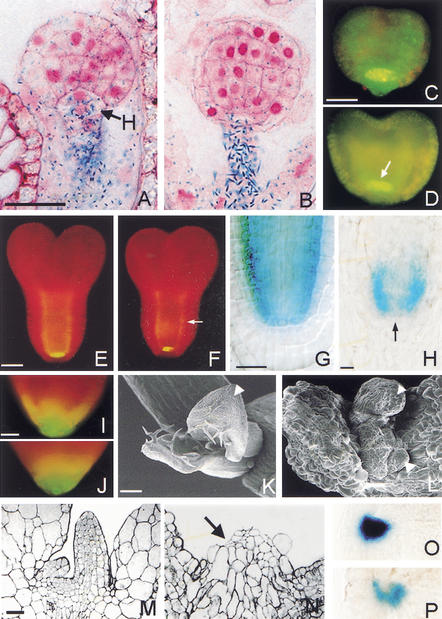
Studies on a series of EMS-induced recessive alleles previously established that the HBT gene is required for correct orientation of embryonic cell divisions in the embryonic root primordium and for maintenance of cell division and cell differentiation in the postembryonic root meristem (Willemsen et al. 1998). The spatial correlation of the early and late defects suggested that the gene might be required for root primordium specification during early pattern formation. However, analysis of markers expressed in the root primordium at different stages of embryogenesis revealed no significant differences between hbt and wild-type embryos. The suspensor and hypophyseal cell marker 267-1 is expressed in wild-type and hbt hypophyseal cells at the globular stage, when the first cell division defects become apparent in hbt mutant embryos (Fig. 1A,B). SCARECROW (SCR) gene expression was detected using promoter fusions and in situ hybridization, at the early heart stage in the incipient quiescent center of wild-type embryos and the equivalent position in hbt embryos, and later in the ground tissue (Fig. 1C–F; data not shown). After embryogenesis, SCR expression was reduced in the ground tissue of hbt mutants and reduced or absent at the position corresponding to the QC (Fig. 1G,H). Enhancer trap J1092, marking the lateral root cap and to a lesser extent the central root cap in wild-type embryos from the torpedo stage onward, is also distally expressed in hbt embryos until the mature embryo stage (Fig. 1I,J). J1092 expression marks the same root cap domain in wild-type seedlings but is absent in postembryonic root cells of hbt mutants (data not shown).
Figure 1.
The HBT gene is required for progression of cell differentiation. Marker gene expression and shoot apical meristem development in wild-type (A,C,E,G,I,K,M,O) and hbt mutant (B,D,F,H,J,L,N,P) plants. (A,B) Sections of globular-stage embryos with GUS staining of promoter trap 267-1 in suspensor and at position of hypophysis. (C,D) Whole-mount view of early-heart-stage embryos expressing GFP driven by SCR promoter at QC position (arrow). (E,F) Idem, early torpedo stage, expression extends in ground tissue layers (arrow). (G,H) Whole-mount view of seedlings expressing GUS driven by SCR promoter, strongest reduction at QC position (arrow). (I,J) Whole-mount view of GFP expression from J1092 enhancer trap in mature embryos. (K,L) Scanning electron micrographs show arrested leaf primordia in hbt with bloated, incorrectly differentiated epidermal cells (arrowheads) and the absence of a shoot apical meristem. (M,N) Astra-blue-stained sections of seedlings reveal the absence of a shoot apical meristem and aberrant cell division planes (arrow) in young leaf primordia of hbt. (O,P) Whole-mount view of seedlings expressing GUS driven by KNAT2 promoter. Bars: A–J, 20 μm; K,L, 100 μm; M,N, 25 μm.
Although the embryonic shoot apical meristem (SAM) does form in hbt mutants, its postembryonic development is defective (Willemsen et al. 1998). Closer inspection reveals that cell division ceases after an initial proliferation phase, and epidermal cells of leaf primordia and cotyledons become irregular in shape (Fig. 1K,L). Furthermore, whereas the wild-type SAM and leaf primordia display regular anticlinal cell division patterns in the outer layers (Fig. 1M), hbt mutants display unordered divisions in these layers (Fig. 1N, arrow) and give rise to vacuolated cells that do not differentiate appropriately. A reporter driven by the promoter of the KNAT2 homeobox gene marks the SAM in wild-type and hbt embryos (data not shown), continues to be expressed in wild-type, but disappears in the vacuolated cells at the position of the hbt SAM (Fig. 1O,P).
Our results indicate that root and shoot meristem primordia are appropriately patterned in hbt embryos, but that postembryonic progression of differentiation of meristematic cell types and maintenance of gene expression patterns is affected. Cells in the SAM have a limited capacity to divide postembryonically, but ultimately cell division ceases in both meristems.
