Abstract
The RecA family of proteins mediates homologous recombination, an evolutionarily conserved pathway that maintains genomic stability by protecting against DNA double strand breaks. RecA proteins are thought to facilitate DNA strand exchange reactions as closed-rings or as right-handed helical filaments. Here, we report the crystal structure of a left-handed Sulfolobus solfataricus RadA helical filament. Each protomer in this left-handed filament is linked to its neighbour via interactions of a β-strand polymerization motif with the neighbouring ATPase domain. Immediately following the polymerization motif, we identified an evolutionarily conserved hinge region (a subunit rotation motif) in which a 360° clockwise axial rotation accompanies stepwise structural transitions from a closed ring to the AMP–PNP right-handed filament, then to an overwound right-handed filament and finally to the left-handed filament. Additional structural and functional analyses of wild-type and mutant proteins confirmed that the subunit rotation motif is crucial for enzymatic functions of RecA family proteins. These observations support the hypothesis that RecA family protein filaments may function as rotary motors.
INTRODUCTION
The RecA family of proteins mediates homologous recombination, a ubiquitous mechanism for repairing DNA double strand breaks. Homologous recombination is involved in the recovery of collapsed DNA replication forks, in generating genetic diversity in meiosis and in establishing mitochondrial homoplasmy. Members of the RecA family include bacterial RecA, archaeal RadA or Rad51, eukaryotic Rad51 and meiosis-specific Dmc1. At double strand break sites, these proteins associate with single-stranded DNA (ssDNA) to form a right-handed helical nucleoprotein filament called the presynaptic complex. The presynaptic complex has both ATPase and DNA strand exchange activities: in the presence of Mg2+ and ATP, the DNA strand exchange activity ensures the formation of heteroduplex DNA (hDNA) between ssDNA and its complementary strand in the double-stranded DNA (dsDNA). The hDNA formation can be conveniently monitored and quantified on an agarose gel using the D-loop formation assay. The molecular mechanism underlying the rather slow DNA-dependent ATPase activity is still not clear. It has been proposed that ATP hydrolysis promotes product release, recycling of recombinase monomers and/or a rotary mechanism that could relieve the DNA topological problem in the strand exchange reaction (1–3).
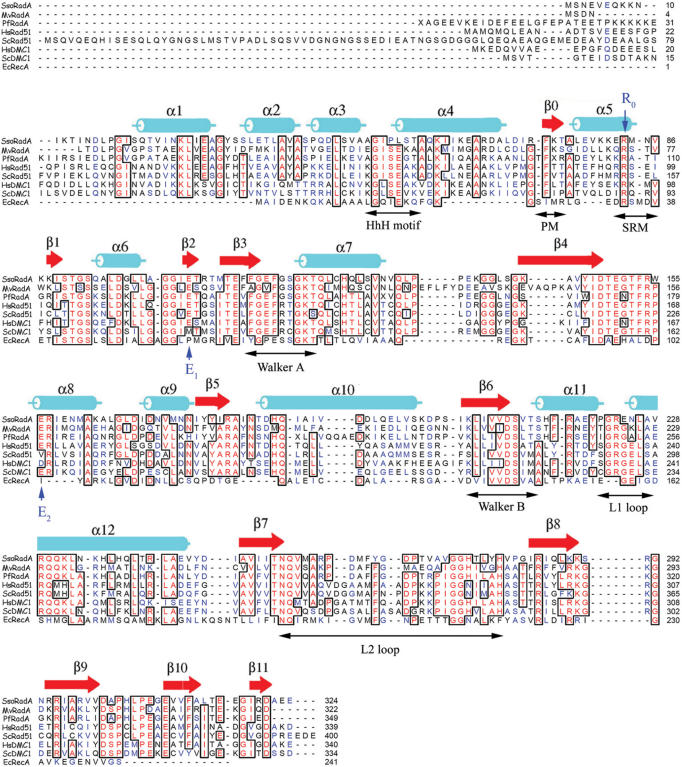
Members of the RecA family share a core ATPase domain containing the putative ssDNA binding motifs, the L1 and L2 loops (4). The ATPase domain is preceded by a short β-strand polymerization motif (5). RadA, Rad51 and Dmc1 have an additional, smaller N-terminal domain (NTD) that may interact with dsDNA (6,7), whereas RecA has a small C-terminal domain with a similar proposed function. The helix-hairpin-helix (HhH) motif in the NTD (Figure 1) mediates dsDNA binding (6,7). The crystal structures of several RecA family members have revealed that the polymerization motif is responsible for assembly of helical filaments and toroidal rings (4,8–13). A hydrophobic residue (phenylalanine in RadA, Rad51 and Dmc1, and isoleucine in RecA) in the polymerization motif docks into a hydrophobic pocket on the neighbouring core ATPase domain. This interaction was also observed in the structure of a fusion construct combining the human Rad51 monomer and a peptide from BRCA2 (5,8).
Figure 1.
Sequence alignment of RecA family proteins from S.solfataricus (Sso RadA), M.voltae (MvRadA), P.furiosus (PfRad51), H.sapiens (HsRad51 and HsDmc1), S.cerevisiae (ScDmc1 and ScRad51) and E.coli (EcRecA). All these RecA-like strand exchange proteins have similar N-terminal domains. The C-terminal RecA domains have been removed for clarity. Secondary structural features of the left-handed SsoRadA helical filament are indicated in cyan (α helices) and red (β strands). Functional motifs are indicated under their corresponding amino acid sequences: the putative dsDNA binding HhH motif, the putative ssDNA binding L1 and L2 loops, the ATP binding Walker A and B motifs, the polymerization motif (PM), the subunit rotation motif (SRM), and others. Positions of the R0–E1–E2 triad are indicated using blue arrows.
For RecA proteins, the transition from closed rings to helical filaments could be flexible or regulated. Archaeal RadA proteins form both closed rings and right-handed helical filaments (11,13–16). Saccharomyces cerevisiae Dmc1 (ScDmc1) and human Dmc1 proteins, in the presence of ssDNA, switch from rings to helical filaments in a Ca2+-dependent manner (17–19). Conceivably, all members of the RecA family form similar right-handed nucleoprotein filaments in which the ssDNA lies close to the filament axis. These nucleoprotein filaments are thought to be catalytically active in mediating strand exchange reactions (14,15,17–20). In this study, we report that both Sulfolobus solfataricus (Sso) RadA and ScDmc1 can exist as left-handed filaments. Using a structure-based approach, we studied the assembly mechanism and functional relevance of the SsoRadA left-handed protein filament.
MATERIALS AND METHODS
Protein expression and purification
The wild-type and mutant SsoRadA cDNAs were cloned into the pET32 expression vector (Novagen, USA), and transformed into BL21(DE3) competent cells. For each protein, an overnight cell culture (100 ml) was grown at 37°C in the presence of 100 mg/l ampicillin. After transferring the cell culture to 6 l LB medium, the cell suspension was allowed to reach an OD600 of about 0.5–0.6 before IPTG (1 mM) was added. Cells were grown for 15 h at 25°C and centrifuged at 12 000 × g for 30 min. The cell pellet was resuspended in 100 ml lysis buffer (50 mM Tris–HCl, pH 8.0, 200 mM NaCl, 5 mM imidazole) and disrupted using a French press (30 000 psi; Sim-Aminco, USA). The total cell lysate was centrifuged at 29 300 × g for 40 min and the soluble protein fraction was loaded onto a 20 ml Ni2+-chelating Sepharose column (Amersham, USA). After the column was washed with 150 ml lysis buffer plus 20 mM imidazole, pH 8.0, a linear gradient (20–300 mM) of imidazole was used to elute thioredoxin-His6-RadA proteins. The 100 ml solution containing the thioredoxin-His6-RadA protein was dialyzed against cleavage buffer (50 mM Tris–HCl, pH 8.0, 100 mM NaCl). Ten units of Factor Xa (Novagen, USA) were added to remove thioredoxin-His6. The resulting RadA protein solution was re-loaded onto a Ni+2-chelating Sepharose column to remove uncleaved proteins. The flow-through was collected and dialyzed against buffer Q (30 mM Tris–HCl pH 7.6, 10% glycerol, 0.2% 2-mercaptoethanol), then loaded onto a Heparin affinity column (Amersham, USA). The RadA proteins were eluted with 0.3 M NaCl, and subsequently run through a desalting column that had been equilibrated with 30 mM Tris–HCl pH 8.0. Purified SsoRadA was lyophilized for long-term storage at −80°C.
