Abstract
For catalysis by bacterial type B RNase P, the importance of a specific interaction with p(recursor)tRNA 3′-CCA termini is yet unclear. We show that mutation of one of the two G residues assumed to interact with 3′-CCA in type B RNase P RNAs inhibits cell growth, but cell viability is at least partially restored at increased RNase P levels due to RNase P protein overexpression. The in vivo defects of the mutant enzymes correlated with an enzyme defect at low Mg2+ in vitro. For Bacillus subtilis RNase P, an isosteric C259-G74 bp fully and a C258-G75 bp slightly rescued catalytic proficiency, demonstrating Watson–Crick base pairing to tRNA 3′-CCA but also emphasizing the importance of the base identity of the 5′-proximal G residue (G258). We infer the defect of the mutant enzymes to primarily lie in the recruitment of catalytically relevant Mg2+, with a possible contribution from altered RNA folding. Although with reduced efficiency, B. subtilis RNase P is able to cleave CCA-less ptRNAs in vitro and in vivo. We conclude that the observed in vivo defects upon disruption of the CCA interaction are either due to a global deceleration in ptRNA maturation or severe inhibition of 5′-maturation for a ptRNA subset.
INTRODUCTION
Ribonuclease P (RNase P), an essential ribonucleoprotein enzyme, catalyzes the 5′-end maturation of tRNAs in all Kingdoms of life (1,2). The bacterial RNase P holoenzyme consists of an RNA subunit (P RNA) of approximately 400 nt and a small basic protein (P protein) of approximately. 13 kDa. In vitro, RNA subunits of bacterial RNase P enzymes are catalytically active in the absence of the protein subunit (3), but the protein is essential in vivo (4,5). Two major architectural subtypes of bacterial P RNA are known, type A (for ancestral, prototype Escherichia coli) and type B (represented by Bacillus subtilis) (6), each containing several secondary structural elements not present in the other. A major difference between type A and B structures is the L15 loop of the catalytic domain, which is an internal one in type A but apical in type B RNAs (Figure 1A–C). For E. coli P RNA, the model system of type A structures, it has been shown that two conserved G residues (G293 and G292) in L15 form Watson–Crick base pairs with the two cytosines of tRNA 3′-CCA (C74 and C75) termini (8). Several in vitro studies focussing on the E. coli RNA-alone reaction have indicated that the CCA contact increases substrate affinity (9–11) and supports selection of the correct cleavage site (8,12) as well as catalysis (10,11,13). Recently, disruption of this interaction was demonstrated to be lethal for E. coli cells (14). Analysis of processing of 4.5S RNA, a non-tRNA substrate of RNase P in E. coli, revealed that restoration of base pairing by an isosteric C293-G74 base pair, but not a C292-G75 pair, fully rescued catalytic performance in vivo. In vitro activity assays of E. coli wild type (wt) and C292/C293 mutant RNase P holoenzymes at magnesium concentrations as low as 2 mM Mg2+ suggested a defect in the recruitment of catalytically active Mg2+ ions as the major cause for the lethal phenotype of the mutant P RNAs (14).
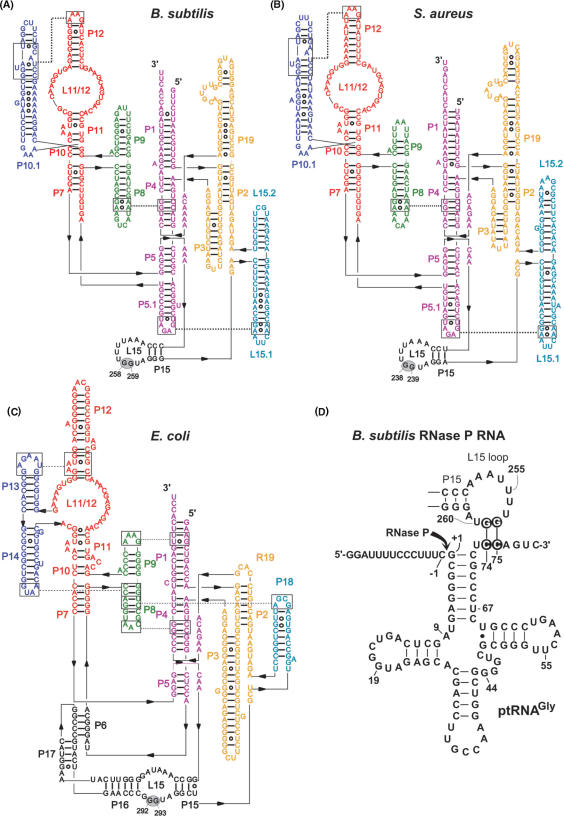
Figure 1.
Secondary structure illustrations of (A) Bacillus subtilis (type B), (B) Staphylococcus aureus (type B) and (C) Escherichia coli (type A) RNase P RNA according to (7). The two G residues in L15, known (E. coli) or suspected (B. subtilis, S. aureus) to be involved in the interaction with tRNA 3′-CCA, are highlighted by gray ovals. (D) Proposed interaction of a canonical bacterial ptRNA (Thermus thermophilus ptRNAGly) with the L15 loop of B. subtilis RNase P RNA. Highlighted nucleotides mark the sites of mutation investigated in this study. The arrow indicates the canonical RNase P cleavage site (between nucleotide –1 and +1).
Type B RNase P RNAs, represented by those from B. subtilis and Staphylococcus aureus, also encode a conserved GG dinucleotide in their L15 loop (Figure 1A and B, G258/G259 in B. subtilis and G238/G239 in S. aureus P RNA). Photo-crosslinking of E. coli and B. subtilis P RNAs to tRNAs with photoreactive groups at the 3′-end has indicated that in enzyme–substrate complexes of both P RNA types the tRNA 3′-CCA terminus is positioned close to the L15 loop region (15). G258 and G259 of B. subtilis P RNA were further shown to be protected from kethoxal modification upon complexation with mature tRNA or precursor tRNA (ptRNA), and this protection was reduced with ptRNA lacking 3′-CCA (16). Potential base pairing to 3′-CCA was addressed by testing cleavage-site selection in L15 mutants of two type A RNAs (from E. coli and Mycobacterium tuberculosis) and of the type B RNA from Mycoplasma hyopneumoniae. With M. hyopneumoniae P RNA, some of the observed mutational effects were consistent with base pairing to 3′-CCA, but the results were not as conclusive as for the type A RNAs (12).
The physiological role of a CCA contact has remained unclear in view of the observation that B. subtilis P RNA, in contrast to its E. coli counterpart, is rather insensitive to 3′-CCA mutations in terms of cleavage-site selection (17). Furthermore, the P protein of B. subtilis RNase P increases the affinity for the ptRNA more than 104-fold (18,19), raising the question whether a contribution of the CCA interaction to substrate affinity of type B enzymes would be significant. Also, the Michaelis constant Km in multiple turnover reactions of the B. subtilis RNase P holoenzyme remained unaffected by the absence of CCA (15).
Furthermore, one-third of tRNAs (27 out of 86) in B. subtilis do not have the CCA motif encoded by their genes. RNase Z removes the 3′-trailers of such CCA-less tRNA primary transcripts by endonucleolytic cleavage 3′ of the discriminator nucleotide as a prerequisite for CCA addition by tRNA nucleotidyl-transferase (CCase); since RNase Z is blocked by the presence of 3′-CCA, it acts specifically on tRNA transcripts lacking the 3′-CCA motif (20). However, B. subtilis RNase Z activity inversely correlates with the length of tRNA 5′ extensions, which led to the proposal that processing by RNase P precedes that of RNase Z in the case of tRNA transcripts with larger 5′ extensions (>30 nt) (20). This would suggest that in vivo B. subtilis RNase P might be able to process certain substrates in the absence of a 3′-CCA moiety.
Here we have analyzed the role of the CCA interaction by mutational analysis in vitro and in vivo in type B bacteria. We demonstrate that B. subtilis rnpBC258 and rnpBC259 mutant alleles carrying a G to C258 or G to C259 mutation in L15 are lethal in the B. subtilis rnpB mutant strain SSB318. We also included a second type B RNA from S. aureus in our complementation study. Corresponding S. aureus rnpBC238 and rnpBC239 mutant alleles also decreased cell viability of B. subtilis SSB318 cells, although not being lethal. Even though the growth defects could be (partially) rescued by parallel overexpression of the B. subtilis P protein, we found no evidence for defects of the mutant P RNAs in P protein or substrate binding. Rather, processing assays with in vitro assembled holoenzymes revealed a defect of the C258/C259 mutant enzymes at free Mg2+ concentrations assumed to be close to those in vivo. We infer that the defect lies primarily in the recruitment of catalytically important Mg2+. An isosteric C259-G74 base pair fully and a C258-G75 pair slightly restored catalytic performance in vitro, which supports the notion that G258/G259 form Watson–Crick base pairs with ptRNA 3′-CCA, but also indicates that the base identity of G258 is important for enzyme function. This is remarkably similar to what has been found for E. coli RNase P (8,11,12,14). Native PAGE experiments further unveiled some differences in RNA-folding equilibria between the wt and mutant P RNAs at low [Mg2+], but folding remained unaffected by the presence of the P protein. This raises the possibility that a folding defect of the C258/C259 mutant RNAs in vivo contributed to their phenotype. We finally addressed the question if RNase P can act on CCA-less ptRNAs in B. subtilis by 5′-RACE experiments in B. subtilis SSB320, an RNase Z-deficient strain that accumulates ptRNA transcripts with CCA-less 3′-extensions. We could demonstrate 5′-end maturation of such ptRNAs by RNase P in vivo, documenting that the presence of 3′-CCA is not an absolute requirement for catalysis by B. subtilis RNase P. We conclude that the in vivo defects of type B rnpB mutant alleles result from a global deterioration of tRNA 5′-end maturation; alternatively, a disruption of the CCA interaction may affect a subset of tRNAs more severely than the majority of tRNAs.
