Abstract
To address the role of N-myc in neurogenesis and in nervous system tumors, it was conditionally disrupted in neuronal progenitor cells (NPCs) with a nestin-Cre transgene. Null mice display ataxia, behavioral abnormalities, and tremors that correlate with a twofold decrease in brain mass that disproportionately affects the cerebellum (sixfold reduced in mass) and the cerebral cortex, both of which show signs of disorganization. In control mice at E12.5, we observe a domain of high N-Myc protein expression in the rapidly proliferating cerebellar primordium. Targeted deletion of N-myc results in severely compromised proliferation as shown by a striking decrease in S phase and mitotic cells as well as in cells expressing the Myc target gene cyclin D2, whereas apoptosis is unaffected. Null progenitor cells also have comparatively high levels of the cdk inhibitors p27Kip1 and p18Ink4c, whereas p15Ink4b, p21Cip1, and p19Ink4d levels are unaffected. Many null progenitors also exhibit altered nuclear morphology and size. In addition, loss of N-myc disrupts neuronal differentiation as evidenced by ectopic staining of the neuron specific marker βTUBIII in the cerebrum. Furthermore, in progenitor cell cultures derived from null embryonic brain, we observe a dramatic increase in neuronal differentiation compared with controls. Thus, N-myc is essential for normal neurogenesis, regulating NPC proliferation, differentiation, and nuclear size. Its effects on proliferation and differentiation appear due, at least in part, to down-regulation of a specific subset of cyclin-dependent kinase inhibitors.
Keywords: N-myc, nervous system, cerebellum, cdk inhibitors, conditional knockout, cyclin D2
The N-myc proto-oncogene was identified nearly two decades ago as a DNA region, frequently amplified in metastatic human neuroblastomas, which possessed homology to the retroviral oncogene v-myc and its cellular ortholog c-myc (Kohl et al. 1983; Schwab et al. 1983). Together, N-myc, c-myc, and L-myc comprise the myc family of proto-oncogenes, which are transcription factors of the basic-helix-loop-helix-zipper (bHLHZ) class and form sequence-specific DNA-binding heterodimers with the small bHLHZ protein, Max (for review, see Facchini and Penn 1998; Grandori et al. 2000).
Considerable evidence indicates that Myc family proteins are involved in fundamental cellular processes including proliferation, growth, apoptosis, and differentiation, most likely through activation and repression of specific sets of target genes (for review, see Dang 1999; Grandori et al. 2000; Eisenman 2001). Although the detailed functional pathways through which Myc proteins mediate their effects have yet to be rigorously delineated, a number of recent studies have identified genes whose expression is modulated by c-Myc. Putative targets include the cyclin-dependent kinase CDK4 (Hermeking et al. 2000), the Cdc25A phosphatase that activates CDKs (Galaktionov et al. 1996), cyclin D2 (Bouchard et al. 1999, 2001; Perez-Roger et al. 1999) and the E2F family (Leone et al. 2001). Myc may further stimulate cyclin E/Cdk2 activity by directly or indirectly abrogating the function of the CDK inhibitor p27Kip1 (Bouchard et al. 1999; Perez-Roger et al. 1999; O'Hagan et al. 2000), and by inducing the Rb antagonist Id2 (Lasorella et al. 2000). Myc also represses genes that function to inhibit proliferation such as gas1 (Lee et al. 1997), gadd45 (Marhin et al. 1997), p15Ink4b (Staller et al. 2001), and p21Cip1 (Gartel et al. 2001). In addition to target genes involved in cell cycle progression, Myc has been found to stimulate expression of multiple genes that control cell size and growth, including those encoding ribosomal proteins, translation factors, and metabolic enzymes (Rosenwald et al. 1993; Coller et al. 2000; Guo et al. 2000; Boon et al. 2001; Schuhmacher et al. 2001). The idea that Myc regulates cell growth is consistent with studies in Drosophila (Johnston et al. 1999) as well as the effects of c-Myc overexpression in mammalian cells (Iritani and Eisenman 1999; Schuhmacher et al. 1999; Beier et al. 2000; Kim et al. 2000). However, murine knockouts of Myc genes have produced conflicting conclusions regarding whether hematopoietic cell size is affected (de Alboran et al. 2001; Douglas et al. 2001; Trumpp et al. 2001).
In the early 1990s, several laboratories produced targeted deletions of c-myc and N-myc in mice. Single knockouts of either gene led to embryonic lethality at mid-gestation (Sawai et al. 1991; Charron et al. 1992; Stanton et al. 1992; Davis et al. 1993) implying that myc genes are required for normal development at the onset of organogenesis. Hypomorphic mutations in N-myc produced a delayed lethality (Moens et al. 1992, 1993; Nagy et al. 1998), suggesting that the absolute levels of expression are important. Deletion of L-myc was more recently shown to produce no observable developmental phenotype (Hatton et al. 1996).
Overall, myc family gene expression appears to be predominantly associated with the expansion of cell populations within a wide range of tissues. However, an understanding of the exact roles that these genes play in development has been hampered by the relatively early embryonic lethal phenotypes of constitutive null mutations. Recently, a number of laboratories have undertaken tissue-specific conditional deletions of c-myc, primarily in hematopoietic cells, in which they showed a profound defect in proliferation, but differed in their determination of whether loss of c-myc influenced cell size (de Alboran et al. 2001; Trumpp et al. 2001). In murine T cells, constitutive loss of c-myc was found to reduce both cell size and proliferation (Douglas et al. 2001). To date, however, there have been no reports on the consequences of conditional loss of any Myc family gene on the development of a specific organ system.
Here, we describe the phenotype of mice in which the N-myc gene has been specifically disrupted in neuronal progenitor cells (NPCs). We focused on the nervous system because of the elevated and dynamic patterns of myc expression in this tissue and because of the link between N-myc and nervous system tumors. Our results show that N-myc is required for development of a complete nervous system, and that loss of N-myc function specifically disrupts the ability of NPCs to expand, differentiate, and populate the brain.
Results
Generation of conditionally targeted N-myc mice
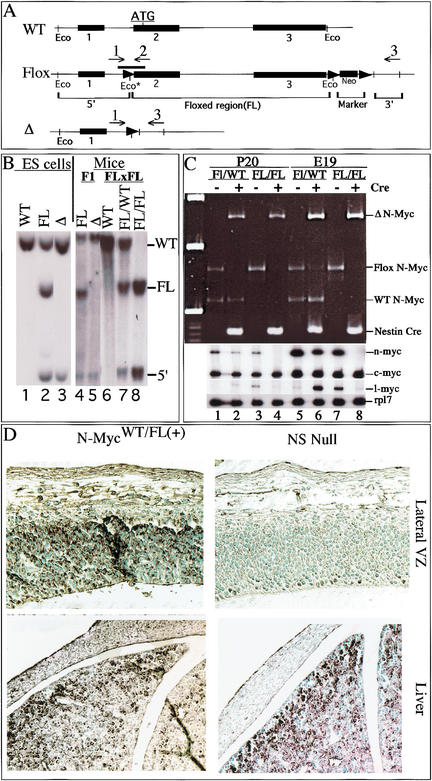
We produced an N-myc-targeting vector by inserting loxP sites 5′ of exon 2 and 3′ of exon 3 (Fig. 1A) of a newly isolated full-length N-myc genomic clone. Because the translational initiation codon of N-myc is in the second exon, the entire coding region of N-myc is floxed, and thus a target for deletion by the Cre recombinase. Six clonal G418R-targeted ES cell lines were produced, and appropriate integration was verified by genomic PCR (primers indicated in Fig. 1A; typical results, Fig. 1C) and Southern blot (Fig. 1B). Expression of Cre in these N-mycFL/WTES cells converted the genotype to N-mycΔ/WT, showing the floxed allele can be deleted appropriately (Fig. 1B, lanes 2–3). Chimeric mice produced from these N-mycΔ/WTES cells transmitted the Δ allele to their F1 offspring (Fig. 1B, lane 5), but extensive intercrosses of N-mycΔ/WT F1 mice failed to produce N-mycΔ/Δ F2 offspring, recapitulating the embryonic lethality seen in earlier nonconditional N-myc knockout mice (Charron et al. 1992; Moens et al. 1992; Stanton et al. 1992; Sawai et al. 1993).
