Abstract
To investigate the feasibility of conducting a genomic-scale protein labeling and localization study in Escherichia coli, a representative subset of 23 coding DNA sequences (CDSs) was selected for chromosomal tagging with one or more fluorescent protein genes (EGFP, EYFP, mRFP1, DsRed2). We used λ-Red recombination to precisely and efficiently position PCR-generated DNA targeting cassettes containing a fluorescent protein gene and an antibiotic resistance marker, at the C-termini of the CDSs of interest, creating in-frame fusions under the control of their native promoters. We incorporated cre/loxP and flpe/frt technology to enable multiple rounds of chromosomal tagging events to be performed sequentially with minimal disruption to the target locus, thus allowing sets of proteins to be co-localized within the cell. The visualization of labeled proteins in live E. coli cells using fluorescence microscopy revealed a striking variety of distributions including: membrane and nucleoid association, polar foci and diffuse cytoplasmic localization. Fifty of the fifty-two independent targeting experiments performed were successful, and 21 of the 23 selected CDSs could be fluorescently visualized. Our results show that E. coli has an organized and dynamic proteome, and demonstrate that this approach is applicable for tagging and (co-) localizing CDSs on a genome-wide scale.
INTRODUCTION
Until relatively recently, it was thought that bacteria lacked much of the complexity and compartmentalization found in eukaryotic cells: essentially being a membranous sack of biomolecules. However, this simplistic view of bacterial cell biology has been dispelled over the past decade or so, largely due to advances in fluorescence labeling and microscopy, and through the global in silico analysis of genome sequences. It is now known that many bacteria have dynamic cytoskeletal structures analogous to those formed by the actin and tubulin proteins in eukaryotes, and exhibit high levels of intracellular organization, asymmetry and coordination of activities (1–6).
As proteins often form dynamic complexes and perform multiple biological or biochemical roles, establishing a protein's interaction partners and localization patterns is essential in order to understand its full range of functions within the cell. To date, global proteome localization and interaction studies have been conducted most comprehensively in the model eukaryote Saccharomyces cerevisae through labeling coding DNA sequences (CDSs) with green fluorescence protein (GFP) and epitope tags, and by two hybrid experiments (7–10). In bacteria, the majority of fluorescent protein tagging and localization studies have been conducted in E. coli, Bacillus subtilis and Caulobacter crescentus. To date, the most comprehensive large-scale protein analyses performed in E. coli have included a tandem affinity tag (TAP)-based protein interaction study (11), as well as several biophysical protein fractionation and localization studies e.g. (12–14). Mori and colleagues have recently constructed and studied the properties of a large collection of E. coli open reading frames (ORFs) expressed from inducible plasmids (15,16) [GenoBase database, http://ecoli.aist-nara.ac.jp/index.html] that have been fused to N-terminal hexa-histidine and C-terminal GFP tags. There have also been many other more focused studies, the majority of which have concentrated on proteins related to cell morphology and cytokinesis, DNA replication and repair or chemotaxis and motility [e.g. see (1–4,6,17,18)]. Various molecular approaches have been used in these investigations, including the use of inducible and native promoters, as well as episomal and chromosome-based copies of the gene of interest fused to the relevant fluorescent protein gene. This has complicated the direct comparison of findings from different studies, as protein-fusions may not have been expressed at levels that are physiologically relevant, or have been functioning in competition with the endogenous un-labeled homolog. With this in mind, we explored the feasibility and utility of conducting a genomic-scale, chromosomal CDS tagging and fluorescent fusion-protein localization study in the model prokaryote E. coli. To this end, we designed and implemented a versatile strategy utilizing the efficient λ-Red homologous recombination system to efficiently tag CDSs at their C-terminus with fluorescent protein genes, leaving the resultant fusions under the control of their native promoters.
MATERIALS AND METHODS
Sequence data, gene annotations and associated information were obtained from the PEC (http://www.shigen.nig.ac.jp/ecoli/pec) (19), Colibri (http://genolist.pasteur.fr/Colibri/) (20), UniprotKB/Swiss-Prot (http://www.ebi.ac.uk/swissprot/) (21) and EchoBase (http://www.ecoli-york.org/) (22) databases.
Bacterial strains and CDS targeting procedures
E. coli DH10B (F− mcrA Δ(mrr–hsdRMS–mcrBC) ϕ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara-leu)7649 galU galK rspL nupG); DY380 (DH10B [λcl857 (cro–bioA)<>tet]); EL250 (DH10B [λcl857 (cro–bioA)<>araC–PBADflpe]; EL350 (DH10B [λcl857 (cro–bioA)<>araC–PBADcre]); DY330 (W3110 ΔlacU169 gal490 [λcl857Δ(cro–bioA)])
Plasmid construction and primers used
See Supplementary Data S1 for primer sequences and details of plasmids used as templates for PCR-amplification of the DNA targeting cassettes. The sources of the fluorescent genes were: EGFP (pEGFP), EYFP (pEYFP-C1), ECFP (pECFP-C1) and DsRed2 (pDsRed2), from BD Biosciences Clontech; mRFP1 (pRSETB-mRFP1, from RY Tsien) (23); mRFPmars (24) (pBsrH-mRFPmars, from A Müller-Taubenberger); RFP (constructed for this study: mRFP1 gene codon optimized for E. coli). Additional plasmids used: pBS246 (Invitrogen), pLysS (Novagen), pFRT2 (25) (from SM Dymecki). All oligonucleotides were purchased from Proligo (Singapore) as the salt-free form. Linear dsDNA targeting cassettes were synthesized by PCR using the appropriate pair of primers (see Table in Supplementary Materials) and plasmid templates were constructed (pEGFP-loxP-CmR-loxP; pEYFP-loxP-CmR-loxP; pECFP-loxP-CmR-loxP; pEGFP-frt-CmR-frt; pDsRed2-frt-CmR-frt; pRSETB-mRFP1-frt-kan-frt; pmRFPmars-frt-kan-frt; pRSETB-RFP-frt-kan-frt).
