Abstract
Many biochemical, physiological, and behavioral processes display daily rhythms generated by an internal timekeeping mechanism referred to as the circadian clock. The core oscillator driving this clock is located in the ventral part of the hypothalamus, the so called suprachiasmatic nuclei (SCN). At the molecular level, this oscillator is thought to be composed of interlocking autoregulatory feedback loops involving a set of clock genes. Among the components driving the mammalian circadian clock are the Period 1 and 2 (mPer1 and mPer2) and Cryptochrome 1 and 2 (mCry1 and mCry2) genes. A mutation in the mPer2 gene leads to a gradual loss of circadian rhythmicity in mice kept in constant darkness (DD). Here we show that inactivation of the mCry2 gene in mPer2 mutant mice restores circadian rhythmicity and normal clock gene expression patterns. Thus, mCry2 can act as a nonallelic suppressor of mPer2, which points to direct or indirect interactions of PER2 and CRY2 proteins. In marked contrast, inactivation of mCry1 in mPer2 mutant mice does not restore circadian rhythmicity but instead results in complete behavioral arrhythmicity in DD, indicating different effects of mCry1 and mCry2 in the clock mechanism
Keywords: Circadian clock, Per, Cry, nonallelic suppressor
The mammalian Period (mPer) and Cryptochrome (mCry) genes are major components of the circadian pacemaker (King and Takahashi 2000; Albrecht 2002). mCRY proteins are part of the negative limb in the transcriptional/translational feedback loop, whereas mPER2 is thought to act positively on Bmal1 expression (Shearman et al. 2000). In vitro studies point to multiple physical interactions between all mPER and mCRY proteins and posttranslational modifications such as phosphorylation and ubiquitylation, thereby offering a variety of putative regulation points for timed accumulation and nuclear appearance of clock proteins (Griffin et al. 1999; Kume et al. 1999; Field et al. 2000; Shearman et al. 2000; Yagita et al. 2000; Lee et al. 2001; Miyazaki et al. 2001; Vielhaber et al. 2001; Zheng et al. 2001; Yu et al. 2002). In particular there is evidence that mPER2 (GFP-tagged) shuttles between cytoplasm and nucleus and is ubiquitylated and degraded by the proteasome unless it is retained in the nucleus by mCRY proteins (Yagita et al. 2002). These findings implicate a regulatory effect of mCRY proteins on mPER2. However, the time course of protein availability, modification, and localization is difficult to resolve in model systems such as cell or slice cultures (Jagota et al. 2000; Hamada et al. 2001; Lee et al. 2001). In bacteria, yeast, Caenorhabditis elegans, and Drosophila, interactions of proteins have frequently been found through nonallelic supressor screens, that is, the restoration of a phenotype by introducing a mutation in another gene (Maine and Kimble 1989; Amin et al. 1999; Nakano et al. 2000; Grandin and Charbonneau 2001; LaJeunesse et al. 2001; Roy et al. 2002). We decided to further elucidate the functional relationship between mPer and mCry genes by studying their genetic interactions in the living animal. To this end, we inactivated the mCry1 or mCry2 gene in an mPer2 mutant mouse strain.
Results and Discussion
Per2Brdm1 mice, carrying a mutant mPer2 gene with a deletion in the PAS domain thought to be important for protein–protein interactions (Zheng et al. 1999), were crossed with mCry1−/− or mCry2−/− mice (van der Horst et al. 1999). The double-heterozygous offspring was intercrossed to produce wild-type and homozygous mutant animals. Per2Brdm1/mCry2−/− and Per2Brdm1/mCry1−/− mice (representative genotyping shown in Fig. 1a) were obtained at the expected Mendelian ratios, appeared fertile, and were morphologically indistinguishable from wild-type animals.
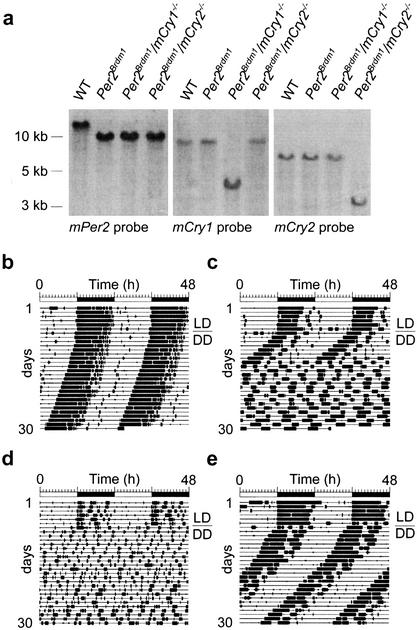
Figure 1.