HOBBIT encodes a CDC27/Nuc2 homolog that partially complements cell division defects in S. pombe nuc2 mutants
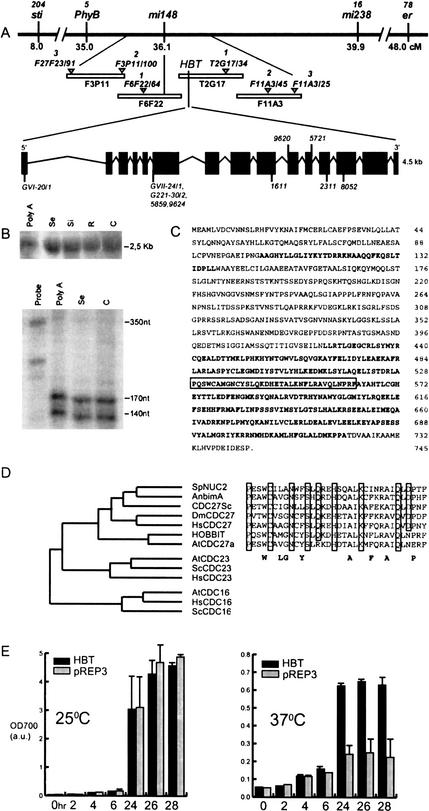
To analyze the molecular basis of the requirement of HBT for cell division and progression of cell differentiation in both root and shoot meristems, we cloned the gene using a map-based approach. Fine mapping located the HBT gene on Chromosome 2 in an area of 120 kb between CAPS markers F6F22/64 and T2G17/34 on the overlapping BACs F6F22 and T2G17 (Fig. 2A). We amplified and sequenced genomic DNA of the hbt2311 and hbt9620 alleles spanning 30 annotated open reading frames (ORFs; Lin et al. 1999), and identified point mutations only in ORF T2G17.20. Eight more hbt alleles revealed sequence changes in the same ORF (Table 1). A 9.2-kb genomic fragment spanning the HBT locus and containing no other predicted ORFs fully complemented hbt2311 homozygotes (data not shown). We conclude that this transcription unit represents the HBT gene.
Figure 2.
The HBT gene encodes a CDC27/Nuc2 homolog that can act in the Schizosaccharomyces pombe anaphase promoting complex. (A) Map-based cloning and structure of the HBT gene. The position of the markers sti, phyB, mi148, mi238, and er are given in centimorgans. The HBT gene location is shown relative to a contig of four BAC clones (F3P11, F6F22, T2G17, and F11A3). Triangles represent CAPS markers (from left to right, F27F23/91, F3P11/100, F6F22/64, T2G17/34, F11A3/45, and F11A3/25). The numbers in italics above the markers correspond to the recombinants that have a recombination breakpoint between the marker and the HBT locus. Structure of the HBT gene: The black boxes represent the exons. Nucleotide sequence changes have been described for 10 hbt alleles (see Table 1). (B) Expression analysis of the HBT gene. Northern blot (upper panel) with RNA from seedling (Se), siliques (Si), roots (R), and cotyledons (C). PolyA, mRNA purified from seedling RNA. RNAse protection analysis (lower panel) showing two potential transcription starts. Probe, probe used for RNAse protection without hybridization with the plant RNAs. (C) Amino acid sequence of the HBT gene-encoded protein. The sequences of the TPR domains are represented in bold letters. The TPR domain specific for CDC27 orthologs is boxed. (D) Protein similarity tree of the TPR-containing APC components and alignment of CDC27-specific TPR domain (boxed in C). The predicted sequences of the TPR-containing APC components from S. pombe (Sp), Aspergillus nidulans (An), Saccharomyces cerevisiae (Sc), Homo sapiens (Hs), and A. thaliana (At) were used to generate a phylogenic tree using Megalign software. Conserved TPR domains characteristic of CDC27 orthologs are aligned for different species. Identical amino acids are boxed, and the consensus of the TPR domain is shown in bold. (E) S. pombe strain nuc2ts complementation with the coding region of HBT cDNA. Growth at permissive (25°C) or restrictive (37°C) temperature of a nuc2ts strain carrying an empty vector as control (pREP3) or expressing the HBT gene (HBT) is followed by optical density at 700 nm (O.D.700) measurements (in arbitrary units, a.u.).
Table 1.
Molecular lesions in hobbit alleles
| Allele
|
Ecotype
|
Mutation
|
Predicted
effect on protein
|
|---|---|---|---|
| GVI-20/1 | L-er | C to T | AA 21: A to V |
| GVII-24/G221-30/2 | L-er | G to A | AA 145: G to S & splicing: intron 5 translated in frame |
| 5859, 9624 | Col-0 | G to A | AA 145: G to S & splicing: intron 5 translated in frame |
| 1611 | L-er | G to A | AA 421: G to V |
| 9620 | Col-0 | G to A | AA 434–441: Splicing: exon 10 absent & frameshift, stop at AA 441 |
| 5721 | Col-0 | G to A | AA 456–495: Splicing: exon 11 absent & frameshift, stop at AA 495 |
| 2311 | Col-0 | C to T | AA 519: Q to stop at AA 519 |
| 8052 | Col-0 | C to T | AA 537: A to V |
HBT cDNAs were isolated from a cDNA library and by RT-PCR. Comparison of the genomic and cDNA sequences revealed that the HBT gene contains 15 exons and 14 introns (Fig. 2A). RNase protection analysis showed two transcriptional start sites (Fig. 2B), both with putative upstream TATA and CAAT boxes. Northern blot analysis confirmed the 2.5-kb size of the transcript (Fig. 2B). The predicted HBT protein of 744 residues contains 10 tetratrico peptide repeats (TPRs; Fig. 2C,D), thought to be involved in protein–protein interactions (Lamb et al. 1995). The HBT protein has the highest homology to the CDC27/BimA/Nuc2 class of proteins found in such evolutionarily distant organisms as yeast, insects, and mammals (Fig. 2D). These proteins are components of the APC, involved in the control of cell cycle progression (King et al. 1995; Zachariae and Nasmyth 1999).