Crystallization, data collection and structural determination
Wild-type SsoRadA (24 mg/ml) in 30 mM Tris–HCl (pH 8.0) was crystallized using the hanging drop vapour diffusion method. Hampton Research and Emerald Biostructure crystallization kits were used for initial screening. The protein solution: reservoir ratio was 2 μl : 2 μl. Protein crystals of the left-handed helical filaments were obtained in about 1 week with the reservoir containing 500 μl of 3% PEG4000 and 50 mM sodium acetate (pH 4.0). X-ray diffraction experiments were performed at the Photon Factory in Japan and the National Synchrotron Radiation Research Center in Taiwan (Table 1). Before flash cooling, crystals were briefly soaked in mother liquor containing 20–25% glycerol (by volume) as a cryoprotectant. The diffraction data were processed and scaled using the HKL package (21). The space group (SG) of the left-handed filament crystals was P4x with typical unit cells of a = b = 55.2 Å and c = 125.6 Å. Each asymmetric unit comprised one RadA molecule. The structure of this tetragonal crystal was determined by molecular replacement (MR) using only the C-terminal domain of Methanococcus voltae (Mv) RadA (PDB = 1T4G) as a search model. A good MR solution was obtained in the SG of P43, with an initial R-value of 0.45. Searches in the other SGs did not yield a correct solution; the best R-value in P41 was 0.54. After bulk solvent and overall anisotropic B factor correction, we refined this model by repeated cycles of positional minimization, simulated annealing and restrained B factor refinement with CNS (22), and manual model building with O (23). The refinement was carried out with tight stereochemical restraint and resulted in acceptable geometry. The electron density maps produced at later stages allowed relocation of the N-terminal domain. The Ramachandran plot (non-Gly and non-Pro residues) with PROCHECK for the final model was 81.2% in the most favored regions, 18.8% in the additionally allowed regions and no residues in the disallowed regions. Data collection and refinement statistics are summarized in Table 1. All of the molecular figures were generated using PyMol (http://pymol.sourceforge.net).
Table 1.
Data collection and refinement statistics for the P43 tetragonal RadA crystals
| Data collection | ||
| X-ray source (beamline) | PF (BL5) | NSRRC (17B2) |
| Unit cell a, b, c (Å) | 55.18, 55.18, 125.58 | 55.23, 55.23, 125.71 |
| Resolution (Å) | 50–2.88 (2.98–2.88) | 50–3.01 (3.12–3.01) |
| Number of observations | 35438 (3593) | 35379 (3560) |
| Unique reflections | 8521 (841) | 7575 (759) |
| Completeness (%) | 99.8 (100.0) | 99.9 (100.0) |
| Average I/σ(I) | 27.8 (2.1) | 24.0 (2.2) |
| Rmerge (%) | 5.3 (72.3) | 6.4 (61.8) |
| Refinement | ||
| Resolution (Å) | 30–2.9 (3.0–2.9) | |
| Number of reflections | 7951 (668) | |
| Rwork (95% data) | 0.217 (0.326) | |
| Rfree (5% data) | 0.288 (0.467) | |
| R.m.s.d. bond distance (Å) | 0.011 | |
| R.m.s.d. bond angle (deg) | 1.8 | |
| Ramachandran plot (% non-Gly & non-Pro residues) | ||
| In most favored regions | 81.2 | |
| In additional allowed regions | 18.8 | |
| Average B (Å2)/No. of non-H atoms | ||
| Protein | 87.3/2289 | |
| Water | 70.8/234 |
Two data sets from PF and NSRRC were merged with an Rmerge of 7.5%. All positive reflections are used in the refinement. Numbers in parentheses are for the highest resolution shell.
The CCP4 program (24) was also used to compare the interface areas of different archaeal RadA and Rad51 crystal structures. In the SsoRadA left-handed filament, the total reduction of solvent accessible area by polymerization was estimated to be 2555 Å2, which corresponds to 17% of the total surface of the monomer. In the Pyrococcus furiosus (Pf) Rad51 ring crystal structure (8) and the MvRadA–AMP-PNP right-handed filament (25), the total buried areas were 3095 Å2 (22%) and 2769 Å2 (19%), respectively. The interface between the adjacent ATPase domains in the left-handed filament was 1584 Å2, smaller than those of the PfRad51 ring structure (2149 Å2) and the MvRadA–AMP–PNP-right-handed filament (1948 Å2).
Protein Data Bank accession code
Atomic coordinates and structure factors have been deposited in the PDB under accession code 2DFL.
DNA binding, ssDNA-stimulated ATPase activity and the strand assimilation assay
The DNA binding assay was carried out according to a previously described protocol (26), except that SsoRadA was used. The ssDNA-stimulated ATPase activity assay was performed as described previously (16). For strand assimilation or D-loop formation assays, different RadA proteins (2 μM) were preincubated for 5 min at 65°C with 3 μM (in nucleotides) 32P-labeled P1655 in the presence of 1 mM magnesium acetate and 2 mM ATP. Assimilation reactions were initiated by the addition of an equal volume of a solution containing supercoiled plasmid (GW1; 20 μM in base pairs). The reactions were incubated at 65°C for 0, 5, 10, 20 or 40 min, then stopped by incubating at 37°C for 5 min with SDS (0.25%) and proteinase K (0.5 mg/ml) in order to remove the proteins. DNA from the reaction mixtures was resolved by electrophoresis for 4 h at 4 V/cm on a 0.8% agarose gel in Tris–acetate–EDTA buffer (40 mM Tris, 1 mM Na2–EDTA, and 20 mM acetic acid, pH 8.0). A phosphorimage of an agarose gel shows D-loop reactions carried out in the presence of wild-type and mutant SsoRadA proteins at different time points (16).
Yeast strain, plasmids and MMS sensitivity assay
The SK1 rad51 null mutant strain used in this study was previously described (26). The expression vectors for the wild-type and SRM point mutant ScRad51 proteins are available upon request. The rad51 cells (WHY2262) bearing control vector pYC2, or wild-type or various mutant pYC2-RAD51 plasmids, were cultured in uracil-deficient selective medium to a density of 1 × 107 cells/ml. Cells were diluted 10-fold; 2 µl of cells was taken from each dilution and spread on uracil-deficient selective medium plates containing either 0% or 0.01% (v/v) MMS, cultured at 30°C for 3 days and observed for cell growth. Plates were photographed using an Alpha ImagerTM 2200 (Alpha Imager, USA).