MATERIALS AND METHODS
Bacteria
Bacillus subtilis strain SSB318 (17) was used for complementation studies and SSB320 (20) for 5′-RACE experiments.
Complementation studies in strain SSB318
Recombinant rnpB-coding pHY300 derivatives were introduced into SSB318 using the LS/HS-medium protocol as described (17). Residual IPTG was removed by centrifugation (1 min, 7000 g) and resuspension of the cells in LB medium. Cells were plated in parallel on LB agar plates with or without 1 mM IPTG and containing 0.5 µg/ml erythromycin, 12.5 µg/ml lincomycin and 30 µg/ml tetracycline (to select for the presence of pHY300). In cases of poor transformation efficiency, cells were first plated exclusively in the presence of IPTG. For plasmids encoding rnpB or rnpA genes under control of the Pxyl promoter, the medium was additionally supplemented with 2% (w/v) glucose or 2% (w/v) xylose. For the construction of complementation plasmids, see Supplementary Data.
Growth curve monitoring
Bacillus subtilis SSB318 cells were grown overnight at 37°C in the presence of the appropriate antibiotics and 1 mM IPTG. IPTG was then washed out by repeated centrifugation and resuspension of the cell pellet in LB without IPTG; the final cell pellet was resuspended in LB, adjusted to a starting OD578 of 0.05–0.1 in 50 ml LB supplemented with the respective antibiotics and grown at 37°C under shaking.
In vitro transcription and 5′-endlabeling
T7 runoff transcription and 5′-endlabeling was performed as described (11). For transcription of ptRNAGly (and G74 and G75 variants thereof) as well as E. coli P RNAs, see (14). For transcription of B. subtilis wt and mutant P RNAs, as well as other mutant substrates (Table 5), see Supplementary Data.
Table 5.
Cleavage rates for the B. subtilis wild-type RNase P holoenzyme acting on substrates with 3′-end variations at low magnesium concentrations
| ptRNAGly | Enzyme concentration | [Mg2+] | kobs | krel |
|---|---|---|---|---|
| U73 | 10 nM P RNA/ 37 nM RnpA | 4.5 | 4.3 ± 0.08 | 0.36 |
| U73UAAAUA | 10 nM P RNA/ 37 nM RnpA | 4.5 | 3.3 ± 0.05 | 0.28 |
| U73CCAAUA | 10 nM P RNA/ 37 nM RnpA | 4.5 | 11.8 ± 0.15 | 1.0 |
| U73 | 10 nM P RNA/ 37 nM RnpA | 2.0 | 1.5 ± 0.01 | 0.21 |
| U73UAAAUA | 10 nM P RNA/ 37 nM RnpA | 2.0 | 1.4 ± 0.01 | 0.2 |
| U73CCAAUA | 10 nM P RNA/ 37 nM RnpA | 2.0 | 7.0 ± 0.04 | 1.0 |
| U73 | 50 nM P RNA/ 185 nM RnpA | 2.0 | 9.5 ± 0.08 | 0.19 |
| U73UAAAUA | 50 nM P RNA/ 185 nM RnpA | 2.0 | 6.8 ± 0.03 | 0.13 |
| U73CCAAUA | 50 nM P RNA/ 185 nM RnpA | 2.0 | 49.9 ± 0.43 | 1.0 |
For assay conditions, see legend to Table 4; krel is defined as the ratio of kobs obtained with the mutant versus canonical (U73CCAAUA) substrate under the respective conditions; values are based on at least four independent experiments.
Preparation of recombinant RNase P protein
Bacillus subtilis RNase P protein carrying an N-terminal His-tag (His-tagged peptide leader: MRGSHHHHHHGS, encoded in plasmid pQE-30 in E. coli JM109) was prepared as described for the E. coli RNase P protein (14).
Processing assays
RNase P holoenzyme kinetics were performed in buffer F (2 or 10 mM MgCl2, 50 mM Mes pH 6.1, 100 mM KCl) or buffer KN (20 mM HEPES-KOH, pH 7.4, 2 or 4.5 mM Mg(OAc)2, 150 mM NH4OAc, 2 mM spermidine, 0.05 mM spermine and 4 mM β-mercaptoethanol) (21). In vitro reconstitution of RNase P holoenzymes was performed as follows: P RNAs were incubated in the respective buffer for 5 min at 55°C and 50 min at 37°C, after which RNase P protein was added, followed by another 5 min at 37°C before addition of substrate. Processing reactions were started by combining enzyme and substrate solutions (final volume 10 µl) and assayed at 37°C.
Folding analysis by native PAGE
Folding analyses were conducted exactly as described (14).
5′-RACE
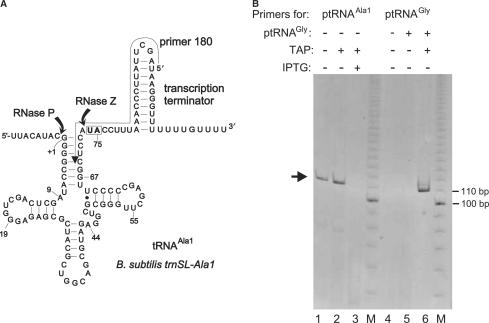
SSB320 cells were grown in parallel either in the presence or absence of IPTG to an OD578 of 0.6. Total RNA was isolated using the TRIzol (Invitrogen) method (here we did not use the RNeasy Mini/Midi Kit from Qiagen employed below, because its cutoff of about 200 nt would have resulted in a substantial loss of smaller RNAs, such as the tRNA transcripts of interest in this experiment), followed by DNase I treatment (Turbo DNase, Ambion). In order to include primary transcripts, one half of total RNA was treated with tobacco acid pyrophosphatase (TAP, Epicentre). Afterwards, the adapter oligonucleotide A1 (5′-GTC AGC AAT CCC TAA GGA G; the three 3′-terminal residues were RNA, the remainder DNA) was ligated to the 5′-monophosphates of RNA molecules. Specific primers were designed to map 3′-unprocessed transcripts of trnSL-Ala1 (primer 180, 5′-TCG AAT AAG GGT TTA AAG GTA TGG AG) and trnSL-Val2 (primer 178, 5′-CTG CGC AAG GGT TTA AGC TAT GAT T). These primers were used in cDNA synthesis and in combination with primer 5′-GTC AGC AAT CCC TAA GGA G (matches the sequence introduced by the adapter oligonucleotide) in the following PCR reaction. PCR products were identified by native 8% PAGE, gel-purified and cloned into pCR2.1-TOPO for sequencing. Five clones were sequenced for each tRNA. For the ptRNAGly control (Figure 5, lanes 4–6), the reverse transcriptase primer was 5′-GAC TGG AGC GGG AGA CGG, yielding an RT-PCR product with an expected length of 112 bp.
Figure 5.
5′-RACE analysis of B. subtilis trnSL-Ala1 3′-precursors. (A) Secondary structure of tRNAAla1 primary transcripts with RNase P and Z cleavage sites indicated; the arrow aligning the transcription terminator and tRNA acceptor stem indicates the position of the primer (‘180’) used for the 5′-RACE experiment; the boxed nucleotides (U74, A75) are replaced with the two C residues after RNase Z cleavage and CCA addition. (B) Analysis of 5′-RACE RT-PCR products on a native 8% PAA gel; M, 10-bp ladder as size marker; in lanes 1–3, primer ‘180’ (see above) and in lanes 4–6 a primer (see the Materials and Methods section) specific for T. thermophilus tRNAGly (Figure 1D) was used. The arrow on the left indicates the main amplification product obtained with primer ‘180’; its size was expected for a tRNAAla1 processing intermediate with a mature 5′-end but carrying the entire 3′-extension as in the primary transcript; this type of product was only detected in total RNA isolated from SSB320 cells grown in the absence of IPTG (lanes 1 and 2), but not in the presence of IPTG (lane 3); before reverse transcription, total RNA preparations were treated (+TAP) or not treated (− TAP) with tobacco acid pyrophosphatase (TAP). In lanes 5 and 6, total RNA from SSB320 cells grown in the absence of IPTG was supplemented with in vitro transcribed ptRNAGly (Figure 1D; 0.05 A260 units ptRNAGly added to 1.0 A260 unit of total cellular RNA) starting with 5′-pppG. A product corresponding to the size of ptRNAGly was only detected after TAP treatment (lane 6), but not without TAP treatment (lane 5) or without addition of in vitro transcribed tRNAGly (lane 4), thus confirming that TAP was active.
