Abstract
The genome of the human hepatitis delta virus (HDV) harbors a self-cleaving catalytic RNA motif, the genomic HDV ribozyme, whose crystal structure shows the dangling nucleotides 5′ of the cleavage site projecting away from the catalytic core. This 5′-sequence contains a clinically conserved U − 1 that we find to be essential for fast cleavage, as the order of activity follows U − 1 > C − 1 > A − 1 > G − 1, with a >25-fold activity loss from U − 1 to G − 1. Terbium(III) footprinting detects conformations for the P1.1 stem, the cleavage site wobble pair and the A-minor motif of the catalytic trefoil turn that depend on the identity of the N − 1 base. The most tightly folded catalytic core, resembling that of the reaction product, is found in the U − 1 wild-type precursor. Molecular dynamics simulations demonstrate that a U − 1 forms the most robust kink around the scissile phosphate, exposing it to the catalytic C75 in a previously unnoticed U-turn motif found also, for example, in the hammerhead ribozyme and tRNAs. Strikingly, we find that the common structural U-turn motif serves distinct functions in the HDV and hammerhead ribozymes.
INTRODUCTION
Underlying the ability of ribozymes to accelerate site-specific phosphodiester transfer 107-fold are many of the same catalytic strategies that protein enzymes employ (1–3). For example, general acid–base catalysis (4–11), an energetic contribution of the 5′-sequence to lowering the energetic barrier of catalysis (12–14), and global and local conformational changes around a strained cleavage site backbone to drive the reaction forward (9,14–17) have all been proposed to contribute to hepatitis delta virus (HDV) ribozyme catalysis, essential for double-rolling circle replication of the human pathogenic HDV (18).
The genomic and antigenomic forms of the HDV ribozyme catalyze the self-(cis-)cleavage of a specific phosphodiester bond by a transesterification reaction, which requires deprotonation of a specific 2′-OH group and its nucleophilic attack on the adjacent scissile phosphate. After stabilizing the negative charge of the 5′-oxyanion leaving group by protonation, the reaction generates two products, the 5′-sequence with 2′,3′-cyclic phosphate and the 3′-product with 5′-OH terminus (2,19,20). C75 of the trefoil turn motif in joiner J4/2 (Figure 1) is crucial for ribozyme activity as determined by mutagenesis studies (21–23). Crystallographic studies of the self-cleaved 3′-product form of the genomic ribozyme supported the notion of a direct catalytic function of C75 (4,7). A role of the corresponding nucleotide in the antigenomic ribozyme form, C76, as a general base catalyst to activate the 2′-OH group of the −1 nucleotide (N − 1) was suggested by rescue of cleavage in a C76U mutant by imidazole with an apparent pH dependence consistent with base catalysis (5). In contrast, the slow cleavage with inverted pH profile observed for the genomic ribozyme in the absence of Mg2+ and presence of molar Na+ concentrations supported a model in which C75 acts as the general acid to protonate the 5′-oxyanion leaving group (6). This residual activity at low pH in the presence of Na+ was later, however, attributed to the structurally essential protonation of C41 in a quadruple interaction buttressing the catalytic core (24).
Figure 1.
Sequence and structure of the cis-acting genomic HDV ribozyme used in this study. (A) Secondary structure of the genomic HDV ribozyme with the set of 5′-sequences (green) immediately upstream of the cleavage site (open arrow) as employed in our cleavage and footprinting experiments. Nucleotides in color correspond to important structural elements in the catalytic core. Red dashed lines, functionally relevant tertiary interactions. (B) Secondary structure of the truncated sequence used in our MD simulations, color coded as in (A). (C) Backbone ribbon representation of the precursor crystal structure (9), color-coded as in (A and B).
In contrast to the conformation of the catalytic pocket in the product crystal structure, the precursor crystal structure depicts base 75 retracted from the 5′-oxygen leaving group by ∼2 Å (9). Interpreting their new structure in light of divergent mechanistic models, Doudna and co-workers proposed that catalysis occurs when U − 1 undergoes a rotation, bringing its 2′-OH into hydrogen-bonding distance with N3 of C75. According to this model, C75 acts as the general base catalyst to position and activate the 2′-OH for in-line attack on the scissile phosphate. Molecular dynamics (MD) simulations are consistent with this model as they observe the necessary conformational change (25,26). Additional mechanistic data obtained from activity assays on a hyperactivated external RNA substrate carrying a 5′-phosphorothiolate, however, have recently again contradicted this model (10). The substrate specifically suppressed the deleterious effects of C75 mutations and pH changes, thereby linking the protonation of C75 to leaving-group stabilization by general acid catalysis. As a chemical modification was introduced at the scissile phosphate group to accelerate leaving group departure, however, it cannot be ruled out that the reaction pathway and/or active site architecture may be affected in this relatively slowly cleaving trans-acting model system compared to the naturally cis-acting ribozymes. Clearly, more studies are needed to understand structure, dynamics and ultimately catalysis in the HDV ribozyme.
Most small catalytic RNAs, such as the hammerhead, hairpin and VS ribozymes, carry cleavage sites that are constrained by flanking upstream and downstream helices. The cleavage site in the HDV ribozyme is unique in that it is located at the junction of the dangling, single-stranded, so-called 5′-sequence and a G1:U37 wobble pair that closes the 7-bp P1 helix (Figure 1) (27). There is a specific requirement for the presence of a G1:U37 wobble pair since changes to either base reduce activity in the genomic (28) and antigenomic sequences (23,29), a A1:C37 wobble combination is the most effective substitution for the G1:U37 pair (23), and both stacking energy and placement of the 5′-leaving group resulting from the wobble pair are essential for activity (30). In addition, the cleavage reaction requires, at a minimum, the presence of a single dangling nucleotide 5′ of the scissile phosphate of G1 (31). It was shown that this 5′-sequence impacts catalysis by lowering the energetic barrier for cleavage in trans-acting HDV ribozyme model systems (12–14). A plausible explanation for the observed ground-state destabilization came from observations by fluorescence resonance energy transfer (FRET), NMR and terbium(III)-mediated footprinting, indicating that local and global conformational changes occur upon cleavage and 5′-sequence dissociation; such changes may facilitate catalysis by better positioning the catalytic C75 toward the reactive groups to an extent that depends on the 5′-sequence (14–17,32,33). More specifically, we showed that varying the length and composition of the 5′-sequence results in relatively subtle differences in catalytic core conformation, which translate into significant changes in global structure in the trans-acting HDV ribozyme (33). Hence, the identity of the 5′-sequence modulates HDV ribozyme structure at local and global levels, influencing the active conformation and resulting in distinct catalytic activities.
Crystallographic studies revealed details of the structural impact of the 5′-sequence on the precursor form of the cis-acting genomic HDV ribozyme. The 5′-sequence is wedged between the P1|P1.1|P4 and P2|P3 stacks, thus widening the gap between the stacks in comparison to the 3′-product structure (9). In addition, the U − 1 nucleotide is bent away from G1 in a sharp ∼180° turn about the scissile phosphate, packing the substrate between P1 and P3 (Figure 1). This bend may destabilize the reaction precursor in a way to favor a transition-state configuration of the scissile phosphate and decrease the entropic barrier for cleavage. From this notion follows the hypothesis that structure and dynamics of the scissile backbone and the adjacent 5′-sequence, particularly N − 1, may impact catalysis of the genomic HDV ribozyme as it does for trans-acting HDV ribozymes. Neither the extent nor the structural origin of such an impact on genomic HDV ribozyme catalysis has been determined yet, prompting us to test our hypothesis.
To this end, here we have used complementary site-directed mutagenesis, terbium(III) footprinting and MD simulations to show that U − 1, conserved in all clinical isolates of HDV, is essential for fast cleavage, with an order of activity of U − 1 > C − 1 > A − 1 > G − 1. Conformations of the P1.1 stem, the cleavage site wobble pair and the A-minor motif of the catalytic trefoil turn depend on the identity of the N − 1 base, with the most tightly folded conformation of the catalytic core, resembling the 3′-product crystal structure, found in the U − 1 wild-type. Furthermore, we find that the propensity of the 5′-sequence to form interactions with adjacent residues impacts the molecular dynamics of the backbone around the cleavage site. U − 1 leads to the most robust kink around the scissile phosphate, exposing the cleavage site to the catalytic C75 in a previously unnoticed U-turn motif found also in the hammerhead ribozyme catalytic core, in tRNAs, in HIV-1 genomic RNA and in large structured RNAs. Our results demonstrate that a common structural motif, the U-turn, is used in distinct ways to accelerate catalysis in both the HDV and hammerhead ribozymes, and they provide an improved structural framework for understanding HDV ribozyme dynamics and function.