PCR amplification of linear dsDNA targeting cassettes
Each PCR reaction included: polymerase and appropriate buffer; 300 μM each dNTP; 5 ng plasmid template; 300 nM of each primer. Typical PCR program used: 95°C for 60s; 5 cycles of: 95°C for 20 s, 56°C for 20 s, 72°C for 180 s; 25 cycles of 95°C for 15 s, 52°C for 15 s, 72°C for 120 s; 72°C for 720 s. Expand Hi-Fidelity DNA polymerase (Roche) was used for the amplification of targeting cassettes containing the kanamycin (Kan, neo) gene, as Amplitaq (Applied Biosystems), pfu (Stratagene) and other DNA polymerases gave poor product yields (data not shown). Amplitaq or taq polymerase purified in-house was used for the amplification of targeting cassettes containing the chloramphenicol (Cm, cat) resistance gene. PCR products were excised from 1.0% agarose gels (in ×1 TAE) and purified using a Qiagen Gel Extraction kit (Qiagen Gmbh, as recommended by the manufacturer), eluting with 32 μl of 5 mM Tris-HCl pH 8.5. DpnI digestion prior to gel purification did not lead to any significant increases in targeting efficiency (data not shown), and was not generally performed. A 100 μl PCR reaction typically yielded enough DNA for 5–10 chromosomal targeting experiments (transformations).
λ-Red mediated dsDNA recombination protocol
The procedure used was similar to those described by Lee et al. (26) and Yu et al. (27) with minor modifications. About 400 μl from overnight cultures [2–3 ml L medium (containing antibiotic where applicable) inoculated from single colonies, grown at 32°C for 18 h] was expanded into 45 ml of L medium in a 250 ml Erlenmeyer flask, and incubated at 32°C for 2–2½ h (until OD600 of ca. 0.4–0.6). Flasks were transferred to a shaking water bath at 42°C and incubated for 14–15 min, before cooling to 0°C as rapidly as possible in iced water. After 15–20 min, cells were harvested by centrifugation at 0°C (4000 g, 9 min). Cell pellets were carefully washed three times with sterilized ice-cold water (2 × 50 ml, then 1 × 1.5 ml) then re-suspended in 100–200 μl of ice-cold water. Competent cells (50 μl) were transformed with 50–200 ng of (gel purified) linear dsDNA targeting cassette using a BioRad electroporator (1.8 kV, 25 mF, 200 W). The L medium (1 ml) was added to the transformed cell mixture, which was incubated at 32°C, for 2–2½ h. Cells were collected by centrifugation, ca. 900 μl of supernatant media was discarded, and then the resuspended cells were plated onto LB agar containing the appropriate antibiotic to select for resistant colonies.
PCR screening of recombinant clones
Colony PCR was used to confirm the correct integration of the fluorescent protein gene and adjacent ‘floxed’ or ‘flirted’ antibiotic-resistance module, using two pairs of primers to separately amplify the 3′- and 5′- junctions with the chromosome. Each PCR reaction included: Taq polymerase and buffer; 300 μM each dNTP; 300 nM of each primer, using the program: 95°C for 120 s; 8 cycles of: 95°C for 20 s, 56°C for 20 s, 72°C for 120 s; 28 cycles of 95°C for 15 s, 52°C for 15 s, 72°C for 90 s; 72°C for 720 s.
Cre-or flpe-mediated removal of antibiotic resistance modules
Cre- (in EL350) or Flpe- (in EL250) mediated removal of ‘floxed’ or ‘flirted’ kanamycin or chloramphenicol resistance modules was performed essentially as described by Buchholtz et al. (28) and Lee et al. (26) with slight modifications. Plasmids 705-cre (for ‘floxed’ Kan modules) or 705-flp (for ‘flirted’ Kan modules) were electroporated into the constructed strains, then plated onto LB-agar containing chloramphenicol (35 μg/ml) and incubated at 32°C overnight. About 50 μl from overnight cultures (in L medium, inoculated from single colonies) was expanded into 2–3 ml of L medium containing 0.1% arabinose and incubated at 32°C for 3–4 h. Aliquots were streaked onto LB-agar plates, and incubated overnight at 32°C. Individual colonies were screened by PCR, and re-streaked onto plates containing kanamycin (20 μg/ml) or chloramphenicol (10 μg/ml), to confirm that the antibiotic resistance modules had been excised from the chromosome. Typically >90% of all colonies had the antibiotic resistance modules correctly excised (data not shown).
Fluorescence microscopy
Aliquots of cells cultured in L Broth (USB) were removed during the stationary or mid-log phases and immobilized on glass slides coated with a partially dehydrated aqueous agarose pad immediately prior to analysis. Fluorescent microscopy was performed with an Nikon ECLIPSE 80i microscope using an oil immersion ×100 objective lens, which was equipped with a Spot CCD camera and Spot Advance software. EYFP/EGFP fluorescence: excitation/emission 450–490/520 nm; all red fluorescence: excitation/emission 510–560/590 nm. When applicable, aliquots were treated with DAPI (0.2 μg/ml) or FM4–64 (0.15 μg/ml) just prior to immobilization. Confocal microscopy was performed using a Biorad Radiance 2100 laser scanning system equipped with a 2-line KR-3 laser and Nikon Eclipse TE300 oil immersion microscope (×100 objective). EGFP/EYFP fluorescence: excitation/emission 488/500–530; FM4-64 red fluorescence excitation/emission 569/580–620 nm. Confocal images were captured and processed using Lasersharp 2000 Acquisition software (Biorad). All fluorescence images were minimally processed using Adobe Photoshop v7.0 (Adobe).
RESULTS
CDS selection and λ-Red mediated tagging strategy
For this pilot study, we selected 23 CDSs of diverse, putative or unknown function (adk; dxs; era; ftsZ; map; metK; msrA; nrdB (ftsB); nusA; pyrH (umk); recG; rpoD; trmE (thdF); yaeL (rseP); yaeN (mesJ); yeaA (msrB); yfgK (engA); yfjB (nadK); yfjP; ygeH; ygjD (gcp); yihA (engB); yqiK; see Supplementary Data S1, Tables 1 and 2) for C-terminal tagging with one or more of the commonly used fluorescent proteins: EGFP; EYFP; ECFP; DsRed2 (29); mRFP1 (23) and mRFPmars (24) [reviewed in (30,31)]. This was to establish which fluorescent fusions were the most suitable for genome-wide tagging, and which (if any) significantly altered protein localization, activity or tertiary complex formation in E. coli. The CDSs were chosen essentially at random from lists of those that are highly conserved (32) or ‘persistent’ (33) in prokaryotes, and were distributed throughout the chromosome on both the replicative leading and lagging strands. The CDSs of known function were involved in diverse cellular functions that included: DNA replication; transcription; translation; metabolism; oxidative repair; proteolytic processing; the cell cycle and cell division. We included CDSs that were located within (putative) operons as well as 13 that are classed as being essential in E. coli (http://www.shigen.nig.ac.jp/ecoli/pec) (19), to investigate whether our tagging strategy had severe genetic polar effects, or significantly affected protein function.
