Abstract
Motor proteins that couple ATP hydrolysis to movement along nucleic acids play a variety of essential roles in DNA metabolism. Often these enzymes function as components of macromolecular complexes, and DNA translocation by the motor protein drives movement of other components of the complex. In order to understand how the activity of motor proteins is regulated within multi-protein complexes we have studied the bacterial transcription-repair coupling factor, Mfd, which is a helicase superfamily 2 member that binds to RNA polymerase (RNAP) and removes stalled transcription complexes from DNA. Using an oligonucleotide displacement assay that monitors protein movement on double-stranded DNA we show that Mfd has little motor activity in isolation, but exhibits efficient oligonucleotide displacement activity when bound to a stalled transcription complex. Deletion of the C-terminal domain of Mfd increases the ATPase activity of the protein and allows efficient oligo-displacement in the absence of RNAP. Our results suggest that an autoinhibitory domain ensures the motor activity of Mfd is only functional within the correct macromolecular context: recruitment of Mfd to a stalled transcription complex relieves the autoinhibition and unmasks the motor activity.
INTRODUCTION
Helicase proteins are essential components of most of the cellular systems that rely on the manipulation of DNA or RNA, including replication, repair, recombination and transcription. Classically helicases were identified as enzymes that couple the free energy of hydrolysis of ATP to the unwinding of double-stranded nucleic acids by translocating along a single strand in either 5′ to 3′ or 3′ to 5′ direction. Such proteins are characterized by a number of conserved amino acid motifs, and three ‘superfamilies’ and two families of helicase proteins have been defined on the basis of sequence and structural homologies (1). The term ‘helicase’ has broadened in meaning, as many of the helicases identified on the basis of sequence homology turned out to be incapable of separating double-stranded nucleic acids, but instead couple ATP hydrolysis to some other mechanical motion such as movement along double-stranded DNA without strand separation, or remodelling of protein–nucleic acid complexes (2).
The Mfd protein is a helicase superfamily 2 member that functions as a transcription elongation factor in bacteria. It was first characterized on the basis of its role in transcription-coupled DNA repair (3). Bulky DNA lesions such as the photoproducts induced by UV irradiation cannot be transcribed by prokaryotic RNA polymerase (RNAP), and so cause transcription elongation complexes to stall (4–6). The stalled transcription complex shields the lesion from detection by DNA repair complexes, but Mfd (the transcription-repair coupling factor, also referred to as TRCF) removes RNAP from the DNA and recruits nucleotide excision repair proteins to the site of damage (3,7). As a result of this transcription-coupled DNA repair pathway, bulky or non-coding lesions in the transcribed strand of an active gene are repaired ∼10-fold more rapidly than similar lesions in the non-transcribed strand or in untranscribed regions of the genome (8,9). In addition to its role in DNA repair, Mfd also participates in processes that regulate transcription, including catabolite repression in Bacillus subtilis (10) and transcription termination by the bacteriophage HK022 Nun protein (11). In these systems, Mfd displaces transcription complexes that have been stalled by regulatory proteins bound to the DNA template (in the case of catabolite repression) or the nascent RNA (in the case of Nun-mediated termination).
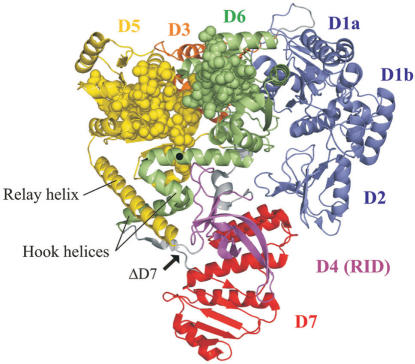
Escherichia coli Mfd is a 130-kDa monomeric protein comprised of eight domains (Figure 1) (3,12). The three most N-terminal domains of the protein (1a, 2 and 1b) show a high degree of structural homology with the UvrB protein, which is a central component of the prokaryotic nucleotide excision repair apparatus (12,13). This region of Mfd interacts with UvrA, the ‘molecular matchmaker’ that is responsible for recruiting UvrB to DNA damage during nucleotide excision repair, and this Mfd–UvrA interaction is thought to be responsible for the enhanced rate of repair that is observed during transcription-coupled repair (14). Domain 4 (the RNAP-interaction domain: RID) interacts with the β subunit of RNAP (12,15). Domains 5 and 6 contain the seven conserved superfamily 2 helicase motifs responsible for the ATPase activity of the protein. These domains are homologous in both sequence and structure to the DNA translocase domains of RecG, a bacterial motor protein involved in the interplay between DNA replication, recombination and repair. In addition to the helicase motifs this region of homology also contains a DNA translocation motif that is unique to RecG and Mfd, termed the TRG motif (for translocation in Rec G) (16,17). Mfd domains 3 and 7 are novel folds with no well-established function. However, it has been proposed that domain 7 may regulate Mfd–UvrA interactions, as in the crystal structure of Mfd it occludes the surface of domain 2 that is thought to interact with UvrA (12).