Generation of mPer/mCry double mutant mice and representative locomotor activity records. (a) Southern blot analysis of wild-type (WT), Per2Brdm1, Per2Brdm1/mCry1−/−, and Per2Brdm1/mCry2−/− tail DNA. The Per2 probe hybridizes to a 12-kb wild-type and a 10-kb mutant fragment of BamHI-digested genomic DNA. The mCry1 probe detects a 9-kb wild-type and a 4-kb NcoI-digested fragment of the targeted locus. In mCry2 mutants, the wild-type allele is detected by hybridization of the probe to a 7-kb EcoRI fragment, whereas the mutant allele yields a 3.5-kb fragment. The left panel indicates size of DNA fragments. (b) Representative locomotor activity records of wild-type (WT), Per2Brdm1, Per2Brdm1/mCry1−/−, and Per2Brdm1/mCry2−/− mice. All animals were kept in a 12-h light:12-h dark cycle (LD) for at least 7 d before release into constant darkness (DD, indicated by the line over the DD). Activity is represented by black bars (three plot heights, >1, >10, and >20 wheel revolutions per 5-min period) and is double-plotted. The top bar indicates light and dark phases in LD. For the first 5 d in DD, wheel rotations per day were 20,021 ± 2,524 (n = 22) for wild-type animals, 17,656 ± 3,301 (n = 17) for Per2Brdm1 mutants, 16,025 ± 3,201 (n = 12) for Per2Brdm1/mCry1−/− mutants, and 17,859 ± 2,703 (n = 15) for Per2Brdm1/mCry2−/− mutants.
To determine the influence of inactivation of either mCry1 or mCry2 on circadian behavior of Per2Brdm1 mice, mutant and wild-type animals were individually housed in circadian activity-monitoring chambers (Albrecht and Oster 2001) for detection of wheel-running activity, an accurate measure of circadian rhythmicity. Mice were kept in a 12-h light:12-h dark cycle (LD 12:12, or LD) for several days to establish entrainment, and were subsequently kept in constant darkness (DD). Under LD conditions, homozygous Per2Brdm1, Per2Brdm1/mCry2−/− and Per2Brdm1/mCry1−/− animals displayed activity patterns similar to that of wild-type mice (Fig. 1b–e). In constant darkness, Per2Brdm1 mutant animals lost circadian rhythmicity after a few days (Fig. 1c), as described previously (Zheng et al. 1999, 2001; Bae et al. 2001). In contrast, Per2Brdm1/mCry1−/− mice lost circadian rhythmicity immediately upon release into DD (Fig. 1d), as also observed for mPer1−/−/Per2Brdm1 and mCry1−/−/mCry2−/− double mutant mice (van der Horst et al. 1999; Vitaterna et al. 1999; Bae et al. 2001; Zheng et al. 2001). Surprisingly, Per2Brdm1/mCry2−/− animals maintained a circadian rhythm in DD (Fig. 1e). Determination of the period length (τ) by χ2 periodogram analysis (using activity record intervals in which the circadian periodicity appeared stable on the activity record) revealed an average circadian period length of 23.8 ± 0.1 h (mean ± S.D., n = 22) for wild-type mice, 22.1 ± 0.3 h (n = 17) for Per2Brdm1 mice and 23.4 ± 0.2 h (n = 15) for Per2Brdm1/mCry2−/− animals. Interestingly, the period length of Per2Brdm1/mCry2−/− animals is the average of that of mCry2−/− mice (24.6 ± 0.1 h, van der Horst et al. 1999) and Per2Brdm1 mice. However, this might be coincidental. Wild-type and mutant mice all showed comparable levels of total wheel-running activity (see Fig. 1b legend), indicating that circadian period measurements were not influenced by aberrant running behavior.