MATERIALS AND METHODS
Preparation of RNA
The cis-acting precursor variants of the genomic HDV ribozyme (Figure 1A) were generated by run-off transcription from a double-stranded, PCR amplified template that encodes an upstream T7 promoter. Transcription reactions contained 40 mM Tris-HCl (pH 7.5), 15 mM MgCl2, 5 mM dithiothreitol, 2 mM spermidine, 4 mM each rNTP, 5 units/ml inorganic pyrophosphatase and 0.1 mg/μl T7 RNA polymerase and were incubated at 10°C (to avoid extensive self-cleavage) for 16 h. The enzyme was removed by phenol–chloroform extraction and the RNA was concentrated using Centricon YM-3 (3 kDa cutoff) ultrafiltration. The full-length precursor transcripts were isolated after denaturing, 8 M urea, 8% (w/v) polyacrylamide gel electrophoresis by UV shadowing, diffusion elution of small gel slices into a chelated 1 mM EDTA (pH 8.0) solution overnight at 4°C, from which they were recovered by ethanol precipitation, then diluted into chelated 0.1 mM EDTA (pH 8.0) and stored at −20°C. The chelated EDTA solution had been passed over the sodium form of Chelex100 to remove residual metal ions.
For cleavage reactions, radiolabeled precursors were transcribed as described above, except that 0.4 mCi of [α-32P] GTP were added to the reaction mixture. The transcription reaction was incubated at 10°C for 24 h and the RNA was fractionated by electrophoresis on denaturing, 8 M urea, 8% (w/v) polyacrylamide gels. Uncleaved precursor RNA was located by autoradiography, excised, eluted into chelated 1 mM EDTA (pH 8.0) overnight at 4°C and recovered by ethanol precipitation. The radiolabeled precursors were stored in chelated 0.1 mM EDTA (pH 8.0) at −20°C.
Cleavage assays
To ensure proper folding, radiolabeled precursor forms of the cis-cleaving HDV ribozyme (Figure 1A) were heated to 90°C for 2 min in a buffer containing 5 mM Tris-HCl (pH 7.5), 0.5 mM spermidine and 1 mM EDTA. Each precursor was then pre-incubated at 37°C for 10 min, after which the reactions were adjusted to the final pH with a buffer containing 25 mM acetic acid, 25 mM MES, 50 mM Tris-HCl (pH 7.5). These mixtures were incubated for an additional 5 min at room temperature (22°C), after which cleavage was initiated with the addition of magnesium dichloride (final concentration) at room temperature using two different mixing techniques. For fast-cleaving ribozymes, 4 μl aliquots of the Mg2+-free reaction mix were distributed to the wells of a microplate. Cleavage at room temperature was initiated by addition of an equal volume of a solution containing 22 mM MgCl2 (to a final concentration of ∼10 mM free magnesium), 0.2 mM spermidine and 0.4 mM EDTA. Cleavage kinetics were followed by quenching individual aliquots at specified times with 8 μl of 80% (v/v) formamide, 0.025% (w/v) xylene cyanol, 0.025% (w/v) bromophenol blue and 50 mM EDTA. In the second mixing technique used for slower cleaving ribozymes, a larger volume of reaction mix was initiated with the addition of an equal volume of 22 mM MgCl2, 0.2 mM spermidine and 0.4 mM EDTA. Here, 8-μl reaction aliquots were taken at appropriate time intervals and the reaction quenched with 8 μl of 80% (v/v) formamide, 0.025% (w/v) xylene cyanol, 0.025% (w/v) bromophenol blue and 50 mM EDTA. In both cases, the reaction product was separated from the precursor by denaturing gel electrophoresis on 8 M urea, 8% (w/v) polyacrylamide gels, and was quantified and normalized to the sum of the precursor and product bands using a PhosphorImager Storm 840 instrument with Image Quant software (Molecular Dynamics). Time traces of product formation were fit with the single-exponential first-order rate equation y = y0 + A1 (1 - e-t/ τ1, employing Marquardt–Levenberg nonlinear least-squares regression (Microcal Origin 7.0), where A1 is the amplitude of the fraction cleaved and 1/τ1 is the reported cleavage rate constant kcleav. Errors were obtained from the standard deviation of at least three independent measurements. To derive apparent magnesium-binding affinities, cleavage assays were performed as described above with the MgCl2 concentration varying between 0 and 11 mM. The concentration of free magnesium cation (Mg2+) was calculated by subtraction of the EDTA concentration present in the reaction mixture from the added Mg2+ concentration. The dependency of the rate constant on free Mg2+ was fit with the cooperative binding equation:
 |
yielding the metal ion half-titration points, Mg1/2, reported in Figure 2C with cooperativity coefficients of n = 1.
Figure 2.
Self-cleavage activity of the cis-acting genomic HDV ribozyme. (A) Cleavage time courses under standard conditions (see Materials and methods section) measured for ribozyme variants with a pyrimidine in the −1 position (U − 1, wild-type with 5′-sequence GAU; C − 1 with 5′-GAC). Data were fit with single-exponential functions to yield the reported rate constants kcleav. (B) Cleavage time courses under standard conditions for ribozyme variants with a purine in the −1 position (A − 1 with 5′-GAA; G − 1 with 5′-GAG). (C) Mg2+ dependence of activity of the HDV ribozyme variants of (A) and (B). Data were fit with a binding equation to yield the indicated Mg2+ half-titration points Mg1/2 and a cooperativity coefficient of 1 (see Materials and methods section). (D) Cleavage time courses under standard conditions for ribozyme variants with the indicated 5′-sequences, found in clinical isolates of HDV. Data were fit with single-exponential functions to yield the reported rate constants kcleav.
Terbium(III)-mediated footprinting
The highest purity terbium(III) chloride (99.9%) was purchased from Sigma-Aldrich. TbCl3 stock solutions at 100 mM were prepared in 5 mM sodium cacodylate (pH 5.5) and stored in small aliquots at −20°C to prevent formation of insoluble hydroxide species (34). To observe the slow backbone scission mediated by Tb(OH)(aq)2+, purified cis-acting precursor forms of the genomic HDV ribozymes (Figure 1A) were (3′-32P)-phosphorylated with T4 RNA ligase and [γ-32P]pCp at 4°C overnight. The labeling reaction was terminated by addition of EDTA (pH 8.0) to a final concentration of 50 mM. Since partial self-cleavage occurred during the labeling reaction, both reaction precursor and product were re-purified by denaturing, 8 M urea, 8% (w/v) polyacrylamide gel electrophoresis, followed by diffusion elution into chelated 1 mM EDTA (pH 8.0) and ethanol precipitation, as described previously (17). The radiolabeled RNA (1 00 000 c.p.m. per 10 μl reaction volume) was pre-annealed in 5 mM Tris-HCl (pH 7.5), 0.5 mM spermidine, denatured at 90°C for 2 min and incubated at 37°C for 10 min. The final pH was adjusted with a buffer containing 25 mM acetic acid, 25 mM MES and 50 mM Tris-HCl (pH 7.5). To fold the ribozyme into an active conformation, a mixture of Mg2+ (11 mM final concentration) with spermidine (0.1 mM) and Tb3+ (5 mM) was added and incubated for 30 min at room temperature (22°C). Backbone scission was slow under these conditions so that only a small fraction of the RNA is cut by terbium(III), avoiding secondary hits on an RNA molecule already cut. The scission reaction was stopped by addition of EDTA (pH 8.0) to a final concentration of 50 mM, followed by ethanol precipitation overnight at −20°C. The precipitated RNA was redissolved in urea loading buffer [80% formamide, 0.025% xylene cyanol, 0.025% bromophenol blue, 9 M urea, 50 mM EDTA (pH 8.0)] and analyzed on a wedged 8 M urea, 15% (w/v) polyacrylamide sequencing gel, alongside sequencing ladders from partial digestion with G-specific RNase T1 and from alkaline hydrolysis as described (15,17,33,35). Product bands were directly visualized using autoradiography and quantified using a PhosphorImager Storm 840 with ImageQuant software (Molecular Dynamics).
Initial structures for MD simulations
Trajectories were analyzed from MD simulations of initial structures based on the precursor crystal structure of the HDV ribozyme, PDB ID 1SJ3, that contains a Mg2+ cation in the active site and a C75U mutation (9). To obtain our initial structures, loop L4 was removed and U75 replaced in the PDB file by C75. The crystal structures 1SJ3 and 1SF3 were overlapped in order to add the nucleotide A − 2, contained only in 1SF3. The average structure after 100 ps of simulation Pre-5AU MD was used to construct the initial structure for the Pre-5GAU simulation (with 5′-sequence 5′-GAU-3′), where nucleotide G − 3 was modeled using the program InsightII. Mutations were manually introduced into the –1 position of the equilibrated structure of wild-type simulation Pre-5GAU using InsightII to yield simulations Pre-GAC, Pre-5GAA and Pre-5GAG. C41(N3) was protonated to assure the formation of the structurally critical A43:C41H+:G73:C44 base quadruple in the J1.1/4 junction (25,26).