Figure 1.
Structure of E. coli Mfd. The three most N-terminal domains (domains 1A, 2 and 1B: blue) show a high degree of structural homology with UvrB. Domain 4 (magenta) is the RNA polymerase interaction domain (RID), which interacts with a region of the β subunit of RNAP. Domains 5 and 6 (yellow and green) contain the seven conserved superfamily 2 helicase motifs (shown in space-fill representation), and are structurally homologous to the DNA translocation domains of RecG. Domains 3 (orange) and 7 (red) are novel folds with as yet no clearly defined function. The location of R953 within the TRG motif is indicated by a black circle, and the point of truncation in MfdΔD7 is indicated by an arrow. The figure was produced using Pymol software (Delano Scientific) and is based on pdb file 2EYQ (12).
Mfd dissociates stalled transcription complexes by pushing RNAP forward along the DNA in an ATP-dependent fashion (15,18). This is thought to destabilize the transcription complex by unwinding the DNA–RNA hybrid and rewinding the single-stranded transcription bubble (18). RNAP displacement requires the interaction of Mfd with the DNA immediately upstream of the stalled RNAP (15), and, by analogy to RecG, Mfd is thought to act as an ATP-dependent double-stranded DNA translocase. RecG consists of two translocase domains attached to a ‘wedge’ domain that binds specifically to branched DNA structures (19,20). As the translocase domains hydrolyse ATP they move along double-stranded DNA and pull the DNA across the attached wedge domain, which strips the strands apart (20). Mfd lacks the wedge domain of RecG, but instead binds to the β subunit of RNAP (15,21): once recruited to the stalled transcription complex via the RID–RNAP interaction, it is thought that domains 5 and 6 engage with the upstream DNA and the DNA translocation activity of Mfd pushes RNAP forward.
Previous studies have shown that Mfd has no detectable strand-separating helicase activity on either double-stranded DNA or on DNA–RNA hybrids (3). In this work, we have investigated the motor activity of Mfd using an oligonucleotide displacement assay that is able to detect DNA translocation in the absence of strand separation. We show that a derivative of Mfd that lacks domain 7 (MfdΔD7) is an efficient DNA-based motor that can remove a triplex-forming oligonucleotide (TFO) from double-stranded DNA. This activity is ATP dependent, and requires the TRG DNA translocation motif of the protein. Full-length Mfd shows little TFO displacement activity in isolation, but displaces a TFO efficiently when able to interact with a stalled transcription elongation complex positioned adjacent to the TFO-binding site. Our results indicate that domain 7 of Mfd is a cis-acting regulatory element that inhibits the motor activity of isolated wild-type Mfd, and suggest that interaction of Mfd with stalled transcription elongation complexes removes this inhibitory effect to unmask the motor activity necessary for RNAP displacement.
MATERIALS AND METHODS
Plasmid construction
The Mfd1–997 (ΔD7) expression plasmid, pETMfd1–997 T7, contains a truncated mfd gene under the control of the T7 promoter with a stop codon and a HindIII site immediately downstream of codon 997. It was constructed as follows: the NdeI-HindIII fragment (encoding aa 911–997) from pETMfd21–997 (12) was inserted into pET21a (Novagen) to create an intermediate construct. The mfd gene was amplified by PCR using a primer that introduced an NdeI site upstream of the start codon. pETMfd1–997 T7 was produced by ligating the NdeI-digested PCR fragment (encoding aa 1–910) into the NdeI site of the intermediate construct. A derivative of pETMfd1–997 T7 encoding an RA953 substitution was constructed using the QuikChange site-directed mutagenesis kit (Stratagene). The plasmid encoding the His-tagged RNAP βIA117 KA118 EA119 subunit is a derivative of the wild-type expression plasmid, pETLRpoBHis (21) and was constructed by site-directed mutagenesis.