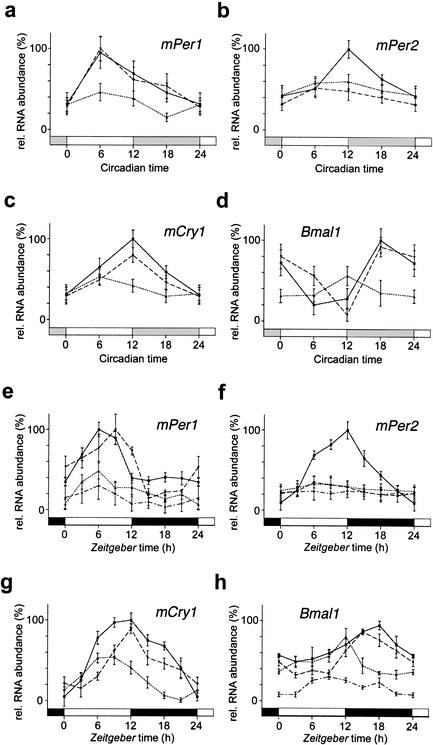
To unravel whether the rescue of circadian rhythmicity in Per2Brdm1 mutant animals by additional inactivation of the mCry2 gene was reflected at the molecular level, we examined the expression patterns of the mPer1, mPer2, mCry1, and Bmal1 clock genes in the SCNs of wild-type and mutant animals under DD (Fig. 2 a–d) and LD (Fig. 2 e–h) conditions. In situ hybridization experiments revealed that mPer1 expression in wild-type mice peaks at CT6, which is in line with previous reports (Sun et al. 1997; Tei et al. 1997). In Per2Brdm1 mutant animals, this rhythmicity is severely blunted (Fig. 2a). As might be expected from the behavioral data, mPer1 gene expression was back to a normal amplitude, reaching a maximum at CT6 in Per2Brdm1/mCry2−/− mutants (Fig. 2a). Thus, circadian expression of mPer1 in Per2Brdm1 mutants is rescued by inactivation of mCry2. A similar rescue was observed for mCry1 expression: both amplitude and timing of mCry1 oscillation in Per2Brdm1/ mCry2−/− mutant animals was not significantly different from that observed in wild-type animals (Fig. 2c). Per2Brdm1 mutant mice displayed abnormal Bmal1 mRNA rhythms, as evident from the reduced amplitude and shift of maximal expression levels to earlier CT times (Fig. 2d; Shearman et al. 2000). In the Per2Brdm1/ mCry2−/− mutants, however, both the phase and amplitude of Bmal1 expression were comparable to those of the wild-type animals (Fig. 2d). Oscillation of mutant mPer2 expression was not rescued in Per2Brdm1/ mCry2−/− mutants (Fig. 2b). Taken together, these data strongly indicate that normal circadian behavior and core oscillator performance is possible in the absence of functional mPer2 and mCry2 genes (Fig. 1e).
Figure 2.
In situ hybridization profiles of cycling clock genes in the SCN of wild-type (solid line), Per2Brdm1 (pointed line), Per2Brdm1/mCry2−/− (dashed line), and Per2Brdm1/mCry1−/− (point/dash line) mice kept in DD (a–d) or in a 12-h:12-h LD cycle (e–h). Each value is the mean ± S.D. (n = 3). Data at circadian time (CT) and Zeitgeber time (ZT) 0/24 are double-plotted. Bars on X-axis indicate light and dark phase. (a) mPer1 expression in DD; (b) mPer2 expression in DD; (c) mCry1 expression in DD; (d) Bmal1 expression in DD; (e) mPer1 expression in LD; (f) mPer2 expression in LD; (g) mCry1 expression in LD; (h) Bmal1 expression in LD.
We also found a rescue of the amplitude of mPer1, mCry1, and Bmal1 expression profiles in Per2Brdm1/mCry2−/− animals kept under LD conditions (Fig. 2 e,g,h), whereas mPer2 mRNA levels remained low as in the Per2Brdm1 mice (Fig. 2f). These results are comparable to the expression patterns observed under DD conditions (Fig. 2 a–d), except that in the Per2Brdm1 mutant mice, the amplitude of Bmal1 expression was almost as high as in wild-type animals (there is no statistically significant difference between both maxima; Fig. 2h; Shearman et al. 2000). The phase advance of Bmal1 expression in Per2Brdm1 mice compared to wild-type mice, which might explain the frequently observed early onset of wheel running activity before the beginning of the dark phase (Zheng et al. 1999), is also lost in Per2Brdm1/mCry2−/− animals (Fig. 2d). These data further underline the correction of the circadian phenotype of Per2Brdm1 mice by inactivation of mCry2.