We first constructed a set of plasmids (Supplementary Data S1) that contained one of the fluorescent protein genes (EGFP, EYFP, ECFP, DsRed2, mRFP1, mRFPmars or RFP), as well as a ‘floxed’ or ‘flirted’ (34) kanamycin (Kan) or chloramphenicol (Cm) resistance module as a selectable marker. These were used as templates for the PCR amplification of DNA ‘targeting cassettes’, to create C-terminal fluorescent fusion proteins of the CDS of interest. PCR primers each included 45–50 bases homologous to the chromosome immediately upstream (forward primer, sense strand) and downstream (reverse primer, antisense strand) of the desired insertion point (which was just 5′- to the stop codon of the CDS), as well as 17 bases homologous to the plasmid template. Forward primers also encoded a short peptide linker (-LEGSG-) (35) to separate the CDS and fluorescent protein domains.
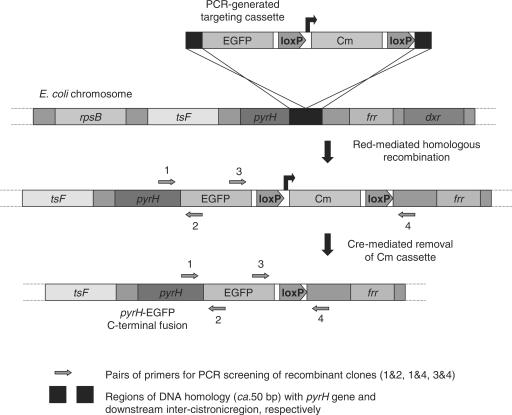
Contemporary to the studies of Butland and co-workers (11) who recently reported the large-scale analysis of protein complexes in E. coli DY330 (27) using a TAP-tag-based approach, we also used strains containing the λ-Red recombination system, namely DY380, EL250 and EL350 (26) for our chromosomal targeting. In these strains (derived from DH10B), exogenous DNA (e.g. targeting cassettes) may be efficiently introduced by electroporation, and integrated into the chromosome via a homologous recombination-based process mediated by the Beta and Exo proteins, which are located in a defective cI857 λ-prophage (27). A schematic illustrating the targeting procedure (as exemplified by the construction of pyrH-EGFP) is shown in Figure 1. In addition to the λ-Red genes, strains EL250 and EL350 also contain arabinose-inducible copies of the flpE and cre site-specific recombinases, respectively, which enables efficient removal of ‘flirted’ or ‘floxed’ Kan or Cm resistance cassettes. This allows additional rounds of CDS-tagging to be performed in the same strain, enabling multiple proteins to be co-localized within the same cell. To confirm that the DNA-targeting cassettes had been accurately inserted (and antibiotic resistance modules removed, when applicable), colonies were screened by PCR, using pairs of primers that flanked the junctions created with the chromosome (see Figure 1, Supplementary Data S1). For each targeting experiment, between 8 and 12 colonies were analyzed. Cells from all the constructed strains were cultured in Luria broth to the log or stationary phase, immobilized on agarose pads on glass slides, and analyzed by fluorescence microscopy.
Figure 1.
Diagram illustrating the strategy used to tag CDSs on the E. coli chromosome with fluorescent protein genes, as exemplified by the construction of pyrH-EGFP.
Efficiency of CDS tagging and observed fluorescence patterns in live cells
All 52 of the independent CDS targeting experiments performed (in DY380, EL250 and EL350) were deemed successful as determined by initial PCR colony screening (i.e. there were >3 individual clones for each constructed strain that produced DNA fragments of the predicted sizes, data not shown). The total number of false positives, i.e. colonies that had antibiotic resistance, but displayed an incorrect pattern upon PCR screening, was low. The majority of these contained a targeting cassette that was correctly inserted at only one end, with the remainder containing an antibiotic cassette that was integrated non-specifically at some other genetic locus. When the recombination-competent cells were transformed with 50–200 ng of gel-purified DNA targeting cassette, 85% of all recombinant clones (n = 430) selected by antibiotic resistance were correctly tagged, as determined by colony PCR (data not shown). When a number of the targeting experiments were repeated with the gel-purification step omitted, the efficiency dropped to 60–70% (n = 120).
Fluorescence patterns for the C-terminal fusion proteins broadly fell into three non-exclusive categories: (a) diffuse and distributed evenly throughout the cytoplasm, (b) concentrated in foci or clusters and (c) associated with the membrane (membrane proximal). Results are summarized in Supplementary Data S1 Table 2, and representative examples shown in Figure 2. An even cytoplasmic distribution was the most commonly observed protein fluorescence pattern. In strains where the fluorescently tagged protein localized in foci, or was associated with the membrane, there was always also a (variable) proportion of the protein distributed throughout the cytoplasm. Significant cell-to-cell variations in fluorescence intensities and protein localization patterns were often encountered. With the exception of the ftsZ strains (discussed below), all the other constructed strains displayed essentially normal growth rates and cell morphologies (see Supplementary Data S1).
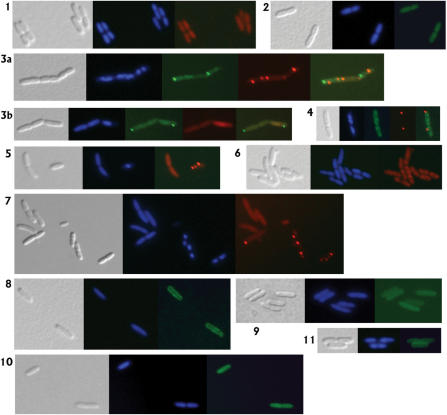
Figure 2.
Representative fluorescence microscopy images of live, immobilized E. coli cells containing C-terminal fluorescent fusions to one or two CDSs on the chromosome. All panels are at the same magnification, and show a phase contrast image of cells (gray); DAPI fluorescent staining of (nucleoid) DNA (blue); mRFP1 fluorescence (red), EGFP or EYFP fluorescence (green). Panel 1: DY380 adk-mRFP1; AdK distributed diffusely throughout cytoplasm. Panel 2: DY380 yeaA-EYFP; YeaA located evenly throughout cytoplasm. Panel 3a and 3b: DY380 metK-mRFP1/pyrH-EGFP; cells containing MetK and PyrH in varying proportions of diffuse cytoplasmic and punctate forms (foci). Panel 4: DY380 yfjP-EYFP/pyrH-mRFP1; PyrH located in one membrane-associated foci per cell, NadK distributed irregularly throughout the cytoplasm, with some clusters formed. Panel 5: DY380 metK-mRFP1; cells containing almost exclusively diffuse cytoplasmic MetK, or foci. Panel 6: DY380 nusA-mRFP1; NusA located predominantly in 2 clusters associated with each nucleoid. Panel 7: DY380 pyrH-RFP; PyrH distributed diffusely throughout cytoplasm, and in 1–4 foci per cell. Panel 8: DY380 yaeL-EYFP; YaeL generally associated with the membrane. Panel 9: DY380 yqiK-EGFP; YqiK distributed diffusely throughout cytoplasm. Panel 10: DY380 yihA-EGFP; YihA distributed diffusely throughout cytoplasm. Panel 11: DY380 ygeH-EGFP; YgeH located predominantly beside the membrane.