Electron microscopy
The wild-type SsoRadA proteins (1 μM) were first incubated at 65°C for 5 min in D-loop formation reaction buffer [1 mM AMP–PNP, 2 mM MgCl2, 0.5 mM DTT, 10 mM HEPES pH 7.0, 5 μM linear ssDNA (500 nucleotides in length), 5 μM dsDNA (2000 base pairs in length), 50 mg/ml bovine serum albumin], and then chilled on ice to stop the reaction. The reaction product was diluted 100-fold with EM sample dilution buffer (1 mM AMP–PNP, 2 mM MgCl2, 0.5 mM DTT, 10 mM HEPES pH 7.0). Alternatively, the wild-type or mutant SsoRadA protein (0.83 μM protein, 2 mM AMP-PNP and 2 mM MgCl2) were mixed with an ssDNA substrate (2.49 μM nucleotides) and then imaged by transmission EM using the negative staining technique. A droplet (4 μl) was placed for 1 min at room temperature on a copper grid (300 mesh, Pelco, USA) coated with fresh carbon. The excess buffer was then carefully blotted away from the edge of the grid with Whatman #1 filter paper (Whatman Inc., USA). After staining for 4 min with 2.5% uranyl acetate, excess liquid was removed and samples were dried at room temperature. Bio-transmission electron microscopy (EM) was performed with a Tecnai™ G2 Spirit Bio TWIN (FEI Co., The Netherlands) using 120 kV acceleration voltage. Images were recorded with a slow-scan CCD camera (Gatan MultiScan™ 600, USA) at a resolution of at least 1024 × 1024 pixels.
RESULTS
Visualization of the SsoRadA left-handed helical filaments by atomic force microscopy and EM
Archaeal RadA proteins were previously observed mainly as monomers or ring structures in the absence of DNA or nucleotide cofactor, as octameric rings bound to DNA in the absence of nucleotide cofactor and as helical filaments in the presence of DNA and ATP (or ATP analogue) (14,15,27). Using the atomic force microscopy (AFM) imaging method, we previously showed (16) that SsoRadA proteins could spontaneously polymerize into long, fine helical filaments in a slightly basic buffer (pH 8.0). A closer look at those AFM images revealed that there were both right- and left-handed helices. In addition, both right- and left-handed filaments displayed more than 20 helical periodicities (16). We then carried out cross-section scanning analysis with better AFM calibration, and determined that the average helical pitches of these left- and right-handed filaments were 137 ± 5 Å and 98 ± 3 Å, respectively (data not shown). The right-handed helical filament observed here is similar to that observed in a crystal structure of an overwound filament with three subunits per helical rise (L.-Z. Chen, unpublished results, 13).
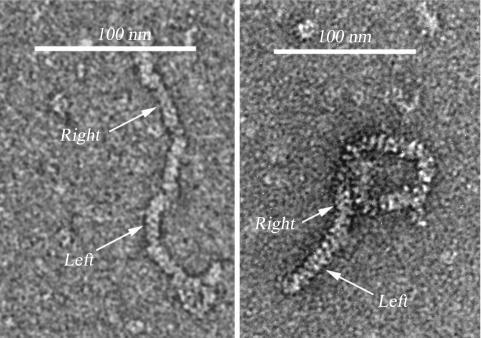
Using an EM imaging approach, we found that SsoRadA protein, after incubation with homologous ssDNA and dsDNA in the presence of Mg2+ and AMP–PNP (pH 7.0), could form a single filament with both right- and left-handed helices (Figure 2). This result confirms our AFM findings that SsoRadA proteins form both right- and left-handed helical filaments. Because the left-handed filaments were more frequently observed in the D-loop formation reaction buffer (pH 7.0; Figure 2), it is intriguing to speculate that they might represent an intermediate during the strand assimilation reaction.
Figure 2.
EM images of the wild type SsoRadA left-handed helical filaments. SsoRadA protein (1 mM) was incubated at 65°C for 5 min in a D-loop formation reaction buffer [1 mM AMP–PNP, 2 mM MgCl2, 0.5 mM DTT, 10 mM HEPES pH 7.0, 5 μM linear ssDNA (500 nucleotides in length), 5 μM dsDNA (2000 base paris in length), 50 mg/ml bovine serum albumin], and chilled on ice to stop the reaction. The reaction product was diluted 100-folds with EM sample dilution buffer (1 mM AMP–PNP, 2 mM MgCl2, 0.5 mM DTT, 10 mM HEPES pH 7.0). The right- and left-handed helices are indicated by arrow, respectively.
To see if the left-handed filamentous structure was unique to SsoRadA or a general feature common to RecA proteins, we imaged another RecA family protein using the AFM-CNT method. We previously found that ScDmc1 proteins assembled into closed-rings in the absence of Ca2+ ion (17) and formed a right-handed helical filament with ssDNA upon addition of Ca2+ ion (18). Here, we found that Ca2+ also promotes ScDmc1 protein self-polymerization into both right- and left-handed helical filaments at a pH near neutrality (Supplementary Figure S1A). Like SsoRadA helical filaments, ScDmc1 helical filaments could be 0.1 μm or greater in length. Cross-section scanning analysis further revealed that the helical pitches (∼165 Å) of both filaments were very similar to each other (Supplementary Figure S1B). Intriguingly, a previous AFM imaging study also indicated that Escherichia coli RecA proteins might form left-handed helices with a circular dsDNA substrate (28). We conclude that the ability to form a left-handed filament may be a general property for most, if not all, RecA family proteins.
Crystal structure of the SsoRadA left-handed helical filament
To further investigate the existence of the left-handed helical filament, we crystallized and determined the protein structures of both a left-handed SsoRadA helical filament and a right-handed SsoRadA filament. The latter, packed in the space group P31 (data not shown), is similar to a P3121 overwound right-handed SsoRadA filament reported previously (13). Although this left-handed SsoRadA filament structure was crystallized in an acidic solution (pH 4.0), low pH is not a prerequisite for formation of left-handed SsoRadA filaments: as described above, left-handed SsoRadA filaments also have been observed at near-neutral pH in our AFM and EM imaging experiments.
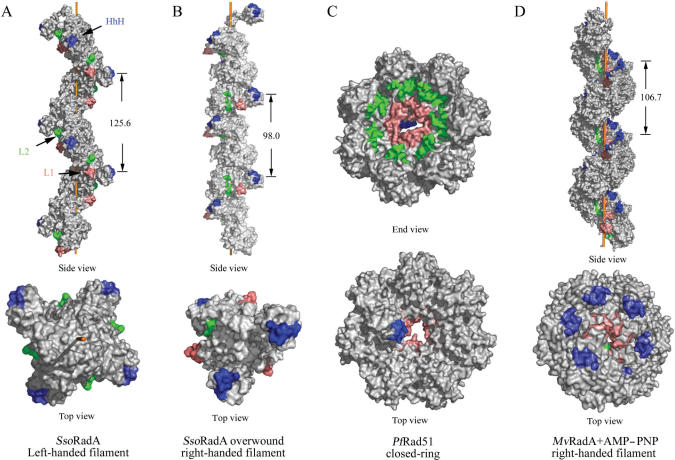
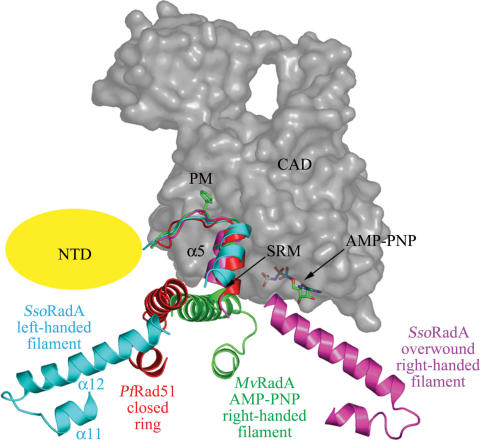
The left-handed filament crystal structure is packed in the space group P43 (Table 1). There are four monomers per helical turn. Its 125.6 Å helical pitch is close to the average pitch of 137 ± 5 Å that was determined by AFM cross-section analysis. The crystal structures show that all three putative DNA binding motifs (L1, L2 and HhH) are on surfaces of the left-handed filament (Figure 3A) and the P31 overwound right-handed filament (Figure 3B). This spatial arrangement is distinct from those in two other crystal structures: for example, the L1 and L2 motifs are clustered in the central channels of PfRad51 rings (Figure 3C) and mammalian Dmc1 rings (8,9); they are located near the filament axes of the right-handed filaments of E.coli RecA and MvRadA–AMP–PNP (Figure 3D) (25,29). Like the polymerization motifs in the right-handed filaments or protein rings (8–10,13,25), the primary polymerization motif in the left-handed SsoRadA filament consists of the same β-strand (amino acid residues 73–75; designated β0 in Figure 1) that binds to the neighbouring core ATPase domain, on which the phenyl group of SsoRadA Phe73 is docked (Supplementary Figure S2). Phe73 is evolutionarily conserved among all archaeal and eukaryotic DNA strand exchange proteins (Figure 1). To avoid model bias, we generated a simulated annealing omit map of residues 70–76 using the CNS program (22). The phenyl group of SsoRadA Phe73 is docked in a hydrophobic pocket formed by residues of β4 (Val145, Ile147), β5 (Tyr177, Ile179) and α10 (Ile190, Asp193, Leu194, Leu197) on the neighbouring core ATPase domain (Supplementary Figure S2). This arrangement gives Phe73 the greatest buried surface area (156 Å2) of any interface residue.