Overexpression of rnpA—effect on P RNA levels
To determine rnpB expression levels in strain SSB318 complemented with S. aureus or B. subtilis rnpBwt or mutant alleles as a function of B. subtilis rnpA overexpression, cells were grown overnight at 37°C in 3 ml LB medium supplemented with 1 mM IPTG, 12.5 µg/ml lincomycin, 0.5 µg/ml erythromycin and 30 µg/ml tetracycline. IPTG was then washed out by repeated centrifugation/resuspension of the cell pellet in LB without IPTG; the final cell pellet was then resuspended in LB, and adjusted to a starting OD578 of 0.05–0.1 in 50 ml LB; antibiotics (see above) and 2% xylose (w/v) were added and cells were grown at 37°C under aeration to an OD578 of 0.5–0.6. Aliquots of the cell suspensions were then diluted and plated in parallel on four plates (+xylose and +IPTG;+xylose and −IPTG;+glucose and +IPTG; +glucose and −IPTG) supplemented with antibiotics (all concentrations as in liquid media) to verify the original growth phenotype (retarded growth with mutant rnpB alleles in the absence of Pxyl rnpA, Table 2). Total RNA from the cultures was prepared using the RNeasy Mini/Midi Kit from Qiagen according to the protocol provided by the manufacturer, followed by DNase I treatment (Turbo DNase, Ambion). RT-PCR was performed with the Access RT-PCR System (Promega). PCR was stopped after 12 cycles within the exponential phase of amplification. Primers specific for S. aureus rnpB were 5′- GGG TAA TCG CTA TAT TAT ATA GAG G combined with either 5′- ATT TGG ATT GCT CAC TCG AGG G (5′-endlabeled in Figure 3) or 5′- CTA GTA GTG ATA TTT CTA TAA GCC ATG (5′-endlabeled in Figure S1); primers specific for B. subtilis rnpB were 5′-CTT AAC GTT CGG GTA ATC GC and 5′- AAG TGG TCT AAC GTT CTG TAA GCC (5′-endlabeled in Figure S2); primers specific for B. subtilis ribosomal protein S18 were 5′- GCA GAG GCG GTC GTG CGA AA and 5′-ACG TGC GCG TTT GAT CGC TGC A (5′-endlabeled in Figures 3, S1 and S2); RT-PCR reactions contained the normal amounts of unlabeled primers, and, in addition, trace amounts of the respective 5′-endlabeled primer.
Table 2.
Heterologous complementation of B. subtilis RNase P mutant strain SSB318 by S. aureus (type B) rnpB alleles
| rnpB variants in pHY300 | + IPTG | −IPTG | Aldose |
|---|---|---|---|
| S. aureus PBs rnpB rnpBwt | + + + | + + + | None |
| S. aureus PBs rnpB rnpBC238 | + + + | + | None |
| S. aureus PBs rnpB rnpBC239 | + + + | + | None |
| S. aureus PBs rnpB rnpBwt + Pxyl rnpA | + + + | + + + | Xylose |
| S. aureus PBs rnpB rnpBC238 + Pxyl rnpA | + + + | + + + | Xylose |
| S. aureus PBs rnpB rnpBC239 + Pxyl rnpA | + + + | + + + | Xylose |
Growth of mutant strain SSB318 transformed with wild-type S. aureus (rnpBwt) and respective mutant alleles on plasmid pHY300; promoter types: PBs rnpB, native B. subtilis rnpB promoter; Pxyl, inducible xylose promoter; B. subtilis rnpA was overexpressed in parallel from the same plasmid. Cell growth was analyzed on LB plates in the presence (1 mM) or absence of IPTG; the aldose xylose was added to 2% (w/v) for pHY300 derivatives encoding B. subtilis rnpA.
+ + +: growth with equal numbers of colonies on the corresponding + and − IPTG plates; +: retarded cell growth, but equal numbers of colonies on + and − IPTG plates
Figure 3.
Radioactive reverse transcription PCR (RT-PCR) analysis of strain SSB318 complemented with S. aureus rnpBwt or rnpBC238/C239. PCR products were analyzed on a 10% polyacrylamide/8 M urea gel. Lanes 1–30: total RNA from SSB318 complemented with S. aureus rnpBwt (lanes 1–4 and 13-16), rnpBC238 (lanes 5–8, 17–20 and 25–30) or rnpBC239 (lanes 9–12 and 21–24) grown at 37°C in the absence of IPTG and in the presence of 2% xylose (w/v); amounts of total RNA were 200 ng in lanes 1–24, 26 and 29, 100 ng in lanes 25 and 28, and 400 ng in lanes 27 and 30. Pxyl rnpA: presence (+) or absence (−) of a xylose-inducible plasmid-encoded B. subtilis rnpA gene. Lanes 1–12 and 25–27: primers specific for S. aureus rnpB (rnpB); lanes 13–24 and 28–30: primers specific for the mRNA encoding B. subtilis ribosomal protein S18 (S18). AMV: presence (+) or absence (−) of reverse transcriptase. For details on RT-PCR, see the Material and Methods section. Lanes 25–30 document that the amount of RT-PCR product was sensitive to RNA template concentration. The figure illustrates a representative experiment, but the results shown here were reproduced in five individual experiments using three independent total RNA preparations.
RESULTS
Complementation studies in B. subtilis
The B. subtilis rnpB mutant strain SSB318 was employed to investigate the in vivo role of the tRNA 3′-CCA interaction with type B RNase P RNA. In strain SSB318, the chromosomal rnpB gene is under the control of an IPTG-inducible Pspac promoter (17). In the absence of IPTG, cell growth depends on the presence of a functional rnpB gene provided on a plasmid (pHY300 derivatives in our study). Complementation was assayed for B. subtilis wt P RNA (rnpBwt), and the C258 or C259 mutant alleles (rnpBC258, rnpBC259). B. subtilis rnpBwt rescued cell growth in the absence of IPTG, as expected, no matter if expressed from the native B. subtilis rnpB promoter (17) or the xylose-inducible Pxyl promoter (Table 1). Even basal expression of Pxyl under repressed conditions (medium supplemented with glucose instead of xylose) sufficed for cell growth. In contrast, with complementation plasmids encoding B. subtilis rnpBC258 or rnpBC259 under control of the native B. subtilis rnpB promoter we could not get a single transformant in strain SSB318. For the rnpBC259 allele, some transformants were obtained after substituting the native for the Pxyl promoter; for some clones, we could indeed confirm the mutant rnpBC259 genotype by sequencing, whereas others turned out to be revertants to rnpBwt. However, cells expressing rnpBC259 had a lethal phenotype in the absence of IPTG, which may explain our severe cloning problems. We could not isolate a single colony expressing C258 P RNA.
Table 1.
Homologous complementation of B. subtilis RNase P mutant strain SSB318 by B. subtilis rnpB wild type and mutant alleles
| rnpB variants in pHY300 | + IPTG | − IPTG | Aldose |
|---|---|---|---|
| B. subtilis PBs rnpB rnpBwt | + + + | + + + | None |
| B. subtilis PBs rnpB rnpBC258 | a | None | |
| B. subtilis PBs rnpB rnpBC259 | a | None | |
| B. subtilis Pxyl rnpBwt | + + + | + + +b | Xylose or glucose |
| B. subtilis Pxyl rnpBC258 | a | ||
| B. subtilis Pxyl rnpBC259 | + + + | − | Xylose |
| B. subtilis Pxyl rnpBwt + Pxyl rnpA | + + + | + + + | Xylose |
| B. subtilis Pxyl rnpBC258 + Pxyl rnpA | + + + | + + c | Xylose |
| B. subtilis Pxyl rnpBC259 + Pxyl rnpA | + + + | + + +d | Xylose |
| B. subtilis Pxyl rnpBwt + Pxyl rnpA-stop | + + + | + + + | Xylose |
| B. subtilis Pxyl rnpBC258 + Pxyl rnpA-stop | + + +e | − | Xylose |
| B. subtilis Pxyl rnpBC259 + Pxyl rnpA-stop | + + + | − | Xylose |
| pHY300 | + + + | − | Xylose |
| pHY300 + Pxyl rnpA | + + + | − | Xylose |
| pHY300 + Pxyl rnpA-stop | + + + | − | Xylose |
Growth of mutant strain SSB318 transformed with wild-type B. subtilis rnpB (rnpBwt) and mutant (rnpBC258, rnpBC259) alleles on plasmid pHY300; promoter types: PBs rnpB, native B. subtilis rnpB promoter; Pxyl, inducible xylose promoter; B. subtilis rnpA was overexpressed in parallel from the same plasmid; rnpA-stop designates the B. subtilis rnpA gene with two stop codons in the 5′-coding region. Cell growth was analyzed on LB plates (with appropriate antibiotics) in the presence (1 mM) or absence of IPTG; an aldose (xylose or glucose) at 2% (w/v) was added where indicated.
+ + +: growth with equal numbers of colonies on the corresponding + and − IPTG plates; ++: somewhat retarded growth, but equal numbers of colonies on + and − IPTG plates.; −: no growth;
aUnable to clone.
bBasal expression of xylose promoter (in the presence of 2% glucose) is sufficient for cell growth in the absence of IPTG.
cBasal expression of xylose promoter (in the presence of 2% glucose) is not sufficient for cell growth in the absence of IPTG.
dBasal expression of xylose promoter (in the presence of 2% glucose) is sufficient for cell growth in the absence of IPTG, but retarded cell growth.
eHere we succeeded to isolate a single colony with a stable rnpBC258 genotype; however, the majority of SSB318 cells transformed with rnpBC258 were still unstable and tended to form revertants to the wild type, most likely by recombination with the chromosomal rnpBwt gene.