PyrH and MetK formed discrete protein foci within the cell
PyrH exhibited the most striking and dynamic protein localization patterns (see Figure 2, panels 4 and 7 (red fluorescence), 3a and 3b (green fluorescence); Figures 3 and 4; Supplementary Figures S2–S4). It localized diffusely and uniformly throughout the cytoplasm, as well as in 1–4 foci (typically 1 or 2) per cell that were often mobile over the timescale of the microscopic observation (tens of seconds to minutes). These foci moved around regions of the cytoplasm in a circular motion, generally spending the majority of their time beside the membrane, especially within the polar regions (Figure 4, panel 1). Sequential confocal microscopic images of the motion of PyrH-EYFP foci are shown in Figure 3, top panel. As well as catalyzing the phosphorylation of uridine monophosphate (UMP), PyrH regulates the transcription of carbamoyl phosphatase (36), and is thought to play a key role in chromosome partitioning and cell division (37). Immunogold labeling has previously indicated that PyrH localizes predominantly near the inner membrane (38), and a protein fractionation/mass spectrometry analysis has suggested that it is mainly cytoplasmic (12). This may reflect the differences between dynamic in vivo analysis and electron microscopy ‘snapshots’ of cells that have been fixed by a complex procedure, possibly altering the intracellular protein organization.
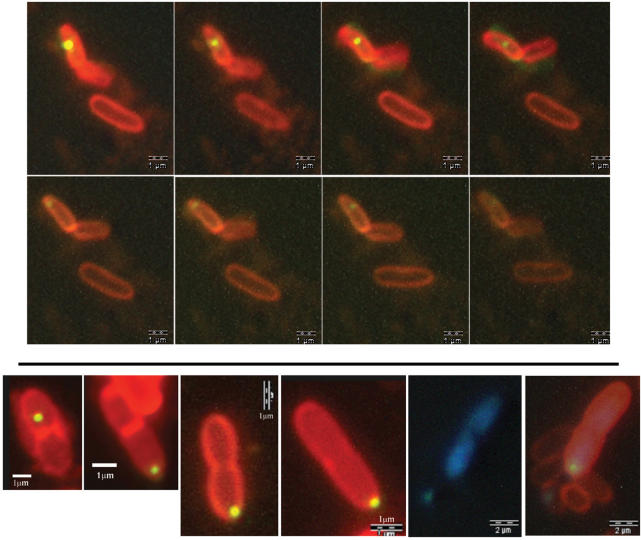
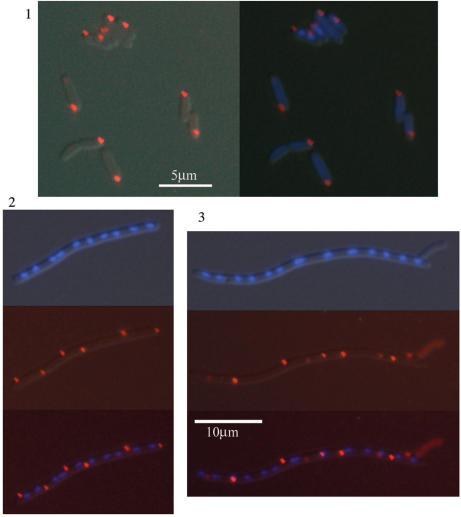
Figure 3.
Confocal fluorescence microscopy images of PyrH-EYFP foci (yellow) in live E. coli DY380 pyrH-EYFP cells stained with the membrane-specific FM4-64 dye (red). Top panel: the 8 frames (left to right, top to bottom) were taken sequentially over ca. 2 min. The PyrH focus moved around the cytoplasm in a generally circular motion. Bottom panel: images 1–4 (left to right) are snapshots showing asymmetrical segregation of PyrH-EYFP foci in different cells undergoing septation and division. Images 5 and 6 show the same cell: image 6 is a composite of the DAPI, EYFP and FM4-64 fluorescence; image 5 shows only DAPI (blue) staining of DNA.
Figure 4.
Fluorescence microscopy images of PyrH fluorescence within live E. coli cells. Panel 1: Preferential localization of PyrH-RFP foci (red) at the old cell pole in DY380 pyrH-RFP. Left image: merge of phase contrast and red fluorescence; right: same image including DAPI staining (blue) of DNA. Panels 2 and 3: Typical images of PyrH-mRFP1 fluorescence (red) and nucleoids (DAPI stained blue) in filamentous DY380 pyrH-mRFP1/ftsZ-Cext cells (both at the same magnification), showing PyrH protein existing predominantly in foci beside the membranes, located at the poles and at positions of potential cell division.
The metK gene is essential in E. coli, with ‘leaky’ mutants exhibiting slow growth and defects in cell division (39). Fluorescent fusions of MetK, which catalyses the synthesis of S-adenosylmethionine (SAM), the main intracellular methylation agent, were localized diffusely throughout the cytoplasm, as well as in 1–4 (typically 1 or 2) apparently static foci (see Figure 2, panels 3a, 3b and 5 (red fluorescence); Supplementary Figures S2, S4). Analogous to the localization of PyrH, we found that cells usually contained a mixture of diffuse and punctate forms of MetK: however, some cells contained almost exclusively one or the other. Both the pyrH and metK genes were tagged with each of the EYFP, EGFP, mRFP1 and DsRed2 fluorescent genes to test whether the different fluorescent fusions exhibited significantly altered protein distribution. Localization patterns for the EYFP, EGFP, DsRed2 and mRFP1 fusions were essentially indistinguishable from one another (see Supplementary Figures S2–S4). In addition, we observed little difference in the intensity of red fluorescence for pyrH fusions to mRFPmars, mRFP1 and RFP (mRFP1 codon optimized for E. coli, data not shown).