As shown above, Per2Brdm1/mCry1−/− mice are arrhythmic in DD. To investigate whether these animals lack a circadian clockwork, we studied the expression patterns of the mPer1, mPer2, and Bmal1 clock genes under LD conditions. We found that in Per2Brdm1/mCry1−/− mutant animals, none of these genes is rhythmically expressed and mRNA levels are very low (Fig. 2e,f,h). This indicates that these animals lack a functional clock and that their diurnal behavioral activity under LD conditions (Fig. 1d) is most likely driven by the light/dark cycle. The loss of cyclic Bmal1 expression in the Per2Brdm1 mutant mice following inactivation of mCry1 suggests that mCry1 might play a role in transcriptional regulation of Bmal1.
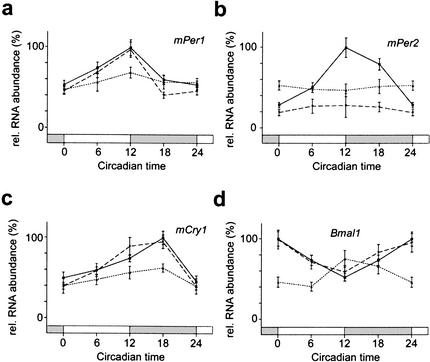
The experiments described above indicate that an inactivation of mCry2 in Per2Brdm1 mice rescues circadian rhythmicity at the behavioral level as well as at the molecular level in the SCN. To determine whether this is also valid for peripheral clocks, we performed Northern blot analysis on kidney tissue. The expression profiles of mPer1, mPer2, mCry1, and Bmal1 (Fig. 3a–d) under DD conditions are comparable to those observed in the SCN (Fig. 2 a–d), except that peak expression of these genes is delayed by several hours in the kidney (Zheng et al. 2001). Thus, rescue of circadian gene expression and probably clock function is also manifest in the periphery.
Figure 3.
Quantificaion of Northern blots identifying clock genes in the kidney of wild-type (solid line), Per2Brdm1 (pointed line), and Per2Brdm1/Cry2−/− (dashed line) mice kept in DD. Each value is the mean ± S.D. of two (mutants) or four (wild-type) different experiments. Data at circadian time (CT) 0/24 is double-plotted. Bars on X-axis indicate subjective night (gray) and day (white). (a) mPer1 expression in DD; (b) mPer2 expression in DD; (c) mCry1 expression in DD; (d) Bmal1 expression in DD.
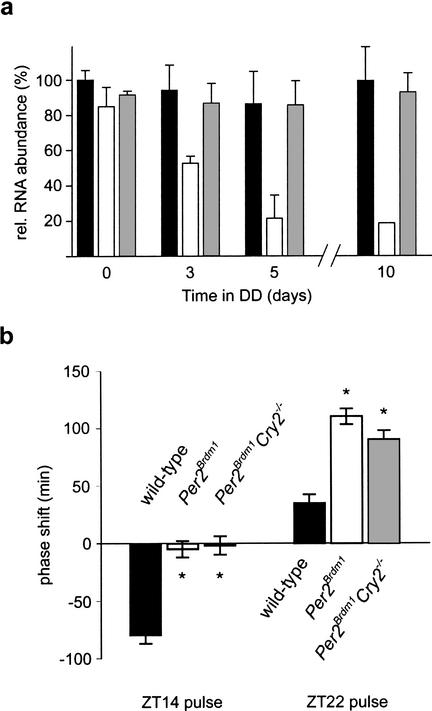
Inactivation of Bmal1 causes immediate arrhythmicity, indicating that its gene product is crucial for circadian rhythmicity (Bunger et al. 2000). Since in Per2Brdm1 mutant animals, Bmal1 mRNA cycling seems to be dampened under DD conditions (Figs. 2d, 3d), we investigated expression levels of the latter transcript at CT18 (wild-type and Per2Brdm1/mCry2−/− animals) and CT12 (Per2Brdm1 mice ) at 0, 3, 5, and 10 d after transfer of animals to constant darkness conditions. We found that in wild-type and Per2Brdm1/mCry2−/− mice, Bmal1 expression is maintained, whereas in Per2Brdm1 animals, this mRNA rhythm decreases gradually until it disappears after 5–10 d (Fig. 4a). This observation parallels the gradual loss of circadian rhythmicity of mPer2 mutant mice in DD (Fig. 1c; Zheng et al. 1999, 2001; Bae et al. 2001) and further supports the idea that the mPER2 protein is involved in regulating Bmal1 expression (Shearman et al. 2000). The mCRY1 and mCRY2 proteins were recently reported to activate the Bmal1 promoter in vitro (Yu et al. 2002). Because only mCry1−/−/mCry2−/− and mPer1−/−/Per2Brdm1 double mutant mice, but not the corresponding single mutant animals, display an immediate loss of circadian rhythmicity in constant darkness, neither mCRY1 and mCRY2 proteins alone nor mPER1 and mPER2 