Figure 3.
MvRadA-AMP-PNP, PfRad51 and SsoRadA quaternary structures. Side and top views are shown as indicated. The helical pitches of the right-handed MvRadA-AMPPNP filament, the left-handed SsoRadA filament and the overwound right-handed SsoRadA filament are 106.7 Å, 125.6 Å and 98.0 Å, respectively. The putative dsDNA binding regions or HhH motifs are highlighted in blue. The putative ssDNA binding L1 and L2 motifs are highlighted in pink and green, respectively. The helical filament axis is depicted as an orange rod running through the filament.
Rotation between two neighbouring SsoRadA protomers accounts for different quaternary structures
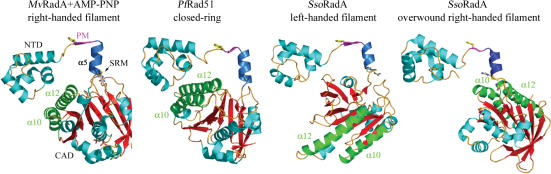
Comparative structure analysis of the SsoRadA left-handed filament, the PfRad51 ring (8), the MvRadA–AMP–PNP right-handed filament (25) and the overwound SsoRadA right-handed filament (13) revealed that their monomers had high similarity in the structural domains (including the NTD, polymerization motif and core ATPase domains) (Figure 4). When the NTDs and polymerization motif of the monomers were superimposed, we identified a hinge region (amino acid residues 83–90), located immediately after the α5 helix (amino acid residues 76–82), which we designated the subunit rotation motif (SRM). This hinge region exhibited sequential conformational changes corresponding to structural transitions from to a closed ring (8), to the AMP–PNP right-handed filament (25), to the overwound right-handed filament (13) and finally to the left-handed filament (Figure 5). These conformational changes appear to be caused mainly by the progressive rotations of their polypeptide backbones, as the torsion angles in this hinge region change stepwise during this clockwise axial rotation (Supplementary Figure S3). Such gross changes in torsion angles were not observed in other regions of SsoRadA (data not shown). These Rad51/RadA structures represent four stepwise intermediates of a 360° rotating protein filament; it is particularly intriguing to speculate that the SRM might be flexible enough to allow Rad51/RadA filaments to carry out 360° axial rotation.
Figure 4.
MvRadA-AMP-PNP, PfRad51 and SsoRadA monomer structures. The N-terminal domain (NTD, in cyan), the conserved phenylalanine residues (in yellow), polymerization motif (PM, in pink) and α5 helix (in blue) are indicated. The subunit rotation motif (SRM) is located at the C-terminal of the α5 helix. Two α helices (α10 and α12) at the core ATPase domain (CAD) are shown in green to illustrate subunit rotation.
Figure 5.
Subunit rotation influences the quaternary structures of archaeal RadA and Rad51 proteins. The structures of the left-handed SsoRadA helical filament (in cyan), the PfRad51 closed ring (in red), the right-handed MvRadA-AMPPNP helical filament (in green) and the overwound right-handed SsoRadA helical filament (in pink) are superimposed by fixing the N-terminal domain (NTD, in yellow) and polymerization motif (PM). The side chain of phenylalanine residue of PM is indicated in green. The Core ATPase domain (CAD) of neighboring protomers is indicated in gray. The subunit rotation motif (SRM) locates immediately after the α5 helix. AMP-PNP in the right-handed MvRadA-AMPPNP helical filament is also marked by arrow, respectively. The disposition of adjacent α11 and α12 helices are shown to illustrate subunit rotation.
Subunit rotation affects the protomer–protomer interface for ATP binding
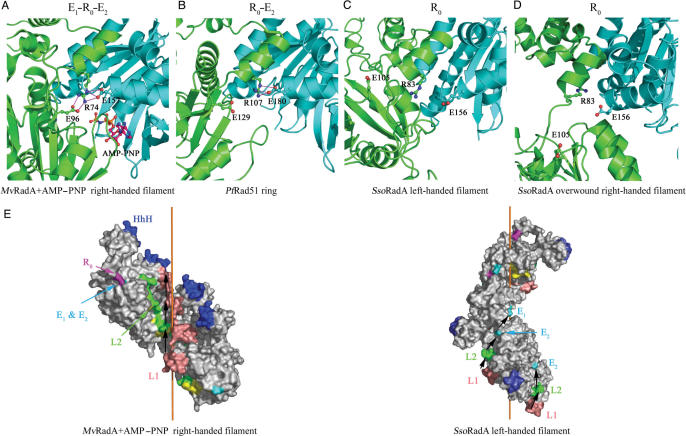
The SRM does not appear to act only as a flexible inter-domain linker. Arg83, a critical amino acid in the SRM of SsoRadA, is evolutionarily conserved not only in archaeal RadA and Rad51, but also in other RecA family members, including E.coli RecA, eukaryotic Rad51 and Dmc1 proteins (Figure 1). The equivalent amino acid residues of MvRadA and PfRad51 are Arg74 and Arg107, respectively. These Arg residues are referred to here as ‘Ro’. In the MvRadA–AMP–PNP right-handed filament (25), the guanidinium group of Ro forms salt bridges with the carboxylic groups of two glutamate residues: Glu96 (denoted ‘E1’) of the same protomer and Glu157 (denoted ‘E2’) of the neighbouring promoter (Figure 6A). Importantly, residues in this E1–R0–E2 triad are also conserved in the archaeal and eukaryotic proteins (Figure 1), except that the E1 glutamate of ScDmc1 is replaced with methionine, and the E2 glutamate of ScRad51 is replaced with valine.
Figure 6.
Subunit rotation influences ATP and DNA binding. (A) Two interacting core ATPase domains are shown as ribbons (in cyan or green) along with the ball-and-stick models of R0, E1, E2 and AMP–PNP. The E1–R0–E2 triad acts as a clip to fasten two AMP–PNP binding surfaces at the neighbouring core ATPase domains. The E1–R0–E2 triad gradually vanishes in the PfRad51 ring (B), in the SsoRadA left-handed filament (C) and in the overwound SsoRadA right-handed filament (D). Oxygen (red) and nitrogen (blue) atoms are shown. Selected hydrogen bonds are shown in pink lines in (A) and (B). (E) Effect of subunit rotation on DNA binding. R0 is shown in deep pink. E1 and E2 are shown in cyan. The HhH (blue), L1 (pink) and L2 (green) motifs are indicated. The putative ssDNA binding paths, connecting all L1 and L2 motifs along the helical filaments, are indicated by black arrows. In the SsoRadA left-handed filament, all dsDNA binding HhH motifs are located at the outermost surfaces.