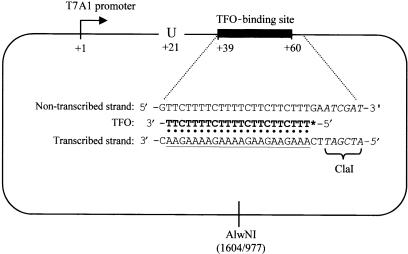
Plasmid pSRT7A1 is a derivative of the in vitro transcription vector pSR (22) carrying residues −91 to +28 of the T7A1 promoter between the EcoRI and HindIII sites. A fragment carrying the T7A1 promoter was generated by PCR, using pAR1707 (23) as a template and primers that placed an EcoRI site upstream of −91 and a HindIII site downstream of +28. Plasmid pSRTB1 is a derivative of pSRT7A1 containing a binding site for a 22-nt TFO located between +39 and +60 downstream of the T7A1 transcription start site (Figure 2). It was constructed by digesting pSRT7A1 with HindIII and inserting a 35-bp double-stranded oligonucleotide that formed a binding site for the TFO. The nucleotide sequence of the TFO used was 5′-TTTCTTCTTCTTTTCTTTTCTT-3′ and the nucleotide sequence to which it bound was 5′-(C)AAAGAAGAAGAAAAGAAAAGAA(C)-3′. In this construct the TFO forms Hoogsteen base pairs with residues +39 to +60 of the transcribed strand, with the 3′ end of the TFO bound at +39.
Figure 2.
Features of the DNA template used in this work. Plasmid pSRTB1 contains a T7A1 promoter (bent arrow) upstream of a TFO-binding site (filled rectangle). RNAP initiating transcription from the T7A1 promoter can be stalled at +20 by omission of UTP from the transcription reactions. The inset shows the sequence of the DNA surrounding the TFO-binding site, with the TFO-binding site underlined. The TFO (bold type) binds in the major groove of DNA by forming Hoogsteen base pairs to a specific sequence within the transcribed strand. In TFO displacement reactions, the 5′ end of the TFO was labelled with 32P (*). The position of the restriction sites used to make linearized template (ClaI and AlwNI) are shown. The ClaI site is adjacent to the TFO-binding site and is shown in italics. The distances from the centre of the AlwNI site to the ends of the TFO-binding site are given in bp.
Proteins
Wild-type and mutant RNAP holoenzyme with a His-tag at the C-terminus of the β subunit were purified as described (21). Full-length Mfd was purified as described (17). MfdΔD7 and MfdΔD7 RA953 were purified from BL21DE3 cells transformed with pETMfd1–997 T7 or its derivative. Cells were grown at 37°C in 1-l of LB medium containing 100 μg/ml ampicillin. Protein expression was induced by the addition of 1 mM IPTG at an OD600 of ∼0.5 and cells were grown at 30°C for a further 3 h before harvesting by centrifugation. The proteins were then purified following the protocol used for full-length Mfd (17).
ATPase assays
ATPase activity was measured using an ATP–NADH-coupled ATPase assay (24). The assays were carried out at 37°C in a 200-μl reaction volume containing 50 nM Mfd, MfdΔD7 or MfdΔD7 RA953. λ DNA was present at a final concentration of 150 μM (moles of base pairs) where indicated (addition of higher concentrations of λ DNA did not increase the ATPase activity of any of the three proteins further: data not shown). Assays were carried out in repair buffer (40 mM HEPES pH 8.0, 100 mM KCl, 8 mM MgCl2, 4% glycerol (v/v), 5 mM DTT and 100 μg/ml BSA) containing 4.4 units pyruvate kinase, 5.7 units lactate dehydrogenase, 500 μM phosphoenolpyruvate and 400 μM NADH. The reactions were started by the addition of ATP at a final concentration of 2 mM and the absorbance at 340 nm was measured every 15 s for 1 h in a VERSAmax 96-well plate reader (Molecular Devices). The accompanying software (SOFTmax Pro) was used to obtain linear fits and the rate of ATP hydrolysis was calculated from the change in absorbance as described (24).