Simultaneous overexpression of B. subtilis rnpA
We recently reported increased viability of E. coli cells expressing the E. coli rnpBC293 mutant gene when simultaneously overexpressing the homologous P protein (14). We thus transformed B. subtilis SSB318 cells with plasmids encoding the B. subtilis rnpA gene and either rnpBwt, rnpBC258 or rnpBC259, with both rnpA and rnpB alleles under control of the xylose promoter. In the presence of xylose, conditions which fully induced rnpA expression, cell viability in the absence of IPTG could indeed be substantially improved for cells expressing rnpBC258 or rnpBC259. For rnpBC259, basal expression of the Pxyl promoter (in the absence of xylose) supported retarded growth of SSB318 cells in the absence of IPTG. In contrast, for rnpBC258 full induction of the xylose promoter was required, with growth still somewhat retarded relative to rnpBwt and rnpBC259 (Table 1). In conclusion, P protein overexpression restored growth of bacteria expressing the rnpB mutant alleles, but fitness did not fully reach that of bacteria expressing the rnpBwt allele. To demonstrate that rnpA-related rescue effects were due to higher expression levels of the B. subtilis P protein and not to unspecific effects caused by the presence of additional copies of the rnpA gene, we also tested complementation with an rnpA gene with two stop codons in its 5′-portion (rnpA-stop), thus directing expression of a truncated inactive protein (5). As expected, rnpA-stop expression was unable to restore growth of SSB318 bacteria expressing rnpBC258 or rnpBC259 (Table 1). Our findings document a lethal phenotype associated with point mutations at G258 and G259 in L15 of B. subtilis P RNA. However, concomitant overexpression of the P protein (also verified by Western-blot analysis, data not shown) largely rescued cell viability. Furthermore, the phenotype was more severe for the C258 than C259 mutant RNA.
Complementation with S. aureus P RNA
The heterologous type B RNA from S. aureus was included in our complementation analysis to broaden the general significance of our findings on the CCA interaction of type B RNase P RNAs. Residues G238 and G239 in S. aureus P RNA correspond to G258 and G259 of B. subtilis P RNA (Figure 1A and B). Whereas S. aureus rnpBwt was fully functional in B. subtilis SSB318, the mutant alleles rnpBC238/C239 resulted in a significant growth defect, though not being lethal (Table 2). This was also confirmed by monitoring growth curves in liquid culture (Figure 2). Here, the phenotypes of rnpBC238 and rnpBC239 were very similar, thus deviating from the observation that the B. subtilis rnpBC258 allele was more deleterious than rnpBC259 (see above). The growth defect caused by the S. aureus rnpBC238/C239 mutant alleles could be completely rescued by overexpression of B. subtilis rnpA (Table 2).
Figure 2.
Growth curves of SSB318 cells complemented with pHY300 derivatives, carrying S. aureus rnpBwt (squares) or rnpBC238 (triangles with apex at the top) or rnpBC239 (triangles with apex at the bottom) mutant alleles in the presence (+) or absence (−) of IPTG. The better growth of SSB318 bacteria expressing S. aureus rnpBwt (squares) relative to SSB318 carrying the empty vector and grown in the presence of IPTG (open circles) can be explained by the finding that IPTG-induced expression of the chromosomal rnpB gene in the SSB318 mutant strain is weaker than rnpB expression from the native promoter in the original strain W168 used to construct SSB318 ((17), Figure 3 therein). Improved growth was also observed when we expressed B. subtilis rnpBwt from pHY300 in strain SSB318 (data not shown), showing that this effect is not specific for S. aureus rnpB. We conclude that plasmid-borne expression of S. aureus or B. subtilis rnpBwt saturates the cellular RNase P levels in SSB318 bacteria and thereby restores wild-type-like growth.
Complementation with E. coli P RNA
We have recently shown that E. coli P RNA (type A) can functionally replace B. subtilis P RNA (type B) in vivo, no matter if E. coli rnpBwt was expressed from its own native or from the B. subtilis rnpB promoter, and even a single copy of E. coli rnpBwt integrated into the amyE site of the B. subtilis chromosome was sufficient for cell viability (17). Since the S. aureus rnpBC238/C239 mutant alleles displayed a somewhat relaxed phenotype relative to B. subtilis rnpBC258/C259, we also tested function of the corresponding E. coli mutant alleles (rnpBC292/C293) in the B. subtilis background of strain SSB318. The E. coli mutant alleles were unable to restore growth under non-permissive conditions (without IPTG, Table S1). Even when co-expressed with B. subtilis rnpB (in the presence of IPTG), the E. coli rnpBC292/C293 mutant alleles impaired cell growth; overexpression of B. subtilis rnpA did not rescue this growth defect (Table S1).
Overexpression of rnpA—effect on P RNA levels
The complementation results shown in Tables 1 and 2 revealed a rescue of the B. subtilis rnpBC258/C259 and S. aureus rnpBC238/C239 mutant phenotypes when the B. subtilis P protein was overexpressed. One explanation may be that higher levels of P protein increase the steady-state level of RNase P holoenzyme, thereby alleviating the processing defects caused by the mutant alleles. To test this possibility, we initially used SSB318 bacteria expressing the S. aureus rnpB mutant alleles, because here the rescue by rnpA overexpression was complete, reducing the risk that spontaneous mutant bacteria might have dominated the liquid cultures. PCR reactions were limited to 12 cycles, because under these conditions product yields were essentially linearly dependent on RNA template amounts (Figure 3, lanes 25–30). The mRNA for ribosomal protein S18 served as the control to be able to assess experimental fluctuations. Overexpression of rnpA caused, on average, a 3- to 5-fold increase in the yields for the P RNA-specific product (based on five individual experiments), independent of whether bacteria expressed the rnpBwt or one of the mutant alleles (Figure 3, lanes 1–12). No increase in the S18-specific RT-PCR product was observed upon overexpression of rnpA (lanes 13–24). Essentially the same results were obtained when the RT-PCR analysis was performed with a primer pair covering the 5′- and 3′-terminal regions of S. aureus P RNA in order to detect full-length P RNAs (Figure S1). This finding ruled out the possibility that partially degraded P RNA molecules might have contributed to the RT-PCR products in Figure 3. We also analyzed the levels of full-length B. subtilis P RNA as a function of rnpA overexpression. For the B. subtilis rnpBC258/C259 alleles, we could not include the control experiment (without rnpA overexpression), as SSB318 bacteria expressing these alleles did not grow under non-permissive conditions (−IPTG; Table 1); also, growth was impaired for rnpBC258, despite rnpA overexpression (Table 1), and this defect was even more pronounced in liquid culture (data not shown). For the above reasons, we compared P RNA levels for B. subtilis wt P RNA in the absence of a plasmid-borne rnpA gene with those for wt and C259 P RNAs in the presence of overexpressed P protein. As for S. aureus P RNA, we detected increased B. subtilis P RNA levels due to rnpA overexpression (Figure S2); intensity of the RT-PCR product for the C259 mutant RNA was reproducibly found to be somewhat lower than the signal for the wt P RNA, suggesting that the C259 mutation (and, by inference, the C258 mutation) may result in somewhat decreased P RNA levels. Overall, our RT-PCR results demonstrate that overexpression of the P protein causes an increase in the steady-state levels of P RNA, and by inference of the RNase P holoenzyme, which explains the observed rescue effects (Tables 1 and 2).
In vitro processing of ptRNA by B. subtilis RNase P holoenzymes
To shed light on the nature of the in vivo defect of the C258/C259 mutant P RNAs, we analyzed the kinetics of B. subtilis RNase P holoenzymes assembled in vitro. The rescue effect observed when the B. subtilis P protein was overexpressed (Table 1) pointed, among several possibilities, to a protein affinity defect of the mutant P RNAs. We thus tested the activity of B. subtilis wt and mutant holoenzymes as a function of P protein concentration. We applied a buffer system which has been used for B. subtilis holoenzyme kinetics, containing 10 mM MgCl2, 50 mM Mes pH 6.1 and 100 mM KCl (referred to as buffer F10; adapted from (22)). Surprisingly, activities were indistinguishable for the wt and mutant holoenzymes at all P protein concentrations (Figure S3). We then decreased the Mg2+ concentration to 2 mM (buffer F2), assuming that this more closely mimics the intracellular free Mg2+-concentration (1–2 mM) (23). Now the two mutant holoenzymes were more than 100-fold less active than wt RNase P (Table 3) when acting on ptRNAGly with a canonical 3′-CCA terminus (termed ptRNAwt in the following). However, combining the C259 enzyme with the mutant substrate ptRNAG74, restoring base pairing to the C259 P RNA, resulted in cleavage efficiencies essentially identical to those for the wt enzyme acting on ptRNAwt, whereas the wt and C258 enzymes cleaved ptRNAG74 at a more than 100-fold decreased rate. Likewise, the C258 mutant enzyme performed best with ptRNAG75 as the substrate, but the improvement in catalytic performance was only 2- to 6-fold (Table 3). This indicated that nt258 and 259 indeed form Watson–Crick base pairs with C75 and C74 of tRNA 3′-CCA. However, only the isosteric C259-G74 pair, but not the C258-G75 pair, in the E–S complex fully restored activity to that of wt complexes, illustrating the importance of G258 identity. A rescue of activity in the presence of the ptRNAG74 substrate was also observed for chimeric holoenzymes composed of the B. subtilis P protein and the S. aureus C239 mutant P RNA partially purified from SSB318 bacteria (data not shown).