Intracellular distributions of other notable proteins
In actively dividing cells, mRFP1 and EGFP fusions of the transcription elongation factor NusA were found to co-localize with the nucleoids, predominantly in two ‘loose’ clusters associated with each nucleoid (see Figure 2, panel 6). A very similar finding has been reported for NusA-GFP in B. subtilis, which during rapid growth, primarily co-localized with RNA polymerase and the ribosomes in transcription foci (TF), revealing the sites of ribosomal RNA synthesis (40,41). Our results suggest that in the Gram-negative E. coli bacterium, NusA regulates transcriptional pausing and anti-termination with a similar spatial and mechanistic arrangement to that which occurs in the Gram-positive B. subtilis.
Although distributed evenly throughout the cytoplasm in most cells, NadK-EGFP (yfjB, NAD kinase) was occasionally found to localize in a number of clusters at various positions within the cytoplasm that were not co-incident with the nucleoids (Figure 2, panel 4, green fluorescence; Supplementary Data S2, S4). EGFP fusions of the hypothetical protein YgeH localized throughout the cytoplasm, but preferentially at the inner membrane interface (Figure 2, panel 11). EGFP fusions of another hypothetical protein, YqiK were distributed throughout the cytoplasm (Figure 2, panel 9), even though its extreme N-terminus is predicted to be periplasmic (UniprotKB/Swiss-Prot, entry P77306). We could not visualize the mRFP1 fusion of the yaeL-encoded inner membrane protease (RseP), and EYFP fusions localized as a mixture of cytoplasmic and membrane-associated forms (Figure 2, panel 8). This is partially consistent with the previous reports (42), and suggests that the C-terminus (including the EYFP fusion) may be partially localized within the periplasm, and consequently largely non-fluorescent (43), with a percentage of the protein possibly mis-localized in the cytoplasm, but fluorescent.
Dual CDS fluorescent tagging
To test the practical applicability and efficiency of our methodology for double CDS tagging in E. coli, eight pairs of proteins were selected for fluorescent labeling and co-localization (e.g. see Figure 2; Supplementary Data, Table 2; Supplementary Figure S4). PyrH was included in seven of these, to investigate whether there were any significant differences in its observed localization patterns between the strains. Seven of these strains were successfully constructed, and fluorescence was observed for every tagged protein except for NrdB in the DY380 nrdB-mRFP1/pyrH-EGFP strain. In these doubly tagged strains, the observed localization patterns for the labeled proteins were essentially identical to those found in the corresponding singly labeled strains (see Supplementary Figures S2, S3, S4).
Whole cell protein extracts from eight representative constructed strains were analyzed by western blotting using a polyclonal anti-GFP (Abcam, #ab290) antibody (see Supplementary Data S1, Supplementary Figure S5), to confirm that the C-terminal fluorescent fusions had been accurately created, and that detectable amounts of the protein were expressed during growth in liquid culture. Using this antibody, it was possible to analyze the expression of EGFP, EYFP and ECFP tagged proteins. With the exception of ftsZ, all the strains analyzed specifically expressed one fluorescently tagged protein species with the correct apparent molecular mass, which had not been noticeably degraded (proteolyzed) in vivo. However, the western blots revealed that the FtsZ-EGFP fusion was not expressed: with only EGFP detectable (Mr ca. 27kDa). Even though DNA fragments of the correct size were produced during the initial PCR screening, chromosomal sequencing of the target loci revealed that two separate single nucleotide deletions were present in the linker regions of all the (independently constructed) clones of the DY380 ftsZ-EGFP and DY30 ftsZ-EGFP/pyrH-mRFP1 strains (see Supplementary Data S1). Both of these deletions encoded for short FtsZ C-terminal extensions of six amino acids and not the intended full-length FtsZ-EGFP fusions, and consequently the strains were renamed ftsZ-Cext. Further efforts to tag the ftsZ gene in the DY380 and DY330 strains using a new batch of (PAGE-purified) targeting oligonucleotides were similarly unsuccessful.
The bacterial tubulin homolog FtsZ polymerizes into a ring-like structure (Z-ring) at the mid-cell, and plays an essential role in septum formation and cell division (4,44–46). Cells from both the DY380 ftsZ-Cext and DY30 ftsZ-Cext/pyrH-mRFP1 strains were highly elongated (some more than 20 times normal cell length; e.g. see Figure 4, panels 2 and 3; Supplementary Figures S2, S4) and exhibited a slower growth rate (Supplementary Data S1), indicative of impaired FtsZ function. In these strains, (un-tagged) EGFP protein was still expressed, and localized predominantly throughout the cytoplasm, as well as in small membrane-associated clusters in some cells (Supplementary Figure S2).
DISCUSSION
Experimental rationale and significance of results obtained
We conducted this pilot study for two main reasons: (1) to establish which protein localization patterns are commonly found in E. coli, and by inference, in other similar, symmetrically dividing bacteria; and (2) to determine optimal experimental parameters for future large-scale fluorescent protein localization, or co-localization analyses. We chose the EGFP, EYFP and mRFP1 fluorescent proteins, as these have been the ones used most extensively in localization studies in bacteria, yeast and other higher organisms (23,30,31,47). Of the 21 successfully tagged CDSs, 13 were distributed evenly throughout the cytoplasm under all conditions employed (Adk, Dxs, Era, Map, MsrA, NrdB, RecG, YaeN, YeaA, YfgK, YgjD, YihA and YqiK); MetK, NusA, NadK, PyrH and TrmE formed foci or clusters; and NadK (YfjB), TrmE, YfjP, YgeH associated with the membrane, in all or a proportion of the cells analyzed. NrdB-EGFP and Era-EGFP fluorescence was barely detectable (although an exhaustive screen of growth conditions, was not performed). Only YaeL(RseP)-mRFP1 and RpoD-EGFP could not be visualized, which may be attributable to sequence errors in the long targeting oligonucleotides: as YaeL-mRFP1 has previously been localized to the membrane in E. coli (42), and the expression of a C-terminal RpoD-TAP tag fusion has similarly been detected (11).
The combination of λ-Red and cre/loxP or flpe/frt technologies creates a highly efficient yet versatile strategy by which specific genetic modifications may be made on the E. coli chromosome. Although performed on a smaller scale, our success rate compares favorably with those obtained for analogous large-scale studies in yeast (8–10), and with the TAP-tagging study performed in E. coli DY330 (11). Whilst it is impossible to say with certainty of which 21 successfully tagged CDSs had their function or localization compromised, as 12 of them are putatively essential in E. coli (http://www.shigen.nig.ac.jp/ecoli/pec) (19), this implies that these C-terminal fusion proteins must retain at least a proportion of their native functionality. Only the YaeL-mRFP1/-EYFP and YqiK-EGFP proteins did not exhibit the expected or predicted cellular distributions. This suggests that it may be prudent to verify localization results by constructing N-terminal fusions, or by using other microscopic or proteomic fractionation techniques.