Figure 1.
Nestin-Cre(+).N-mycFL/FL mice are nervous system-specific N-myc nulls. (A) Wild-type (WT), Floxed (Flox), and deleted (Δ) N-myc alleles. The thick line above the Flox allele is the Southern probe. (Eco*) The introduced EcoRI site just 5′ of the loxP site. loxP sites are represented as arrowheads. Numbers and arrows above the alleles represent PCR primers. (B) Southern blotting of EcoRI-digested DNA from ES cells (left) and the mice generated from the ES cells (right). (Lane 1) Wild-type ES cells. FL/WT ES cells (lane 2) were used to generate chimeric mice that were subsequently used to produce FL/WT F1 mice (lane 4). FL/WT mice were then interbred, producing F2 mice: WT (lane 6), FL/WT (lane 7) and FL/FL (lane 8). Cre was introduced into FL/WT ES cells to produce true Δ/WT ES cells (lane 3). (C) RNA and DNA samples were isolated simultaneously from whole brains of P20 pups (lanes 1–4) and E19 embryos (lanes 5–8). Genomic PCR was conducted on DNA samples (top), FL/WT with (lanes 2,6) and without nestin-Cre (lanes 1,5) as well as FL/FL with (lanes 4,8) and without nestin-Cre (lanes 3,7). RT–PCR for N-myc, L-myc, and c-myc as well as Rpl7 (ribosomal protein gene used as control) was conducted on total RNA samples from the same brains (bottom). (D) N-Myc staining of E12.5 embryos.
Chimeric mice were also produced from N-mycFL/WT ES cells (Fig. 1B, lane 4). Crosses of N-mycFL/WT F1 mice produced the appropriate ratio of N-mycFL/FL homozygous F2 pups (Fig. 1B, lanes 6–8). N-mycFL/FL mice were then crossed with hemizygous nestin-Cre transgenic mice (Tronche et al. 1999), producing N-mycFL/WT mice minus and plus nestin-Cre (referred to henceforth as N-mycFL/WT(−) and N-mycFL/WT(+), respectively). Nestin-Cre was chosen because it is expressed as early as E9.5–E10.5, specifically in NPCs, neural crest cells, and their progeny, an expression pattern quite similar to that of N-myc. The nestin-Cre transgene contains the rat nestin promoter and intronic enhancer elements (Tronche et al. 1999) and its specific expression in embryonic neuronal progenitors has been verified (Graus-Porta et al. 2001). The N-mycFL/WT(+) mice were then crossed once more with N-mycFL/FL animals, producing the four genotypes used throughout this project, N-mycFL/FL(−), N-mycFL/FL(+), N-mycFL/WT(−), and N-mycFL/WT(+). The ratios of pups produced with these four genotypes were non-Mendelian. N-mycFL/FL(+) and N-mycFL/WT(−) pups were approximately twofold under-represented compared with both N-mycFL/FL(−) and N-mycFL/WT(+), likely due to a genetic linkage between loci comprising the N-mycWT allele and the nestin-Cre transgene, with the few N-mycFL/FL(+) and N-mycFL/WT(−) pups produced probably being the result of meiotic recombination.
N-mycFL/FL(+) mice are null for N-myc in the nervous system
To verify that N-mycFL/FL(+) animals are null for N-myc in the nervous system, PCR (Fig. 1C, top) and Southern blotting (data not shown) for N-myc were conducted on brain genomic DNA from null and control littermates. N-mycFL/WT(−) mice have the expected WT and FL products and N-mycFL/WT(+) mice have WT and Δ bands (Fig. 1C, top, lanes 1–2,5–6). N-mycFL/FL(−) mice have a single FL allele band, whereas N-mycFL/FL(+) mice have instead a Δ band, showing efficient deletion of the N-myc gene by Cre (Fig. 1C, top, lanes 3,4,7,8).
N-myc mRNA levels were quantitated by RT–PCR on mRNA isolated from the same brain samples from which genomic DNA was isolated as described above (Fig. 1C, bottom). N-mycFL/WT(+) mice exhibit a twofold decrease in N-myc levels in brain compared with N-mycFL/WT(−) littermates (Fig. 1C, lanes 1,2,5,6). Although N-mycFL/FL(−) mice retain neo 3′ of the N-myc gene (Fig. 1A), they exhibit only moderately (25%–30%) reduced N-myc expression (Fig. 1C, lanes 1,3,5,7) and develop normally. N-mycFL/FL(+) mice as both pups and late embryos (E19) have no detectable N-myc mRNA in their brains, and do not exhibit compensatory increases in c-myc and L-myc (Fig. 1C, bottom), although there is some variability in L-myc expression among the other three genotypes.
The nestin-Cre transgene is also effective earlier, as N-mycFL/FL(+) E12.5 embryos have very little detectable N-Myc protein in the nervous system (Fig. 1D, top). Control littermates have high levels of N-Myc protein in the ventricular zone (VZ) as reported previously for N-myc mRNA expression (Hurlin et al. 1997), whereas only very few VZ cells (1%–2%) in N-mycFL/FL(+) embryos retained N-Myc staining. N-Myc protein levels in other organs of nulls such as liver are indistinguishable from that of control littermates, demonstrating the tissue specificity of the nestin-Cre transgene (Fig. 1D, bottom). Thus, on the basis of evidence from assays of N-myc genomic DNA, mRNA, and protein, it is clear that N-mycFL/FL(+) mice are null for N-myc in the nervous system and so will be referred to as N-myc NS nulls.
N-myc NS null mice exhibit tremors, ataxia, and behavioral abnormalities along with a twofold reduction in brain mass
From birth onward, N-myc NS null animals exhibit moderate growth retardation (Fig. 2A), but surprisingly, null and control pups survive weaning at the same rate. Although from the age of 4 wk onward, their general health improved and their rate of weight gain paralleled their littermates, other abnormalities became more apparent in the nulls. In addition to tremors, the null animals also exhibited ataxia and behavioral abnormalities such as constantly raised tails as well as excessive time spent digging and grooming themselves. Importantly, animals of the other three genotypes did not exhibit these or any other phenotypic characteristics, indicating that the null phenotype is specifically related to deletion of N-myc, and that the presence of neo has no apparent effect on the control mice.
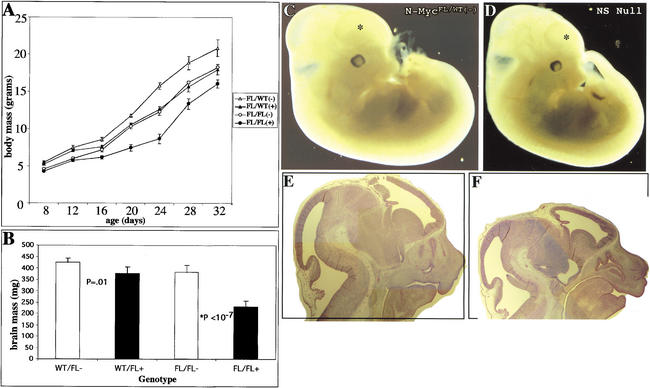
Figure 2.
N-myc deficiency results in reduced brain size. (A) Total body mass of animals from P8 to P32. (B) Total brain mass of animals from 8 to 16 wk of age. (C,D) External views of E12.5 control and null littermate embryos, respectively. Asterisks indicate forebrain area. (E,F) H&E stain of control and null E12.5 head regions, respectively.
The brains of null and littermate controls were examined. N-myc NS null animals aged 8–16 wk exhibited profound microencephaly, with small heads and eyes, and a significant reduction (twofold absolute) in total brain mass relative to total body weight (reduced only <25%) when compared with all three controls (Fig. 2B). Gross pathological analysis of the null brain indicated that the cerebellum and cerebral cortex were especially diminutive. To trace the origin of the diminution of the nervous system in N-myc NS nulls, embryos were examined. Nulls as early as E12.5 had very small heads and CNS (Fig. 2C–F). In contrast, the rest of the null embryo was only slightly smaller than that of control littermates. Histological analysis indicated that the N-myc NS null cerebellum primordium and the developing cerebral cortex were particularly small and many regions of VZ were noticeably reduced in thickness. Thus, we decided to concentrate on analyzing cerebellar and cerebral cortex development, beginning with an analysis of pups.