Looking more closely at the crystal structure in this region, we find that the E1–R0–E2 triad functions as a clip to fasten the AMP–PNP binding between two protomers (Figure 6A). Intriguingly, sequential removal of the E1–R0 and R0–E2 interactions correlates very well with counter clockwise subunit rotation. In the PfRad51 closed-ring (8), R0 interacts only with E2 but not with E1 (Figure 6B). Both interactions disappear in the SsoRadA left-handed filament. Meanwhile, the ATP-binding interfaces between the two neighbouring promoters also separate in a stepwise manner filament (Figure 6C). E1–R0 and R0–E2 interactions also do not form in the P31 overwound SsoRadA right-handed filament; however, the ATP-binding interface between the two neighbouring promoters in this filament become closer compared to those in the left-handed filament (Figure 6C and D). These results indicate that the subunit rotation mediated by the SRM most likely plays an important role in controlling ATP binding and/or hydrolysis. Alternatively, inclusion of ATP or non-hydrolysable ATP analogues (AMP–PNP or ATPγS) into a close-ring or a left-handed filament may induce subunit rotation resulting in the ‘active’ right-handed filament.
Subunit rotation and DNA binding
Additional structural analysis reveals that the E1–R0–E2 triad may control DNA binding as well as DNA release. E1 and E2 are located on the protein surface in the SsoRadA left-handed filament. It is likely that their acidic carboxylic groups would interfere with ssDNA binding by electrostatically repelling the phosphate groups in DNA, because E1 and E2 are both localized in the right-handed spiraling path (L1–L2–E2–E1–L1) that connects all L1 and L2 motifs in the left-handed SsoRadA filament (Figure 6E, right panel). On the other hand, in the MvRadA–AMP–PNP right-handed filament (Figure 6A), these two acidic carboxyl groups not only are electrically neutralized but also buried in the protein interior by the guanidinium group of R0. Moreover, E1 and E2 are also excluded from the filament axis, in which the L1, L2 and HhH motifs are located (Figure 6E, left panel). Therefore, establishing the E1–R0–E2 triad during this left-to-right structural transition can promote ssDNA or dsDNA binding. In contrast, a reverse transition from right to left may repel ssDNA from SsoRadA. Because all HhH motifs are exposed on the outermost surfaces of the left-handed filament, this filament may still have the capability to bind dsDNA. We speculate that the left-handed helical filament represents a conformation that forms during or at the end of the DNA strand exchange reaction. In this scenario, the right-to-left subunit rotation might promote the formation of hDNA by ssDNA exclusion. This speculation is consistent with our finding in the EM imaging experiments that left-handed filaments were more frequently observed if SsoRadA proteins were incubated with D-loop formation reaction buffer (Figure 2).
Point mutant analysis indicates that the SRM is crucial for SsoRadA function in vitro
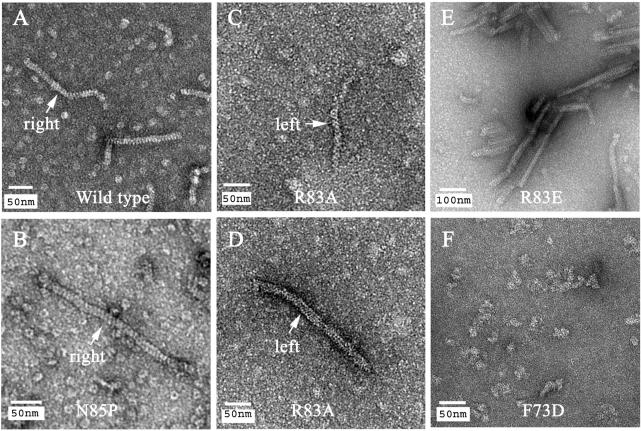
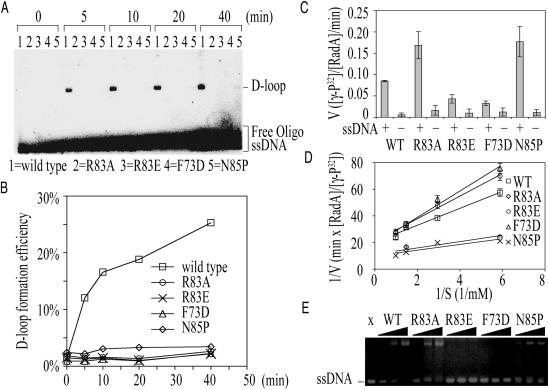
To determine whether the SRM could affect SsoRadA's enzymatic activity in vitro, we constructed three SRM point mutants (R83E, R83A and N85P) in which Arg83 (R0) of SsoRadA was replaced by Glu or Ala, and Asn85 by Pro. Because the dihedral angle (ϕ) of proline is restricted to approximately −60° by the five-membered ring, the N85P mutation might affect normal rotation or movement of the SRM. Time course analysis of D-loop formation was carried out first to determine if these SRM point mutants could catalyze the homologous strand assimilation reaction. As described previously (16), wild-type SsoRadA catalyzed homology-dependent D-loop formation between a 32P-labeled oligonucleotide P1655 (50-mer) and a supercoiled plasmid, GW1. The F73D mutant was used here as a negative control: EM imaging analysis found that this mutant failed to form filamentous structures in the presence of ssDNA (Figure 7F), and filamentous assembly of SsoRadA with ssDNA is a prerequisite for strand assimilation activity. We found that none of the three SRM point mutants (R83A, R83E and N85P) was capable of promoting the D-loop formation (Figure 8A and B). However, EM imaging analysis (Figure 7) revealed point mutations in Arg83 or Asn85 did not affect SsoRadA's capability to form filamentous structures, indicating that their D-loop formation phenotypes are unlikely to be due to a defect in polymerization. The wild-type protein (Figure 7A) and N85P mutant (Figure 7B) often form helical filaments with right-handed helices in the presence of ssDNA. In contrast, left-handed helices were observed in the R83A mutant protein filaments (Figure 7C and D). The R83E proteins form filamentous structures with variable lengths (Figure 7E). The polymerization-defective F73D mutants form protein aggregates with irregular shapes (Figure 7F).
Figure 7.
EM images of the wild-type and mutant SsoRadA proteins. Negative staining EM images of SsoRadA wild-type, R83A, N85P, R83E and F73D proteins in the presence of AMP-PNP, Mg2+ and a ssDNA substrate (872 nucleotides in length). The results indicate that wild-type, R83A, N85P and R83E proteins are capable of forming filamentous structures. The wild-type (left-top panel) and N85P (left-middle panel) protein filaments often exhibited right-handed helices. In contrast, left-handed helices were often observed in the R83A protein filaments (right-top and right-middle panels). R83E proteins form filamentous structures with different lengths (left-bottom panel). The F73D point mutant proteins, defective in protein polymerization, form only protein aggregates with irregular morphology (right-bottom panel). Right- and left-handed helices are marked using white arrows.
Figure 8.