TFO displacement assays
TFO displacement assays were carried out essentially as described (25). Linear templates were generated from pSRTB1 either by digestion with AlwNI (triplex flanked by ∼1000- and 1600-bp dsDNA) or by digestion with ClaI and treatment with Klenow DNA polymerase plus dNTPs to generate blunt ends (triplex flanked by ∼2600- and 6-bp dsDNA). TFO was 5′ end-labelled using T4 polynucleotide kinase and [γ-32P] ATP. Triplex substrates were produced by incubating 25 nM labelled TFO with 50 nM linear or supercoiled pSRTB1 template in buffer MM (10 mM MES pH 5.5, 12.5 mM MgCl2) at 20°C overnight. Immediately prior to use the triplex was diluted 1 in 10 in TR buffer (50 mM Tris-HCl pH 8.0, 10 mM MgCl2, 1 mM DTT) unless otherwise indicated. Displacement reactions thus contained 5 nM template DNA. For standard displacement reactions Mfd proteins were added at 250 nM, ATP was added at 2 mM unless indicated otherwise, and reactions were incubated at 20°C. At appropriate time intervals after the addition of ATP (or after the addition of protein in reactions not started by the addition of nucleotide) reactions were quenched by the addition of 1/4 volume GSMB buffer (15% (w/v) glucose, 3% (w/v) SDS, 250 mM MOPS pH 5.5, 0.4 mg/ml bromophenol blue). Samples were run on 1% (w/v) agarose gels (20 mM Tris-acetate, 5 mM sodium acetate, 5 mM MgCl2, 0.1 mM EDTA, pH 5.5) and analysed using a Molecular Dynamics Typhoon PhosphorImager and ImageQuant software as described (25). Transcription initiation complexes were formed where indicated by incubating the diluted triplex with 20 nM RNAP holoenzyme for 15 min at 20°C prior to the addition of Mfd and 2 mM ATP. Stalled transcription elongation complexes were formed where indicated by incubating the diluted triplex with 20 nM RNAP holoenzyme, 10 μM ATP, 10 μM GTP, 10 μM CTP and 100 μM ApU for 15 min at 20°C prior to the addition of Mfd and 2 mM ATP (when transcription is initiated at the T7A1 promoter under these conditions the transcription elongation complex stalls at +20 because the reaction mixture does not contain the UTP necessary for extension to +21 (26)).
RNAP displacement assays
Displacement of stalled transcription complexes by Mfd and MfdΔD7 was analysed in vitro by electrophoretic mobility shift assays (EMSAs) essentially as described in (21). RNAP was stalled by nucleotide starvation on the 308-bp EcoRI-BamHI fragment of pSRTB1, which contains the T7A1 promoter, transcribed region and transcription terminator. The fragment was 5′ end-labelled using T4 polynucleotide kinase and [γ-32P] ATP. RNAP holoenzyme (10 nM) was incubated in repair buffer (40 mM HEPES, pH 8.0, 100 mM KCl, 8 mM MgCl2, 4% glycerol (v/v), 5 mM DTT, 100 μg/ml BSA) with 0.4 nM end-labelled fragment for 15 min at 37°C, and then heparin (10 μg/ml) was added. ATP (2 mM), CTP (10 μM), GTP (10 μM) and ApU (100 μM) were added and reactions were incubated for a further 15 min at 37°C. Mfd or MfdΔD7 (250 nM) was added and the reactions were incubated at 37°C. At timed intervals aliquots were removed and loaded onto a 4.5% acrylamide/1 × TBE gel. Gels were run at 4°C. Radiolabelled bands were detected and quantified as described above.
RESULTS
Removal of Mfd domain 7 stimulates DNA translocation and ATPase activity of Mfd
Several lines of evidence indicate that Mfd pushes stalled RNAP forward using an ATP-dependent motor activity, and that this forward motion results in removal of RNAP from DNA if transcription cannot reinitiate (15,18). To study the DNA translocation activity of Mfd we used a triplex displacement assay, in which protein movement on DNA is reported by the displacement of a labelled TFO (25). The basis of this assay is that a pyrimidine-rich TFO can bind in a sequence-specific manner to the major groove of a DNA duplex via Hoogsteen base pairing. The TFO requires acidic conditions in order to bind, but once formed the resulting DNA triplex is stable for several hours at physiological pH. If a TFO is displaced from its binding site by the action of an enzyme that moves along DNA it is unable to rebind under non-acidic conditions.
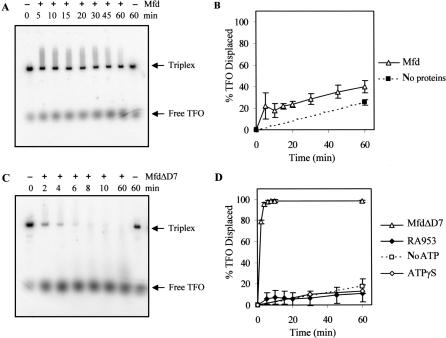
We measured the ability of purified E. coli Mfd to displace a 32P-labelled TFO from a 2603-bp linear template DNA. Binding of the TFO to the template, and its subsequent displacement, was monitored by EMSAs and the proportion of TFO displaced at set time intervals was quantified (Figure 3A and B). Purified Mfd had little effect on the stability of the triplex in these assays: the amount of TFO displaced after 60-min incubation with Mfd in the presence of ATP was only slightly greater than that displaced by spontaneous triplex dissociation in the absence of proteins.