Loss of N-myc severely disrupts cerebellar development
N-myc NS null pups had a number of specific histologically apparent defects with the most obvious being a small cerebral cortex and a striking failure of cerebellar development (Fig. 3). Relative to control littermates, the N-myc NS null cerebellum was significantly smaller and disorganized, exhibiting severe defects in foliation (Fig. 3A–F). Although all three primary mature cerebellar layers—molecular layer (ML), Purkinje cell layer (PCL), and granular cell layer (GCL)—were present to some degree in P20 null animals, there were significant abnormalities with the rostral portion of each layer most severely affected. The GCL was thin and in places completely absent (Fig. 3A–F). In any given area of equivalent position within folia, the N-myc NS null had a granular cell density that was reduced three- to sixfold compared with all three controls. We estimate the reduction in total granular cell numbers in the whole P20 null cerebellum to be ∼30-fold, given both the small total foliar area and the reduced cellular density therein. The null ML appears relatively less affected by N-myc deficiency. However, because of the severe defect in foliation, the ML has failed to expand and the total cerebellar area and cell number of the ML is significantly reduced. The PCL is disrupted in the null animals and this phenotype is characterized by several defects including breaks in the layer and ectopic Purkinje cells (Fig. 3E,F). The total number of cerebellar Purkinje cells is significantly reduced in the NS null cerebellum because of the limited expansion and reduced number of folia, however, within existing NS null folia, the number of Purkinje cells in the vicinity of the GCL is near normal.
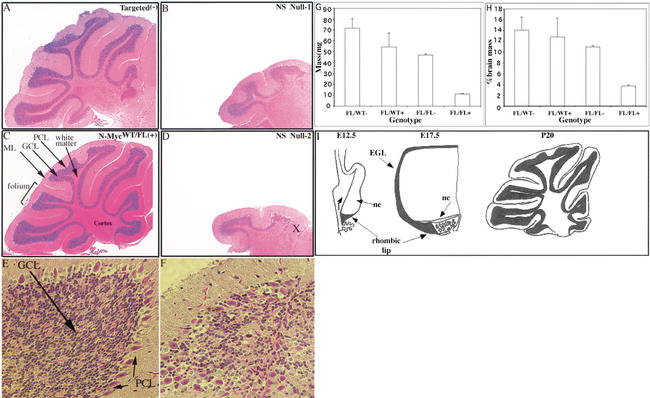
Figure 3.
Lack of N-myc severely disrupts cerebellar development. (A–F) H&E staining of P21 cerebella. (A,C) Control cerebella. (B,D) Null cerebella. (E,F) Folia of control and null cerebella. (G) Absolute cerebellar mass in milligrams from more than four animals of each genotype. (H) Cerebellar mass relative to total brain mass. In D, X is a reference spot to refer to the equivalent position in Figure 4B. (I) Diagram of murine cerebellar development [based on data in Altman and Bayer (1997)]. (Top) Rostral; (bottom) caudal. (NE) neuroepithelium; (EGL) external germinal layer; (ML) molecular layer; (PCL) Purkinje cell layer; (GCL) granular cell layer. Magnification: A–D, 5×; E,F, 40×.
To more accurately quantitate the diminution of the NS null cerebellum at P20, cerebella of NS null and control mice were excised and weighed and the weight compared with the total brain weight. The mean weight of the N-myc NS null cerebellum was just 11 mg and represented ∼3% of the total brain weight (Fig. 3G,H). In contrast, the cerebella of N-mycFL/FL(−) mice had a mean weight of ∼45 mg, representing ∼11% of total brain mass, a percentage roughly equivalent to that seen in wild-type mice (Airey et al. 2001). N-mycFL/WT(−) (70 mg) and N-mycFL/WT(+) (54 mg) cerebella were not statistically different in weight. Thus, the NS null cerebellum is four- to sixfold reduced both in absolute weight and percent of total brain weight.
The null E17.5 cerebellum primordium is small and deficient in germinal cells
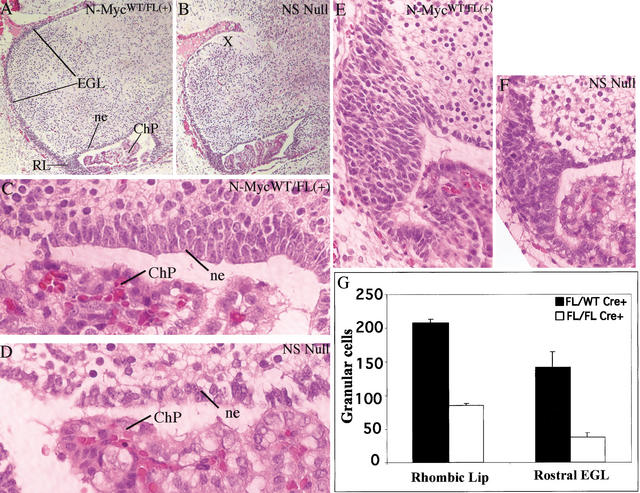
During late embryonic and early postnatal cerebellar development, there are two cerebellar germinal populations. Granular cell precursors are present in the external germinal cell layer (EGL), which emanates from the rhombic lip, a ventral, triangular-shaped pool of germinal cells (Hatten and Heintz 1995; Figs. 3I, 4A), whereas the cerebellar neuroepithelium contains Purkinje, cortical, and molecular layer cell precursors (Figs. 3I, 4A). Although the N-myc NS null cerebellum primordium was only mildly reduced in overall size (Fig. 4A,B), it exhibited a pronounced deficiency in both cerebellar progenitor cell pools (Fig. 4C–G). The deficiency in EGL cells was most pronounced in the most rostral portion (marked X in Fig. 4B), which is the most distant area to which these cells ultimately migrate. The relative severity of the embryonic phenotype at this specific location correlates well with the loss of the rostral portion of the GCL (also marked by X in Fig. 3D) observed later in null P20 animals. In the rhombic lip and the rostral portion of the EGL of the cerebellum primordium, N-myc NS null animals had 2.5- and almost 4-fold fewer cells, respectively, per section (Fig. 4G), whereas the most rostral portion of the EGL is completely absent, suggesting the germinal cells of the EGL prematurely terminate their migration. The N-myc NS null neuroepithelium also appears strikingly thin (only 1–2 cells thick vs. 3–5 cells in the controls) and exhibits breaks in places as well as evidence of disorganization (Fig. 4C,D).
Figure 4.
The N-myc NS null E17.5 cerebellum primordium is small and exhibits germinal cell deficiency. (A,B) H&E-stained control and null cerebella primordia, respectively. (C,D) Control and null neuroepithelium. (E,F) Control and null rhombic lip. (G) Cell counts, the mean of three sections, from the rhombic lip and EGL. RL, rhombic lip; ChP, choroid plexus; EGL, external germinal layer; ne, neuroepithelium. Magnification: A,B, 5×; C,D, 100×; E,F, 40×.
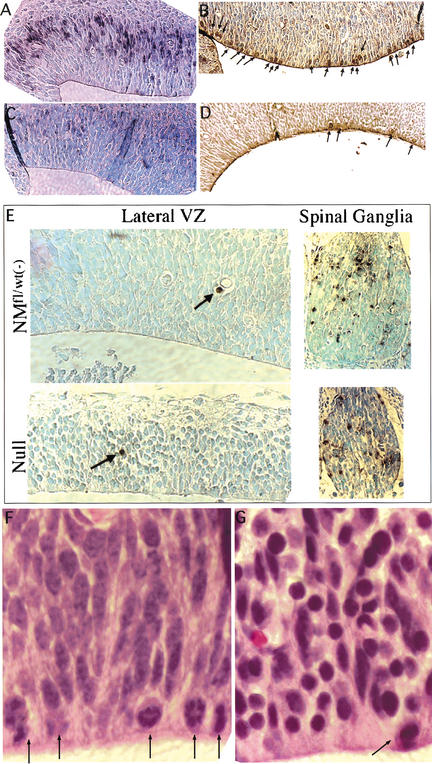
A subset of the null rhombic lip NPCs were more rounded and more densely packed than control NPCs, strongly suggesting reductions in cytoplasmic volume. Similar, but more severe changes were also observed in the lateral VZ of E17.5 null embryos (Fig. 6E, left side, and F,G, see below). Many of the N-myc-deficient NPCs therein have nuclei that are quite small and rounded with some regions exhibiting alterations in cellular packing. The small round nuclei exhibit rather intense H&E (Fig. 6F,G, see below) and DAPI staining (data not shown), suggestive of altered nuclear structure. There are also many cytoplasmic gaps between these NPCs, indicating the cells may have reduced cytoplasmic volume (most evident in Fig. 6E, bottom left, and F,G, see below).