proteins alone are likely to be responsible for rhythmic Bmal1 transcriptional activation. Rather, Bmal1 gene expression appears to be regulated by a combination of mPer and mCry gene products. Analogous to mCry1−/−/mCry2−/− (van der Horst et al. 1999; Vitaterna et al. 1999) and mPer1−/−/Per2Brdm1 double mutant mice (Bae et al. 2001; Zheng et al. 2001), inactivation of both mPer2 and mCry1 leads to immediate loss of circadian rhythmicity (Fig. 1d). This implies that Per2Brdm1/mCry1−/− mutant mice are lacking a circadian clock and that mPER1 and mCRY2 proteins are not sufficient for maintenance of circadian rhythmicity and expression of clock genes such as Per1, Cry2, and Bmal1 (Figs. 2a,b,d; 3a,b,d). However, mice with inactivated mPer2 and mCry2 genes display stable circadian rhythmicity and normal Bmal1 mRNA cycling, indicating that mPER1 and mCRY1 proteins can maintain circadian rhythmicity and rhythmic Bmal1 expression in the absence of functional mPer2 and mCry2 genes. These data suggest that in Per2Brdm1 single mutant mice, the functional mCry2 gene product interferes with mPER1 and/or mCRY1, which leads to a gradual loss of circadian rhythmicity. Thus, mPer1 only in combination with mCry1 and in the absence of a functional mCry2 gene seems to be able to sustain the circadian clock in vivo. However, the resetting defect observed in Per2Brdm1 single mutant mice (Albrecht et al. 2001) was not rescued in Per2Brdm1/mCry2−/− mice (Fig. 4b). This indicates that in Per2Brdm1/mCry2−/− mice, PER1 can substitute PER2 in the core clock mechanism but not in the light input signaling pathway. As a note of caution, one should keep in mind that the phenotypic effect of genetic modifications of genetically altered animals in a nonhomogeneous genetic background are prone to epigenetic effects. However, we tried to minimize this risk by using double heterozygous animals throughout the crossings from which the wild-type and mutants were derived. Thus the wild-type control animals in this study have a mixed background similar to that of the mutants.
Figure 4.
(a) Bmal1 maximal expression in the SCN of wild-type (black column), Per2Brdm1 (white column), and Per2Brdm1/mCry2−/− (gray column) mice released into DD. Mice were adapted to a 12-h:12-h LD cycle and subsequently put into DD for 3, 5, or 10 subjective days. Only rhythmic animals were sacrificed at time points of maximal Bmal1 expression (CT18 for wild-type and Per2Brdm1/mCry2−/−; CT12 for Per2Brdm1 mice). Each value is the mean ± S.D. of three animals (except for day 10 in Per2Brdm1 mice with only one rhythmic animal). (b) Light-induced activity phase shifts in wild-type (black column), Per2Brdm1 (white column), and Per2Brdm1/mCry2−/− (gray column) mice. After entrainment to a 12-h:12-h LD cycle, animals were exposed to a 15-min light pulse (400Lux) at the beginning (ZT14) or at the end (ZT22) of the night and subsequently relased into DD. Negative values represent phase delays, and positive values phase advances. Each value is the mean ± S.D. of 10–14 animals. Only animals which remained rhythmic for at least 7 consecutive days in DD were used for quantification. Significance as indicated by asterisks was determined by one-way ANOVA with subsequent Bonferroni posttest (p < 0.05).