Figure 3.
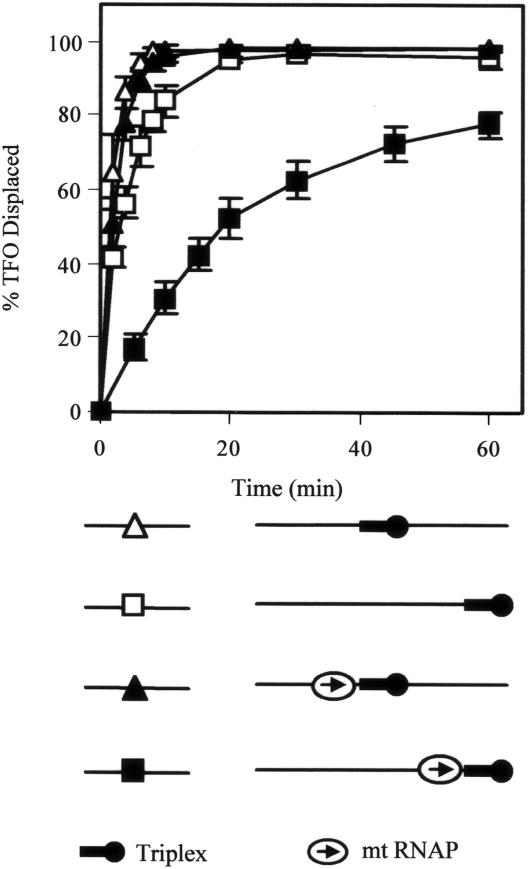
TFO displacement by Mfd and MfdΔD7. 5 nM linear pSRTB1 DNA (linearized with AlwNI) containing a triplex was incubated with 250 nM Mfd or derivatives. Reactions were initiated by the addition of 2 mM ATP or ATPγS where indicated. Aliquots were removed at the indicated time points, quenched and analysed by gel electrophoresis. (A) TFO displacement by wild-type Mfd in the presence of ATP. (B) Quantification of TFO displacement by wild-type Mfd (open triangles) and spontaneous dissociation of TFO (filled squares), both in the presence of ATP. (C) TFO displacement by MfdΔD7 in the presence of ATP. (D) Quantification of TFO displacement by MfdΔD7 in the presence of ATP (open triangles), in the absence of nucleotide (open squares) and in the presence of ATPγS (open diamonds), and TFO displacement by MfdΔD7 RA953 in the presence of ATP (filled diamonds). The graphs show the percentage of TFO displaced at each time point, normalized for the amount of triplex present at t = 0. Data points are the average of at least three independent experiments, with standard deviation.
We also measured the TFO displacement activity of a truncated form of Mfd that lacks the C-terminal domain (D7). D7 is structurally linked to the translocase domains via two helices that form a ‘hook’ around the relay helix joining the RID and domain 5 (Figure 1), and it has been suggested that conformational changes in the translocase domains during ATP hydrolysis may be transmitted via the hook to D7 (12). We have previously purified a derivative of Mfd that lacks this domain (MfdΔD7) and demonstrated that it retains the ability to displace stalled transcription elongation complexes from DNA (12). Like the full-length protein, MfdΔD7 binds DNA stably in the presence of a poorly hydrolysable ATP analogue ATPγS with an apparent KD of less than 50 nM, but does not form a stable complex with DNA in the presence of ATP or in the absence of nucleotide (data not shown). To determine whether removal of D7 affected the DNA translocation activity of Mfd, we performed TFO displacement assays with MfdΔD7 using the same TFO and linear template used in the experiments with the full-length protein. In contrast to the results obtained with the full-length protein, MfdΔD7 was able to rapidly and efficiently displace the TFO from the double-stranded template (Figure 3C and D). Similar results were obtained when the experiments were repeated with the TFO bound to circular supercoiled template DNA, indicating that the TFO displacement activity of MfdΔD7 does not require the presence of free DNA ends and is not significantly affected by template supercoiling (supplementary Figure 1). These results suggest that D7 inhibits the ability of full-length Mfd to catalyse TFO displacement, and deletion of D7 removes this inhibitory effect.