Table 3.
Cleavage activity of B. subtilis wt and mutant RNase P holoenzymes acting on ptRNAGly wt and G74 or G75 mutant substrates at 2 mM Mg2+ and two protein concentrations
| P RNA | ptRNAGly | B. subtilis P protein [nM] | kobs | krel |
|---|---|---|---|---|
| B. subtilis wt | wt | 37.5 | 0.18 ± 0.03 | 1.0 |
| B. subtilis wt | wt | 62.5 | 0.20 ± 0.04 | 1.1 |
| B. subtilis C258 | wt | 37.5 | (4.0 ± 2.0) × 10−4 | 2.2 × 10−3 |
| B. subtilis C258 | wt | 62.5 | (1.3 ± 0.5) × 10−3 | 7.2 × 10−3 |
| B. subtilis C259 | wt | 37.5 | (5.0 ± 1.0) × 10−4 | 2.8 × 10−3 |
| B. subtilis C259 | wt | 62.5 | (1.6 ± 0.9) × 10−3 | 8.9 × 10−3 |
| B. subtilis wt | G74 | 37.5 | (7.0 ± 1.0) × 10−4 | 3.9 × 10−3 |
| B. subtilis wt | G74 | 62.5 | (1.3 ± 0.3) × 10−3 | 7.2 × 10−3 |
| B. subtilis C258 | G74 | 37.5 | (2.0 ± 1.0) × 10−4 | 1.1 × 10−3 |
| B. subtilis C258 | G74 | 62.5 | (3.0 ± 2.0) × 10−4 | 1.7 × 10−3 |
| B. subtilis C259 | G74 | 37.5 | 0.15 ± 0.006 | 0.8 |
| B. subtilis C259 | G74 | 62.5 | 0.30 ± 0.04 | 1.7 |
| B. subtilis wt | G75 | 37.5 | (3.0 ± 1.0) × 10−4 | 7.2 × 10−3 |
| B. subtilis wt | G75 | 62.5 | (4.0 ± 3.0) × 10−4 | 2.2 × 10−3 |
| B. subtilis C258 | G75 | 37.5 | (6.0 ± 4.0) × 10−4 | 3.3 × 10−3 |
| B. subtilis C258 | G75 | 62.5 | (1.8 ± 0.2) × 10−3 | 1.0 × 10−2 |
| B. subtilis C259 | G75 | 37.5 | (1.4 ± 0.8) × 10−4 | 0.6 × 10−3 |
| B. subtilis C259 | G75 | 62.5 | (5.0 ± 0.1) × 10−4 | 2.8 × 10−3 |
Assay conditions: 50 mM Mes pH 6.1, 100 mM KCl, 2 mM MgCl2; P RNA concentration was 20 nM, protein concentration as indicated and substrate concentration was 200 nM; 5′-endlabeled substrate was added in trace amounts (<1 nM). kobs is given in pmol substrate converted per pmol of P RNA per min; for krel, individual kobs values were divided by kobs for the wt P RNA at 37.5 nM P protein and acting on ptRNAwt (first line); values are based on at least three independent experiments.
In vitro processing of ptRNA by B. subtilis holoenzymes in a buffer assumed to closely mimic intracellular conditions
We recently showed (14) for E. coli P RNA that folding of the catalytic RNA may be incomplete in low Mg2+ buffers, such as buffer F2 (Table 3). However, a buffer originally optimized for ribosome function (buffer KN, see Material and Methods) and containing spermine and spermidine appears to favor formation of a native fold (14). We therefore tested activity of the wt and mutant B. subtilis holoenzymes in buffer KN at two different Mg2+ concentrations (2 and 4.5 mM, KN2 and KN4.5). At 4.5 mM Mg2+, the mutant enzymes cleaved ptRNAwt at almost identical (C259) or 2-fold reduced (C258) rates. However, at 2 mM Mg2+, the rate dropped 2-fold for the wt enzyme, but about 6-fold for the mutant enzymes (Table 4). Thus, the minor shift from 4.5 to 2 mM Mg2+ exacerbated the defect of the mutant enzymes. To analyze if the defect was on the level of substrate affinity or holoenzyme stability, we tested processing activity under the same conditions but at a 5-fold higher holoenzyme concentration (Table 4). If substrate binding or holoenzyme stability were the major defect in reactions catalyzed by the C258/C259 mutant enzymes at 2 mM Mg2+, one would have expected that the ratio of cleavage rates for the mutant enzyme versus wt enzymes (krel, Table 4) improves at the higher enzyme concentration. However, krel values rather showed a tendency to further decrease (0.12 and 0.13 versus 0.17 and 0.30, Table 4).
Table 4.
Cleavage rates for B. subtilis holoenzymes reconstituted in vitro at low Mg2+ concentrations
| P RNA | ptRNAGly | Enzyme concentration | [Mg2+] | kobs | krel |
|---|---|---|---|---|---|
| B. subtilis wt | wt | 10 nM P RNA/ 37 nM RnpA | 4.5 | 9.7 ± 0.08 | 1.0 |
| B. subtilis C258 | wt | 10 nM P RNA/ 37 nM RnpA | 4.5 | 4.4 ± 0.08 | 0.45 |
| B. subtilis C259 | wt | 10 nM P RNA/ 37 nM RnpA | 4.5 | 8.4 ± 0.12 | 0.87 |
| B. subtilis wt | wt | 10 nM P RNA/ 37 nM RnpA | 2.0 | 4.6 ± 0.09 | 1.0 |
| B. subtilis C258 | wt | 10 nM P RNA/ 37 nM RnpA | 2.0 | 0.8 ± 0.01 | 0.17 |
| B. subtilis C259 | wt | 10 nM P RNA/ 37 nM RnpA | 2.0 | 1.4 ± 0.03 | 0.30 |
| B. subtilis C259 | G74 | 10 nM P RNA/ 37 nM RnpA | 2.0 | 6.5 ± 0.08 | |
| B. subtilis wt | wt | 50 nM P RNA/ 185 nM RnpA | 2.0 | 47.7 ± 0.68 | 1.0 |
| B. subtilis C258 | wt | 50 nM P RNA/ 185 nM RnpA | 2.0 | 5.9 ± 0.07 | 0.12 |
| B. subtilis C259 | wt | 50 nM P RNA/ 185 nM RnpA | 2.0 | 6.2 ± 0.05 | 0.13 |
Assay conditions: 20 mM Hepes pH 7.4 (37°C), 2 mM Mg(OAc)2, 150 mM NH4OAc, 2 mM spermidine, 0.05 mM spermine, 4 mM β-mercaptoethanol, and P RNA and B. subtilis P protein (RnpA) concentrations as indicated; the substrate concentration was 100 nM; 5′-endlabeled substrate was added in trace amounts (<1 nM); kobs is given in pmol substrate converted per pmol of P RNA per min; krel is defined as the ratio of kobs obtained with the mutant versus wt holoenzyme under the respective conditions; values are based on at least four independent experiments.
We also analyzed cleavage of ptRNAG74 by the C259 mutant enzyme in buffer KN2 (Table 4). This fully restored the cleavage rate to that of the wt enzyme, in line with the results of Table 3.
Activity assays performed with chimeric holoenzymes consisting of E. coli P RNA (wt and the corresponding C292/C293 mutant variants) and the B. subtilis P protein (Table S2) resulted in essentially the same findings as with the B. subtilis holoenzymes.