Figure 6.
Analysis of mitosis, S phase, and apoptosis in the E13 lateral VZ. (A,C) BrdU staining of a control and null. (B,D) Anti-phosphoH3 staining of a control and null. (E) TUNEL staining. (F,G) The dorsal portion of the lateral VZ in a control and null embryo, respectively. Arrows point to mitotic figures (A,F,G) or apoptic cells (E). Magnification: E, 40×; F,G, 100×.
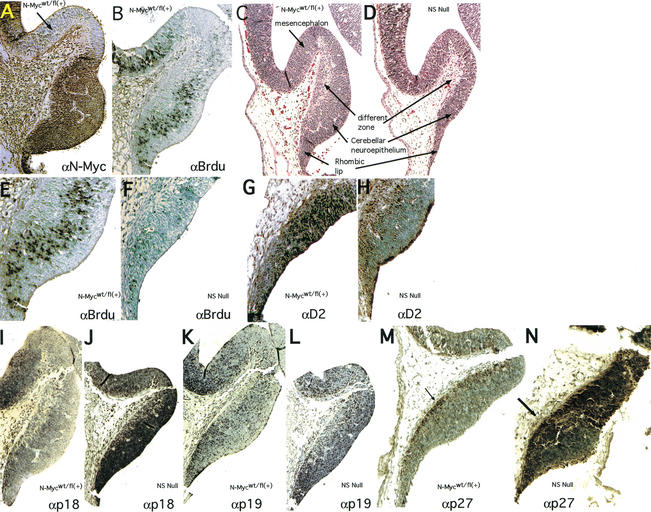
The N-myc NS null E12.5 cerebellum primordium, normally a site both of high N-Myc levels and BrdU incorporation, has decreased numbers of NPC
To further trace the defects in cerebellar development in the N-myc-deficient brain, earlier embryos were analyzed histologically. Interestingly, the E12.5 cerebellar neuroepithelium and the rhombic lip, which together appear as a bulge histologically (Fig. 5C), and contain all of the germinal cells necessary for making a cerebellum (Hatten and Heintz 1995; Goldowitz and Hamre 1998) have very high levels of N-Myc protein (Fig. 5A). The same region has a high percentage of cells in S phase as determined by BrdU staining (Fig. 5B). We note that essentially all BrdU-positive cells are also N-myc positive. Precisely in this zone, N-myc NS null embryos have a substantial reduction in NPC and the interior differentiating zone is also reduced in size (Fig. 5C,D). Interestingly, the top of the control cerebellum primordium, a mesencephalic region that does not contribute to the development of the cerebellum itself (Altman and Bayer 1997), is relatively deficient in N-Myc, BrdU incorporation, and cyclin D2 (arrow in Fig. 5A; data not shown), suggesting N-Myc is differentially regulated within, and may demarcate, distinct developmental domains. Our findings from E12.5–E17.5 embryos indicate that the more profound cerebellar defects seen in P20 animals may be caused by a deficiency in embryonic cerebellar germinal cells.
Figure 5.
The N-myc NS null E12–E13 cerebellum primordium exhibits NPC deficiency, loss of cyclin D2 expression, and ectopic CDKI expression. Sections of cerebella primordia were stained as indicated. The arrow in A identifies the domain at the apex of the cerebellum primordium that lacks significant N-myc, cyclinD2, and BrdU staining.
Loss of N-myc inhibits NPC proliferation without increasing apoptosis
To examine the hypothesis that disruption of N-myc may cause NPC deficiency by reducing cellular proliferation, BrdU incorporation was monitored in control and null E13 embryos. The N-myc null CNS generally exhibited a striking reduction in the percentage of cells in S phase as determined by the extent of BrdU incorporation, and the cerebellum primordium was an area of particular contrast between controls and nulls (Fig. 5E,F). Whereas control embryos exhibited significant anti-BrdU staining as a band through the interior of the cerebellum primordium, only a tiny fraction of nuclei in the same region of the N-myc NS null were BrdU positive. Similarly, loss of N-myc in the VZ of the developing telencephalon, an area of normally high BrdU incorporation, resulted in a notable decrease in the number of S-phase NPCs (Fig. 6A,C). Thus, disruption of N-myc results in a nearly complete loss of S-phase NPCs at E13 in several distinct germinal regions of the brain. To further examine the effects of loss of N-myc on the cell cycle, sections were stained with an antibody against phosphorylated histone H3 (phosphoH3), a marker for mitotic cells (Gurley et al. 1978). Consistent with a substantial reduction in mitotic figures evident by H&E and DAPI staining (Fig. 6F,G; data not shown), very few N-myc NS null NPCs of the cerebellum primordium (data not shown) and lateral VZ (Fig. 6B,D) were phosphoH3 positive (mitotic).
Because the NPC deficiency linked to loss of N-myc could also be the result of an increase in apoptosis, TUNEL staining was conducted on E13 sections of control and N-myc NS null embryos (Fig. 6E). The normal level of apoptosis in the E13 brain is extremely low (<0.01% in the lateral ventricle and undetectable in the cerebellum primordium) and loss of N-myc causes no detectable increase in this rate throughout the CNS, including the cerebellum primordium (data not shown) and the lateral VZ (Fig. 6E). Furthermore, N-myc did not modulate apoptosis in regions of the CNS with normally high apoptotic rates, such as the spinal ganglia (5%–10% TUNEL positive; Fig. 6E). Thus, N-myc does not appear to regulate cell survival at E13, and the NPC deficiency observed in nulls is not the result of increased cell death.
Loss of N-myc disrupts cyclin D2 expression
To further explore the mechanism by which loss of N-myc affects the cell cycle in NPCs, the expression of cyclin D2 was examined in E13 control and N-myc NS null embryos. Cyclin D2 is a particularly attractive potential mediator of N-myc function in this context because it is a well-established c-Myc target gene (Bouchard et al. 1999, 2001; Perez-Roger et al. 1999), and targeted disruption of the cyclin D2 gene in mice results in a diminutive cerebellar phenotype (Huard et al. 1999). High levels of cyclin D2 are broadly distributed throughout the most proliferative regions of the control E13 CNS. For example, ∼90% of the germinal cells of the E13 cerebellum primordium are cyclin D2 positive (Fig. 5G). In contrast, in the N-myc NS null cerebellum primordium, <10% of the cells are cyclin D2 positive (Fig. 5, cf. G and H). Thus, loss of N-myc results in a profound down-regulation of cyclin D2 in cells within the cerebellar primordium.
Null progenitors exhibit an altered expression pattern of cdk inhibitors
Because Myc function has also been linked to modulation of cdk inhibitors (CDKI's), we examined, by immunostaining, the expression of CDKI's associated previously with Myc function or neurogenesis. In the developing CNS at E13, the normal expression patterns of p27Kip1, p18Ink4c, and p15Ink4b are very restricted, limited to the differentiating zone in which cells have begun to exit the cell cycle (Fig. 5M, arrow; data not shown) and where there is little detectable N-Myc, BrdU, and cyclin D2 staining. Although p18Ink4c and p27Kip1 expression is restricted to these differentiation zones in controls, null embryos, in contrast, exhibit widespread, intense expression of both proteins throughout the cerebellum primordium, and to a lesser extent, the lateral VZ (Fig. 5, cf. I and J, M and N; data not shown). In contrast, loss of N-Myc did not affect other CDKI's as p19Ink4d was unaffected, p21Cip1 was undetectable in both control and null cerebellar primordia (Fig. 5K,L; data not shown), and p15Ink4b levels were also unchanged by disruption of N-myc (data not shown), despite the fact that the latter two are considered Myc target genes (Bouchard et al. 2001; Staller et al. 2001). Therefore, regulated expression of a distinct group of CDKI's in the developing nervous system may represent a unique cellular program that is controlled by N-Myc (see Discussion).