To investigate whether the Arabidopsis HBT protein can act as a component of the APC, the full-size cDNA was cloned in a yeast vector with a thiamine-repressible promoter and transformed into an S. pombe nuc2ts strain. HBT expression partially rescued the nuc2 phenotype at the restrictive temperature, reproducibly restoring growth to approximately fourfold higher density compared with the empty vector control. Complementation was not complete as growth rates and final density were, respectively, 5- and 10-fold less compared with the permissive temperature (Fig. 2E). We concluded that HBT protein can restore defects in its homologous yeast APC component and hence may act as a component of the plant APC.
Mutations in the hbt9620, hbt5721, and hbt2311 alleles introduce premature stop codons and should result in truncated proteins lacking major portions of the TPR domains (Table 1). In S. pombe, constructs carrying versions of the Nuc2 protein with truncations in the TPR domains cannot complement nuc2-663, which has no detectable Nuc2 function (Hirano et al. 1988, 1990). Thus, the presumptive absence of many TPR domains in three strong hbt alleles suggests that these alleles may be nulls.
Notably, a second CDC27-related sequence is present in the Arabidopsis genome, whose homology to yeast CDC27/Nuc2 is comparable to that of HBT (36% vs. 32%). The deduced AtCDC27a protein (accession no. BAB01271) shares an overall similarity of 47% with HBT, which increases to 65% when comparing the TPR domains. On the contrary, AtCDC16 and AtCDC23 homologs, encoding two other putative components of the APC with TPR domains, are present as single-copy genes in the Arabidopsis genome and group in distinct clades (Fig. 2D). An insertion in the AtCDC16 gene has been expected to lead to gametophytic lethality (V. Sundaresan, pers. comm.). However, phenotypes that point to an early essential role of the HBT protein in the APC complex, that is, metaphase arrest and gametophytic lethality, are not observed in the strong hbt alleles. This lack of early cell division arrest in hbt mutants may be explained by redundancy with AtCDC27a or by a divergence of HBT gene function from an ancestral role in the APC.
HBT gene expression is cell cycle regulated
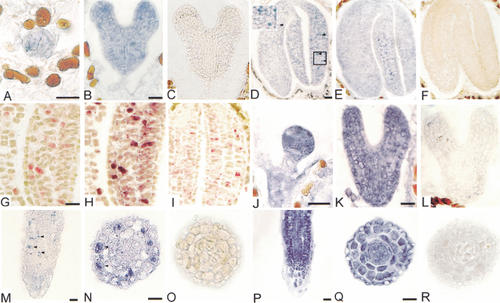
We analyzed whether redundant gene action and differences in transcription of the HBT and AtCDC27a genes could explain the region-specific mitotic defects in hbt mutants. On Northern blots, HBT transcripts were detected at similar levels in all organs tested (Fig. 2B). Antisense probes corresponding to unique 5′ sequences were used for in situ hybridization experiments. The distribution of HBT transcript varies during different stages of embryogenesis and also among cells within one embryo (Fig. 3A–D). HBT expression remains low in all cells until early heart stage onward, where particular cells express the gene at higher levels than others. The stochastic and punctuate distribution of these cells with higher HBT mRNA levels suggested that expression is elevated in a specific phase of the cell cycle. Colocalization of HBT and AtCYCB2;2 transcripts (Mironov et al. 1999) on single sections of late-stage embryos revealed high HBT transcript accumulation in the same cells that accumulate AtCYCB2;2 message, thus showing that the HBT message is cell cycle regulated with a peak around the G2/M phase (Fig. 3G–I). AtCDC27a is expressed ubiquitously at high levels during all embryonic stages (Fig. 3J–L), and this expression pattern contrasts with the low and punctuate expression pattern of HBT. AtCDC23 and AtCDC16 display similar high and ubiquitous transcript levels in the embryo (data not shown).
Figure 3.
The HBT transcript is cell cycle-regulated in contrast to transcripts from other APC-related genes. HBT expression during embryogenesis in wild-type (A–D) and in hbt2311 mutant (E,F) plants, and postembryonically (M–O) in wild-type roots using antisense (A,B,D,E,M,N) or sense (C,F,O) probe. (D, insert) Detail of boxed region. Double labeling of AtCYCB2;2 and HBT in a single section: Embryos were hybridized first with AtCYCB2;2 antisense (G) and then with HBT antisense (H) probes. As control, a double labeling was done with AtCYCB2;2 antisense and HBT sense probes in the same section (I). In both cases, the AtCYCB2;2 antisense probe was detected using the fast red substrate, the alkaline phosphatase was inactivated, and HBT transcripts were detected using NBT/BCIP. AtCDC27 expression in wild-type embryos (J–L) and seedlings (P–R) with antisense (J,K,P,Q) or sense (L,R) probes. Arrowheads show punctuate HBT expression. Bars: A–F,J–L, 25 μm; G–I, 50 μm; M,P, 10 μm; O,Q, 20 μm.