MD simulations
All MD simulations (20 ns for each system) were carried out using the AMBER7.0 program package (36) with the parm99 Cornell et al. force field (37–39). The RNA was solvated in a rectangular box of TIP3P waters (40) extended to a distance of ≥10 Å from any solute atom. The single resolved Mg2+ cation at the active site was excluded from the initial structures to avoid possible artifacts from the imperfect description of divalents by the force field and uncertainty in the exact coordination of the ion after replacing the crystal U75 with C75 (25,26). The simulated system was neutralized with Na+ cations initially placed by the LeaP module at points of favorable electrostatic potential close to the RNA (41). This corresponds to an ion concentration of ∼0.2 M. The Sander module of AMBER7.0 was used for the equilibration and production runs using our standard protocols (25,26,42). The particle mesh Ewald method (43) was applied with a heuristic pair list update, using a 2.0-Å nonbonded pair list buffer and a 9.0 Å cutoff. A charge grid spacing of close to 1 Å and a cubic interpolation scheme were used. The production runs were carried out at 300 K with constant-pressure boundary conditions using the Berendsen temperature coupling algorithm (44) with a time constant of 1.0 ps. SHAKE (45) was applied in the simulations with a tolerance of 10−8 to constrain bonds involving hydrogen.
Analysis of MD trajectories
MD trajectories were analyzed using the carnal and ptraj modules of the AMBER7.0 and 8.0 packages, and the ptraj module of AMBER8.0 was used to obtain cross-correlation matrices, which were plotted using Mathematica5.2. Structures were visualized using the program VMD (46). Time trajectories of heavy-atom distances and angles were monitored using Microcal Origin 7.0. The occupancy criterion for a hydrogen bond was defined as the time in percentage during which the hydrogen bond distance between the heavy atoms (donor (D) and acceptor (A)) was ≤3.0 Å and the hydrogen (H) bond angle of D-H-A was ≥120°.
RESULTS
The nucleotide immediately 5′ of the active site affects catalysis in the cis-acting genomic HDV ribozyme
Experimental studies have provided evidence for ground-state destabilization associated with the 5′-sequence in trans-acting ribozyme–substrate complexes (12,13,33). FRET, NMR and crystallographic studies have reported differences in conformation between precursor and product ribozymes that vary only in their 5′-sequence (4,7,9,14,16,17,32,33,35). To investigate the impact of the 5′-sequence on conformational dynamics and function of the cis-acting ribozyme, here we have designed several sets of cis-cleaving genomic HDV ribozymes with short 5′-sequences and varying N − 1 residue (Figure 1) and have studied them by a combination of experimental and simulation techniques.
Cleavage assays carried out on the simplest set of four constructs carrying U − 1, A − 1, C − 1 and G − 1 in the context of the 3-nt 5′-sequence 5′-GAN-3′ (Figure 1A) under standard conditions (25 mM acetic acid, 25 mM MES, 50 mM Tris-HCl (pH 7.5), 11 mM MgCl2, 0.1 mM spermidine, at 22°C) revealed an order of activity of U − 1 > C − 1 > A − 1 > G − 1, with the fastest construct, U − 1, cleaving at (19.9 ± 1.9) min−1 and the slowest construct, G − 1, cleaving at (0.75 ± 0.03) min−1 (Figure 2A and B). We find that a majority of molecules in our cleavage assays fold into an active conformation as indicated by the generally high cleavage extent of ∼78% for all four variants (Figure 2A and B). The cleavage rate constant of the U − 1 ribozyme, the wild type, is with ∼20 min−1 comparable to previously reported values for the wild type genomic ribozyme (24,47). Interestingly, the cleavage rate constants of the least active A − 1 [(1.5 ± 0.1) min−1] and G − 1 variants ((0.75 ± 0.03) min−1) are rather similar to those of wild-type trans-acting ribozymes, in which removal of typically the J1/2 joiner and/or the capping loop on P4 lowers the catalytic activity by about an order of magnitude, presumably by depopulating the catalytic conformation ∼10-fold (12,14,16,48).
Higher Mg2+ affinities of purines in the −1 position coincide with lower catalytic activities
Doudna and co-workers identified six functional groups within the outer coordination sphere of a presumably catalytically involved metal ion in the active site of the genomic HDV ribozyme precursor, including one of the non-bridging phosphate oxygens of U − 1 (9). To ask whether mutation of U − 1 simply alters the affinity towards this catalytic metal ion as a possible mechanism for the observed mutagenesis effects, we determined the dependence of the cleavage rate constant on the Mg2+ concentration (Figure 2C). We chose reaction conditions for which Bevilacqua and co-workers previously demonstrated that they specifically probe the catalytic metal-binding site, since the higher affinity structural sites are saturated under these conditions. As Na+ ions compete for the structural sites, these conditions require a low ionic strength background with no added monovalents (49,50). As shown in Figure 2C, the U − 1, C − 1, A − 1 and G − 1 5′-sequences yield binding isotherms that are well fit with a cooperativity coefficient of n = 1 (i.e. no cooperativity between metal ions), resulting in Mg2+ half-titration points, Mg1/2, of 1.8, 3.5, 0.6 and 0.5 mM, respectively. The value for U − 1 is in accord with prior studies of the wild-type conducted under similar solution conditions (6,50). Our results lead to two conclusions: (i) Slow mutants (particularly A − 1 and G − 1) cannot be activated to wild-type levels by high Mg2+ concentrations, suggesting that the mechanism of mutagenic interference is not simply a weakened affinity for the catalytic metal ion. (ii) The purines A − 1 and G − 1, which carry an additional N7 imino group as a well-known metal ion chelator, exhibit up to 7-fold higher Mg2+ affinities than their pyrimidine counterparts U − 1 and C − 1, which lack this metal chelator. These higher Mg2+ affinities of the purine derivatives, however, coincide with catalytic deficiency.
The clinically conserved U − 1 is obligatory for efficient catalysis by the cis-acting genomic HDV ribozyme
Phylogenetic comparison of 22 clinically isolated HDV sequences reveals that the only nucleobase found at the −1 position is uracil. The −2 and −3 positions vary, with the most prevalent sequences, aside from the 5′-GAU-3′ wild-type, being 5′-UGU-3′, 5′-UUU-3′ and 5′-CGU-3′ (27). We therefore constructed an additional set of three cis-acting genomic ribozymes with these trinucleotides in the 5′-sequence to further test the functional role of U − 1 in the HDV genome; we added a common G − 4 nucleotide for efficient radioactive in vitro transcription (see Materials and methods section). We find that the constructs carrying the 5′-UGU-3′, 5′-UUU-3′ and 5′-CGU-3′ 5′-sequences are all fast-cleaving ribozymes, with rate constants of (24.9 ± 1.1) min−1, (18.7 ± 1.2) min−1 and (12.2 ± 0.4) min−1, respectively (Figure 2D), supporting the notions that U − 1 was naturally selected as the catalytically most effective N − 1 and that the nucleotides in positions −2 and −3 are far less critical. Taken together, our observations indicate an important functional role in the cis-acting genomic HDV ribozyme for the physicochemical properties contributed by a U − 1. A slight reduction in the extent of cleavage from 76 to 66% observed for the UUU 5′-sequence in comparison to the UGU and CGU 5′-sequences (Figure 2D) suggests that the UUU trinucleotide enhances ribozyme misfolding.
No hydrogen-bonding contact between U − 1 and G25 as basis for their conservation
The helical stack of P3 is continued by the U20 and G25 bases (Figure 1), which form different types of wobble interactions in the precursor and product forms of the ribozyme (4,7,9,26). Interestingly, a G25A mutation that would stabilize standard Watson–Crick base pairing with U20 reduces HDV ribozyme activity ∼3000-fold (22), consistent with the notion that hydrogen bonding between positions 20 and 25 must be sufficiently weak for catalysis to occur. We therefore tested the possibility that U − 1 may be forming a functionally important, transient contact with G25, a configuration we observed in one of our MD simulations wherein U − 1 and G25 formed a wobble pair (data not shown). A similar interaction between, for example, G − 1 and G25 would not be feasible, which could explain the lower catalytic activity associated with G − 1. We therefore designed a G25U mutant, which is expected to form wobble interactions with G − 1, but not U − 1; the presence of an interaction between U − 1 and G25 would be supported by the observation of a switched preference of the G25U mutant, with higher activity in the context of the GAG than the GAU 5′-sequence. However, this was not observed (data not shown), providing no support for a catalytically relevant transient formation of hydrogen bonds between U − 1 and G25.