Our observations would be compatible with a hypothesis that there is a hierarchy of “activity potentials” for the four members of the negative limb (mPer1, mPer2, mCry1, mCry2) of the clock mechanism. It seems that CRY1 is a stronger repressor than CRY2, and PER2 is a more potent repressor than PER1. The activity potentials on the negative limb will have effects on the positive limb of the clock mechanism as well. Thus one could envisage a similar hierarchy of activity potentials for the positive limb or more precisely on activation of Bmal1. Assuming that all four CRY and PER proteins can be assembled into a protein complex with two CRY and two PER proteins, the activity potential of this complex is dependent on its composition. For example, a complex of two PER1 proteins and two CRY2 proteins would have a lower repressor potential than a complex composed of two PER2 and two CRY1 proteins. In turn, a complex that only forms in a wild-type animal composed of PER1, PER2, CRY1, and CRY2 would have an intermediate repressor potential. Assuming that circadian clocks are based on limit cycles of feedback-transcription to generate circadian rhythms (Glass and Mackey 1988; Leloup et al. 1999), too-low or too-high repressor potentials will destabilize the limit cycle, and the system will fall into equilibrium and become arrhythmic. Such a model might be further supported by the gene dosage effects observed in mCry double mutant mice. For instance, knocking out one mCry1 allele in mCry2−/− mice normalizes the behavioral rhythm, and knocking out one mCry2 allele in mCry1−/− mice further disturbs rhythmicity (van der Horst et al. 1999). In mPer1−/− animals, both PER subunits would be PER2 proteins with a high repressor potential, which accelerates the feedback loop, as has been observed (Zheng et al. 2001). However, the overall repressor potential is still compatible with the parameters of the limit cycle, and therefore the mPer1−/− mice are still rhythmic. In Per2Brdm1 mutant animals, both PER subunits are PER1 proteins that are weak repressors, and therefore the feedback loop is slowed down to a degree that is no longer compatible with the limit cycle; thus, Per2Brdm1 mice become arrhythmic. In Per2Brdm1/mCry2−/− mice, however, the repressor complex is composed of two CRY1 proteins (strong repressors) and two PER1 proteins (weak repressors). The overall repressor potential of this complex would approach that of the wild-type complex and is thus compatible with the parameters of the limit cycle. Per2Brdm1/mCry2−/− mice would therefore be rhythmic, which is compatible with our observations.
Taken together, our findings suggest that mCry2 can act as a nonallelic suppressor of mPer2 and that there might exist PER/CRY complexes with different transcriptional activity potentials, some of which are compatible with the parameters of a limit cycle whereas others are not.
Materials and methods
Generation of mPer and mCry mutant mice
We crossed mPer2Brdm1 mice (Zheng et al. 1999) with mCry1 and mCry2 knockout animals (van der Horst et al. 1999). The genotype of the offspring was determined by Southern blot analysis as described (Ramirez-Solis et al. 1993). Hybridization probes were for mPer2 as described by Zheng et al. (1999) and for mCry1 and mCry2 as described by van der Horst et al. (1999). Matching wild-type control animals were produced by back-crossing heterozygous animals.
Locomotor activity monitoring and circadian phenotype analysis
Mice housing and handling were performed as described (Albrecht and Oster 2001). For LD–DD transitions, lights were turned off at the end of the light phase and not turned on again the next morning. Activity records are double plotted so that each day cycle's activity is shown both to the right and below that of the previous day cycle. Activity is plotted in threshold format with three different thresholds set to >1, >10, and >20 wheel revolutions per 5-min period (Fig. 1). For activity counting, we used the ACS program of Simon Fraser University. For actograms and period determination, we used the Circadia program of Simon Fraser University. Period length was assessed by χ2 periodogram analysis using mice running in constant darkness for at least 10 d (5 d for Per2Brdm1).