N-Myc inhibits neuronal differentiation
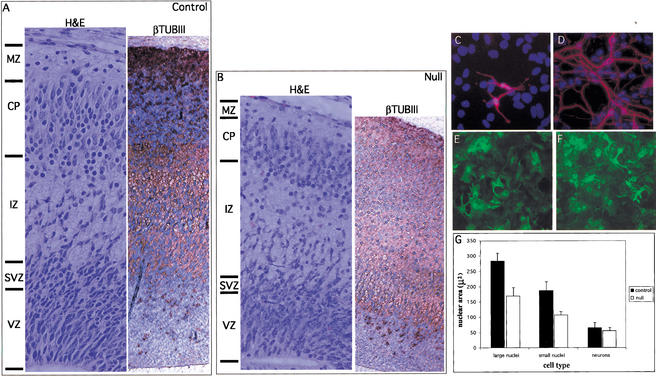
To examine the hypothesis that N-Myc functions to maintain a progenitor cell state, we examined differentiation of neural progenitors both in vivo (Fig. 7A,B,) and in vitro in primary progenitor cell cultures derived from E12.5 embryos (Fig. 7C–F). Generally, staining for βTubIII in the control E17.5 cerebrum revealed a clearly recognizable pattern of layers of βTubIII-negative and positive cells that correlated with layering evident by H&E staining. In sharp contrast, little layering was evident in the null cerebrum, which was characterized by expansive βTubIII staining. Whereas the VZ of both the nulls and controls was almost uniformly βTubIII negative, the null subventricular zone (SVZ) was markedly abnormal. The control SVZ is characterized by its predominantly βTubIII-negative progenitors with large nuclei (recognizable as counterstaining bright blue) radially streaming into the intermediate zone (IZ). In contrast, the null SVZ was largely βTubIII positive and the relatively few migrating cells possessed small nuclei. Similarly, the null IZ, which appears to constitute a disproportionately increased fraction of the cerebral wall compared with that of the control, contained many cells with small, abnormally round nuclei. Although it was difficult to determine the boundary between the cortical plate (CP) and IZ by βTubIII staining in the null, it was clearly evident by H&E staining (Fig. 7B, left), again suggesting significant ectopic expression of βTubIII in the null. Thus, the null cerebral wall generally exhibits a greatly increased proportion of βTubIII-positive cells, suggesting precocious differentiation in vivo in the absence of N-myc.
Figure 7.
N-Myc inhibits neuronal differentiation. (A,B) H&E (left) and βTubulinIII (right) staining of the control and null E17.5 cerebral cortex in the lateral ventricle region. Cells positively stained for β-tubulinIII are brown. (C,D) Anti-βTubulinIII immunofluorescence of control and null E12.5 neurosphere cultures stimulated to differentiate for 5 d. (E,F) Anti-GFAP immunofluorescence of control and null E12.5 neurosphere cultures stimulated to differentiate for 5 d. (G) Nuclear size in microns of the same neurosphere cultures as above. VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate; MZ, marginal zone.
Primary neurosphere progenitor cell cultures were established from control and null E12.5 whole brain (Palmer et al. 1995; see Materials and Methods section). Null progenitors proliferated at an approximately twofold decreased rate compared with controls (data not shown). Furthermore, they exhibited strongly enhanced neuronal differentiation (induced by growth factor withdrawal and the addition of serum and retinoic acid) as judged by a significant increase in βTubIII-positive neurons, extensions, and connections compared with littermate controls (Fig. 7C,D). In contrast, gliogenesis appeared relatively unaffected by N-myc status as judged by αGFAP immunofluorescence (Fig. 7E,F). Thus, disruption of N-myc leads to enhanced neuronal differentiation both in vivo (Fig. 7A,B) and in vitro (Fig. 7C,D), suggesting N-Myc normally suppresses neuronal differentiation.
Null primary neuronal progenitor cells exhibited morphological abnormalities. In agreement with our observations that a subset of nuclei in saggital sections of null embryos are significantly smaller, N-myc-deficient progenitor cells [i.e., staining negatively for both neuron (βTubIII) and glial (GFAP) markers] also exhibit up to a 40% reduction in nuclear volume as determined by DAPI staining and measurement of nuclear area (Fig. 7G). Surprisingly, whereas null glia (GFAP+) cells also exhibit reduced nuclear volume, the nuclei of N-myc-deficient neurons (βTubIII+) are not significantly reduced in volume (Fig. 7G), suggesting that the relationship between N-myc and nuclear volume is specific to progenitors, and that differentiation may dictate ultimate nuclear size. Thus, we have shown both in vivo and in vitro that disruption of N-myc enhances neuronal differentiation, indicating a normal role for N-Myc as an inhibitor of differentiation.
Discussion
Here, we present the first systematic study of the function of a Myc family gene in organogenesis with a specific focus on the function of N-Myc in the development of the nervous system. Disruption of N-myc in the nervous system during murine embryogenesis causes neurological dysfunction manifested postnatally by tremors, ataxia, and behavioral abnormalities. Although these mice survive to adulthood, they exhibit a twofold reduction in total postnatal brain mass with particularly severe phenotypic changes in the cerebellum and cerebral cortex evident as early as E12.5. Because cerebellar defects are frequently associated with tremors and ataxia (Hirano and Dembitzer 1973; Sheldon et al. 1997), there is likely a causal relationship between the cerebellar abnormalities and the tremors and ataxia we observe in the N-myc NS null animals. We chose to focus our analysis primarily on the cerebellum, the most severely affected region of the null nervous system, but it is critical to emphasize that the changes observed in the cerebellum are consistent with relatively less severe, yet similar, alterations observed throughout the null CNS.
Our findings with conditional N-myc NS null mice are consistent with previously published reports using constitutive knockouts of N-myc, which were limited to histological studies of embryos of E11.5 or earlier and therefore precluded analysis of both cerebellar development and the mid-late stages of neurogenesis (Sawai et al. 1991, 1993; Stanton et al. 1992; Charron et al. 1992). These earlier studies found that disruption of N-myc generally reduced the size of both the CNS and PNS at E11, but were unable to investigate the causes or the ultimate consequences of this reduction in size. Our conditional knockout shows that the effects in the nervous system following constitutive loss of N-myc are specifically due to disruption of NPC function and not a secondary consequence of a more general failure of embryogenesis. In addition, our work shows that the consequences of N-myc loss are manifested throughout the full course of neural development. We also specifically identify a group of genes, including cyclin D2, p27Kip1 and p18Ink4c, that likely mediate the effects of disruption of N-myc on proliferation and possibly differentiation.