Terbium(III) footprinting highlights structural differences in the catalytic core in dependence of the identity of N − 1
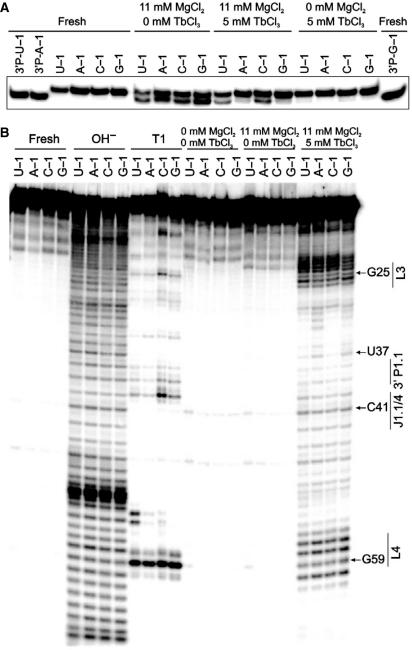
The lanthanide terbium(III) is a straightforward and useful probe for secondary and tertiary structure in RNA (35,51,52). High (millimolar) concentrations of Tb3+ ions bind relatively non-specifically to RNA and thus result in backbone scission in a sequence-independent manner, preferentially cutting solvent accessible, single-stranded or non-Watson–Crick base-paired regions. By applying terbium(III) footprinting to our U − 1, A − 1, C − 1 and G − 1 containing cis-acting genomic HDV ribozyme variants, we tested whether the conformation of the catalytic pocket changes depending on the identity of the base in the −1 position; we then sought to correlate observed conformational changes with changes in cleavage activity. As a control, we first performed standard cleavage assays in the presence of 5 mM terbium(III) and found that the lanthanide ion strongly inhibits activity of the genomic ribozyme, as previously shown for the hairpin (34) and antigenomic HDV ribozymes (17). [Please note that self-cleavage of the properly annealed ribozyme in 11 mM Mg2+, in the absence of Tb3+, is already slowed down under these (low temperature) conditions (Figure 3A)]. Yet inhibition is incomplete, as all four genomic HDV ribozyme variants still self-cleave to different (small) extents over the footprinting incubation period (30 min) when both 11 mM Mg2+ and 5 mM Tb3+ are added (to ∼37, 18, 34, and 20% in case of the U − 1, A − 1, C − 1 and G − 1 ribozymes, respectively; lanes labeled ‘11 mM MgCl2, 5 mM TbCl3’ in Figure 3A). In contrast, inhibition is complete in the absence of Mg2+ (lanes labeled ‘0 mM MgCl2, 5 mM TbCl3’ in Figure 3A). We therefore probed and compared the structure of each precursor variant with 5 mM Tb3+ in both the presence and absence of 11 mM Mg2+. Each precursor displayed a different terbium(III) scission pattern, identical in the presence and absence of Mg2+ (data not shown) and clearly distinct from that of the reaction product (see below), suggesting that: (i) The footprinting pattern under the strongly inhibited plus-Mg2+ condition is largely dominated by the excess of uncleaved precursor and thus represents the precursor footprint. (ii) Secondary and tertiary structure of the cis-acting genomic HDV ribozyme do not significantly differ in 5 mM Tb3+ in the presence and absence of 11 mM Mg2+.
Figure 3.
Terbium(III)-mediated footprinting of the four 3′-32P-labeled cis-acting genomic HDV ribozyme variants. (A) Control experiment of background cleavage in the presence of Tb3+. The HDV ribozyme N − 1 variants were incubated for 30 min at 22°C under the indicated ionic conditions (see also Materials and methods section). We then separated the 88-nt reaction precursor from the 85-nt, faster migrating 3′-product, as identified by comparison with purified 3′-product from self-cleavage of the U − 1, A − 1 and G − 1 variants (3′P-N − 1). ‘Fresh’ indicates that the material was not incubated before loading onto the gel. (B) Terbium(III)-mediated footprinting of genomic HDV ribozyme variants. As in (A), the HDV ribozyme N − 1 variants were incubated for 30 min at 22°C under the indicated ionic conditions and then analyzed on a sequencing gel alongside alkaline hydrolysis (OH−) and G-specific RNase T1 ladders for sequence identification (see also Materials and methods section). ‘Fresh’ indicates that the material was not incubated before loading onto the gel.
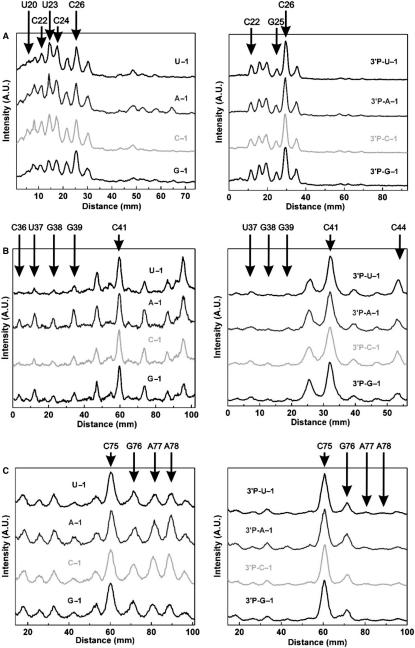
The raw and analyzed data in Figures 3B and 4, respectively, were therefore derived after incubation in the presence of both 11 mM Mg2+ and 5 mM Tb3+. Terbium(III) footprinting profiles of the precursor and product generated for each −1 variant are consistent with the expected secondary structure of the genomic HDV ribozyme, revealing strong scission 5′ to nucleotides in the single-stranded regions of loops L3 and L4 as well as joiners J1.1/4 and J4/2 (Figure 4). We find that the scission patterns of loop L3 (U20–U27), including the 5′-segment of the P1.1 stem (C21 and C22), show strong similarities between the ribozyme variants U − 1, A − 1 and C − 1 (Figure 4A, left). In contrast, the scission intensities 5′ to U23 and C24 are relatively lower for the G − 1 variant, resembling somewhat the product footprinting pattern shown on the right side of Figure 4A. This suggests that residues U23 and C24 are more accessible to and/or less protected from backbone scission by hydrated terbium(III) ions in the U − 1, A − 1 and C − 1 variants compared to the G − 1 mutant.
Figure 4.
Quantitative analysis of terbium(III) footprinting patterns of the four cis-acting genomic HDV ribozyme variants (left line graphs, derived from vertical lines through the middle of the gel lanes) and their respective 3′-products (3′P, right line graphs) in the presence of 11 mM Mg2+ and 5 mM Tb3+. The relative scission intensity is given in arbitrary units (A.U.), with important nucleotides indicated 5′ to which Tb3+ cuts. The backbone scission observed in loop L3 reported is (A); stem P1.1 and joiner J1.1/4 (B); and joiner J4/2 (C).
As a control, we carried out terbium(III) footprinting on the 3′-product forms derived from the four ribozyme variants, 3′P-U − 1, 3′P-A − 1 3′P-C − 1 and 3′P-G − 1, which were generated by self-cleavage during transcription. Similar terbium scission patterns for the L3 loop residues are expected among all four 3′-product forms, considering that they all are missing the 5′-sequence and thus have identical sequences. Indeed, we observe indistinguishable scission patterns (Figure 4A, right), underscoring the significance of the considerably distinct profiles of the precursors with different 5′-sequences and thus supporting the notion that loop L3 adopts distinct conformations or conformational equilibria depending on the N − 1 base identity.
Pyrimidines in the −1 position stabilize the P1.1 platform and the G1:U37 wobble pair of the catalytic pocket
Analysis of terbium(III) footprinting patterns of the P1.1 stem and the joiner J1.1/4 reveals that G38 and G39 of the P1.1 stem as well as U37, the wobble pair partner of the cleavage site G1, are less susceptible to terbium(III) scission in the U − 1 and C − 1 compared to the A − 1 and G − 1 variants (Figure 4B, left). This observation indicates that the conformation of residues forming the catalytic pocket depends on the identity of the −1 nucleotide. In fact, the P1.1 stem forms the platform of the binding pocket on which the cleavage site G1:U37 wobble pair stacks (Figure 1) (9). The fact that the pyrimidine derivatives of N − 1 lead to stronger protection of this region against terbium(III) scission than the purine derivatives suggests that this region is more strongly base paired in the pyrimidine derivatives (17,35,52). This observation correlates with the higher cleavage activity of the pyrimidine compared to the purine derivatives (Figure 2). We note that the scission patterns for joiner J1.1/4 (encompassing G40–A42), in contrast, are similar among all four variants (Figure 4B, left) and that again the P1.1 and J1.1/4 footprinting profiles of the various 3′-products are identical, as expected (Figure 4B, right). Finally, residues U37, G38 and G39 are more strongly protected from terbium(III) scission in the 3′-product than in any of the precursors, although the precursor variants with pyrimidine in position N − 1 lead to nearly as strong a protection (Figure 4B).
Formation of the A-minor motif anchoring the catalytic C75 depends on the identity of N − 1
We find that the A-minor motif, which anchors the catalytic C75 in the core (Figure 1), shows stronger terbium(III) scission in the A − 1 and C − 1 variants 5′ of A77 and A78 than does the A-minor motif in the U − 1 and G − 1 variants (Figure 4C, left). Generally, A77 and A78, and to some extent G76, are less protected in all four precursor variants than in the 3′-product form (Figure 4C, right), in agreement with terbium(III)-footprinting data obtained previously on a trans-acting HDV ribozyme (33). Again, the patterns for the four products derived from self-cleavage of each of the precursors are essentially identical, as expected, further supporting the notion that the relative protections in the U − 1 wild-type and the G − 1 variant are significant and likely represent more tightly formed tertiary interactions in the A-minor motif involving A77 and A78. Notably, we find one of the strongest terbium(III) hits in both the precursors and the 3′-product 5′ of the catalytic C75 in the trefoil motif, suggesting that this site represents a strong Tb3+-binding site and/or exhibits a specific geometry particularly amenable to rapid scission by the Tb(OH)(aq)2+ species and does not change significantly in this regard upon self-cleavage (17,35,52).