For light-induced phase shifts, we used the Aschoff Type II protocol as described (Albrecht et al. 2001). We chose this protocol because mPer2Brdm1 mice become arrhythmic in constant darkness, precluding the determination of circadian times. Animals were entrained to an LD cycle for at least 7 d before the light administration [15 min bright white light (400Lux) at Zeitgeber time (ZT)14 or ZT22] and subsequently released into DD. The phase shift was determined by eye-fitting a line through at least 7 consecutive d of onset of activity in LD before the light pulse and in DD after the light pulse. The difference between the two lines on the day of the light pulse determined the value of the phase shift.
In situ hybridization
Mice were sacrificed by cervical dislocation under ambient light conditions at ZT6 and ZT12 and under a 15W safety red light at ZT18 and ZT0/24 as well as at circadian time (CT) 0, 6, 12, and 18. For DD conditions, animals were kept in the dark for 3 d before decapitation. Specimen preparation, 35S-UTP labeled riboprobe synthesis, and hybridization steps were performed as described (Albrecht et al. 1998). The probes for mPer1 and mPer2 were as described (Albrecht et al. 1997). The mCry1 probe was made from a cDNA corresponding to nucleotides 190–771 (accession no. AB000777) and the Bmal1 probe corresponding to nucleotides 654–1290 (accession no. AF015953). The mPer2 probe is located outside the region deleted in the mutant. Quantification was performed by densitometric analysis of autoradiograph films (Amersham Hyperfilm MP) using the NIH Image program after conversion into the relative optical densities by the 14C-autoradiographic microscale (Amersham). Data from the SCN were normalized with respect to the signal intensities in an equal area of the lateral hypothalamus. Three sections per SCN were analyzed. “Relative mRNA abundance” values were calculated by defining the highest value of each experiment as 100%.
Northern blot analysis
Rhythmic animals were sacrificed at the specified circadian time points on the third day in DD. Total RNA from kidney was extracted using RNAzol B (WAK Chemie). Northern analysis was performed using denaturing formaldehyde gels (Sambrook and Russel 2001) with subsequent transfer to Hybond-N membrane (Amersham). For each sample, 20μg of total RNA was used. cDNA probes were the same as described for in situ hybridization. Labeling of probes was done using the random prime labeling kit (Pharmacia) incorporating [P32]dCTP to a specific activity of 108 cpm/μg. Blots were hybridized using QuickHyb solution (Stratagene) containing 100 μg/mL salmon sperm DNA. The membrane was washed at 60°C in 0.1× SSC and 0.1% SDS. Subsequently, blots were exposed to phosphoimager plates (Bio-Rad) for 20 h, and signals were quantified using Quantity One 3.0 software (Bio-Rad). Signal intensities were normalized by comparison of the 18S ribosomal RNA bands' methylene blue staining on the blotted membrane. The highest clock gene signal was determined as 100% for each experiment.
Acknowledgments
We thank E. Maronde for comments on the manuscript. This work was supported by the Max-Planck-Society and grants from the Deutsche Forschungsgemeinschaft, the Swiss National Science Foundation, the Novartis Foundation (U.A.), and a SPINOZA premium from the Netherlands Organization for Scientific Research (NWO; G.T.J. v. d. H.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL urs.albrecht@unifr.ch; FAX 41-26-300-9735.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.233702.