In vitro enzymatic activities of wild-type SsoRadA and SRM point mutants. (A) Time course analysis of D-loop formation was carried out as described in ‘Materials and Methods’ (16). (B) Quantitation of the D-loop time course experiments shown in (A). The amount of joined molecule or D-loop was quantified as the ratio of counts co-migrating with GW1 plasmid DNA to the total counts in each lane. A small faction (∼1/200) of total reaction mixture was used to determine the total counts. The efficiency of D-loop formation was calculated according to the molar ratio of joint molecules over total GW1 plasmid DNA. (C) The ATPase activities of SsoRadA proteins in the presence (+) or absence (−) of ssDNA substrate. The wild-type or mutant SsoRadA protein (2.4 µM) was first incubated with or without ΦX174 ssDNA (1 mM in nucleotides) in the presence of 1 mM Mg2+. ATP hydrolysis was initiated by addition of 1 mM [γ-32P]ATP at 65°C. At different time points, 0.3 µl aliquots were withdrawn and spotted on thin layer chromatography paper as described previously (16). As expected, all five proteins examined here exhibited relatively low ATPase activities in the absence of ssDNA. (D) Kinetic analysis of the ssDNA-stimulated ATPase activities of wild-type or mutant SsoRadA proteins. The concentrations of wild-type, N85P, R83A, R83E and F73D proteins, denoted as [RadA], were 3.0, 2.0, 2.5, 5.0 and 5.0 μM, respectively. ATP hydrolysis was determined by addition of different amounts of [γ-32P]-ATP (0.17, 0.34, 0.68 and 1.02 mM) at 65°C. Kinetic parameters (Km and Vmax) were determined by curve fitting using the Michaelis–Menten equation with a nonlinear least squares algorithm (Microcal Origin software). (E) EMSA analysis of wild-type or mutant SsoRadA proteins. A ssDNA substrate (50-mer; 1 μM) was incubated with increasing amount of protein (2.5, 5, and 10 μM). The resulting products were separated on a agarose gel and visualized by staining with ethidium bromide.
To further investigate these three SRM point mutant proteins, we examined their ssDNA binding activities and ssDNA-stimulated ATPase activities. ATPase activity was determined by monitoring the release of 32P from [γ-32P]ATP. The ssDNA binding activity was assayed using an electrophoresis mobility shift assay (EMSA). The wild-type and F73D point mutant proteins were again used as positive and negative controls, respectively. First of all, we found that all five proteins exhibited relatively low ATPase activity in the absence of ssDNA (Figure 8C). The Km of the wild-type SsoRadA for ATP in the ssDNA-stimulated ATPase activity was determined to be 0.30 ± 0.01 mM, and kcat was 0.051 ± 0.001 min−1. kcat was determined by the ratio of Vmax to E, where E is the concentration of SsoRadA protein used in the ssDNA-stimulated ATPase assay. For the F73D mutant, Km was 0.49 ± 0.03 mM and kcat was 0.051 ± 0.001 min−1 (Figure 8D). The R83E mutant exhibited almost identical ATPase activity as that of the F73D mutant. In the ssDNA-stimulated ATPase assay, the R83E mutant had a Km of 0.46 ± 0.06 mM and a kcat of 0.052 ± 0.004 min−1 (Figure 8D). These results are consistent with our hypothesis that the SRM or the E1–R0–E2 triad play an important role in the use of ATP. The R83A and N85P mutants, on the other hand, were more efficient in hydolysing ATP (Figure 8D). Their Km's for ATP were 0.21 ± 0.01 and 0.20 ± 01 mM, respectively. They also exhibited overactive ATPase activities: for R83A, the kcat was 0.091 ± 0.001 min−1, and for N85P, the kcat was 0.104 ± 0.007 min−1 (Figure 8D). Finally, the results of the EMSA analysis indicated that R83E, like F73D, is defective in ssDNA binding. On the contrary, R83A, N85P and the wild-type protein exhibited similar ssDNA binding affinity (Figure 8E). Because all the three SRM mutants also failed to promote D-loop formation (Figure 8A), these results taken together support the idea that the SRM plays important roles in coupling SsoRadA's ssDNA binding and ATPase activities to its strand assimilation activity.
The SRM is also essential for ScRad51 function in vivo
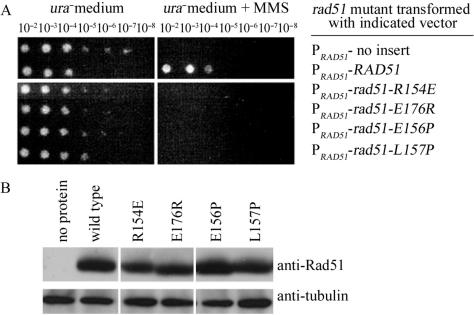
Our structural analyses indicate that the SRM and the E1–R0–E2 triad are evolutionarily conserved among archaeal and eukaryotic RecA family proteins, except the E1 amino acid residue in ScDmc1 protein is replaced by a methionine and the E2 amino acid residue in ScRad51 is replaced by valine (Figure 1). The E1 and R0 residues in the ScRad51 protein are Glu176 and Arg154, respectively. Glu156 and L157 are two amino acid residues located in the SRM of ScRad51. To substantiate the biological relevance of SRM, we constructed a series of yeast CEN/ARS vectors that expressed wild-type yeast Rad51 or its SRM point mutant proteins (R154E, E176R, E156P, L157P). Induction of these proteins was under the control of the RAD51 gene promoter (PRAD51). Each expression vector, and a control vector without the RAD51 insert, was transformed into a rad51 mutant strain. The resulting transformants were tested for sensitivity to a DNA damaging reagent, methyl methanesulfonate (MMS; Figure 9A). As expected, the rad51 cells transformed with the control vector were sensitive to MMS. When the rad51 mutant cells were transformed with the wild-type RAD51 gene, these cells became resistant to MMS. We found that none of the SRM point mutant proteins (R154E, E176R, E156P, L157P) were able to compliment the loss of wild-type ScRad51 protein in the rad51 null mutant (Figure 9A). Western blot analysis confirmed that all Rad51 proteins were expressed in the rad51 cells (Figure 9B). These results indicate that the SRM is indispensable for ScRad51 function in vivo, and support the idea that the SRM is important for RecA family protein function.
Figure 9.
Subunit rotation motif is essential for ScRad51 function in vivo. The yeast rad51 null mutant was transformed either with an empty control vector pYC2, with the pYC2-Rad51 expression vector for the wild-type Rad51, or with point mutant Rad51 proteins as indicated. Induction of these proteins was under the control of the RAD51 gene promoter. (A) MMS sensitivity assay was carried out as described in the ‘Material and Methods’. (B) Western blot analysis of the wild-type and point mutant Rad51 proteins. Total cell lysate was prepared, proteins were separated by SDS-PAGE and analyzed by Western blotting. Anti-Rad51 antibody (Santa Cruz Biotechnology, USA) and anti-tubulin (Invitrogen, USA) was used for detection of Rad51 and tubulin proteins. Tubulin was used here as a protein loading control. Final detection was performed using the ECL detection system. The emitted chemiluminescent light was recorded on X-ray films.
DISCUSSION
In summary, we report that members of the RecA protein family can self-polymerize into left-handed helical filaments in vitro. Subsequent structural and functional analysis led to the identification of a new functional motif, the SRM, in this protein family. We favor the notion that axial rotation of the SRMs may control not only the quaternary structures of RecA family proteins (at least in protein crystals), but also regulate their enzymatic functions. We speculate that the left-handed helical filament, if it forms in a physiological context, may stabilize hDNA at the end of the DNA strand assimilation reaction. Since the left-handed helical filament tends to exclude ssDNA, it may also represent a conformation for uncoupling RecA family proteins from ssDNA when the latter fails to find a homologous dsDNA partner to complete the DNA strand assimilation reaction. In both scenarios, i.e. hDNA stabilization and ssDNA exclusion, the left-handed helical filament may represent an intermediate during or after the strand assimilation reaction. This supposition is consistent with our findings that left-handed filaments were more often observed when the wild-type SsoRadA protein was incubated with the reaction buffer for D-loop formation (Figure 2).
However, we have yet to obtain indisputable evidence confirming that the left-handed helical filament is involved in the strand exchange reaction. It may be critical to identify a mutant defective in left-handed filament formation. It would be interesting to determine if the R83A proteins only form left-handed filaments and if the N85P proteins only form right-handed filaments. Alternatively, the SRM may be flexible enough to promote the strand exchange reaction by adapting alternative conformations or structural intermediates.