HBT again showed a punctuate expression in the actively dividing cells of the postembryonic root meristem (Fig. 3M–O), shoot meristem, flowers, and lateral root initiation sites (data not shown). In contrast, AtCDC27a (Fig. 3P–R), AtCDC23, and AtCDC16 (data not shown) transcript levels are uniformly high in all dividing cells. In lines segregating for the strong hbt2311 allele, no obvious difference in the expression level or pattern between wild-type (WT) and mutant embryos was observed (Fig. 3D,E). This indicated that HBT function is not required for the regulation of its own expression and, taking HBT expression as a marker of the G2/M phase, it also suggested that no dramatic cell cycle phasing defects occur in hbt embryos. As HBT mRNA was always detected in a subset of AtCDC27a-transcribing cells, the late cell division block in hbt appears not to be caused by the postembryonic absence of AtCDC27a transcription in the HBT expression domain. Rather, the distinct expression pattern of HBT in comparison to all other TPR-containing APC subunit homologs tested suggests that the gene serves a specialized function in meristematic cells.
hobbit mutants lack distal auxin reporter gene expression and accumulate a repressor of auxin responses
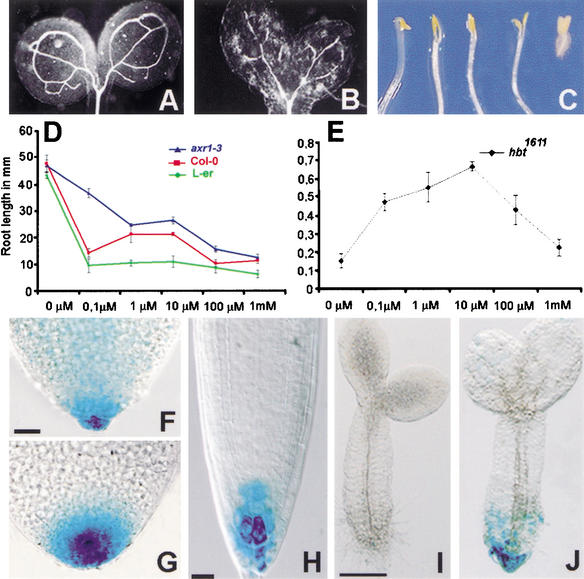
Several auxin-related cell division and cell differentiation processes appear to be affected in the root of hbt seedlings. Furthermore, hbt cotyledons display vascular patterning defects similar to those observed in axr6, bdl, and mp mutants (Fig. 4A,B; Hardtke and Berleth 1998; Hamann et al. 1999; Hobbie et al. 2000). Moreover, dark-grown hbt seedlings fail to maintain an apical hook as do the auxin-response mutants axr1, tir1, and bdl (Fig. 4C). This prompted us to investigate whether the HBT gene was required for auxin responses. A root growth assay revealed that growth of wild-type and the axr1-3 auxin-response mutant was inhibited from 0.1 μM IAA onward (Fig. 4D). The limited growth of the root of the weak hbt1611 allele was stimulated at IAA concentrations that already inhibited growth of wild type (Fig. E) and, to a lesser extent, the axr1-3 mutant. Notably, root growth stimulation also occurs in wild-type plants, but at very low IAA concentrations (Evans et al. 1994; data not shown). Only high concentrations acted as inhibitory on hbt suggesting that hbt seedlings displayed a shifted biphasic response to IAA that indicates a lower sensitivity (Fig. 4E). To investigate further the auxin responsiveness of hbt seedlings, we used a synthetic auxin-responsive promoter, DR5 :: GUS (Ulmasov et al. 1997). hbt2311 embryos displayed a basal DR5 :: GUS maximum after heart stage similar to that of wild-type embryos, and hence the basal auxin transport and response machinery appears to be active (Fig. 4F,G). hbt2311 seedlings, however, lack the DR5 :: GUS maximum that is present in the columella of wild-type roots (Fig. 4H,I). After growing wild-type and hbt seedlings for 3 d on media containing 5 × 10−7 M the synthetic auxin 2,4-D, intense ectopic DR5 :: GUS expression was detected in the wild-type root (data not shown), and a basal DR5 :: GUS peak appeared in the hbt root pole (Fig. 4J), suggesting that the signal transduction machinery to perceive auxin in the root tip is still present at the appropriate location in hbt mutants but less active.
Figure 4.
Reduced auxin sensitivity and auxin reporter gene expression in hbt seedlings. Vascular pattern is abnormal in hbt as xylem elements are not complete compared with wild-type (A,B). hbt, axr1, bodenlos, and tir1 do not form an apical hook (C; from left to right: WT, bodenlos, axr1-3, tir1-1, and hbt1611). hbt1611 root growth is induced by IAA. (D) Root growth is inhibited in wild-type and axr1-3 plants, whereas an initial stimulation is negated only at higher IAA concentrations in hbt mutants (E). DR5 :: GUS expression in wild-type (F) and hbt2311 (G) embryos, and in wild-type roots (H) and hbt2311 seedlings (I,J). Three d.p.g. seedlings were transferred to 1/2 GM medium (H,I) and to medium containing 5 × 10−7 M 2,4-D (J) and incubated for another 3 d. Bars: B, 50 μm; F,G, 10 μm; H, 20 μm; I,J, 100 μm.
Auxin-responsive transcription factors that activate DR5 :: GUS can dimerize with short-lived repressor proteins of the Aux/IAA class, encoded by early auxin-response genes, which reduce auxin responses (Kim et al. 1997; Ulmasov et al. 1997). Dominant mutations in several Aux/IAA genes stabilize the protein and reduce auxin sensitivity (e.g., Rouse et al. 1998; Ouellet et al. 2001). The axr3-1 mutant carries such a dominant mutation in the IAA17/AXR3 protein, and axr3-1 roots have reduced DR5 :: GUS responses and lack properly differentiated distal cell types (Sabatini et al. 1999). The axr3-1 mutant defect in the root tip reminded us of the stronger but similar defects in hbt mutants. Moreover, the possibility that HBT functioned in an APC-like complex that targets proteins for degradation prompted us to investigate whether IAA17/AXR3 turnover could depend on HBT activity.