To examine the mechanism by which MfdΔD7 catalyses TFO displacement we determined the nucleotide-dependence of the process, and investigated the effect of disrupting the TRG motif in the translocase domains. We repeated the TFO displacement assays in the absence of ATP, in the presence of the poorly hydrolysable ATP analogue ATPγS, and using an MfdΔD7 derivative containing an RA953 substitution that disrupts the TRG motif (17). In each case, TFO displacement was decreased to background levels (Figure 3D). The requirements for TFO displacement by MfdΔD7 thus mirror known requirements for RNAP displacement by Mfd (and by implication, DNA translocation): the RNAP displacement activity of Mfd is dependent on ATP hydrolysis and requires an intact TRG motif, whereas in the presence of ATPγS, or when the TRG motif has been disrupted, Mfd is able to bind stably to DNA but cannot displace RNAP (14,17).
Deletion of D7 also enhanced the ATPase activity of Mfd. We measured the ATPase activity of MfdΔD7 in the absence of DNA, and found it to be more than 5-fold higher than the activity of full-length wild-type Mfd (Table 1). The ATPase activity of Mfd has previously been found to be unaffected by the addition of DNA (3), in contrast to RecG whose ATPase activity is dependent on the presence of double-stranded DNA (27). We found that the ATPase activity of MfdΔD7 was stimulated more than 2-fold by double-stranded DNA. We also observed a slight DNA-dependent enhancement of ATP hydrolysis by the full-length wild-type Mfd. Finally, we examined the ATPase activity of MfdΔD7 RA953, in which the TRG motif is disrupted. The mutant protein showed the same elevated ATPase activity as MfdΔD7 in the absence of DNA. However, disruption of the TRG motif reduced the DNA-dependent stimulation of ATPase activity.
Table 1.
Rates of ATP hydrolysis by Mfd proteins
| Mfd protein | +/− λ DNA | Rate (min−1) | Av. Stim. |
|---|---|---|---|
| Full length Mfd | − | 17.1 ± 2.0 | 1.4× |
| + | 24.5 ± 1.9 | ||
| MfdΔD7 | − | 111.5 ± 2.6 | 2.1× |
| + | 238.8 ± 14.6 | ||
| MfdΔD7 RA953 | − | 102.0 ± 1.4 | 1.4× |
| + | 139.3 ± 7.2 |
Rates were measured at 37°C using an ATP–NADH-coupled assay. Assays were carried out in repair buffer and contained 50 nM Mfd or its derivatives, 2 mM ATP and 150 μM λ DNA where indicated. Rate values (ATP/min) were calculated from the decrease in absorbance by NADH and the data shown are the average of three independent experiments with standard deviation. Average fold stimulation by DNA (Av. Stim.) is also indicated.
RNAP displacement by MfdΔD7remains dependent on Mfd–RNAP interactions
As MfdΔD7 was able to displace a TFO from DNA we considered the possibility that it may also be able to remove RNAP from DNA without the need for specific protein–protein interactions between Mfd and RNAP. Domain 4 of Mfd (the RID) interacts with a region in the β subunit of RNAP (12,14,15,21). Amino acid substitutions in either Mfd or the β subunit that disrupt this interaction abolish the ability of full-length Mfd to displace RNAP from DNA, presumably because wild-type Mfd must be recruited to the stalled transcription complex in order to exert its effect (12,21). If TFO displacement by MfdΔD7 is the result of the protein acting as a ‘snow plough’ to push aside obstacles during random DNA translocation events, might the same random process be sufficient to displace any transcription complexes encountered?
To answer this question we tested the ability of MfdΔD7 to displace RNAP βIA117 KA118 EA119, an RNAP derivative in which the Mfd contact patch was disrupted. We have shown earlier that alanine substitutions at positions 117–119 of the RNAP β subunit specifically disrupt the Mfd–RNAP interaction and abolish RNAP displacement by wild-type Mfd (12,21). We formed stalled transcription complexes using either wild-type RNAP or RNAP βIA117 KA118 EA119, incubated the complexes with wild-type Mfd or MfdΔD7, and monitored transcription complex dissociation by EMSA as described earlier (21). In order to obtain gels in which the complexes were sufficiently well resolved to allow quantitation of RNAP displacement the reaction conditions used for these experiments varied from those used in the earlier TFO displacement assays. Control experiments showed that MfdΔD7 was able to displace the TFO from DNA under the conditions used for these experiments (Supplementary Figure 2), and that qualitatively similar results were obtained when RNAP displacement assays were performed under the conditions optimized for TFO displacement (data not shown).