Analysis of B. subtilis P RNA folding equilibria
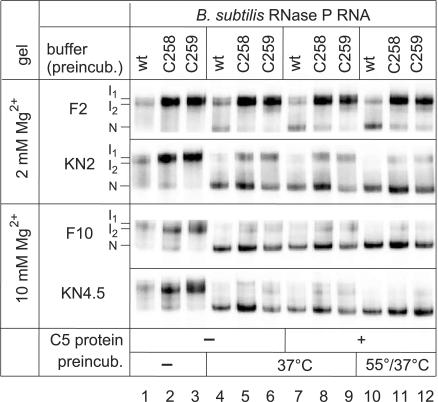
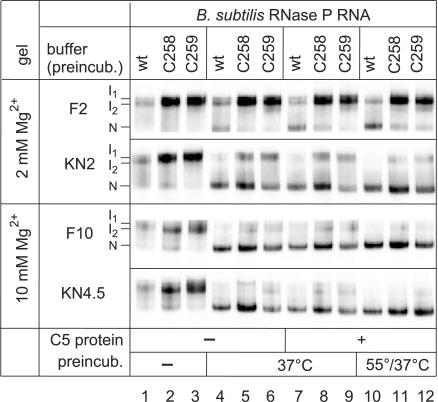
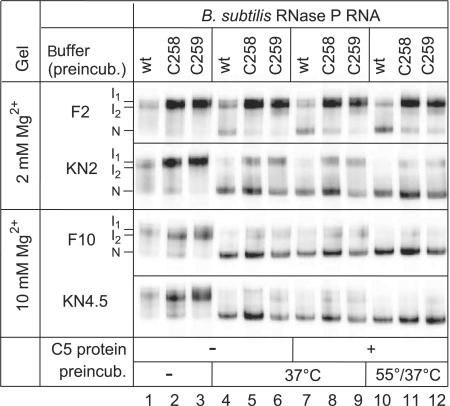
Incomplete (24) or aberrant folding might have contributed to the in vivo and in vitro phenotypes of the C258/C259 mutant P RNAs. To address this possibility, we performed a native PAGE analysis using the same buffer systems as for the kinetic experiments. When 5′-endlabeled wt, C258 or C259 P RNAs were directly loaded (after gel purification and ethanol precipitation) onto the native gel without a preincubation step, under all four tested conditions two major bands appeared, which we termed I1 and I2 (Figure 4, lanes 1–3). We are aware that I1 and I2 observed in the presence of 2 mM Mg2+ during electrophoresis may not necessarily be identical to those seen at 10 mM Mg2+. Upon P RNA preincubation for 55 min at 37°C (37°C) or for 5 min at 55°C followed by 50 min at 37°C (55/37°C), a faster migrating conformer appeared, which we assign to the native fold (termed ‘N’), in line with a previous study (19). Remarkably, in buffer F2 (2 mM Mg2+) the N conformer was much less populated with the two mutant than with the wt P RNA (Figure 4, lanes 4–12). This difference was largely reduced in buffer KN2. When preincubation was performed in buffers F10 and KN4.5 (10 and 4.5 mM Mg2+, respectively) and native PAGE at 10 mM Mg2+, differences between wt and mutant P RNAs were further mitigated (lanes 4–9) and essentially abolished after preincubation at 55/37°C (lanes 10–12). No substantial changes were seen when the P protein was added to the preincubation mix (lanes 4–6 versus 7–9). In summary, our native PAGE analysis indicates that (i) the KN2 buffer containing spermine and spermidine favors formation of the native fold relative to the F2 buffer and (ii) folding differences can be considered insignificant in buffer KN4.5 combined with our standard preincubation (5 min at 55°C, 50 min at 37°C). The data in Figure 4 illustrate that the folding equilibria of wt and C258/C259 mutant RNase P RNAs are sensitive to the buffer conditions. As we do not know the exact ionic milieu in vivo, a contribution from impaired folding of the mutant P RNAs to their growth defect in vivo cannot be excluded.
Figure 4.
Analysis of folding equilibria for wt and C258/C259 mutant B. subtilis P RNAs by native PAGE. RNAs (50 fmol, 5′-endlabeled) were preincubated either in buffer F containing 2 mM (F2) or 10 mM (F10) Mg2+, or in buffer KN containing 2 mM (KN2) or 4.5 mM (KN4.5) Mg2+ in a volume of 4–5 µl (for buffer F and KN compositions, see Materials and Methods). Lanes 1–3: no preincubation (kept at 4°C); lanes 4–6: preincubation of P RNAs for 70 min at 37°C; lanes 7–9: preincubation of P RNAs for 55 min at 37°C, addition of 1 µl B. subtilis P protein (final concentration 37 nM) and further incubation for 15 min at 37°C; lanes 10–12: as in lanes 7–9, but preincubation of P RNAs for 5 min at 55°C and 50 min at 37°C. Samples were run on 11.25% polyacrylamide gels in 1× THE buffer supplemented with 100 mM NH4OAc and either 2 mM (F2, KN2) or 10 mM (F10, KN4.5) MgCl2.
Processing of CCA-less ptRNAs
Since in B. subtilis only two-thirds of the tRNA genes encode 3′-CCA, the question arose whether tRNA transcripts not encoding 3′-CCA have to undergo 3′-end processing and CCA addition before 5′-end maturation by RNase P can occur. To address this question, we constructed ptRNAGly 3′-variants, either encoding 3′-U73CCAAUA, 3′-U73UAAAUA or ending with the discriminator nucleotide (3′-U73 = ΔCCA); UAA instead of CCA downstream of the discriminator base is found in the native B. subtilis trnI-Thr precursor (20). Processing of these substrates by wt RNase P was tested in buffer KN, either at 4.5 or 2 mM Mg2+, and for 2 mM Mg2+ also at a 5-fold higher P RNA and P protein concentration (Table 5). The obtained pattern of relative cleavage rates roughly mirror-imaged what we had seen with the C258/C259 mutant enzymes in Table 4: (i) the cleavage rate was reduced for the UAA and ΔCCA variant, and the ratio of rates for the mutant versus canonical substrate worsened when the Mg2+ concentration was reduced from 4.5 to 2 mM; (ii) a 5-fold increase in P RNA and protein concentration did not improve performance of the mutant substrates relative to the canonical ptRNA (Table 5), suggesting that substrate affinity is not the major cause of the defect.
5′-RACE experiment of trnSL-Ala1 and trnSL-Val2
In vitro processing of the CCA-less substrates was impaired but not abolished, raising the question whether this residual activity would suffice for RNase P cleavage of CCA-less tRNA transcripts in vivo. This was addressed by 5′-RACE experiments. For this purpose, we used strain SSB320, in which the chromosomal rnz gene (encoding RNase Z) is put under control of the IPTG-inducible Pspac promoter. In B. subtilis, RNase Z is involved in the 3′-maturation of tRNA primary transcripts lacking the CCA motif. We chose two native tRNAs (trnSL-Ala1 and trnSL-Val2), which were shown to accumulate as 3′-precursors in strain SSB320 when rnz expression is decreased (20). Both tRNAs are transcribed from monocistronic genes with expected 5′-precursor segments of 8 nt (trnSL-Ala1) and 38 nt (trnSL-Val2), and both are 3′-extended by their transcription terminator stem-loop (Figure 5A) (20). To ensure that only tRNAs with unprocessed 3′-ends were reverse transcribed during 5′-RACE, we designed a primer for reverse transcription whose 5′-proximal and central portion targeted the 3′-precursor sequences (Figure 5A). A control experiment performed with RNA from SSB320 cells grown in the presence of IPTG (normal RNase Z expression) did not produce RT-PCR products (Figure 5B, lane 3), demonstrating that our primer was specific for the 3′-extended tRNA. Using total RNA isolated from SSB320 cells grown in the absence of IPTG we obtained a prominent 5′-RACE product, roughly corresponding to the size of 5′-matured 3′-precursor tRNA (ca. 110 nt; Figure 5B, lane 1). An additional treatment with tobacco acid pyrophosphatase (TAP; to permit RT-PCR of primary transcripts with 5′-triphosphates) did not give rise to an additional RT-PCR product (Figure 5B, lane 2). We further controlled for TAP activity by adding a T7 primary transcript of ptRNAGly (Figure 1D, carrying a 5′-triphosphate) to the total cellular RNA pool (Figure 5B, lanes 5 and 6). In the case of inefficient 5′-end processing we would have expected a product extended by 8 nt for tRNAAla1 (20). To ensure that RT-PCR products corresponded to 5′-matured transcripts, we cloned the main band of Figure 5B (lanes 1 and 2, indicated by the arrow) into the pCR2.1-TOPO vector (Invitrogen) and sequenced 5 clones (the same was done for trnSL-Val2; data not shown). Sequencing analysis confirmed mature 5′-ends for both tRNAs. This was surprising as we would have anticipated, based on our kinetic in vitro results (Table 5), to detect at least some 3′-extended tRNA with immature 5′-ends. In any case, these results clearly demonstrated that RNase P processing can proceed in vivo in the absence of the CCA interaction.