The biochemistry data in this report suggests that SRM is important for coupling SsoRadA's ssDNA binding and ATPase activities to its strand exchange activity. Similar to the rotary motor F1-ATPase, the use of ATP in SsoRadA might involve three sequential states: an ATP-bound state (TP), an intermediate (DP), and a state in which the nucleotide binding site is empty (E) (30). The R83A and N85P point mutants may carry out the TP to DP transition, but are incapable of transmitting these allosteric effects to strand assimilation or exchange. On the other hand, the R83E point mutant may be defective in the transition from E to TP. Accordingly, the SRM may control the TP-DP-E transition. This model is consistent with a previous report that the use of ATP in E.coli RecA occurs sequentially in three steps (31). In addition, a recent study indicated that the transition from TP to DP by MvRadA involved extensive allosteric effects spanning the ATPase site and the L2 putative DNA-binding loop (12).
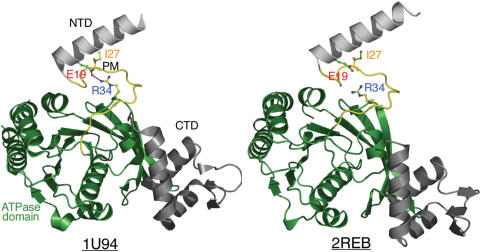
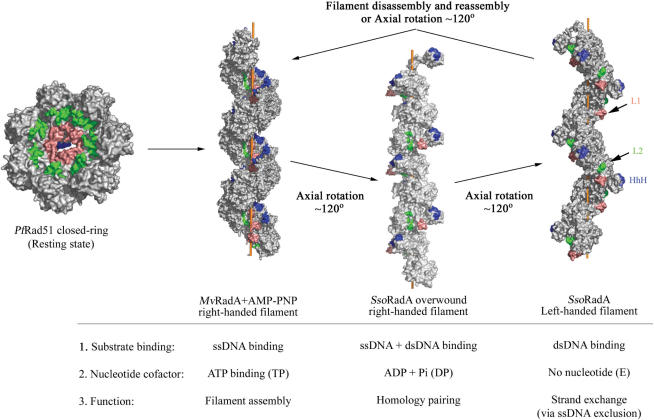
Another important finding in this report is that the R0 of the SRM is evolutionarily conserved in almost all RecA family proteins, including archaeal RadA and Rad51, eukaryotic Rad51 and Dmc1 and E.coli RecA (Figure 1). Arg34, the R0 of E.coli RecA, is also located immediately after a single β-strand which functions as the polymerization motif (Ile27) for RecA filament assembly (10). The polymerization motif of the E.coli RecA protein resides between its small NTD (amino acid residues 1–22) and the core ATPase domain (amino acid residues 36–353) (29). A recent structural study reported that changes in the dihedral angles of two amino acid (Lys23 and Gly24) residing in the polymerization motif were responsible for rotation of the RecA filament between compressed and stretched conformations (32). We noticed that Arg34 formed a salt bridge with Glu19 in a compressed RecA helical filament (74 Å helical pitch; 33), and this salt bridge fell apart in a relaxed RecA helical filament (83 Å helical pitch; 29) (Figure 10). Accordingly, this Arg34–Glu19 dyad may be functionally similar to the E1–R0–E2 triads of the archaeal and eukaryotic proteins. It would be interesting to determine whether Arg34 or Glu19 is essential for RecA enzymatic activity, or for RecA to form left-handed nucleoprotein filaments (28). Conservation of R0 in all RecA family proteins also indicates that the SRM does not simply function as a flexible inter-domain linker. This is consistent with our findings that the R83E mutant is defective in ssDNA-stimulated ATPase activity and that the R83A and R83E mutants are defective in the strand exchange reaction. Therefore, R0 may be directly involved in coupling ATP hydrolysis to the strand exchange reaction. In this scenario, R0 is functionally similar to Arg242 in the E.coli F1F0 ATPase γ subunit. Arg242 forms a salt-bridge with Glu381 of the catalytic βDP subunit, and this β–γ interaction plays an important role in turnover and coupling (34). Intriguingly, a rotary motor model had been proposed for RecA family proteins to explain the coupling of ATP hydrolysis to DNA strand exchange activity (3). Since the comparative structural analysis in this report (Figure 5) led to the speculation that the SRM may be flexible enough to allow the archaeal RadA and Rad51 protein filaments to rotate 360°, we propose the following rotary motor model for RadA or Rad51 proteins (Figure 11). First, RadA/Rad51 proteins exist as protein rings in the resting state (left panel). After association with ATP or AMP–PNP, the RadA proteins form a P61 right-handed helical filament, which represents the TP intermediate state. Second, ssDNA binding induces ATP hydrolysis (by disrupting the E1–Ro–E2 triad and slightly widening the ATP binding pocket) in conjunction with an ∼120° axial rotation into the P31 overwound right-handed filament. This overwound right-handed filament may represent or resemble the DP intermediate state. Intriguingly, the P61 to P31 structural transition is accompanied by concerted movements of the L1, L2 and HhH motifs from filament centre to filament surface. Together, these three DNA binding motifs constitute a 25 Å open pocket (L.-T. Chen et al., unpublished results) that is wide enough to capture dsDNA and mediate homologous pairing between ssDNA and dsDNA. Third, release of ADP and inorganic phosphate (Pi) induces an additional ∼120° axial rotation and results into the P43 left-handed helical filament or the E intermediate state, because the ATP binding pocket of the left-handed filament is wide open. The strand exchange reaction between ssDNA and dsDNA likely occurs during this structural transition, since the HhH dsDNA binding motifs not only move to the most exterior surfaces, but also depart from the L1 and L2 ssDNA binding motifs. Further, the two negatively charged residues of the E1–Ro–E2 triad are exposed to the filament surface and conceivably function to exclude ssDNA (Figure 6E). Finally, the left-handed filaments may dissociate from the heteroduplex DNA either via disassembly into RadA monomers or by carrying out a third ∼120° axial rotation to form P61 right-handed filaments.
Figure 10.
Subunit rotation accounts for the structural transition of RecA right-handed filament from a compressed conformation (left panel) to a relaxed conformation (right panel). The N-terminal domain (NTD, in gray), the polymerization motif (PM, in yellow), ATPase domain (in green) and C-terminal domain (in gray) are shown as indicated, respectively. Ball-and-stick models of Ile27, Arg34 (R0), Glu19 are shown along with oxygen (red) and nitrogen (blue) atoms. The side chain of Ile27 (in yellow) is responsible for filament assembly via interacting with a hydrophobic site in the ATPase domain of neighboring promoter. Arg34 forms a salt bridge with Glu19 in a compressed RecA helical filament (74 Å helical pitch, 33), and this salt bridge falls apart in a relaxed RecA helical filament (83 Å helical pitch, 29). The protein databank accession numbers of these two filaments are 1U94 and 2REB, respectively.
Figure 11.
The rotary motor hypothesis for RadA or Rad51 protein filaments.