N-myc regulates the cell cycle of neuronal progenitors
The dramatically decreased size of the CNS in N-myc NS null mice could be due to a reduction in cell number, cell size, or both. Our data suggest that decreases in both cell number and cell size are occurring. We observed significantly reduced numbers of NPCs in diverse germinal regions of the brain. For example, the reduction in overall cerebellum size, weight, number of folia, and density of the granular cell layer in N-myc NS null mice at P21 is reflected in the substantially decreased progenitor cell numbers in the cerebellum primordium at E17.5 and E12.5 (Figs. 4, 5). This NPC deficiency, in turn, appears to be due in large part to attenuated proliferation, as evidenced by substantially reduced numbers of S- and M-phase NPCs, sharply diminished expression of cyclin D2, and elevated levels of p27Kip1 and p18Ink4c, whereas p19Ink4d and p15Ink4b are unaffected. It was reported previously that during neurogenesis p18Ink4c levels, but not those of p19Ink4d, only begin to increase during the transition in progenitors from rapid to a slower proliferation rate that coincides with increased differentiation at E13.5–E15.5 (Zindy et al. 1997). Thus, our observation that disruption of N-myc specifically increases p18Ink4c levels, but not those of p19Ink4d, early in neurogenesis, supports the idea that N-Myc may normally sustain an early progenitor identity. At E11, cell cycle duration in the cerebral cortex is remarkably rapid, as short as 8 h, mainly due to an extremely brief G1 phase (for review, see Caviness et al. 1995). As neurogenesis proceeds, the duration of G2+M and S phases remain essentially unchanged, but G1 becomes progressively longer, more than doubling by E15. Interestingly, N-myc levels decline during the same period (Stanton et al. 1992). In our N-myc NS null mice early (e.g., E11), rapidly cycling NPCs that normally have high levels of N-myc expression may be reprogrammed to behave biologically as though they were later (e.g., E15–P0), more slowly replicating, or even quiescent NPCs in which N-Myc levels are lower. N-myc, perhaps through its induction of target genes like cyclin D2 and through suppression of CDKIs is, therefore, a good candidate for maintaining the extraordinarily short cell cycle duration of NPCs in early neurogenesis. This work suggests a specific subset of CDKIs whose regulation may be responsible for neural cell cycles.
Interestingly, targeted disruption of the cyclin D2 gene produces a phenotype of cerebellar diminution that is considerably milder than what we observe in the N-myc NS null animals (Huard et al. 1999). The cyclin D2 null phenotype is quite consistent with it being a transcriptionally up-regulated target of Myc (Bouchard et al. 1999, 2001; Perez-Roger et al. 1999). We find a dramatic reduction in cyclin D2 levels in NPCs in the null mice (Fig. 5G,H). Because the phenotype of our N-Myc disruption is more severe than observed for deletion of cyclin D2, we surmise that targets of N-Myc, in addition to cyclin D2, are crucial for neural development. It seems likely that the increase in p18Ink4c and decrease in cyclin D2 act to inhibit overall D kinase activity, whereas the increase in p27Kip1 acts to inhibit E kinase activity, thereby severely limiting cell cycle progression (Sherr and Roberts 1999). Although the decrease in cyclin D2 levels are consistent with the fact that it is a direct transcriptional target of Myc (Bouchard et al. 2001), it is also possible that the elevated levels of p18Ink4c detected in the absence of N-Myc lead to degradation and further loss of cyclin D function. Our data suggest that p18Ink4c is likely to be a new Myc repression target.
Differentiation, but not death
Reduced progenitor cell numbers in the N-myc NS null mice could also be the result of two other causes, stimulation of programmed cell death or precocious differentiation of progenitors. Elevated levels of apoptosis are unlikely to be significant, as we failed to detect any change in the extent of apoptosis using TUNEL assays. However, we present evidence that disruption of N-myc relieves suppression of neuronal differentiation in the cerebrum, suggesting that N-Myc normally inhibits neuronal differentiation. Precocious terminal differentiation in the absence of N-myc may shift NPCs into a differentiated, nonproliferative state. We did not find evidence of precocious differentiation in the E17.5 cerebellum, in which we observed cell cycle arrest, but it is unclear whether this is because loss of N-Myc differentially affects neuronal differentiation in the cerebrum and cerebellum, or whether altered differentiation in the null cerebellum may occur much later. Very little is known about the mechanistic role of Myc proteins in differentiation, however, two reports using cell lines imply a role for Myc in neuronal differentiation, consistent with our observations in vivo and in vitro (Bernard et al. 1992; Hoshimaru et al. 1996). It remains unclear whether N-Myc directly blocks differentiation or whether it indirectly inhibits differentiation by maintaining a state of rapid proliferation. Nonetheless, our finding that loss of N-Myc induces p27Kip1, a CDKI shown to be linked to differentiation (Casaccia-Bonnefil et al. 1997) as well as p18Ink4c, a CDKI linked to a shift from symmetric to asymmetric progenitor division, suggests that N-Myc normally regulates a switch in progenitor programming from proliferation to differentiation.
Progenitor cell size and morphology
A decrease in cell size may also contribute to the reduced size of the CNS. There is considerable evidence that Myc is a positive effector of cell growth (see Introduction). We observed that a subset of the NPCs (e.g., cells within the rhombic lip, the adjacent neuroepithelium, and the lateral VZ) from N-myc NS null animals contain smaller, more densely spaced nuclei and apparently less cytoplasmic area than those of littermate controls (see Figs. 4, 6). We found that N-myc-deficient nuclei were up to 40% smaller than that of littermate controls in primary progenitor cultures. However, it remains unclear whether reductions in cell or nuclear size affect the ultimate mass of the fully mature N-myc-deficient CNS, because the only evidence of a reduction in the size of more mature neurons in vivo is a moderate reduction in the spacing between neuronal nuclei in the mature brain, the cause of which remains unclear (data not shown).
N-myc NS null NPCs, even those without small nuclei, also exhibited signs of altered nuclear structure. A subset of N-myc-deficient nuclei are abnormally rounded and display highly dense staining with H&E and DAPI compared with control NPCs (Fig. 6F,G; data not shown), suggestive of more compact chromatin in the N-myc NS null nuclei. There is strong evidence that Myc's transcriptional activity is closely linked to its ability to acetylate nucleosomal histones in the vicinity of Myc–Max-binding sites (McMahon et al. 2000; Frank et al. 2001). Recent studies identifying target genes (Coller et al. 2000) as well as DNA-binding sites (A. Oryan and R.N. Eisenman, unpubl.) indicate that there are likely to be hundreds of chromatin-binding sites for Myc–Max dimers. Furthermore, c-Myc overexpression has been tied to genetic instability (Felsher and Bishop 1999). These considerations point to the interesting possibility that Myc, through histone acetylation, may have a more global effect on chromatin structure than has perhaps been generally appreciated. In this regard, loss of N-Myc function, in addition to down-regulation of specific gene targets (e.g., cyclin D2), might drive chromatin condensation in a way that attenuates DNA replication. It remains to be determined exactly what the consequences of N-Myc loss are for chromatin structure and nuclear morphology.
N-myc control of the normal and abnormal growth of CNS domains
Why is the cerebellum, and to a lesser extent the neocortex, particularly affected by N-myc deficiency? We favor the notion that N-myc is a critical regulator of cerebellar and cerebral cortex development. Support for this idea comes from our data showing sharply delineated areas, or domains, of high-level N-Myc protein expression in the cerebellar and cerebral primordia, which correlate strongly with zones of DNA synthesis. In N-myc null mice, these domains correspond to regions of cells with reduced DNA synthesis, mitoses and cyclin D2 expression, decreased size, elevated levels of cdk inhibitors, and altered nuclear structure. It is also possible that temporal and spatial aspects of Cre expression from the nestin-Cre transgene may influence the specificity and severity of the N-myc NS null phenotype. For example, spatially, the nestin-Cre transgene may be more active or more persistent in a subgroup of NPCs that are destined to form the cerebellum and cerebral cortex. Temporally, a significant proportion of N-myc function during neurogenesis may simply precede its deletion by Cre. The nestin-Cre transgene is known to produce low levels of Cre activity in reporter mice by E9.5 in NPCs (A. Imamoto, pers. comm.) as well as to be strongly active by E10.5, more broadly in the CNS and PNS (Graus-Porta et al. 2001), but significant N-Myc function may precede E10.5.
Our data suggesting that N-myc is involved in driving expansion of distinct cellular populations during brain development points to potential links between N-Myc function and brain evolution as well as the genesis of nervous system tumors. Recent studies have suggested that different taxonomic groups vary in the fraction of total brain volume occupied by parts of the brain, such as the neocortex and cerebellum. However, within a taxonomic group, proportions between parts are maintained, even though the absolute size of the brain can vary considerably (Clark et al. 2001). N-Myc is a highly conserved, and highly regulated protein, which, as we have shown here, promotes expansion of NPCs in different regions of the brain in a manner that influences regional size (Fig. 2). N-Myc is therefore a reasonable candidate for involvement in the molecular mechanism responsible for the coordination or unlinking of regional brain growth.