Conformational dynamics vary among the N − 1 variants in MD simulations
We carried out a total of 80 ns of unconstrained MD based on the available crystal structures of the cis-acting genomic HDV ribozyme to supplement our experimental findings and provide a structurally sound account of the changes in conformational sampling expected in response to alterations in the 5′-sequence. The four simulated sequences contain the same three nucleotides 5′ of the cleavage site with varying N − 1; accordingly, the simulations are termed Pre-5GAU, Pre-5GAA, Pre-5GAC Pre-5GAG (Figure 1B). G − 3 was modeled into the structure as none of the available crystal structures resolved this residue. The final structure of the equilibration run of simulation Pre-5GAU was used to initiate modeling and simulation of Pre-5GAA, Pre-5GAC and Pre-5GAG. All simulations revealed stable trajectories with modest root-mean-square deviations of between 4.25 and 6 Å from their corresponding starting structures.
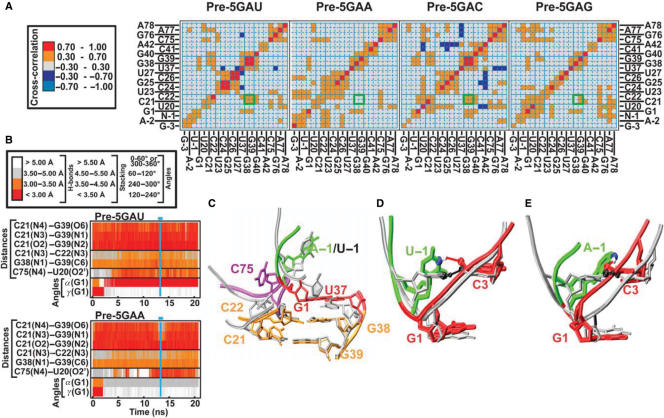
Cross-correlation matrices of MD-simulated motions in individual solute segments provide insight into coupled (correlated or anti-correlated) motions and thereby quickly assess, for example, the occupancy of base pairs in an RNA (42,53). We obtained comparable cross-correlation patterns over the whole and the second half of the simulation trajectory for all four simulations Pre-5GAU, Pre-5GAA, Pre-5GAC and Pre-5GAG, indicating that the simulated structural dynamics converge in each case (data not shown). While the patterns between the four simulations are similar, we find important differences (Figure 5A). In particular, the motions of the P1.1 stem residues C21, C22, G38 and G39 undergo correlated motions, consistent with joint movements as mediated by base pairing, in simulations Pre-5GAU and Pre-5GAC. In contrast, the correlation is weakened in simulation Pre-5GAG and completely lost in simulation Pre-5GAA (Figure 5A, highlighted by green boxes), indicating that the P1.1 stem elements move rather independently from one another. The stronger cross-correlations in the pyrimidine derivatives of N − 1 correlate with the fact that the G38/39 residues are better protected from terbium(III)-mediated footprinting in the presence of U − 1 and C − 1 than A − 1 and G − 1 (Figure 4B); all of these observations consistently suggest stronger base pairing in the P1.1 stem for pyrimidine derivates of N − 1. The P1.1 stem is known to be a functionally important structural platform for the cleavage site G1:U37 wobble base pair (Figure 1) (9,47). Strikingly, the simulated cross-correlation and experimental footprinting patterns mirror the catalytic activities (Figure 2), in that pyrimidines in the −1 position lead to a more stable P1.1 stem as well as faster catalysis than purines.
Figure 5.
Analysis of precursor MD simulations with four different 5′-sequences. (A) Cross-correlation maps of the residues defining the active site, as indicated. Positive cross-correlations are indicated in red and orange, negative cross-correlations in shades of blue as indicated. Intersections of vertical and horizontal lines in cyan and green boxes highlight cross-correlation of the base-paired nucleotides in the P1.1 stem. (B) Time trajectories of heavy-atom distances and torsional angles around the scissile phosphate shown over the entire duration of simulations Pre-5GAU (top panel) and Pre-5GAA (bottom panel). Vertical lines and horizontal bars in cyan indicate the time period used to obtain the representative average structures shown in panels (C–E). (C) Overlay of the representative average structures of the catalytic pocket from simulations Pre-5GAU (silver) and Pre-5GAA (backbone and key nucleotides color-coded as in the Figure 1C). Overlay of the initial crystal-structure-based model (silver) and the representative average structure (color-coded) from simulations Pre-5GAU (D) and Pre-5GAA (E); solid lines depict the hydrogen bonds from U − 1(N3) and A − 1(N6), respectively, to C3(O2P), which are rendered in CPK mode.
Upon closer inspection of the most extreme examples, the wild-type simulation Pre-5GAU and the purine mutant simulation Pre-5GAA, we find the likely reason for the loss of cross-correlation of motions in the P1.1 stem of the Pre-5GAA simulation; in the presence of A − 1, but not the wild-type U − 1, the C21:G39 base pair establishes a fluctuating propeller twist that weakens its hydrogen bonding and limits its stacking with the adjacent C22:G38 base pair of P1.1 (Figure 5B and C). Consistent with the observation of a structurally destabilized P1.1, the ∼13-fold decrease in cleavage activity of the A − 1 variant relative to the U − 1 wild-type (Figure 2A and B) is very similar in magnitude to the loss of cleavage activity in P1.1 mutants with conservative changes that weaken hydrogen bonding, such as mutations of the C22:G38 base pair to U22:A38 or U22:G38 base pairs (47). In addition, striking changes in the cleavage site kink occur during our simulations, where the α torsional angle of N − 1(O3′)–G1(P)–G1(O5′)–G1(C5′) around the scissile phosphate, which is ∼(278 ± 9)° in all simulations immediately after equilibration, changes after 5–10 ns and remains (178.0 ± 10.2)° in wild-type simulation Pre-5GAU and (86 ± 11.6)° in mutant simulation Pre-5GAA until the end of our 20-ns simulations (Figure 5B and Table 1). Coincidentally, the β torsional angle of G1(P)–G1(O5′)–G1(C5′)–G1(C4′) around the O5′–C5′ bond changes from ∼(194 ± 10)° to (174.1 ± 9.4)° and (166.6 ± 7.6)° in simulations Pre-5GAU and Pre-5GAA, respectively, whereas the γ torsional angle of G1(O5′)–G1(C5′)–G1(C4′)–G1(C3′) around the C5′–C4′ bond changes from ∼(181 ± 7)° to (49.0 ± 8.8)° and (53.9 ± 9.1)° in simulations Pre-5GAU and Pre-5GAA, respectively (Figure 5B and Table 1). While the Pre-5GAU wild-type simulation sustains pairwise stacking interactions between its 5′-sequence nucleotides during the entire 20-ns simulation time, the α torsional angle change in the A − 1 mutant simulation is accompanied by unstacking of the three nucleotides in the GAA 5′-sequence. More specifically, the U − 1 nucleotide in simulation Pre-5GAU prominently forms a relatively weak U − 1(N3)–C3(O2P) base–backbone hydrogen bond with C3 of stem P1, with an occupancy of ∼35%, as well as very transient interactions with A − 2, G1, C3 and G28, most of which are backbone–backbone interactions. By comparison, A − 1, C − 1 and G − 1 each establish several base–backbone hydrogen bonds with occupancies significantly above 35%, as well as several significant backbone-backbone contacts (Table 2 and data not shown). As expected, the most flexible residue in the 5′-sequence is the one furthest away from the catalytic core, G − 3 (Table 2). Generally, hydrogen-bonding interactions between the 5′-sequence residues and the P1 helix result in decreased hydrogen bond occupancies in the Watson–Crick base pairs of stem P1 (data not shown).
Table 1.