Our results are to a large extent consistent with the microscopic images obtained by Mori and colleagues, from cultures of E. coli cells expressing C-terminal GFP fusions from plasmids (without deletion of the corresponding wild-type CDSs) (16) (GenoBase database, http://ecoli.aist-nara.ac.jp/index.html). Out of the 21 CDSs visualized here, the localizations of 15 were either entirely or partially consistent with those of Mori et al.; 4 were inconsistent (dxs, recG, yqiK, ygeH); 2 were not observed by Mori and co-workers; (yaeL, msrB); and 2 were not localized by us (rpoD, ftsZ). However, at this point in time it is difficult to draw further conclusions, as no articles have been published that describe precisely how these experiments were performed, or that contain a detailed description or analysis of the intracellular protein localization patterns observed.
For the various nusA, pyrH and metK-fusion strains constructed, there appeared to be no correlation between the type of fluorescent fusion protein used and the proportion of protein that existed condensed in foci or clusters, as opposed to being evenly distributed throughout the cytoplasm. Furthermore, MetK-EGFP, NusA-EGFP and PyrH-EGFP fusions created in E. coli DY330 [derived from W3110 (27)], had essentially identical distribution patterns as those found in DY380, EL250 and EL350 (data not shown), indicating that the localization patterns were not strain specific.
The formation and polar localization of MetK and PyrH foci
In cultures of all the relevant constructed strains, we always observed individual cells that contained almost exclusively either the diffuse or punctate forms of the PyrH and MetK fusion proteins, as well as cells that contained various proportions of both (e.g. see Figure 2, panels 3a, 3b, 5 and 7). As we were unable to directly observe their inter-conversion, it is difficult to suggest exactly why such different protein distributions exist within the cell, and what the underlying factors or processes may be. However, it is tempting to speculate that the two forms may correspond to distinct activities within the cell. MetK and PyrH are involved in a variety of biochemical and biological processes: ones that are primarily metabolic, and others that are regulatory in nature. Both have activities that are essential for successful cell division (37,39). In the absence of additional evidence, it is hard to say whether their tendency to aggregate or dissipate is a cause, or a result of these different activities within the cell, or is due to some environmental or growth-related factor.
Intriguingly, when present within the cell, PyrH and MetK foci tended to localize preferentially within the polar regions of the cell, particularly at the mature cell pole, i.e. the cell pole not formed at the last cell division event (e.g. see Figure 4, panel 1; supplementary Figures S2, S3, S4 for images). Butland et al. (11) found that the C-terminal ‘TAP-tagged’ PyrH and MetK proteins co-purified with 47 and 26 other protein species, respectively in E. coli. The interacting proteins have a variety of reported or predicted intracellular localizations (e.g. see (12); GenoBase, EchoBase and UniprotKB/Swiss-prot databases), and are involved a diverse range of functions. Consequently, it is difficult to speculate exactly which protein interactions (if any) may be involved in the observed distribution patterns. In addition, the solubility of PyrH (<0.1 mg/ml at pH 7.4) is enhanced significantly by the presence of Mg-UTP, as well as by acidic/alkaline pH conditions (48), which suggests that there may be other physical factors underlying the formation/dissipation of foci. Also, we cannot discount the possibility that these foci may correspond to non-functional protein aggregates or inclusion bodies within the cell.
The dynamic, targeted localization of proteins to one or both of the cell poles has been demonstrated to occur in a number of Gram-negative and -positive bacterial species, which undergo symmetrical as well as differential cell division (reviewed in (1,3,6,49). A number of general mechanisms have been postulated to explain this type of behavior, including: specific polar targeting and membrane insertion; ‘diffusion and capture’ by a membrane-anchored protein; and localized or spatially specific proteolysis (50–52). Furthermore, proteins associated with the repair, transcription, replication or partitioning of chromosomal and episomal DNA, are often dynamically and asymmetrically located within the cell (18,35,53,54). The preferential localization at the old pole may also reflect general physical or morphological differences in the peptidoglycan, or membrane composition of the new and old poles, or could be due to aging or oxidative effects (55–58). It is also possible that the two daughter cells are not ‘programmed’ to perform identical overall functions, and may have corresponding differences in protein and RNA composition (5,59,60).
Localization of PyrH foci at sites of potential cell division
In the filamentous DY380 ftsZ-Cext/pyrH-mRFP1 cells (Figure 4, panels 2 and 3), the PyrH protein localized predominantly in foci that were separated by approximately one cell length, and contained regularly spaced nucleoids. This localization pattern was highly reminiscent of that found for the actin filament nucleation protein IcsA, in E. coli cells that had been rendered filamentous through inhibition of the septum-specific cell wall transpeptidase FtsI by treatment with aztreonam (61). Under these conditions, IcsA located to the points of potential cell division (septation zones), as well as the cell poles. As noted earlier, the observed cell morphology is consistent with the short C-terminal extension interfering with FtsZ polymerization, or disrupting the association of the ZipA or FtsA cell division proteins to their normal binding sites in the short, highly conserved C-terminal peptide domain (45,62). This greatly inhibits efficient Z-ring formation, resulting in cell filamentation: somewhat analogous to treatment with aztreonam. As western blotting (Supplementary Data S1, Supplementary Figure S5) indicates that it is not specifically proteolyzed within the cell, the fluorescent images suggest that the PyrH protein may have an affinity for some protein and/or structural feature within the septation zones or cell poles (which are derived from the septum during the previous round of cell division).
Asymmetric segregation of PyrH foci between daughter cells
Closer examination of labeled-protein fluorescence in actively dividing cells revealed that a variable but significant proportion of single PyrH foci were kept localized near the old cell pole during the division process, and were segregated into only one of the daughter cells (Figure 3, bottom panel). This phenomenon has previously been observed in differentiating bacteria such as Caulobacter crescentus, which divide asymmetrically forming two distinct cell types (e.g. see (3,6,17,49) and references contained therein). To the best of our knowledge, this is the first time that the asymmetrical segregation of a predominantly cytoplasmic protein focus or cluster has been directly demonstrated to occur in a symmetrically dividing bacterium. It must be noted however, that the segregation of a PyrH focus into one daughter cell does not correspond to an exclusive division of all the PyrH protein present within the parent cell, as there is still a proportion distributed diffusely within the cytoplasm.