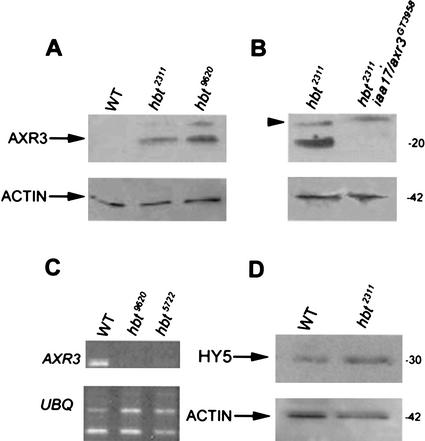
Polyclonal antibodies raised against IAA17/AXR3 do not detect a candidate protein in wild type because of the low abundance of AXR3 (Ouellet et al. 2001), but detected a prominent band close to the expected size in protein extracts of different hbt alleles (Fig. 5A). This band was markedly reduced in double mutants of hbt2311 and the GT3958 gene trap (Fig. 5B), which we determined by PCR to be homozygous for a Ds transposon insertion that upon sequence analysis appeared to reside 130 bp downstream of the start codon in the first exon of IAA17/AXR3 (data not shown). We concluded that the antiserum predominantly detected IAA17/AXR3 protein that accumulates to high levels in hbt seedlings. However, signals of lower intensity accumulate at a molecular weight in the range of the AUX/IAA proteins in hbt iaa17/axr3GT3958 seedlings, which may be caused by IAA17/AXR3-related proteins (Fig. 5B, arrowhead). RT-PCR experiments revealed that AXR3 RNA is less abundant in hbt2311 seedlings compared with wild-type seedlings (Fig. 5C), strongly suggesting that the observed protein accumulation is caused by defective posttranscriptional regulation. Posttranscriptional regulation was not generally defective in hbt as we detected wild-type levels of the proteolytically regulated HY5 protein (Fig. 5D; Osterlund et al. 2000).
Figure 5.
Elevated AXR3/IAA17 protein levels in hbt. Western blot analysis of AXR3/IAA17 expression in wild-type (Col-0) and hbt background (hbt2311 and hbt9620; A); and in the hbt2311/axr3GT3958 double mutant (B). Proteins were extracted from whole plants (10 d.p.g.) and quantified, and 1 μg was separated on a 15% polyacrylamide gel. After transfer, membranes were incubated first with anti-AXR3/IAA17 antibody (upper blot), and then with anti-actin antibody (lower blot) as control of protein loading. The numbers indicate size in kilodaltons. (C) RT-PCR analysis of AXR3expression in wild-type (Col-0) and hbt background (hbt9620 and hbt5722). RNA was extracted from wild-type and mutant seedlings. After cDNA synthesis, PCR was performed using AXR3-specific primers and ubiquitin (UBQ) as constitutive control. (D) Western blot analysis of HY5 expression in wild-type and hbt2311 mutant. The experiment was carried out under the same conditions as A and B.
Although the accumulation of IAA17/AXR3 repressor protein in hbt mutants correlates with the observation that auxin responses are reduced, the hbt root phenotype was not suppressed in the hbt2311 iaa17/axr3GT3958 double mutant (data not shown). Taking into account the residual signals detected by the Aux/IAA antibody in this double mutant, this may be caused by additional Aux/IAA proteins in hbt mutants.
Discussion
We have shown that the HBT gene is required in meristems for postembryonic progression of cell differentiation and maintenance of cell division. The gene encodes a protein homologous to the CDC27/Nuc2/BimA/APC3 subunit of the APC, which regulates M-phase progression in yeast and animals through targeted proteolysis. In line with a cell cycle-related role, HBT protein can partially complement an S. pombe nuc2 mutant, and HBT transcripts mainly accumulate around the G2/M phase in dividing cells. In contrast with mutants in other plant APC homologs, presumptive hbt null mutants are not defective in cell cycle progression during gametophyte development and embryogenesis. Arrest of cell division occurs only at later stages in root and shoot meristems, in both cases associated with aberrant progression of cell differentiation. The defects in auxin-requiring cell types in the hbt root meristem correlate with low auxin sensitivity, and we have obtained evidence that the Aux/IAA proteins may be one class of factors regulated by HBT. This raises the possibility that cell division or cell differentiation defects in hbt mutants arise from differences in auxin responsiveness. It should be interesting to establish whether AUX/IAA proteins are direct targets of a HBT-containing APC-like complex or whether HBT indirectly affects AUX/IAA abundance by modulation of the activity of other factors, for example, the SCFtir1 complex that targets Aux/IAA proteins for destruction (Gray et al. 2001).
The requirements for HBT activity in postembryonic cell division and cell differentiation may be independent, but they can also be connected, and below we discuss possible causal relationships.
HOBBIT as cell cycle regulator
Our findings pose the question whether, in line with its molecular identity and expression pattern, the HBT gene is required primarily for cell cycle regulation. In this case, redundant gene action, for example, by AtCDC27a, of which we detected high transcript levels throughout the embryo, should explain the normal cell division rate in hbt embryos. In postembryonic meristems, higher levels of HBT may be required at a specific phase of the cell cycle, for which AtCDC27a activity can no longer substitute.
Several mechanisms can be proposed by which cell differentiation defects arise from cell cycle progression defects. First, it is possible that the cell types that do not appropriately differentiate in hbt cannot achieve appropriate ploidy levels. However, we consider this unlikely as flow cytometry experiments indicate that hbt nuclei can perform at least three endoreduplication cycles (data not shown).