Backbone torsional angles α [of Ni−1(O3′)–Ni(P)–Ni(O5′)–Ni(C5′)], β [of Ni(P)–Ni(O5′)–Ni(C5′)-Ni(C4′)] and γ [of Ni(O5′)–Ni(C5′)–Ni(C4′)–Ni(C3′)] in degrees (± standard deviation)
| Origin | α | β | γ |
|---|---|---|---|
| U4/G5, U-turn Hammerhead ribozyme crystal, PDB ID 2GOZ | 150.26 | 172.07 | 66.97 |
| U33/2′-OMeG34, anticodon U-turn yeast tRNAPhe crystal, PDB ID 1TN2 | 130.61 | 145.96 | 85.27 |
| U − 1/G1, U-turn HDV ribozyme crystal (PDB ID 1SJ3) | 276.38 | 251.67 | 156.19 |
| U − 1/G1, U-turn HDV ribozyme over 19–20 ns of MD simulation Pre-5GAU | 178.0 ± 10.2 | 174.1 ± 9.4 | 49.0 ± 8.8 |
| A − 1/G1, U-turn HDV ribozyme over 19–20 ns of MD simulation Pre-5GAA | 86.0 ± 11.6 | 166.6 ± 7.6 | 53.9 ± 9.1 |
| Average of C27–A31, PseudoU39–G43, anticodon A-helix yeast tRNAPhe crystal (PDB ID 1TN2) | 284.5 ± 8.2 | 173.6 ± 10.9 | 173.6 ± 10.9 |
Table 2.
Occupancy (in%) of hydrogen bonds between the residues of the 5′-sequence and the remainder of the ribozyme during the course of 20-ns MD simulations
| 5′-Sequence | Hydrogen bond | Pre-5GAU | Pre-5GAA | Pre-5GAC | Pre-5GAG |
|---|---|---|---|---|---|
| N − 1 | U − 1(N3)–C3(O2P) | 35.27 | – | – | – |
| N − 1(O2′)–G1(O1P) | 6.37 | 96.43 | 90.45 | 15.96 | |
| A − 1(N6)–C3(O2P) | – | 43.25 | – | – | |
| N −1(O2P)–U27(N3) | – | 43.28 | – | – | |
| C − 1(N4)–C3(O2P) | – | – | 76.28 | – | |
| G − 1(N1)–C3(O2P) | – | – | – | 91.8 | |
| G − 1(N2)–C3(O2P) | – | – | – | 69.08 | |
| G − 1(O2P)–A77(N6) | – | – | – | 31.85 | |
| A − 2 | A − 2(O4′)–G − 3(O2′) | 4.92 | 26.52 | – | 30.6 |
| A − 2(N6)–C3(O2P) | – | 15.35 | – | – | |
| A − 2(N6)–C33(O2P) | – | – | 67.92 | – | |
| A − 2(N6)–G76(O2P) | – | – | 2.35 | – | |
| A − 2(N1)–C4(N4) | – | – | – | 16.32 | |
| A − 2(N1)–C3(N4) | – | – | – | 5.78 | |
| G − 3 | G − 3(N7)–C32(N4) | 13.67 | – | – | – |
| G − 3(O6)–C33(N4) | 6.75 | – | – | – | |
| G − 3(N2)–C4(O2P) | – | 47.83 | – | – | |
| G − 3(N1)–C4(O2P) | – | 17.42 | – | – | |
| G − 3(N2)–G6(O2P) | – | – | 40.6 | – | |
| G − 3(O5′)–C7(N4) | – | – | 25.93 | – | |
| G − 3(O2′)–G31(O6) | 3.05 | 7.57 | 22.51 | 16.38 | |
| G − 3(N2)–G6(N7) | – | – | 15.13 | – | |
| G − 3(O2′)–A − 2(O4′) | 4.92 | 26.52 | – | 30.6 | |
| G − 3(N2)–G5(O6) | – | – | – | 23.55 | |
| G − 3(N2)–C32(N3) | 3.84 | 4.95 | 9.84 | 13.05 |
U − 1 leads to a stable U-turn kink around the scissile phosphate
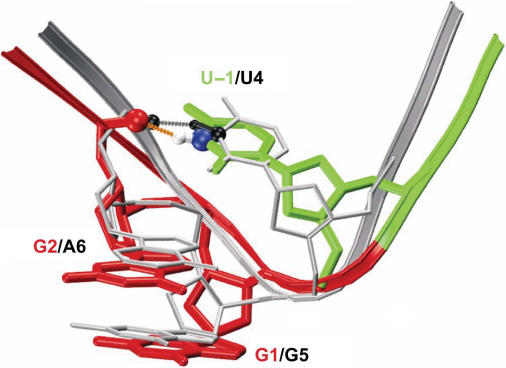
About 9.7 ns into the wild-type simulation Pre-5GAU, the U20–G25 base-pair breaks and does not reestablish throughout the remainder of the simulation. U20 remains in a crystal-structure-like conformation, while G25 undergoes large motions that change its conformation several times, coinciding with substantial unfolding of the L3 loop (data not shown) as previously observed (25,26). Importantly, U20–G25 unpairing followed by L3 unfolding does not have any obvious effects on the positioning of the cleavage site kink relative to the catalytically involved nucleotide C75, which remains stacked below A77, anchored by fluctuating hydrogen bonds of C75(N4) with U20(O2′) of loop L3 (Figure 5B). This robustness of the catalytic core architecture attests to the stable kink facilitated by the GAU 5′-sequence of the wild-type HDV ribozyme compared to, for example, the GAA mutant (Figure 5D and E). In fact, the wild-type sequence of U − 1/G1 with a sharp bend 3′ of U − 1, which in the crystal structure is mediated by an unusually large β torsional angle around the downstream O5′–C5′ bond of G1 and during our MD simulation is replaced by unusually small α and γ torsional angles around the scissile P-O5′ and the C5′–C4′ bonds of G1, respectively (Table 1), is nearly identical in backbone trajectory to that of the U4/G5 U-turn motif in the hammerhead ribozyme (Figure 6). The U-turn is one of the most commonly found RNA structural motifs, leading to strongly bent RNA structures such as those in the hammerhead ribozyme catalytic core (54–56), the TΨC and anticodon hairpin loops of tRNAs (57,58), the A-rich loop in HIV-1 genomic RNA (59) and GNRA tetraloops of large structured RNAs (60,61). In fact, our Pre-5GAU wild-type MD simulation adjusts the α, β and γ torsional angles of the HDV ribozyme crystal structure, which were modeled into a medium-resolution electron density map of this dynamic segment (9) and, except for β, are surprisingly close to standard A-helix values, to values much closer to those found in the canonical U-turns of the hammerhead ribozyme and yeast tRNAPhe crystal structures (Table 1).
Figure 6.
Overlay of the U-turn from the initial crystal-structure-based model from simulation Pre-GAU (backbone and residues color-coded as in Figure 1) with the crystal structure of the hammerhead ribozyme (PDB ID 2GOZ; backbone and residues colored in silver); dashed lines depict the hydrogen bonds between U − 1(N3) and C3(O2P) (HDV ribozyme; orange) and U4(N3) and U7(O2P) (hammerhead ribozyme; gray); key atoms are rendered in CPK mode and colored as follows: HDV ribozyme U − 1(N3) in blue, U − 1(H3) in white and C3(O2P) in red; hammerhead ribozyme U4(N3), U4(H3) and U7(O2P) in black.
DISCUSSION
Here we have shown that U − 1, clinically conserved in the genome of the human HDV, is essential for fast self-cleavage of the HDV ribozyme and thus critical for double-rolling circle replication of the virus. Distinct conformations of the catalytic core, in particular the P1.1 stem, the cleavage site wobble pair and the A-minor motif of the trefoil turn, as revealed via terbium(III) footprinting, provide the structural basis for a >25-fold activity loss across U − 1 > C − 1 > A − 1 > G − 1. Moreover, the most solvent protected and thus tightly folded catalytic core, including the most stable P1.1 stem, cleavage site G1:U37 wobble pair and A-minor tertiary structure motif are found in the most active U − 1 wild-type. Consistent with this picture, we find in complementary MD simulations that a 5′-sequence with U − 1 leads to the most stable P1.1 stem as well as the most robust kink around the scissile phosphate connecting U − 1 and G1. This kink exposes the cleavage site to the catalytic C75 in the form of a common U-turn motif, found for example in the hammerhead ribozyme catalytic core, but previously unnoticed in the HDV ribozyme. The architectural function of the U-turn is very distinct in the HDV and hammerhead ribozymes, in that in the genomic HDV ribozyme it forms the bend that presents the scissile phosphate to the catalytic C75, while in the hammerhead ribozyme, in contrast, it molds the catalytic core residues around the cleavage site. Our results provide an improved structural framework for future studies of HDV ribozyme dynamics and function.
Why did nature choose the U-turn motif for such diverse functional roles? The U-turn presents the minimal sequence motif, consisting of only 2–3 nt, that adopts a 180° turn in an RNA backbone trajectory. This property makes it an evolutionarily favored motif for organizing highly structured RNAs such as tRNAs into compact folds. In the HDV ribozyme, this feature of the U-turn additionally helps mark and perhaps prepare a particular phosphodiester bond for site-specific cleavage; in the hammerhead ribozyme it allows for a maximum number of residues to be arranged around the scissile phosphate. The particular use that nature makes of the U-turn motif thus depends, perhaps not surprisingly, on the structural context, making the U-turn motif not only one of the smallest distinct RNA structural motifs, but also one of the most versatile.