Summary of tagging and microscopy results
We believe that the Aequorea-derived EGFP and EYFP, or the Discosoma-derived mRFP1 proteins, would be the best choice for a general large-scale (single CDS) fluorescent localization study in E. coli. The mRFP1 fusion may be preferable for proteins that are predicted to be integrated into the membrane or translocated to the periplasm, as GFP fusions are often non-fluorescent when localized outside the cytoplasm (14,42,43). For straightforward double CDS co-visualization experiments, mRFP1 and EGFP/EYFP-fusions may be individually excited and observed. However, for more detailed spacio-temporal analyses (e.g. using fluorescence resonance energy transfer (FRET)), other pairs of fluorescent fusions may be desirable (31,63).
Although the number of CDSs selected for tagging with fluorescent protein genes represents ca. 0.5% of the predicted genome [4487 genes for strain MG1655 according to EchoBase v1.3, (22) (http://www.ecoli-york.org/)], our results clearly indicate that this approach is applicable for genome-wide CDS expression and protein localization studies in E. coli. Furthermore, we show that such an investigation would prove highly revealing in this symmetrically dividing bacteria, as even within this limited subset of proteins we have observed significant variation, dynamism and polarity in the observed protein localization patterns.
CONCLUSIONS
The microscopic visualization of fluorescently tagged proteins in live E. coli cells revealed a surprising variation in localization patterns. Intracellular protein distributions fall into three general, non-mutually exclusive categories: diffuse cytoplasmic, concentrated in foci or clusters, and membrane associated. Most notably, fluorescent fusions of the PyrH protein formed mobile foci within the cytoplasm that tended to localize at the mature cell pole or points of potential septation, and which were asymmetrically segregated between daughter cells during cell division. It remains to be seen whether the formation of (dynamic) protein foci and preferential polar localization are phenomena commonly found within bacteria. The λ-Red-based CDS tagging procedure we have used here is efficient and specific enough to be applicable for the fluorescent labeling and (co-) visualization of E. coli proteins on a genomic scale. Our investigation has indicated that the future analyses of protein localization patterns in symmetrically dividing bacteria, such as E. coli, have a high probability of revealing many interesting and unexpected results.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by a joint grant from the National Science Foundation (NSF) of China and the Research Grant Council (RGC) of Hong Kong (N_HKU712/02) to J.D.H. and D.L., and by a CRCG seed funding grant (University of Hong Kong) to J.D.H. R.M.W. was supported by the Area of Excellence Scheme of the University Grants Committee of Hong Kong (AoE/P-10/01), and by a Small Project Funding Grant (200507176162) from the University of Hong Kong. Funding to pay the Open Access publication charge was waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gitai Z. The new bacterial cell biology: moving parts and subcellular architecture. Cell. 2005;120:577–586. doi: 10.1016/j.cell.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 2.Lewis PJ. Bacterial subcellular architecture: recent advances and future prospects. Mol. Microbiol. 2004;54:1135–1150. doi: 10.1111/j.1365-2958.2004.04343.x. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro L, McAdams HH, Losick R. Generating and exploiting polarity in bacteria. Science. 2002;298:1942–1946. doi: 10.1126/science.1072163. [DOI] [PubMed] [Google Scholar]
- 4.Carballido-Lopez R, Errington J. A dynamic bacterial cytoskeleton. Trends Cell Biol. 2003;13:577–583. doi: 10.1016/j.tcb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Danchin A, Guerdoux-Jamet P, Moszer I, Nitschke P. Mapping the bacterial cell architecture into the chromosome. Philos. Trans. R. Soc. Lond. B, Biol. Sci. 2000;355:179–190. doi: 10.1098/rstb.2000.0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lybarger SR, Maddock JR. Polarity in action: asymmetric protein localization in bacteria. J. Bacteriol. 2001;183:3261–3267. doi: 10.1128/JB.183.11.3261-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wohlschlegel JA, Yates JR. Proteomics: where's Waldo in yeast? Nature. 2003;425:671–672. doi: 10.1038/425671a. [DOI] [PubMed] [Google Scholar]
- 8.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 9.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 10.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 11.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Campistrous A, Semchuk P, Burke L, Palmer-Stone T, Brokx SJ, Broderick G, Bottorff D, Bolch S, Weiner JH, et al. Localization, annotation, and comparison of the Escherichia coli K-12 proteome under two states of growth. Mol. Cell Proteomics. 2005;4:1205–1209. doi: 10.1074/mcp.D500006-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Lai EM, Nair U, Phadke ND, Maddock JR. Proteomic screening and identification of differentially distributed membrane proteins in Escherichia coli. Mol. Microbiol. 2004;52:1029–1044. doi: 10.1111/j.1365-2958.2004.04040.x. [DOI] [PubMed] [Google Scholar]
- 14.Daley DO, Rapp M, Granseth E, Melen K, Drew D, von Heijne G. Global topology analysis of the Escherichia coli inner membrane proteome. Science. 2005;308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- 15.Arifuzzaman M, Maeda M, Itoh A, Nishikata K, Takita C, Saito R, Ara T, Nakahigashi K, Huang HC, et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 17.Ryan KR, Shapiro L. Temporal and spatial regulation in prokaryotic cell cycle progression and development. Annu. Rev. Biochem. 2003;72:367–394. doi: 10.1146/annurev.biochem.72.121801.161824. [DOI] [PubMed] [Google Scholar]
- 18.Meile JC, Wu LJ, Ehrlich SD, Errington J, Noirot P. Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory. Proteomics. 2006;6:2135–2146. doi: 10.1002/pmic.200500512. [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto M, Ichimura T, Mizoguchi H, Tanaka K, Fujimitsu K, Keyamura K, Ote T, Yamakawa T, Yamazaki Y, et al. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 2005;55:137–149. doi: 10.1111/j.1365-2958.2004.04386.x. [DOI] [PubMed] [Google Scholar]
- 20.Medigue C, Viari A, Henaut A, Danchin A. Colibri: a functional data base for the Escherichia coli genome. Microbiol. Rev. 1993;57:623–654. doi: 10.1128/mr.57.3.623-654.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasteiger E, Jung E, Bairoch A. SWISS-PROT: connecting biomolecular knowledge via a protein database. Curr. Issues Mol. Biol. 2001;3:47–55. [PubMed] [Google Scholar]