A second possibility is that progression through a specific phase of the cell cycle may be required for appropriate cell differentiation. For example, defined Drosophila neurons are specified by the even-skipped transcription factor, whose expression depends on progression through S phase, presumably for temporary removal of chromatin-remodeling factors that inhibit even-skipped expression (Weigmann and Lehner 1995). In analogy, an hbt-mediated cell cycle block in meristems could prevent the occurrence of the appropriate cell cycle phase for activation of cell differentiation factors. Similarly, an inappropriate elevation of mitotic cyclin levels in hbt, normally not associated with the G1 phase, could prevent cell cycle exit and differentiation. It is noteworthy in this respect that APC activity has been reported to remain active in differentiated animal cells (Gieffers et al. 1999). When appropriate antibodies become available, this hypothesis can be tested by the analysis of mitotic cyclin levels in hbt mutants.
HOBBIT as a regulator of cell differentiation
The HBT gene may have acquired new functions in the regulation of cell differentiation. Diverse roles for the APC have recently been suggested, because mutations in APC components disrupt zygote polarity in Caenorhabditis elegans (Rappleye et al. 2002). Here, identical genes are involved in both polarity and cell cycle progression, and the known APC target separin is required for both processes. Notably, HBT is the only TPR-containing APC homolog in the Arabidopsis genome of which a related copy exists. Hence, it is conceivable that the HBT gene encodes a duplicated APC subunit with new functions. The characterization of SCF ubiquitin–protein ligase complexes provides clear precedence for such a functional diversification of factors originally identified as cell cycle regulators (Patton et al. 1998).
The cell division defects in hbt meristems may be a consequence of defective cell differentiation if HBT-dependent upstream factors in the meristems control mitotic activity. Clear examples of regional cell cycle regulation have emerged from studies in Drosophila, where many transcription factors have been shown to act upstream of cell cycle control through regulation of the CDK phosphatase encoded by string(cdc25) (Edgar et al. 1994).
How can HBT cell cycle-specific transcription fit into the regulation-of-differentiation scenario? One possibility is that this cell cycle-specific transcription links cell cycle progression to control of cell differentiation in the meristem. In the meristem, cell differentiation is reversible and continuously dependent on positional cues. HBT-mediated, cell cycle-regulated abundance of response factors, for example, Aux/IAA proteins, may create a window of perception for cell differentiation cues in plant meristems.
Interestingly, cell cycle-regulated modulation of the perception of positional cues has been documented in C. elegans. The competence of vulval precursor cells to respond to signals that select vulval cell fates is coupled to different phases of the cell cycle (Ambros 1999; Wang and Sternberg 1999). The LIN-12 transmembrane receptor is required for this selection. LIN-12 activity appears to be modulated at the end of the G1 phase by SEL-10, an SCF component regulating ubiquitin-mediated turnover (Hubbard et al. 1997).
Although it remains to be shown how direct and functionally important the control of Aux/IAA levels by HBT is, a dependence of abundance of these proteins on cell cycle-expressed HBT activity seems to be an attractive explanation for the restriction of multiple auxin-mediated responses to dividing cells.
Materials and methods
Plant stocks
The hbt alleles were described in Willemsen et al. (1998). Suspensor-expressed promoter trap 267-1 was provided by P. Gallois; pSCR ∷ GUS by P. Benfey (Wysocka-Diller et al. 2000); and J1092 by J. Haseloff (http://www.plantsci.cam.ac.uk/haseloff/CATALOGUES/Jlines/index.html). The axr3 GT3958 gene trap (L-er background) was obtained from the Nottingham Arabidopsis Stock Centre database (http://nasc.nott.ac.uk; stock number N 100023; an IMA line provided by V. Sundaresan). All plants were grown on 1/2 GM (Scheres et al. 1994). The DR5 :: GUS plants have been described previously (Sabatini et al. 1999).
Auxin-response experiments
For apical hook experiments, seedlings were grown in the dark as described in Hamann et al. (1999). For auxin-sensitivity assays, wild-type (Col-0 and L-er) plants and axr1 and hbt1611 mutants were grown as described above for 3 d, transferred to fresh plates containing different IAA concentrations (10−7–10−3 M), and the root length from the tip to the hypocotyl boundary was measured.
Microscopy
Plant material for light microscopy was prepared as described in Scheres et al. (1994). Images were taken on a Zeiss Axioskop 2 microscope with a Nikon DXM1200 digital camera. For scanning electron microscopy, tissues were prepared as in Bowman et al. (1989) and visualized on a Philips XL30Labs SEM.
Map-based cloning
Heterozygotes carrying the Col-0 allele hbt2311 were crossed to L-er stichel, with sti (II,8) and er (II,48) flanking the HBT gene. In the F2 generation, recombinants between sti and er were selected. To identify hbt heterozygotes among these recombinants, F3 progenies were tested for hbt segregation, and DNA was isolated from 204 recombinants between sti and hbt and 78 recombinants between er and hbt, using a CTAB method (Lukowitz et al. 1996). DNA was analyzed with previously described CAPS markers (http://www.arabidopsis.org; Konieczny and Ausubel 1993). ORFs in the interval between the two nearest flanking markers were PCR-amplified from hbt2311 and hbt9620, and sequenced using Big Dye Terminator (Genpak Ltd.) on an ABI PRISM 310 Genetic Analyzer. Among all these ORFs, only ORFT2G17.20 contained mutations, and the sequence of all hbt alleles revealed the presence of mutations. Thus, 9.2 kb of genomic DNA containing this ORF (position 13960–23244 of T2G17 BAC) were cloned in the vector pgreen0226, conjugated into Agrobacterium tumefaciens C58, and this strain was used to transform hbt2311/+ plants. Resistant plants were genotyped for the hbt2311 mutations using an allele-specific CAPS marker (see below), and complemented plants with wild-type appearance but homozygous for hbt2311 were identified.