In the canonical crystal structures of U-turns, including the one described here in the HDV ribozyme, the bend of the turn is stabilized by a hydrogen bond of the Un with the non-bridging O2P phosphate oxygen of the n + 3 nucleotide (Figure 6). In our MD simulation carried out in native aqueous solvent at room temperature, the relative dynamics of the U − 1 nucleotide lead to an only ∼35% occupancy of the U − 1(N3)–C3(O2P) base–backbone hydrogen bond. This observation suggests a surprisingly transient nature of the canonical hydrogen bond in the U-turn motif. Still, it is by far the most highly occupied hydrogen bond of U − 1 in the simulated motif (Table 2), making it plausible that under the cryogenic conditions of the crystallographic experiment it is frozen out as a stabilizing, low-energy interaction. Thus, our MD simulation data are consistent with the crystallographic data, but add an important dynamic dimension. This dynamic nature will have consequences for our understanding of the widespread U-turn structural motif, for example, to explain how certain naturally occurring modifications restrict the conformational space of the U-turn-containing tRNA anticodon loop and thus affect the accuracy of decoding and the maintenance of the mRNA reading frame (58). Introducing such modifications into the U-turn motif of the HDV ribozyme may present a novel approach to modulating active site conformational dynamics and thus self-cleavage activity of the HDV genome.
Despite the conformational flexibility of the 5′-sequence nucleotides, the backbone trajectory around the cleavage site remains strikingly conserved in our U − 1 wild-type MD simulations (Figure 5E) compared to, for example, the GAA mutant (Figure 5D), consistent with the well solvent-protected catalytic core structure of the wild-type HDV ribozyme observed by terbium(III) footprinting (Figures 3 and 4). This architectural robustness of the cleavage site may contribute to enhanced catalytic activity of the wild-type ribozyme by decreasing specifically the entropic penalty associated with reaching the reaction transition state. Such an architectural feature and catalytic strategy are also found in protein enzymes, which often employ substrate strain as a means to lower the transition state barrier (1–3,33,62).
Why is U found in the U-turn? Uracil possesses specific features such as a relatively low diversity of functional groups, a small size and relatively weak stacking interactions that make it stand out among the four natural nucleobases. Consistent with this picture, U − 1 in the HDV ribozyme establishes less of the diverse hydrogen bond interactions with the P1 helix than the other MD-simulated N − 1 variants; these interactions generally result in weakened (lower occupancy) hydrogen bonds of the essential stem P1 Watson–Crick base pairs. In the context of HDV ribozyme self-cleavage, U − 1 thus appears to suppress local misfolding and enhance residency time in functionally relevant conformations, thereby further boosting cleavage activity. In addition, uracil possesses the flexibility needed for rotational motion around the U-turn to establish a catalytically important interaction of its 2′-OH with C75 as proposed for the base catalysis mechanism (9). A future evaluation of the conformational reaction trajectory of this motion using umbrella sampling will likely provide further insight into function of the genomic HDV ribozyme.
In conclusion, we find considerable agreement between our MD simulation and experimental results, as we have previously concluded for results on the HDV and hairpin ribozymes (25,26,42). Such agreement is reassuring in light of potential force-field biases in the description of nucleic acids that may significantly affect, for example, the backbone behavior, in particular over longer MD simulations (63–67). The synergistic power of combining computational with experimental studies seems worth further exploitation. Our finding of a U-turn around the cleavage site provides the basis to frame such future inquiries into genomic HDV ribozyme structure, dynamics and catalytic function.
ACKNOWLEDGEMENTS
This work was supported in part by NIH Grant GM62357 including Supplement S2 for acquisition of a computer cluster to N.G.W., by Wellcome Trust International Senior Research Fellowship in Biomedical Science in Central Europe GR067507, grants GA203/05/0388 and GA203/05/0009, Grant Agency of the Czech Republic, grants 1QS500040581 and IAA400550701 by Grant Agency of the Academy of Sciences of the Czech Republic, by Research center LC512 and research projects AVO Z5 004 0507 and AVO Z4 055 050 by the Ministry of Education of the Czech Republic to J.Š., and by a Margaret and Herman Sokol International Summer Research Fellowship, a NATO Science Fellowship, a Center for the Education of Women Sarah Winans Newman Scholarship and an Eli Lilly Fellowship to J.S. Funding to pay the Open Access publication charge was provided by NIH Grant GM62357.
Conflict of interest statement. None declared.
REFERENCES
- 1.Narlikar GJ, Herschlag D. Mechanistic aspects of enzymatic catalysis: lessons from comparison of RNA and protein enzymes. Annu. Rev. Biochem. 1997;66:19–59. doi: 10.1146/annurev.biochem.66.1.19. [DOI] [PubMed] [Google Scholar]
- 2.Shih IH, Been MD. Catalytic strategies of the hepatitis delta virus ribozymes. Annu. Rev. Biochem. 2002;71:887–917. doi: 10.1146/annurev.biochem.71.110601.135349. [DOI] [PubMed] [Google Scholar]
- 3.Doudna JA, Lorsch JR. Ribozyme catalysis: not different, just worse. Nat. Struct. Mol. Biol. 2005;12:395–402. doi: 10.1038/nsmb932. [DOI] [PubMed] [Google Scholar]
- 4.Ferre-D’Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 5.Perrotta AT, Shih I, Been MD. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- 6.Nakano S, Chadalavada DM, Bevilacqua PC. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 7.Ferre-D’Amare AR, Doudna JA. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J. Mol. Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- 8.Oyelere AK, Kardon JR, Strobel SA. pK(a) perturbation in genomic hepatitis delta virus ribozyme catalysis evidenced by nucleotide analogue interference mapping. Biochemistry. 2002;41:3667–3675. doi: 10.1021/bi011816v. [DOI] [PubMed] [Google Scholar]
- 9.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 10.Das S, Piccirilli J. General acid catalysis by the hepatitis delta virus ribozyme. Nat. Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 11.Perrotta AT, Wadkins TS, Been MD. Chemical rescue, multiple ionizable groups, and general acid-base catalysis in the HDV genomic ribozyme. RNA. 2006;12:1282–1291. doi: 10.1261/rna.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deschenes P, Lafontaine DA, Charland S, Perreault JP. Nucleotides −1 to −4 of hepatitis delta ribozyme substrate increase the specificity of ribozyme cleavage. Antisense Nucleic Acid Drug Dev. 2000;10:53–61. doi: 10.1089/oli.1.2000.10.53. [DOI] [PubMed] [Google Scholar]
- 13.Shih I, Been MD. Energetic contribution of non-essential 5′ sequence to catalysis in a hepatitis delta virus ribozyme. EMBO J. 2001;20:4884–4891. doi: 10.1093/emboj/20.17.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira MJ, Harris DA, Rueda D, Walter NG. Reaction pathway of the trans-acting hepatitis delta virus ribozyme: a conformational change accompanies catalysis. Biochemistry. 2002;41:730–740. doi: 10.1021/bi011963t. [DOI] [PubMed] [Google Scholar]
- 15.Harris DA, Rueda D, Walter NG. Local conformational changes in the catalytic core of the trans-acting hepatitis delta virus ribozyme accompany catalysis. Biochemistry. 2002;41:12051–12061. doi: 10.1021/bi026101m. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka Y, Tagaya M, Hori T, Sakamoto T, Kurihara Y, Katahira M, Uesugi S. Cleavage reaction of HDV ribozymes in the presence of Mg2+ is accompanied by a conformational change. Genes Cells. 2002;7:567–579. doi: 10.1046/j.1365-2443.2002.00541.x. [DOI] [PubMed] [Google Scholar]
- 17.Harris DA, Tinsley RA, Walter NG. Terbium-mediated footprinting probes a catalytic conformational switch in the antigenomic hepatitis delta virus ribozyme. J. Mol. Biol. 2004;341:389–403. doi: 10.1016/j.jmb.2004.05.074. [DOI] [PubMed] [Google Scholar]
- 18.Lai MM. The molecular biology of hepatitis delta virus. Annu. Rev. Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 19.Sharmeen L, Kuo MY, Dinter-Gottlieb G, Taylor J. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J. Virol. 1988;62:2674–2679. doi: 10.1128/jvi.62.8.2674-2679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu HN, Lai MM. Reversible cleavage and ligation of hepatitis delta virus RNA. Science. 1989;243:652–654. doi: 10.1126/science.2492677. [DOI] [PubMed] [Google Scholar]