- 22.Misra RV, Horler RS, Reindl W, Goryanin II, Thomas GH. EchoBASE: an integrated post-genomic database for Escherichia coli. Nucleic Acids Res. 2005;33:D329–333. doi: 10.1093/nar/gki028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer M, Haase I, Simmeth E, Gerisch G, Muller-Taubenberger A. A brilliant monomeric red fluorescent protein to visualize cytoskeleton dynamics in dictyostelium. FEBS Lett. 2004;577:227–232. doi: 10.1016/j.febslet.2004.09.084. [DOI] [PubMed] [Google Scholar]
- 25.Dymecki SM. A modular set of Flp, FRT and lacZ fusion vectors for manipulating genes by site-specific recombination. Gene. 1996;171:197–201. doi: 10.1016/0378-1119(96)00035-2. [DOI] [PubMed] [Google Scholar]
- 26.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 27.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchholz F, Angrand PO, Stewart AF. A simple assay to determine the functionality of Cre or FLP recombination targets in genomic manipulation constructs. Nucleic Acids Res. 1996;24:3118–3119. doi: 10.1093/nar/24.15.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanushevich YG, Staroverov DB, Savitsky AP, Fradkov AF, Gurskaya NG, Bulina ME, Lukyanov KA, Lukyanov SA. A strategy for the generation of non-aggregating mutants of anthozoa fluorescent proteins. FEBS Lett. 2002;511:11–14. doi: 10.1016/s0014-5793(01)03263-x. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 31.Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 32.Koonin EV. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- 33.Fang G, Rocha E, Danchin A. How essential are nonessential genes? Mol. Biol. Evol. 2005;22:2147–2156. doi: 10.1093/molbev/msi211. [DOI] [PubMed] [Google Scholar]
- 34.Branda CS, Dymecki SM. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 35.Lemon KP, Grossman AD. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 36.Kholti A, Charlier D, Gigot D, Huysveld N, Roovers M, Glansdorff N. pyrH-encoded UMP-kinase directly participates in pyrimidine-specific modulation of promoter activity in Escherichia coli. J. Mol. Biol. 1998;280:571–582. doi: 10.1006/jmbi.1998.1910. [DOI] [PubMed] [Google Scholar]
- 37.Yamanaka K, Ogura T, Niki H, Hiraga S. Identification and characterization of the smbA gene, a suppressor of the mukB null mutant of Escherichia coli. J. Bacteriol. 1992;174:7517–7526. doi: 10.1128/jb.174.23.7517-7526.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landais S, Gounon P, Laurent-Winter C, Mazie JC, Danchin A, Barzu O, Sakamoto H. Immunochemical analysis of UMP kinase from Escherichia coli. J. Bacteriol. 1999;181:833–840. doi: 10.1128/jb.181.3.833-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newman EB, Budman LI, Chan EC, Greene RC, Lin RT, Woldringh CL, D'Ari R. Lack of S-adenosylmethionine results in a cell division defect in Escherichia coli. J. Bacteriol. 1998;180:3614–3619. doi: 10.1128/jb.180.14.3614-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis PJ, Thaker SD, Errington J. Compartmentalization of transcription and translation in Bacillus subtilis. EMBO. J. 2000;19:710–718. doi: 10.1093/emboj/19.4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davies KM, Dedman AJ, van Horck S, Lewis PJ. The NusA:RNA polymerase ratio is increased at sites of rRNA synthesis in Bacillus subtilis. Mol. Microbiol. 2005;57:366–379. doi: 10.1111/j.1365-2958.2005.04469.x. [DOI] [PubMed] [Google Scholar]
- 42.Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol. Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- 43.Feilmeier BJ, Iseminger G, Schroeder D, Webber H, Phillips GJ. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 2000;182:4068–4076. doi: 10.1128/jb.182.14.4068-4076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell. Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 47.Southward CM, Surette MG. The dynamic microbe: green fluorescent protein brings bacteria to light. Mol. Microbiol. 2002;45:1191–1196. doi: 10.1046/j.1365-2958.2002.03089.x. [DOI] [PubMed] [Google Scholar]
- 48.Serina L, Bucurenci N, Gilles AM, Surewicz WK, Fabian H, Mantsch HH, Takahashi M, Petrescu I, Batelier G, et al. Structural properties of UMP-kinase from Escherichia coli: modulation of protein solubility by pH and UTP. Biochemistry. 1996;35:7003–7011. doi: 10.1021/bi960062v. [DOI] [PubMed] [Google Scholar]
- 49.Janakiraman A, Goldberg MB. Recent advances on the development of bacterial poles. Trends Microbiol. 2004;12:518–525. doi: 10.1016/j.tim.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Pugsley AP, Buddelmeijer N. Traffic spotting: poles apart. Mol. Microbiol. 2004;53:1559–1562. doi: 10.1111/j.1365-2958.2004.04273.x. [DOI] [PubMed] [Google Scholar]
- 51.Rudner DZ, Pan Q, Losick RM. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8701–8706. doi: 10.1073/pnas.132235899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Charles M, Perez M, Kobil JH, Goldberg MB. Polar targeting of Shigella virulence factor IcsA in Enterobacteriacae and Vibrio. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9871–9876. doi: 10.1073/pnas.171310498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Possoz C, Sherratt DJ. Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev. 2005;19:2367–2377. doi: 10.1101/gad.345305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherratt DJ. Bacterial chromosome dynamics. Science. 2003;301:780–785. doi: 10.1126/science.1084780. [DOI] [PubMed] [Google Scholar]
- 55.Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Pedro MA, Quintela JC, Holtje JV, Schwarz H. Murein segregation in Escherichia coli. J. Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stewart EJ, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS. Biol. 2005;3:295–300. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woldringh CL. Is Escherichia coli getting old? Bioessays. 2005;27:770–774. doi: 10.1002/bies.20271. [DOI] [PubMed] [Google Scholar]
- 59.Rocha EP, Fralick J, Vediyappan G, Danchin A, Norris V. A strand-specific model for chromosome segregation in bacteria. Mol. Microbiol. 2003;49:895–903. doi: 10.1046/j.1365-2958.2003.03606.x. [DOI] [PubMed] [Google Scholar]
- 60.Aldridge P, Hughes KT. How and when are substrates selected for type III secretion? Trends Microbiol. 2001;9:209–214. doi: 10.1016/s0966-842x(01)02014-5. [DOI] [PubMed] [Google Scholar]
- 61.Janakiraman A, Goldberg MB. Evidence for polar positional information independent of cell division and nucleoid occlusion. Proc. Natl. Acad. Sci. U.S.A. 2004;101:835–840. doi: 10.1073/pnas.0305747101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma X, Margolin W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 1999;181:7531–7544. doi: 10.1128/jb.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miyawaki A, Tsien RY. Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Methods Enzymol. 2000;327:472–500. doi: 10.1016/s0076-6879(00)27297-2. [DOI] [PubMed] [Google Scholar]