DISCUSSION
In vivo phenotypes of P RNAs with disrupted CCA interaction
We have investigated the tRNA 3′-CCA interaction with type B RNase P under in vivo conditions, using B. subtilis mutant strain SSB318 which has the chromosomal rnpB gene under control of the IPTG-inducible Pspac promoter (17). For this purpose, complementation efficiencies of B. subtilis rnpBwt relative to rnpBC258/C259 mutant alleles were analyzed in strain SSB318 under non-permissive conditions (−IPTG). The latter two rnpB alleles encode P RNAs with point mutations of the nucleotides thought to be involved in the CCA interaction. We also tested the functionality of S. aureus rnpBwt and its mutant rnpBC238/C239 alleles (as representatives of a heterologous type B P RNA) and that of the corresponding E. coli rnpBC292/C293 alleles representing type A RNase P RNA architecture. All rnpBwt alleles (B. subtilis, S. aureus, E. coli) were functional in SSB318 bacteria (Tables 1, 2 and S1). However, the mutant P RNA alleles resulted either in a severe growth defect (S. aureus rnpBC238/C239; Table 2) or were even lethal (B. subtilis rnpBC258/C259 and E. coli rnpBC292/C293; Tables 1 and S1). Overexpression of the B. subtilis P protein partially (for B. subtilis rnpB alleles) or fully (for S. aureus rnpB alleles) restored cell viability of SSB318 bacteria expressing the mutant rnpB alleles (Tables 1 and 2). The rescue effect was also observed for the E. coli C293 mutant RNA in its natural host (14), but was not scrutinized there. Here we could attribute the rescue to increases in P RNA steady-state levels owing to P protein overexpression (Figures 3, S1 and S2). These findings demonstrate, for the first time, that the CCA interaction is important for the vitality of bacteria encoding a type B RNase P RNA (B. subtilis in this study), but not necessarily essential, as inferred from the complementation results with the mutant rnpB alleles from S. aureus (Table 2). RNase P is able to cleave CCA-less ptRNAs in vivo, as inferred from 5′-RACE in the RNase Z mutant strain (Figure 5). Since RNase Z, which acts on CCA-less tRNA 3′-precursors, has largely impaired activity on ptRNAs with long 5′-leaders, RNase P cleavage was proposed to precede RNase Z cleavage on CCA-less ptRNAs carrying expanded 5′- in addition to 3′-extensions (20). Our results verify this hypothesis. The finding that B. subtilis RNase P is able to process CCA-less ptRNAs then raises the question why the C258/C259 (or C238/C239) mutations caused a lethal (or severe) phenotype in the presence of normal amounts of P protein (Tables 1 and 2). Most likely, the general deceleration in ptRNA maturation kinetics adds up to a collective defect caused by the incapability of the tRNA biosynthesis machinery to meet the demands of the protein synthesis apparatus. This defect is mitigated when the steady-state amounts of the RNase P holoenzyme increase severalfold as a result of P protein overproduction. It also cannot be ruled out that disruption of the CCA interaction may impair 5′-maturation of a subset of ptRNAs more severely than that of the bulk of ptRNAs.
P protein overexpression leads to increases in P RNA steady-state levels (Figures 3, S1 and S2), and, by inference, to increases in cellular RNase P holoenzyme concentrations. This is remarkable, taking into account that the P protein only covers a small surface area of the RNA subunit (2). In a study on P RNA metabolism in E. coli (25), it was found that rnpB primary transcripts can undergo either 3′-end maturation or oligoadenylation resulting in degradation. A knockout of poly(A) polymerase (pcnB) blocked the degradation pathway and consequently increased the level of 3′-precursor P RNA. Since these higher levels of 3′-precursor P RNA did not entail an increase in the level of mature P RNA, the authors proposed that mature RNase P RNA may only stably accumulate when complexed with its protein subunit (25). Our results (Figures 3, S1 and S2) are consistent with this model, suggesting similar mechanisms to regulate cellular RNase P levels in E. coli and B. subtilis despite fundamentally different P RNA maturation pathways in the two organisms. Whereas the mature 5′- and 3′-ends of B. subtilis P RNA appear to be generated by autolytic processing of precursor P RNA (26), E. coli P RNA primary transcripts are mainly intitiated at the mature 5′-end, and 3′-precursor sequences are removed by RNase E followed by exonucleolytic removal of 1 or 2 remaining nucleotides (25,27).
In vitro results
The importance of the CCA interaction in reactions catalyzed by type B RNase P holoenzymes has been underestimated in previous in vitro analyses (15). We could reconcile these earlier findings when we tested activity upon disruption of the CCA interaction at 10 mM Mg2+ (Figure S3), which has been the ‘gold standard’ magnesium concentration for the testing of bacterial holoenzymes. However, the CCA interaction becomes critical in vitro at low Mg2+ concentrations, such as 2 mM (Tables 3 to 5), assumed to better mimic the cellular milieu than 10 mM Mg2+.
Our kinetic data clearly demonstrate that tRNA 3′-CCA ends form Watson–Crick base pairs with two guanosines in the L15 loop of type B RNase P RNAs. Evidence for the base pairing with C74 was first suggested by a mutational study analyzing cleavage fidelity in the reaction catalyzed by the type B RNA from M. hyopneumoniae (12). As found for E. coli (11,14), an isosteric C259-G74 base pair, but not a C258-G75 pair, fully restored catalytic performance of type B RNase P to that of the wt holoenzyme acting on ptRNA with a canonical 3′-CCA end (Tables 3 and 4). This indicates that the base identity of the 5′-proximal G residue (G258 in B. subtilis) is critical for catalytic performance, as observed for G292 of E. coli P RNA (11,14). For E. coli, the importance of G292 identity can be attributed to the finding that the N7 of G292 is sensitive to a c7-deaza modification, which was interpreted to indicate hydrogen bonding with the 6-amino group of A258 in the 5′-portion of L15 (28). By inference, G258 might interact in an analogous manner with one of the adenines (A251–A253, Figure 1D) in L15 of B. subtilis P RNA.
Toward a better understanding of the defect of holoenzymes with a disrupted CCA interaction at low Mg2+, it is instructive to briefly discuss what is known about the metal ion binding properties of the L15 loop. In E. coli, the P15/L15/P16 region has been identified as a central Me2+ binding module in the active site of P RNA (13,29). There are sites of prominent Pb2+ hydrolysis at two locations in the internal L15 loop (sites III and V). Binding of tRNA 3′-CCA suppresses, in vitro and in vivo, Pb2+ hydrolysis at sites III and V and creates a new prominent Pb2+ hydrolysis site (IVb) nearby (30,31). This indicates a structural rearrangement of the P15/L15/P16 region and a concomitant re-coordination of metal ions upon formation of the G292-C75 and G293-C74 intermolecular base pairs. For B. subtilis P RNA, four out of nine prominent Pb2+ hydrolysis sites could also be assigned to its apical L15 loop (32). Combined with the phenotypic similarities of C258/C259 (C292/C293 in E. coli) mutations in the E. coli and B. subtilis systems, this supports the notion that the L15 loop is a central Me2+ binding module in the active site of type B RNAs as well.
Based on the results shown in Figure 4, the low activity of mutant holoenzymes at 2 mM Mg2+ might have been caused by a P RNA folding defect. However, the results of Table 5 argue against this view. When we deleted or mutated 3′-CCA in the substrate and assayed processing by wt RNase P, results were essentially a mirror image of what we had seen with the mutant enzymes acting on ptRNAwt (cf. Tables 4 and 5): catalytic performance for the mutant substrates relative to ptRNAwt deteriorated upon a reduction of [Mg2+] from 4.5 to 2 mM, and the relative catalytic performance for the mutant substrates did not improve at 5-fold higher P RNA and protein concentrations.
The slight reduction in the fraction of correctly folded C258/C259 mutant P RNA molecules relative to the wt P RNA upon a decrease in Mg2+ concentration from 4.5 to 2 mM (Figure 4) is nonetheless remarkable. Though unlikely to be the major cause of the catalytic defect of the mutant P RNAs at 2 mM Mg2+, this finding is surprising as the L15 loop is thought to be quite unstructured in free P RNA based on its low resolution in the X-ray structure of B. stearothermophilus P RNA (33). Although the effect of the two point mutations in L15 on P RNA folding is not understood, this underscores the high sensitivity of P RNA folding to minimal structural changes at low Mg2+ concentrations.
Formally, the defect of the mutant P RNAs may also include contributions from impaired P protein and/or substrate affinity. However, at 10 mM Mg2+ (buffer F10) and increasing P protein concentrations, the reconstituted B. subtilis C258 and C259 mutant holoenzymes had activities indistinguishable from the wt enzyme (Figure S3). Likewise, in buffer KN supplemented with 4.5 mM Mg2+, activity reductions for the mutant holoenzymes were marginal. Only at 2 mM Mg2+, a substantial loss of activity was seen for the mutant enzymes, which correlated with the severe in vivo defects. Considering that a reduction of the Mg2+ concentration from 10 to 2 mM and particularly from 4.5 to 2.0 mM Mg2+ means only a minor decrease in ionic strength, we think it unlikely that the major defect of the mutant P RNAs is related to impaired P protein and/or substrate affinity. We rather infer that the L15 loop mutations lower the affinity for catalytically relevant Mg2+ ions that either directly contribute to the catalytic process or help to fold the L15–CCA interaction module as part of the active site in E–S complexes.
Differences between individual RNase P enzymes
Despite the striking similarities in the CCA interaction of type A and B RNase P RNAs, some details are different. For E. coli RNase P, a 5-fold increase in P RNA and protein concentrations did not increase the cleavage rate for the C293 and C292 mutant enzymes at 2 mM Mg2+ (14). In contrast, the B. subtilis C258 and C259 mutant enzymes cleaved ptRNAwt at a 4-to 7-fold higher rate when P RNA and protein concentrations were increased 5-fold (Table 4). Activity of the chimeric holoenzymes consisting of E. coli P RNA and B. subtilis P protein also improved 2- to 3-fold at the higher RNase P subunit levels (Table S2). This finding, if not due to differences in the quality of the recombinant E. coli and B. subtilis P proteins, may point to a specific effect exerted by the B. subtilis P protein, possibly related to the observation that binding of P protein and substrate to P RNA are cooperative in the B. subtilis system (34).