- 21.Kumar PK, Suh YA, Miyashiro H, Nishikawa F, Kawakami J, Taira K, Nishikawa S. Random mutations to evaluate the role of bases at two important single-stranded regions of genomic HDV ribozyme. Nucleic Acids Res. 1992;20:3919–3924. doi: 10.1093/nar/20.15.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanner NK, Schaff S, Thill G, Petit-Koskas E, Crain-Denoyelle AM, Westhof E. A three-dimensional model of hepatitis delta virus ribozyme based on biochemical and mutational analyses. Curr. Biol. 1994;4:488–498. doi: 10.1016/s0960-9822(00)00109-3. [DOI] [PubMed] [Google Scholar]
- 23.Perrotta AT, Been MD. Core sequences and a cleavage site wobble pair required for HDV antigenomic ribozyme self-cleavage. Nucleic Acids Res. 1996;24:1314–1321. doi: 10.1093/nar/24.7.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wadkins TS, Shih I, Perrotta AT, Been MD. A pH-sensitive RNA tertiary interaction affects self-cleavage activity of the HDV ribozymes in the absence of added divalent metal ion. J. Mol. Biol. 2001;305:1045–1055. doi: 10.1006/jmbi.2000.4368. [DOI] [PubMed] [Google Scholar]
- 25.Krasovska MV, Sefcikova J, Reblova K, Schneider B, Walter NG, Sponer J. Cations and hydration in catalytic RNA: molecular dynamics of the hepatitis delta virus ribozyme. Biophys. J. 2006;91:626–638. doi: 10.1529/biophysj.105.079368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krasovska MV, Sefcikova J, Špacková N, Šponer J, Walter NG. Structural dynamics of precursor and product of the RNA enzyme from the hepatitis delta virus as revealed by molecular dynamics simulations. J. Mol. Biol. 2005;351:731–748. doi: 10.1016/j.jmb.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 27.Wadkins TS, Been MD. Ribozyme activity in the genomic and antigenomic RNA strands of hepatitis delta virus. Cell. Mol. Life Sci. 2002;59:112–125. doi: 10.1007/s00018-002-8409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu HN, Lee JY, Huang HW, Huang YS, Hsueh TG. Mutagenesis analysis of a hepatitis delta virus genomic ribozyme. Nucleic Acids Res. 1993;21:4193–4199. doi: 10.1093/nar/21.18.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu HN, Wang YJ, Hung CF, Lee HJ, Lai MM. Sequence and structure of the catalytic RNA of hepatitis delta virus genomic RNA. J. Mol. Biol. 1992;223:233–245. doi: 10.1016/0022-2836(92)90728-3. [DOI] [PubMed] [Google Scholar]
- 30.Nishikawa F, Fauzi H, Nishikawa S. Detailed analysis of base preferences at the cleavage site of a trans-acting HDV ribozyme: a mutation that changes cleavage site specificity. Nucleic Acids Res. 1997;25:1605–1610. doi: 10.1093/nar/25.8.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perrotta AT, Been MD. The self-cleaving domain from the genomic RNA of hepatitis delta virus: sequence requirements and the effects of denaturant. Nucleic Acids Res. 1990;18:6821–6827. doi: 10.1093/nar/18.23.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luptak A, Ferre-D’Amare AR, Zhou K, Zilm KW, Doudna JA. Direct pK(a) measurement of the active-site cytosine in a genomic hepatitis delta virus ribozyme. J. Am. Chem. Soc. 2001;123:8447–8452. doi: 10.1021/ja016091x. [DOI] [PubMed] [Google Scholar]
- 33.Jeong S, Sefcikova J, Tinsley RA, Rueda D, Walter NG. Trans-acting hepatitis delta virus ribozyme: catalytic core and global structure are dependent on the 5′ substrate sequence. Biochemistry. 2003;42:7727–7740. doi: 10.1021/bi034627g. [DOI] [PubMed] [Google Scholar]
- 34.Walter NG, Yang N, Burke JM. Probing non-selective cation binding in the hairpin ribozyme with Tb(III) J. Mol. Biol. 2000;298:539–555. doi: 10.1006/jmbi.2000.3691. [DOI] [PubMed] [Google Scholar]
- 35.Harris DA, Walter NG. Probing RNA structure and metal-binding sites using terbium footprinting. Curr. Protoc. Nucleic Acid Chem. 2003;6.8:6.8.1–6.8.8. doi: 10.1002/0471142700.nc0608s13. [DOI] [PubMed] [Google Scholar]
- 36.Case DA, Pearlman DA, Caldwell JW, Cheatham TE, III, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, et al. AMBER 7. San Francisco: University of California San Francisco; 2002. [Google Scholar]
- 37.Cheatham TE, Cieplak P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 38.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, et al. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 39.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 40.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 41.Aqvist J. Ion water interaction potentials derived from free-energy perturbation simulations. J. Phys. Chem. 1990;94:8021–8024. [Google Scholar]
- 42.Rhodes MM, Reblova K, Sponer J, Walter NG. Trapped water molecules are essential to structural dynamics and function of a ribozyme. Proc. Natl. Acad. Sci. USA. 2006;103:13381–13385. doi: 10.1073/pnas.0605090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 44.Berendsen H.J.C, Postma J.P.M, van Gunsteren WF, Dinola A, Haak JR. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 45.Ryckaert JP, Ciccotti G, Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Chem. 1977;23:327–341. [Google Scholar]
- 46.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 47.Wadkins TS, Perrotta AT, Ferre-D’Amare AR, Doudna JA, Been MD. A nested double pseudoknot is required for self-cleavage activity of both the genomic and antigenomic hepatitis delta virus ribozymes. RNA. 1999;5:720–727. doi: 10.1017/s1355838299990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shih I, Been MD. Kinetic scheme for intermolecular RNA cleavage by a ribozyme derived from hepatitis delta virus RNA. Biochemistry. 2000;39:9055–9066. doi: 10.1021/bi000499+. [DOI] [PubMed] [Google Scholar]
- 49.Nakano S, Proctor DJ, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry. 2001;40:12022–12038. doi: 10.1021/bi011253n. [DOI] [PubMed] [Google Scholar]
- 50.Nakano S, Cerrone AL, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: classifying the catalytic and structural metal ion sites within a multichannel reaction mechanism. Biochemistry. 2003;42:2982–2994. doi: 10.1021/bi026815x. [DOI] [PubMed] [Google Scholar]
- 51.Sigel R.K.O, Pyle AM. Lanthanide ions as probes for metal ions in the structure and catalytic mechanism of ribozymes. Met. Ions Biol. Syst. 2003;40:477–512. [PubMed] [Google Scholar]
- 52.Harris DA, Walter NG. In: Handbook of RNA Biochemistry. Hartmann RK, Bindereif A, Schön A, Westhof E, editors. 2005. Wiley-VCH, Weinheim, Vol. 1, pp. 205–213. [Google Scholar]
- 53.Ichiye T, Karplus M. Collective motions in proteins: a covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins. 1991;11:205–217. doi: 10.1002/prot.340110305. [DOI] [PubMed] [Google Scholar]
- 54.Pley HW, Flaherty KM, McKay DB. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 55.Scott WG, Finch JT, Klug A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- 56.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quigley GJ, Rich A. Structural domains of transfer RNA molecules. Science. 1976;194:796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- 58.Stuart JW, Koshlap KM, Guenther R, Agris PF. Naturally-occurring modification restricts the anticodon domain conformational space of tRNA(Phe) J. Mol. Biol. 2003;334:901–918. doi: 10.1016/j.jmb.2003.09.058. [DOI] [PubMed] [Google Scholar]
- 59.Puglisi EV, Puglisi JD. HIV-1 A-rich RNA loop mimics the tRNA anticodon structure. Nat. Struct. Biol. 1998;5:1033–1036. doi: 10.1038/4141. [DOI] [PubMed] [Google Scholar]
- 60.Jucker FM, Pardi A. GNRA tetraloops make a U-turn. RNA. 1995;1:219–222. [PMC free article] [PubMed] [Google Scholar]
- 61.Krasilnikov AS, Mondragon A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA. 2003;9:640–643. doi: 10.1261/rna.2202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fersht A. Structure and Mechanism in Protein Science. New York: Freeman; 1999. [Google Scholar]
- 63.Varnai P, Zakrzewska K. DNA and its counterions: a molecular dynamics study. Nucleic Acids Res. 2004;32:4269–4280. doi: 10.1093/nar/gkh765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beveridge DL, Barreiro G, Byun KS, Case DA, Cheatham TE, III, Dixit SB, Giudice E, Lankas F, Lavery R, et al. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. I. Research design and results on d(CpG) steps. Biophys. J. 2004;87:3799–3813. doi: 10.1529/biophysj.104.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fadrna E, Spackova N, Stefl R, Koca J, Cheatham TE, III, Sponer J. Molecular dynamics simulations of Guanine quadruplex loops: advances and force field limitations. Biophys. J. 2004;87:227–242. doi: 10.1529/biophysj.103.034751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barone F, Lankas F, Spackova N, Sponer J, Karran P, Bignami M, Mazzei F. Structural and dynamic effects of single 7-hydro-8-oxoguanine bases located in a frameshift target DNA sequence. Biophys. Chem. 2005;118:31–41. doi: 10.1016/j.bpc.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 67.McDowell SE, Spackova N, Sponer J, Walter G. Molecular Dynamics Simulations of RNA: An In Silico Single Molecule Approach. Biopolymers. 2007;85:169–184. doi: 10.1002/bip.20620. [DOI] [PMC free article] [PubMed] [Google Scholar]