Three aberrant forms of nervous system growth, medulloblastomas, neuroblastomas, and retinoblastomas have been shown to contain amplifications of the N-myc locus or otherwise overexpress N-Myc (Kohl et al. 1983; Schwab et al. 1983; Lee et al. 1984; Badiali et al. 1991). Our observation that N-Myc specifically inhibits neuronal, but not glial differentiation, could in part explain why N-Myc is strongly linked with the above three tumors, which are all of neuronal origin, whereas there is little evidence linking N-Myc to glial-derived tumors. Medulloblastoma, the major malignant brain tumor of childhood, had long been postulated to arise from cerebellar granule cells, an idea that has recently received strong confirmation (Marino et al. 2000). An oligonucleotide microarray analysis also showed a good correlation between desmoplastic medulloblastoma and high N-myc expression (Pomeroy et al. 2002). Our work, showing that cerebellar granule cell progenitors require N-myc during development, suggests that these cells may be especially sensitive to changes in the abundance of N-Myc. Thus, overexpression of N-myc within a specific developmental window may cause medulloblastoma and other tumors by driving germinal cells to maintain their early embryonic short-duration cell cycles and, therefore, favor self-renewal, genetic instability, and cancer.
Materials and methods
Construction of targeting vector and production of mice
A ClaI fragment containing the first 892 bp of the murine N-myc cDNA was used to probe a 129-murine genomic library, identifying a 18–20-kb full-length N-myc genomic clone. Restriction fragments were subcloned into pPGKneobpalox2DTA. Briefly, a 1.45-kb EcoRI fragment was blunted and subcloned 3′ of the neo cassette and the adjacent loxP site into a blunted SalI site. A synthetic loxP site paired with a new EcoRI site just 5′ were created by annealing two complimentary oligos that contained BamHI sticky ends. The N-myc genomic clone was cut with BglII by partial digest and the EcoRI–loxP site fragment was ligated into the BglII site in the first intron. The construct now containing the internal loxP site was cut with BglII and EcoRI, releasing a 7.7-kb fragment that was blunted and cloned into a blunted SacII site 5′ of the loxP site 5′ of neo. Cre was introduced into heterozygously floxed 129 ES cells by electroporation using the pOG231 vector. A total of 129 ES cells were injected into C57BL/6 blastocysts and chimeric mice were bred to C57BL/6 mates to check for germ-line transmission. The targeted mice bred with the nestin-Cre transgenic mice were of a mixed, C57BL/6 and 129, background.
Immunohisto- and cytochemistry
Embryos or tissue samples were fixed overnight in fresh buffered 4% paraformaldehyde. Paraffin-embedded sections (7 μm), sagittally cut in all cases, were mounted for staining. Antibodies used include anti-Ν-Myc (Santa Cruz, 1:200), anti-cyclin D2 (Santa Cruz, 1:200), anti-TubulinβIII (Covance, 1:200, 1:1000 IF), anti-GFAP (Advanced Immunochemicals, 1:500 ), anti-p15Ink4b, as well as anti-p18Ink4c, p21Cip1, and αp19Ink4d (Santa Cruz Biotechnologies, 1:50), anti-p27Kip1 (Neoprobes), anti-Phospho H3 (Upstate, 1:100), and anti-BrdU (Sigma, 1:50). Sections were counterstained with methyl green. Staining was conducted as described by the manufacturer (Vectastain elite ABC kit). αBrdU staining was conducted on embryos from mothers who were injected IP with BrdU (150 μg/g) following a 1-h BrdU incorporation period. TUNEL staining was conducted using the Apotag kit (Intergen) as directed.
RT–PCR
Total RNA isolated from brain was prepared using Trizol as described by the manufacturer. Primers included the following: (c-Myc, 201bp: s-ACCAACAGGAACTATGACCTC and as-AAGGACGTAGCGACCGCAAC), (N-myc, 345 bp: s-CAGCT GCACCGCGTCCACCATGCCGGGGATGATCTGC and as-CATGCAGTCCTGAAGGATGACCGGATTAGGAGTGAG), (L-Myc, 262 bp: s-CTCGTATCAGCACTATTTCTACG and as-GGTCACCACGTCAATCTCTTCAC), (Rpl7, ribosomal protein L7 internal control, 202 bp: s-GAAGCTCATCTATGA GAAGGC and as-AAGACGAAGGAGCTGCAGAAC). All samples were also run at multiple cycles to determine linear range, and minus reverse transcriptase to control for RNA dependence. Total RNA (2 μg) was used for RT with MMLV and a random primer. A total of 1 μL of this RT reaction was subsequently used for the PCR reaction with [32P]CTP. Cycling was conducted as follows 94°C for 2 min initially followed by variable numbers of cycles of the following: 94°C for 30 sec, 62°C for 30 sec, and 72°C for 1 min, except for N-myc, in which an annealing temperature of 65°C was used. Samples were run on an 8% tris-borate-EDTA acrylamide gel.
Genomic PCR
Primers used for genomic PCR included the following: (1) GTC GCGCTAGTAAGAGCTGAGATC, (2) GGCACACACCTAT AATCCCAGCTAG, and (3) CACAGCTCTGGAAGGTGGGA GAAAGTTGAGCGTCTCC. The flox specific band is 260 bp, the wild-type band is 217 bp, and the deletion band is 350 bp.
Neurosphere culture
Whole E12.5 mouse heads were dissociated by rubbing between two frosted slides. Cells were cultured in the following medium: Neurobasal (GIBCO-BRL) containing 1% glutamine, B27 supplement (GIBCO-BRL), and 20 ng/mL EGF and bFGF. For differentiation, cells were cultured in the following medium: Neurobasal (GIBCO-BRL) containing 1% glutamine, B27 supplement (GIBCO-BRL), 100 nm all-trans retinoic acid, and 1% FCS.
Calculation of nuclear volume
Images of DAPI-stained nuclei were captured on a Deltavision microscope at 40× magnification in which a pixel/μ conversion of 0.1357 was determined. Nuclear area in pixels was determined using the Photoshop program and converted to μ. Error bars, S.D.
Acknowledgments
We thank Philippe Soriano for advice and important reagents throughout the course of this work. We also thank Susan McConnell, Theo Palmer, Cecilia Moens, Jon Cooper, Steve Tapscott, Philippe Soriano, and Susan Mendrysa for their insightful comments on the manuscript. We thank Akira Imamoto for the nestin-Cre mice and for sharing unpublished data. We thank Theo Palmer for help with the neurosphere cultures. We also thank Denny Liggit for his advice as a mouse pathologist, Brian Iritani for helpful advice, Nanyan Jiang for her excellent work on the mouse transgenics, Kay Gurley for the p27 staining, and Karen Englehart for her excellent histological work. We thank members of the Eisenman laboratory for their advice and support. P.S.K. was supported by a Jane Coffin Childs Memorial Fund for Medical Research fellowship. R.N.E. is a Research Professor of the American Cancer Society. This work was supported by grants from the National Institutes of Health (to R.N.E).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL eisenman@fhcrc.org; FAX (206) 667-6522.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1021202.