References
- Albrecht U. Invited Review: Regulation of mammalian circadian clock genes. J Appl Physiol. 2002;92:1348–1355. doi: 10.1152/japplphysiol.00759.2001. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Oster H. The circadian clock and behavior. Behav Brain Res. 2001;125:89–91. doi: 10.1016/s0166-4328(01)00288-1. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Lu H-C, Revelli J-P, Xu X-C-, Lotan R, Eichele G. Studying gene expression on tissue sections using in situ hybridization. In: Adolph KW, editor. Human genome methods. NY: CRC Press; 1998. pp. 93–119. [Google Scholar]
- Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC. mPer1 and mPer2 are essential components for normal resetting of the circadian clock. J Biol Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- Amin NS, Tuffo KM, Holm C. Dominant mutations in three different subunits of replication factor C suppress replication defects in yeast PCNA mutants. Genetics. 1999;153:1617–1628. doi: 10.1093/genetics/153.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2 and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MD, Maywood ES, O'Brien JA, Weaver DR, Reppert SM, Hastings MH. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- Glass L, Mackey MC. From clocks to chaos. The rhythms of life. Princeton, NJ: Princeton University Press; 1988. pp. 22–25. [Google Scholar]
- Grandin N, Charbonneau M. Hsp90 levels affect telomere length in yeast. Mol Genet Genomics. 2001;265:126–134. doi: 10.1007/s004380000398. [DOI] [PubMed] [Google Scholar]
- Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Hamada T, LeSauter J, Venuti JM, Silver R. Expression of Period genes: Rhythmic and nonrhythmic compartments of the suprachiasmatic nucleus pacemaker. J Neurosci. 2001;21:7742–7750. doi: 10.1523/JNEUROSCI.21-19-07742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagota A, de la Iglesia HO, Schwartz WJ. Morning and evening circadian oscillations in the suprachiasmatic nucleus in vitro. Nat Neurosci. 2000;3:372–376. doi: 10.1038/73943. [DOI] [PubMed] [Google Scholar]
- King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu Rev Neurosci. 2000;23:713–742. doi: 10.1146/annurev.neuro.23.1.713. [DOI] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- LaJeunesse DR, McCartney BM, Fehon RG. A systematic screen for dominant second-site modifiers of Merlin/NF2 phenotypes reveals an interaction with blistered/DSRF and scribbler. Genetics. 2001;158:667–679. doi: 10.1093/genetics/158.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Leloup JC, Gonze D, Goldbeter A. Limit cycle models for circadian rhythms based on transcriptional regulation in Drosophila and Neurospora. J Biol Rhythms. 1999;14:433–448. doi: 10.1177/074873099129000948. [DOI] [PubMed] [Google Scholar]
- Maine EM, Kimble J. Identification of genes that interact with glp-1, a gene required for inductive cell interactions in Caenorhabditis elegans. Development. 1989;106:133–143. doi: 10.1242/dev.106.1.133. [DOI] [PubMed] [Google Scholar]
- Miyazaki K, Mesaki M, Ishida N. Nuclear entry mechanism of rat PER2 (rPER2): Role of rPER2 in nuclear localization of CRY protein. Mol Cell Biol. 2001;21:6651–6659. doi: 10.1128/MCB.21.19.6651-6659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Zhu Y, Liu J, Reyes DY, Yoshikawa H, Zuber P. Mutations conferring amino acid substitutions in the C-terminal domain of RNA polymerase alpha can suppress clpX an clpP with respect to developmentally regulated transcription in Bacillus subtilis. Mol Microbiol. 2000;37:869–884. doi: 10.1046/j.1365-2958.2000.02052.x. [DOI] [PubMed] [Google Scholar]
- Ramirez-Solis R, Zheng H, Whiting J, Krumlauf R, Bradley A. Hoxb-4 (Hox-2.6) mutant mice show homeotic transformation of a cervical vertebra and defects in the closure of the sternal rudiments. Cell. 1993;73:279–294. doi: 10.1016/0092-8674(93)90229-j. [DOI] [PubMed] [Google Scholar]
- Roy F, Laberge G, Douziech M, Ferland-McCollough D, Therrien M. KSR is a scaffold required for activation of the ERK/MAPK module. Genes & Dev. 2002;16:427–438. doi: 10.1101/gad.962902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular cloning. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee CC. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- Vielhaber EL, Duricka D, Ullman KS, Virshup DM. Nuclear export of mammalian PERIOD proteins. J Biol Chem. 2001;8:8. doi: 10.1074/jbc.M107726200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K, Yamaguchi S, Tamanini F, van Der Horst GT, Hoeijmakers JH, Yasui A, Loros JJ, Dunlap JC, Okamura H. Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes & Dev. 2000;14:1353–1363. [PMC free article] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van Der Horst GT, Okamura H. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Nomura M, Ikeda M. Interactivating feedback loops within the mammalian clock: BMAL1 is negatively autoregulated and upregulated by CRY1, CRY2, and PER2. Biochem Biophys Res Commun. 2002;290:933–941. doi: 10.1006/bbrc.2001.6300. [DOI] [PubMed] [Google Scholar]
- Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]