Subunit rotation may also occur in other helical protein filaments. For example, it has been shown that subunit rotation controls the right-handed to left-handed transition of the Salmonella typhimurium flagellar filament (35). This filament, together with a hook, forms the flagellum, the rotary motor for bacterial motility. Point mutations of flagellin, the building block of the flagellar filament, lock flagellar filaments in either right-handed or left-handed conformations (36), and analysis of the crystal structure of the Salmonella flagellar protofilament revealed a β-hairpin to be the SRM (37). This scenario very closely resembles our identification of the SRM in RecA family proteins. Taking all these data together, we propose that subunit rotation may be a general molecular mechanism for switching and assembling helical protein filaments.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by the Academia Sinica (Investigator Award Grant to TFW and AS93IBC3 to AHJW) and by the National Science Council (NSC94-2311-B-001-005 to TFW). This work was also supported in part by a National Core Facility of High-Throughput Protein Crystallography Grant (NSC 93-3112-B-001-011-Y to AHJW). The authors thank Sue-Ping Lee (Institute of Molecular Biology, Academia Sinica) for EM analysis. TFW thanks Chung Wang (Academia Sinica), Michael Litchen (NIH, USA) and Guido Guidotti (Harvard University, USA) for helpful discussions. Funding to pay the Open Access charge was provided by Academia Sinica.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kowalczykowski SC, Eggleston AK. Homologous pairing and DNA strand-exchange proteins. Annu. Rev. Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 2.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 3.Cox MM. The bacterial RecA protein as a motor protein. Annu. Rev. Microbiol. 2003;57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- 4.Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 5.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 6.Aihara H, Ito Y, Kurumizaka H, Yokoyama S, Shibata T. The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR. J. Mol. Biol. 1999;290:495–504. doi: 10.1006/jmbi.1999.2904. [DOI] [PubMed] [Google Scholar]
- 7.Kinebuchi T, Kagawa W, Kurumizaka H, Yokoyama S. Role of the N-terminal domain of the human DMC1 protein in octamer formation and DNA binding. J. Biol. Chem. 2005;280:28382–28387. doi: 10.1074/jbc.M503372200. [DOI] [PubMed] [Google Scholar]
- 8.Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. Embo. J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinebuchi T, Kagawa W, Enomoto R, Tanaka K, Miyagawa K, Shibata T, Kurumizaka H, Yokoyama S. Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein Dmc1. Mol. Cell. 2004;14:363–374. doi: 10.1016/s1097-2765(04)00218-7. [DOI] [PubMed] [Google Scholar]
- 10.Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, He Y, Moya IA, Qian X, Luo Y. Crystal structure of archaeal recombinase RADA: a snapshot of its extended conformation. Mol. Cell. 2004;15:423–435. doi: 10.1016/j.molcel.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Qian X, Wu Y, He Y, Luo Y. Crystal structure of Methanococcus voltae RadA in complex with ADP: hydrolysis-induced conformational change. Biochemistry. 2005;44:13753–13761. doi: 10.1021/bi051222i. [DOI] [PubMed] [Google Scholar]
- 13.Ariza A, Richard DJ, White MF, Bond CS. Conformational flexibility revealed by the crystal structure of a crenarchaeal RadA. Nucleic Acids Res. 2005;33:1465–1473. doi: 10.1093/nar/gki288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang S, Yu X, Seitz EM, Kowalczykowski SC, Egelman EH. Archaeal RadA protein binds DNA as both helical filaments and octameric rings. J. Mol. Biol. 2001;314:1077–1085. doi: 10.1006/jmbi.2000.5213. [DOI] [PubMed] [Google Scholar]
- 15.McIlwraith MJ, Hall DR, Stasiak AZ, Stasiak A, Wigley DB, West SC. RadA protein from Archaeoglobus fulgidus forms rings, nucleoprotein filaments and catalyses homologous recombination. Nucleic Acids Res. 2001;29:4509–4517. doi: 10.1093/nar/29.22.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee MH, Leng CH, Chang YC, Chou CC, Chen YK, Hsu FF, Chang CS, Wang AH, Wang TF. Self-polymerization of archaeal RadA protein into long and fine helical filaments. Biochem. Biophys. Res. Commun. 2004;323:845–851. doi: 10.1016/j.bbrc.2004.08.163. [DOI] [PubMed] [Google Scholar]
- 17.Chang YC, Lo YH, Lee MH, Leng CH, Hu SM, Chang CS, Wang TF. Molecular Visualization of the Yeast Dmc1 Protein Ring and Dmc1-ssDNA Nucleoprotein Complex. Biochemistry. 2005;44:6052–6058. doi: 10.1021/bi048897q. [DOI] [PubMed] [Google Scholar]
- 18.Lee MH, Chang YC, Hong EL, Grubb J, Chang CS, Bishop DK, Wang TF. Calcium ion promotes yeast Dmc1 activity via formation of long and fine helical filaments with single-stranded DNA. J. Biol. Chem. 2005;280:40980–40984. doi: 10.1074/jbc.M505896200. [DOI] [PubMed] [Google Scholar]
- 19.Bugreev DV, Golub EI, Stasiak AZ, Stasiak A, Mazin AV. Activation of Human Meiosis-specific Recombinase Dmc1 by Ca2+ J. Biol. Chem. 2005;280:26886–26895. doi: 10.1074/jbc.M502248200. [DOI] [PubMed] [Google Scholar]
- 20.Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429:433–437. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr., Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project No. 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Wu Y, Qian X, He Y, Moya IA, Luo Y. Crystal structure of an ATPase-active form of Rad51 homolog from Methanococcus voltae. Insights into potassium dependence. J. Biol. Chem. 2005;280:722–728. doi: 10.1074/jbc.M411093200. [DOI] [PubMed] [Google Scholar]
- 26.Chen YK, Leng CH, Olivares H, Lee MH, Chang YC, Kung WM, Ti SC, Lo YH, Wang AH, Chang CS, et al. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc. Natl. Acad. Sci. USA. 2004;101:10572–10577. doi: 10.1073/pnas.0404195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seitz EM, Brockman JP, Sandler SJ, Clark AJ, Kowalczykowski SC. RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange. Genes Dev. 1998;12:1248–1253. doi: 10.1101/gad.12.9.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi WX, Larson RG. Atomic force microscopic study of aggregation of RecA-DNA nucleoprotein filaments into left-handed supercoiled bundles. Nano Lett. 2005;5:2476–2481. doi: 10.1021/nl051783v. [DOI] [PubMed] [Google Scholar]
- 29.Story RM, Weber IT, Steitz TA. The structure of the E.coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 30.Menz RI, Walker JE, Leslie AG. Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell. 2001;106:331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- 31.Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- 32.Petukhov M, Lebedev D, Shalguev V, Islamov A, Kuklin A, Lanzov V, Isaev-Ivanov V. Conformational flexibility of RecA protein filament: transitions between compressed and stretched states. Proteins. 2006;65:296–304. doi: 10.1002/prot.21116. [DOI] [PubMed] [Google Scholar]
- 33.Xing X, Bell CE. Crystal structures of Escherichia coli RecA in a compressed helical filament. J. Mol. Biol. 2004;342:1471–1485. doi: 10.1016/j.jmb.2004.07.091. [DOI] [PubMed] [Google Scholar]
- 34.Al-Shawi MK, Ketchum CJ, Nakamoto RK. Energy coupling, turnover, and stability of the F0F1 ATP synthase are dependent on the energy of interaction between gamma and beta subunits. J. Biol. Chem. 1997;272:2300–2306. doi: 10.1074/jbc.272.4.2300. [DOI] [PubMed] [Google Scholar]
- 35.Trachtenberg S, DeRosier DJ. A molecular switch: subunit rotations involved in the right-handed to left-handed transitions of Salmonella typhimurium flagellar filaments. J. Mol. Biol. 1991;220:67–77. doi: 10.1016/0022-2836(91)90381-f. [DOI] [PubMed] [Google Scholar]
- 36.Hyman HC, Trachtenberg S. Point mutations that lock Salmonella typhimurium flagellar filaments in the straight right-handed and left-handed forms and their relation to filament superhelicity. J. Mol. Biol. 1991;220:79–88. doi: 10.1016/0022-2836(91)90382-g. [DOI] [PubMed] [Google Scholar]
- 37.Samatey FA, Imada K, Nagashima S, Vonderviszt F, Kumasaka T, Yamamoto M, Namba K. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature. 2001;410:331–337. doi: 10.1038/35066504. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.