References
- Airey DC, Lu L, Williams RW. Genetic control of the mouse cerebellum: Identification of quantitative trait loci modulating size and architecture. J Neurosci. 2001;21:5099–5109. doi: 10.1523/JNEUROSCI.21-14-05099.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman J, Bayer S. Development of the cerebellar system: In relation to its evolution, structure, and functions. Boca Raton, FL: CRC Press; 1997. pp. 82–137. [Google Scholar]
- Badiali M, Pession A, Basso G, Andreini L, Rigobello L, Galassi E, Giangaspero F. N-myc and c-myc oncogenes amplification in medulloblastomas. Evidence of particularly aggressive behavior of a tumor with c-myc amplification. Tumori. 1991;77:118–121. doi: 10.1177/030089169107700205. [DOI] [PubMed] [Google Scholar]
- Beier R, Burgin A, Kiermaier A, Fero M, Karsunky H, Saffrich R, Moroy T, Ansorge W, Roberts J, Eilers M. Induction of cyclin E-cdk2 kinase activity, E2F-dependent transcription and cell growth by Myc are genetically separable events. EMBO J. 2000;19:5813–5823. doi: 10.1093/emboj/19.21.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard O, Drago J, Sheng H. L-myc and N-myc influence lineage determination in the central nervous system. Neuron. 1992;9:1217–1224. doi: 10.1016/0896-6273(92)90079-s. [DOI] [PubMed] [Google Scholar]
- Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, Weis I, Voute PA, Schwab M, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Luscher B. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes & Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Tikoo R, Kiyokawa H, Friedrich V, Jr, Chao MV, Koff A. Oligodendrocyte precursor differentiation is perturbed in the absence of the cyclin-dependent kinase inhibitor p27Kip1. Genes & Dev. 1997;11:2335–2346. doi: 10.1101/gad.11.18.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviness VS, Jr, Takahashi T, Nowakowski RS. Numbers, time and neocortical neuronogenesis: A general developmental and evolutionary model. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, Robertson EJ, Alt FW. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes & Dev. 1992;6:2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- Clark DA, Mitra PP, Wang SS. Scalable architecture in mammalian brains. Nature. 2001;411:189–193. doi: 10.1038/35075564. [DOI] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes & Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- de Alboran IM, O'Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- Douglas NC, Jacobs H, Bothwell AL, Hayday AC. Defining the specific physiological requirements for c-Myc in T cell development. Nat Immunol. 2001;2:307–315. doi: 10.1038/86308. [DOI] [PubMed] [Google Scholar]
- Eisenman RN. Deconstructing myc. Genes & Dev. 2001;15:2023–2030. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- Facchini LM, Penn LZ. The molecular role of Myc in growth and transformation: Recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci. 1999;96:3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes & Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaktionov K, Chen X, Beach D. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Ye X, Goufman E, Shianov P, Hay N, Najmabadi F, Tyner AL. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc Natl Acad Sci. 2001;98:4510–4515. doi: 10.1073/pnas.081074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. Trends Neurosci. 1998;21:375–382. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Graus-Porta D, Blaess S, Senften M, Littlewood-Evans A, Damsky C, Huang Z, Orban P, Klein R, Schittny JC, Muller U. Beta1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron. 2001;31:367–379. doi: 10.1016/s0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee NH, Dang CV, Liu ET. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- Gurley LR, D'Anna JA, Barham SS, Deaven LL, Tobey RA. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur J Biochem. 1978;84:1–15. doi: 10.1111/j.1432-1033.1978.tb12135.x. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- Hatton KS, Mahon K, Chin L, Chiu FC, Lee HW, Peng D, Morgenbesser SD, Horner J, DePinho RA. Expression and activity of L-Myc in normal mouse development. Mol Cell Biol. 1996;16:1794–1804. doi: 10.1128/mcb.16.4.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, O'Connell BC, Mateyak MK, Tam W, Kohlhuber F, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano A, Dembitzer HM. Cerebellar alterations in the weaver mouse. J Cell Biol. 1973;56:478–486. doi: 10.1083/jcb.56.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshimaru M, Ray J, Sah DW, Gage FH. Differentiation of the immortalized adult neuronal progenitor cell line HC2S2 into neurons by regulatable suppression of the v-myc oncogene. Proc Natl Acad Sci. 1996;93:1518–1523. doi: 10.1073/pnas.93.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huard JM, Forster CC, Carter ML, Sicinski P, Ross ME. Cerebellar histogenesis is disturbed in mice lacking cyclin D2. Development. 1999;126:1927–1935. doi: 10.1242/dev.126.9.1927. [DOI] [PubMed] [Google Scholar]
- Hurlin PJ, Queva C, Eisenman RN. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes & Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- Iritani BM, Eisenman RN. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci. 1999;96:13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Li Q, Dang CV, Lee LA. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc Natl Acad Sci. 2000;97:11198–11202. doi: 10.1073/pnas.200372597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl NE, Kanda N, Schreck RR, Bruns G, Latt SA, Gilbert F, Alt FW. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell. 1983;35:359–367. doi: 10.1016/0092-8674(83)90169-1. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- Lee TC, Li L, Philipson L, Ziff EB. Myc represses transcription of the growth arrest gene gas1. Proc Natl Acad Sci. 1997;94:12886–12891. doi: 10.1073/pnas.94.24.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WH, Murphree AL, Benedict WF. Expression and amplification of the N-myc gene in primary retinoblastoma. Nature. 1984;309:458–460. doi: 10.1038/309458a0. [DOI] [PubMed] [Google Scholar]
- Leone G, Sears R, Huang E, Rempel R, Nuckolls F, Park CH, Giangrande P, Wu L, Saavedra HI, Field SJ, et al. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol Cell. 2001;8:105–113. doi: 10.1016/s1097-2765(01)00275-1. [DOI] [PubMed] [Google Scholar]
- Marhin WW, Chen S, Facchini LM, Fornace AJ, Jr, Penn LZ. Myc represses the growth arrest gene gadd45. Oncogene. 1997;14:2825–2834. doi: 10.1038/sj.onc.1201138. [DOI] [PubMed] [Google Scholar]
- Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes & Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Auerbach AB, Conlon RA, Joyner AL, Rossant J. A targeted mutation reveals a role for N-myc in branching morphogenesis in the embryonic mouse lung. Genes & Dev. 1992;6:691–704. doi: 10.1101/gad.6.5.691. [DOI] [PubMed] [Google Scholar]
- Moens CB, Stanton BR, Parada LF, Rossant J. Defects in heart and lung development in compound heterozygotes for two different targeted mutations at the N-myc locus. Development. 1993;119:485–499. doi: 10.1242/dev.119.2.485. [DOI] [PubMed] [Google Scholar]
- Nagy A, Moens C, Ivanyi E, Pawling J, Gertsenstein M, Hadjantonakis AK, Pirity M, Rossant J. Dissecting the role of N-myc in development using a single targeting vector to generate a series of alleles. Curr Biol. 1998;8:661–664. doi: 10.1016/s0960-9822(98)70254-4. [DOI] [PubMed] [Google Scholar]
- O'Hagan RC, Ohh M, David G, de Alboran IM, Alt FW, Kaelin WG, Jr, DePinho RA. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes & Dev. 2000;14:2185–2191. doi: 10.1101/gad.827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer TD, Ray J, Gage FH. FGF-2-responsive neuronal progenitors reside in proliferative and quiescent regions of the adult rodent brain. Mol Cell Neurosci. 1995;6:474–486. doi: 10.1006/mcne.1995.1035. [DOI] [PubMed] [Google Scholar]
- Perez-Roger I, Kim SH, Griffiths B, Sewing A, Land H. Cyclins D1 and D2 mediate myc-induced proliferation via sequestration of p27(Kip1) and p21(Cip1) EMBO J. 1999;18:5310–5320. doi: 10.1093/emboj/18.19.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- Rosenwald IB, Rhoads DB, Callanan LD, Isselbacher KJ, Schmidt EV. Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2 alpha in response to growth induction by c-myc. Proc Natl Acad Sci. 1993;90:6175–6178. doi: 10.1073/pnas.90.13.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai S, Shimono A, Hanaoka K, Kondoh H. Embryonic lethality resulting from disruption of both N-myc alleles in mouse zygotes. New Biol. 1991;3:861–869. [PubMed] [Google Scholar]
- Sawai S, Shimono A, Wakamatsu Y, Palmes C, Hanaoka K, Kondoh H. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development. 1993;117:1445–1455. doi: 10.1242/dev.117.4.1445. [DOI] [PubMed] [Google Scholar]
- Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, Eick D, Kohlhuber F. Control of cell growth by c-Myc in the absence of cell division. Curr Biol. 1999;9:1255–1258. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F, Brodeur G, Goldstein M, Trent J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305:245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- Sheldon M, Rice DS, D'Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell BW, Cooper JA, Goldowitz D, Curran T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes & Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Moroy T, Bartek J, Massague J, Hanel FM, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes & Dev. 1992;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- Zindy F, Soares H, Herzog KH, Morgan J, Sherr CJ, Roussel MF. Expression of INK4 inhibitors of cyclin D-dependent kinases during mouse brain development. Cell Growth Differ. 1997;8:1139–1150. [PubMed] [Google Scholar]