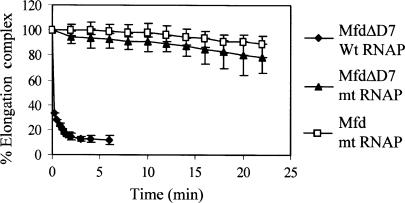
As we have shown earlier (12), MfdΔD7 rapidly displaced stalled wild-type RNAP from DNA (Figure 4). In contrast, RNAP in which the Mfd-binding site in the β subunit was disrupted was displaced very poorly by MfdΔD7, and there was no significant difference between the rate of displacement by MfdΔD7 and wild-type Mfd. These results demonstrate that although removal of D7 from Mfd reveals an activity that allows it to displace a TFO from DNA, the truncated protein remains dependent on an interaction between the Mfd RID and the β subunit of RNAP in order to efficiently displace stalled transcription complexes.
Figure 4.
Displacement of stalled transcription elongation complexes by MfdΔD7 in vitro. Transcription complexes stalled at +20 were formed on an end-labelled fragment of pSRTB1, 250 nM Mfd or MfdΔD7 was added and aliquots were removed and analysed by EMSA. Data points represent the percentage of elongation complex at each time point relative to the amount at t = 0, and are expressed as an average of at least two experiments (shown with data range). The graph shows displacement of wild-type RNAP by MfdΔD7 (filled diamonds) and RNAP βIA117 KA118 EA119 (mt RNAP) by MfdΔD7 (filled triangles) and Mfd (open squares).
DNA flanking the triplex is required for efficient TFO displacement by MfdΔD7
If TFO displacement by MfdΔD7 is the result of translocation of the protein along double-stranded DNA, the process should be dependent on the presence of DNA flanking the triplex. The minimum requirement for the Mfd translocation domains to bind DNA is likely to be ∼26 bp, as this is the amount of DNA that must be accessible upstream of a stalled RNAP in order for it to be displaced by Mfd (15). To determine whether MfdΔD7 requires DNA flanking the TFO-binding site in order to displace the TFO, we initially examined the ability of MfdΔD7 to disrupt triplexes formed by binding a TFO to short double-stranded oligonucleotides of various lengths. However, the triplexes formed using these short substrates proved to be too unstable for unambiguous conclusions to be drawn (data not shown). We therefore took an alternative approach and examined TFO displacement by MfdΔD7 using a substrate in which the DNA on one side of the TFO-binding site was removed and the DNA on the other side of the TFO-binding site was blocked by a protein roadblock.
We digested plasmid pSRTB1 with ClaI and used Klenow DNA polymerase and dNTPs to generate a blunt-ended 2603-bp linear template in which the 5′ end of the bound TFO was located 6 bp from one end. To create a stable protein ‘roadblock’ on the other side of the triplex we used RNAP βIA117 KA118 EA119 to form nucleotide-starved stalled transcription elongation complexes immediately adjacent to the TFO-binding site (as shown in Figure 4, transcription complexes formed with RNAP βIA117 KA118 EA119 are stable, and resistant to displacement by MfdΔD7). Plasmid pSRTB1 contains the T7A1 promoter 39-bp upstream of the 3′ end of the bound TFO, and in these experiments transcription complexes that initiated from this promoter were stalled at +20 because the reaction mixture did not contain the UTP necessary for extension to +21. The ‘footprint’ of transcription elongation complexes in chemical and DNase I protection experiments extends ∼15 bp ahead of the last transcribed base (28), suggesting that the downstream edge of the stalled RNAP and the upstream edge of the DNA triplex would be separated by ∼3 bp on this template.
Removal or blocking of DNA on just one side of the triplex had little effect on the rate of TFO displacement by MfdΔD7 (Figure 5). This is not surprising, as although Mfd would be expected to translocate on DNA with a fixed (though as yet undefined) polarity, both the location and orientation of DNA binding by MfdΔD7 are likely to be random. Translocation events initiating on either side of the triplex could therefore lead to its disruption. When the flanking DNA was simultaneously removed on one side and blocked on the other, however, the rate of TFO displacement by MfdΔD7 was significantly reduced. These results indicate that MfdΔD7 requires a region of flanking DNA on at least one side of the triplex in order to displace the TFO, and support a model in which MfdΔD7 displaces the TFO by translocation along DNA until the triplex is reached.