RNase protection analysis
The HBT transcription start was determined using RNAse protection on 20 μg of total RNA from roots and cotyledons and 1 μg of poly(A) RNA from seedlings. A 350-bp HBT DNA fragment including the putative ATG transcription start was subcloned into a PGEM-T vector (Promega). The RNAse protection experiment was carried out using the Ambion RPA II RNAse protection kit, according to the manufacturer's instructions.
RNA expression analysis
For Northern blot analysis, total RNA from different tissues was isolated (Kay et al. 1987), fractionated in 1% denaturing gels, and transferred to a Hybond-N Nylon membrane (Amersham) as described in Sambrook et al. (1989). Hybridizations were performed at 65°C using the 350-bp probe from the RNAse protection assay. The integrity and equal loading of the RNAs were checked by staining the blot with methylene blue (Sambrook et al. 1989).
For the in situ hybridization experiments, tissues were fixed, dehydrated, and embedded as described in Wilkinson and Nieto (1993). Pretreatment and hybridization were performed according to the method of Lincoln et al. (1994). Hybridizations and washes were carried out under stringent conditions (2× SSC, 50% formamide at 65°C). Double labelings were performed according to Long and Barton (1998). AtCYCB2;2 antisense was labeled with digoxigenin-11-UTP. Fluorescein-12-UTP was used for HBT antisense and sense probes synthesis. AtCYCB2;2 was detected with anti-digoxigenin antibody (1:1250) and fast red substrate (Sigma), and HBTwith anti-fluorescein antibody (1:6000) and NBT/BCIP substrate (Roche).
Yeast complementation
The coding region from HBT cDNA was amplified using the Elongase Amplification System (Life Technologies) and cloned into pGEM-T (Promega). A 2.4-kb SalI–BamHI fragment was transcriptionally fused to the thiamine-repressible promoter (nmt1) in the yeast shuttle vector pREP3, containing the LEU2 selection marker, and transformed into S. pombe strain nuc2-663 (Hirano et al. 1988) as described in Moreno et al. (1991). Leu+ colonies were grown in liquid minimal media (MM) with 5 μM thiamine at 25°C overnight. Cells were washed in MM without thiamine, divided in two flasks, and grown at 25°C to induce the promoter. One flask was shifted to 37°C, the other kept at 25°C, and optical density at 700 nm was measured from samples taken at 2-h intervals.
Genotyping of mutant combinations
The hbt2311 mutations were identified using allele-specific CAPS markers. PCR reactions were carried out using the following primers: HBT-F, GAGTTACTTGGCAGGAACTAATA TCAAC; HBT-R, CATAGCACACCTACATAATTGATTGCT TAT. PCR products were digested with the restriction enzyme MnlI (MBI Fermentas). The primers used for AXR3 wild-type allele genotyping were AXR3-F, TCTTCCCGGTGGAGATA CAG; and AXR3-R, GCCCATGGTAAAAGAGCTGA; and for Ds insertion allele: DS3-1, CGATTACCGTATTTATCCCGT; and AXR3-R.
Western blot analysis
Crude protein extract was produced from whole seedlings using the EZ method as described in Martinez-Garcia et al. (1999). Total proteins were quantified using DC reagent (Bio-Rad). Then 1 μg of extract was separated on a 15% polyacrylamide gel and transferred onto Immobilon-P membranes following the manufacturer's instructions (Millipore). Membranes were blocked and incubated with either anti-AXR3/IAA17 (1/500; Ouellet et al. 2001) or anti-HY5 (1/750; Osterlund et al. 2000) purified antibodies. Secondary anti-rabbit IgG conjugated to HRP (1/5000; Bio-Rad) was used, and blots were revealed using an ECL detection kit (Amersham Pharmacia Biotech). Membranes were then incubated with anti-actin IgG (1/3000; ICN Pharmaceuticals Inc.) and anti-mouse IgG conjugated to HRP (1/5000; Bio-Rad), following the same procedure.
Accession numbers
The cDNA sequence for HOBBIT was submitted to GenBank under accession no. AJ487669.
Acknowledgments
We are grateful to Philip Benfey, Patrick Gallois, and Jim Haseloff for making available promotor fusions/traps and enhancer traps, to Sakis Theologis for making available the AXR3/IAA17 antibody, to Xing Wang Deng and Christian Hardtke for the HY5 antibody, and to Thorsten Hamann for the bodenlos seeds. Frits Kindt and Piet Brouwers are acknowledged for help in preparing the figures. We also thank the members of the Scheres laboratory for the useful comments on the manuscript. F.F. and O.S. have been supported by EC-MCF fellowships HPMF-CT-1999-00013 and HPMF-CT-2001-01349. The Dutch organization of Sciences has supported B.S by Jonge Chemici and Pionier grants and S.F. by an ALW grant. N.E. and P.F. acknowledge CNPq and CAPES for support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL b.scheres@bio.uu.nl; FAX 31-30-251-3655
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.237302.