Also, the in vivo phenotypes of the C258 (B. subtilis) and C238 (S. aureus) type B mutant P RNAs could be partially (B. subtilis; Table 1) or fully (S. aureus; Table 2) rescued by overexpression of B. subtilis rnpA. However, in the E. coli system essentially no rescue was observed for the corresponding E. coli C292 mutant P RNA upon overexpression of E. coli rnpA (14), suggesting that the guanine identity at this position is more critical in type A versus B enzymes. In this context, the better complementation efficiency of mutant S. aureus versus B. subtilis rnpB alleles was surprising. The S. aureus rnpBC238/C239 alleles even enabled some cell growth at normal P protein levels (Table 2). This might be related to the general problem associated with studies of homologous genes especially in B. subtilis which is known to possess an efficient recombination machinery. The presence of homologous rnpB genes carrying point mutations may activate genetic repair systems, especially if the mutant genes cause stress to the cells. This would explain our severe problems to clone the homologous B. subtilis rnpBC258/C259 alleles in SSB318 combined with the high number of wild-type revertants. Finally, the severeness of phenotypes for B. subtilis rnpBC258 and E. coli rnpBC292 exceeded that of B. subtilis rnpBC259 and E. coli rnpBC293, respectively (Table 1) (14). In contrast, the in vivo phenotypes of S. aureus rnpBC238 and rnpBC239 were indistinguishable (Table 2; Figure 2), which we do not understand at present.
Despite some context dependence of L15 mutations, we can conclude from our findings that the Watson–Crick base-pairing interaction with tRNA 3′-CCA is a common feature of the majority of bacterial type A and type B RNase P enzymes. This interaction is important but not necessarily essential for cellular function.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
We like to thank Ciarán Condon for providing B. subtilis strain SSB320, B.M. Fredrik Pettersson and Leif A. Kirsebom for providing genomic DNA of S. aureus, Michal Marszalkowski for preparation of recombinant B. subtilis RNase P protein and Dagmar K. Willkomm for critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (HA 1672/7-4) and the Fonds der Chemischen Industrie. Funding to pay the Open Access publication charge was provided by Deutsche Forschungsgemeinschaft (GK 1384).
Conflict of interest statement: None declared.
REFERENCES
- 1.Hartmann E, Hartmann RK. The enigma of ribonuclease P evolution. Trends Genet. 2003;19:561–569. doi: 10.1016/j.tig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 4.Schedl P, Primakoff P, Roberts J. Processing of E. coli tRNA precursors. Brookhaven Symp. Biol. 1974;26:53–76. [PubMed] [Google Scholar]
- 5.Gößringer M, Kretschmar-Kazemir Far R, Hartmann RK. Analysis of RNase P protein (rnpA) expression in Bacillus subtilis utilizing strains with suppressible rnpA expression. J. Bacteriol. 2006;188:6816–6823. doi: 10.1128/JB.00756-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall TA, Brown JW. The ribonuclease P family. Methods Enzymol. 2001;341:56–77. doi: 10.1016/s0076-6879(01)41145-1. [DOI] [PubMed] [Google Scholar]
- 7.Tsai HY, Masquida B, Biswas R, Westhof E, Gopalan V. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J. Mol. Biol. 2003;325:661–675. doi: 10.1016/s0022-2836(02)01267-6. [DOI] [PubMed] [Google Scholar]
- 8.Kirsebom LA, Svärd SG. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994;13:4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardt WD, Schlegl J, Erdmann VA, Hartmann RK. Kinetics and thermodynamics of the RNase P RNA cleavage reaction: analysis of tRNA 3′-end variants. J. Mol. Biol. 1995;247:161–172. doi: 10.1006/jmbi.1994.0130. [DOI] [PubMed] [Google Scholar]
- 10.Oh BK, Frank DN, Pace NR. Participation of the 3'-CCA of tRNA in the binding of catalytic Mg2+ ions by ribonuclease P. Biochemistry. 1998;37:7277–7283. doi: 10.1021/bi973100z. [DOI] [PubMed] [Google Scholar]
- 11.Busch S, Kirsebom LA, Notbohm H, Hartmann RK. Differential role of the intermolecular base-pairs G292-C(75) and G293-C(74) in the reaction catalyzed by Escherichia coli RNase P RNA. J. Mol. Biol. 2000;299:941–951. doi: 10.1006/jmbi.2000.3789. [DOI] [PubMed] [Google Scholar]
- 12.Svärd SG, Kagardt U, Kirsebom LA. Phylogenetic comparative mutational analysis of the base-pairing between RNase P RNA and its substrate. RNA. 1996;2:463–472. [PMC free article] [PubMed] [Google Scholar]
- 13.Brännvall M, Pettersson BMF, Kirsebom LA. Importance of the +73/294 interaction in Escherichia coli RNase P RNA substrate complexes for cleavage and metal ion coordination. J. Mol. Biol. 2003;325:697–709. doi: 10.1016/s0022-2836(02)01195-6. [DOI] [PubMed] [Google Scholar]
- 14.Wegscheid B, Hartmann RK. The precursor tRNA 3′-CCA interaction with Escherichia coli RNase P RNA is essential for catalysis by RNase P in vivo. RNA. 2006;12:2135–2148. doi: 10.1261/rna.188306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh BK, Pace NR. Interaction of the 3′end of tRNA with ribonuclease P RNA. Nucleic Acids Res. 1994;22:4087–4094. doi: 10.1093/nar/22.20.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaGrandeur TE, Hüttenhofer A, Noller HF, Pace NR. Phylogenetic comparative chemical footprint analysis of the interaction between ribonuclease P RNA and tRNA. EMBO J. 1994;13:3945–3952. doi: 10.1002/j.1460-2075.1994.tb06710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wegscheid B, Condon C, Hartmann RK. Type A and B RNase P RNAs are interchangeable in vivo despite substantial biophysical differences. EMBO Rep. 2006;7:411–417. doi: 10.1038/sj.embor.7400641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurz JC, Niranjanakumari S, Fierke CA. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry. 1998;37:2393–2400. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- 19.Buck AH, Dalby AB, Poole AW, Kazantsev AV, Pace NR. Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J. 2005;24:3360–3368. doi: 10.1038/sj.emboj.7600805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pellegrini O, Nezzar J, Marchfelder A, Putzer H, Condon C. Endonucleolytic processing of CCA-less tRNA precursors by RNase Z in Bacillus subtilis. EMBO J. 2003;22:4534–4543. doi: 10.1093/emboj/cdg435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinos G, Wilson DN, Teraoka Y, Szaflarski W, Fucini P, Kalpaxis D, Nierhaus KH. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site RNA binding. Mol. Cell. 2004;13:113–124. doi: 10.1016/s1097-2765(04)00002-4. [DOI] [PubMed] [Google Scholar]
- 22.Kurz JC, Fierke CA. The affinity of magnesium binding sites in the Bacillus subtilis RNase P x pre-tRNA complex is enhanced by the protein subunit. Biochemistry. 2002;41:9545–9558. doi: 10.1021/bi025553w. [DOI] [PubMed] [Google Scholar]
- 23.Alatossava T, Jutte H, Kuhn A, Kellenberger E. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol. 1985;162:413–419. doi: 10.1128/jb.162.1.413-419.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zarrinkar PP, Wang J, Williamson JR. Slow folding kinetics of RNase P RNA. RNA. 1996;2:564–573. [PMC free article] [PubMed] [Google Scholar]
- 25.Kim KS, Sim S, Ko JH, Lee Y. Processing of M1 RNA at the 3' end protects its primary transcript from degradation. J. Biol. Chem. 2005;280:34667–34674. doi: 10.1074/jbc.M505005200. [DOI] [PubMed] [Google Scholar]
- 26.Loria A, Pan T. The 3' substrate determinants for the catalytic efficiency of the Bacillus subtilis RNase P holoenzyme suggest autolytic processing of the RNase P RNA in vivo. RNA. 2000;6:1413–1422. doi: 10.1017/s1355838200000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundberg U, Altman S. Processing of the precursor to the catalytic RNA subunit of RNase P from Escherichia coli. RNA. 1995;1:327–334. [PMC free article] [PubMed] [Google Scholar]
- 28.Heide C, Feltens R, Hartmann RK. Purine N7 groups that are crucial to the interaction of Escherichia coli RNase P RNA with tRNA. RNA. 2001;7:958–968. doi: 10.1017/s1355838201001753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kufel J, Kirsebom LA. The P15-loop of Escherichia coli RNase P RNA is an autonomous divalent metal ion binding domain. RNA. 1998;4:777–788. doi: 10.1017/s1355838298970923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciesiolka J, Hardt WD, Schlegl J, Erdmann VA, Hartmann RK. Lead-ion induced cleavage of RNase P RNA. Eur. J. Biochem. 1994;219:49–56. doi: 10.1111/j.1432-1033.1994.tb19913.x. [DOI] [PubMed] [Google Scholar]
- 31.Lindell M, Brännvall M, Wagner EG, Kirsebom LA. Lead(II) cleavage analysis of RNase P RNA in vivo. RNA. 2005;11:1348–1354. doi: 10.1261/rna.2590605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zito K, Hüttenhofer A, Pace NR. Lead-catalyzed cleavage of ribonuclease P RNA as a probe for integrity of tertiary structure. Nucleic Acids Res. 1993;21:5916–5920. doi: 10.1093/nar/21.25.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Day-Storms JJ, Niranjanakumari S, Fierke CA. Ionic interactions between PRNA and P protein in Bacillus subtilis RNase P characterized using a magnetocapture-based assay. RNA. 2004;10:1595–1608. doi: 10.1261/rna.7550104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.