References
- Ambros V. Cell cycle-dependent sequencing of cell fate decisions in Caenorhabditis elegansvulva precursor cells. Development. 1999;126:1947–1956. doi: 10.1242/dev.126.9.1947. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla A, Vinardell JM, Kiss E, Olah B, Roudier F, Kondorosi A, Kondorosi E. The mitotic inhibitor ccs52is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 1999;18:4476–4484. doi: 10.1093/emboj/18.16.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft CE, den Boer BG, Healy JM, Murray JA. Cyclin D control of growth rate in plants. Nature. 2000;405:575–579. doi: 10.1038/35014621. [DOI] [PubMed] [Google Scholar]
- Criqui MC, Parmentier Y, Derevier A, Shen WH, Dong A, Genschik P. Cell cycle-dependent proteolysis and ectopic overexpression of cyclin B1 in tobacco BY2 cells. Plant J. 2000;24:763–773. doi: 10.1111/j.1365-313x.2000.t01-1-.x. [DOI] [PubMed] [Google Scholar]
- Doerner P, Jorgensen JE, You R, Steppuhn J, Lamb C. Control of root growth and development by cyclin expression. Nature. 1996;380:520–523. doi: 10.1038/380520a0. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Lehman DA, O'Farrell PH. Transcriptional regulation of string (cdc25): A link between developmental programming and the cell cycle. Development. 1994;120:3131–3143. doi: 10.1242/dev.120.11.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans ML, Ishikawa H, Estelle M. Responses of Arabidopsisroots to auxin studied with high temporal resolution: Comparison of wild type and auxin response mutants. Planta. 1994;194:215–222. [Google Scholar]
- Gieffers C, Peters BH, Kramer ER, Dotti CG, Peters JM. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc Natl Acad Sci. 1999;96:11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;41:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jurgens G. The auxin-insensitive bodenlos mutation affects primary root formation and apical–basal patterning in the Arabidopsisembryo. Development. 1999;126:1387–1395. doi: 10.1242/dev.126.7.1387. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROSencodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly A, Engler J de A, Begounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P. Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 1995;14:3925–3936. doi: 10.1002/j.1460-2075.1995.tb00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Hiraoka Y, Yanagida M. A temperature-sensitive mutation of the Schizosaccharomyces pombe gene nuc2+that encodes a nuclear scaffold-like protein blocks spindle elongation in mitotic anaphase. J Cell Biol. 1988;106:1171–1183. doi: 10.1083/jcb.106.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Kinoshita N, Morikawa K, Yanagida M. Snap helix with knob and hole: Essential repeats in S. pombe nuclear protein nuc2+ Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- Hobbie L, McGovern M, Hurwitz LR, Pierro A, Yang Liu N, Bandyopadhyay A, Estelle M. The axr6 mutants of Arabidopsis thalianadefine a gene involved in auxin response and early development. Development. 2000;127:23–32. doi: 10.1242/dev.127.1.23. [DOI] [PubMed] [Google Scholar]
- Hubbard EJ, Wu G, Kitajewski J, Greenwald I. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes & Dev. 1997;11:3182–3193. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay R, Chan A, Daly M, McPherson J. Duplication of a CaMV 35S promoter sequence creates a strong enhancer for plant genes. Science. 1987;236:1299–1302. doi: 10.1126/science.236.4806.1299. [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. Protein–protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsismutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: To TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, Town CD, Fujii CY, Mason T, Bowman CL, Barnstead M, et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. A knotted1-like homeobox gene in Arabidopsisis expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Barton KD. The development of apical embryonic pattern in Arabidopsis. Development. 1998;125:3027–3035. doi: 10.1242/dev.125.16.3027. [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jurgens G. Cytokinesis in the Arabidopsisembryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Monte E, Quail PH. A simple, rapid and quantitative method for preparing Arabidopsisprotein extracts for immunoblot analysis. Plant J. 1999;20:251–257. doi: 10.1046/j.1365-313x.1999.00579.x. [DOI] [PubMed] [Google Scholar]
- Mironov V, De Veylder L, Van Montagu M, Inze D. Cyclin-dependent kinases and cell division in plants—The nexus. Plant Cell. 1999;11:509–522. doi: 10.1105/tpc.11.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A. IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell. 2001;13:829–841. doi: 10.1105/tpc.13.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Rappleye CA, Tagawa A, Lyczak R, Bowerman B, Aroian RV. The anaphase-promoting complex and separin are required for embryonic anterior–posterior axis formation. Dev Cell. 2002;2:195–206. doi: 10.1016/s1534-5807(02)00114-4. [DOI] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. Changes in auxin response from mutations in an AUX/IAAgene. Science. 1998;279:1371–1373. doi: 10.1126/science.279.5355.1371. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsisroot. Cell. 1999;99:463–472. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P. Embryonic origin of the Arabidopsisprimary root and root meristem initials. Development. 1994;120:2475–2487. [Google Scholar]
- Steeves TA, Sussex IM. Patterns in plant development. Cambridge, UK: Cambridge University Press; 1989. [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Sternberg PW. Competence and commitment of Caenorhabditis elegans vulval precursor cells. Dev Biol. 1999;212:12–24. doi: 10.1006/dbio.1999.9357. [DOI] [PubMed] [Google Scholar]
- Weigmann K, Lehner CF. Cell fate specification by even-skipped expression in the Drosophilanervous system is coupled to cell cycle progression. Development. 1995;121:3713–3721. doi: 10.1242/dev.121.11.3713. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- Willemsen V, Wolkenfelt H, de Vrieze G, Weisbeek P, Scheres B. The HOBBIT gene is required for formation of the root meristem in the Arabidopsisembryo. Development. 1998;125:521–531. doi: 10.1242/dev.125.3.521. [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development. 2000;127:595–603. doi: 10.1242/dev.127.3.595. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes & Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]