Figure 5.
The effect of flanking DNA on TFO displacement by MfdΔD7. 5 nM linear DNA fragment containing a triplex was incubated with or without 20 nM RNAP βIA117 KA118 EA119 (mt RNAP) and nucleotides (10 μM ATP, 10 μM GTP, 10 μM CTP and 100 μM ApU) for 15 min at 20°C. Reactions were initiated by the addition of 250 nM MfdΔD7 and 2 mM ATP. Aliquots were removed and the reaction quenched at the indicated time points. TFO displacement was analysed on a DNA fragment with a triplex in its middle (pSRTB1 DNA linearized with AlwNI: triangles) and a linear fragment with a triplex at its end (pSRTB1 DNA linearized with ClaI: squares). In the schematic of the substrates the 5′ end of the TFO is indicated by a black circle, and the direction in which RNAP was transcribed prior to stalling is indicated by an arrow. The graph shows the percentage of TFO displaced at each time point, normalized for the amount of triplex present at t = 0. Filled symbols indicate reactions in which elongation complexes were stalled adjacent to the triplex. Open symbols indicate reactions in which no stalled elongation complexes were present. The data are the average of at least three independent experiments, with standard deviation.
DNA translocation by wild-type Mfd is stimulated by interaction with transcription elongation complexes
The fact that MfdΔD7 is able to efficiently displace a TFO from DNA suggests that the full-length protein also has the potential to do so, but that this activity is inhibited by the presence of D7. As there is substantial evidence that Mfd is able to catalyse movement of RNAP along DNA when the two proteins interact, it is possible that the motor activity of full-length Mfd is only revealed once it binds to RNAP. To test this hypothesis we measured the ability of full-length Mfd to displace a TFO from a substrate carrying transcription complexes adjacent to the TFO-binding site.
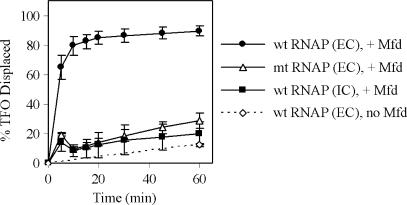
As explained above, the templates used in our experiments allow transcription complexes to be formed adjacent to the triplex. In the absence of nucleotides, RNAP will form a transcription initiation complex at the T7A1 promoter, located 39-bp upstream of the 3′ end of the TFO bound to the transcribed strand. Addition of selected nucleotides allows elongation complexes to be stalled by nucleotide starvation with the leading edge of the stalled RNAP located immediately upstream of the 3′ end of the bound TFO. Mfd can push backtracked RNAP ‘forward’ (i.e. in the direction of transcription) (15), which in our system would result in RNAP being pushed towards the TFO. To determine the effect of RNAP on the DNA translocation activity of Mfd we measured the ability of full-length Mfd to displace a TFO from a linear template on which transcription initiation complexes or stalled transcription elongation complexes had been formed using wild-type RNAP (Figure 6). When Mfd was added to reactions containing stalled transcription elongation complexes the TFO was rapidly and efficiently displaced from the DNA template. The presence of a stalled elongation complex adjacent to the TFO did not destabilize the triplex in the absence of Mfd, and no stimulation of TFO displacement activity was observed when the experiment was performed under conditions that allowed RNAP to form initiation complexes but not elongation complexes. The simplest explanation for these observations is that when Mfd is recruited to the stalled transcription elongation complex its DNA translocation activity results in displacement of the TFO. The TFO might be displaced by the leading edge of RNAP as it is pushed forward by Mfd, or Mfd may continue to translocate along DNA after RNAP has been displaced and displace the TFO directly.
Figure 6.
The effect of stalled transcription elongation complexes on TFO displacement by Mfd. 5 nM linear pSRTB1 DNA (linearized with AlwNI) containing a triplex was incubated for 15 min at 20°C with 20 nM RNAP (IC), or with 20 nM RNAP plus nucleotides (10 μM ATP, 10 μM GTP, 10 μM CTP and 100 μM ApU) (EC) to form transcription initiation or elongation complexes upstream of the triplex. The displacement reaction was initiated by the addition of 250 nM Mfd and 2 mM ATP, aliquots were removed and the reaction quenched at the indicated time points. The graph shows the percentage of TFO displaced at each time point, normalized for the amount of triplex present at t = 0. The data are the average of at least three independent experiments, with standard deviation. The graph shows displacement of the TFO by wild-type Mfd in the presence of a stalled wild-type elongation complex (filled circles), in the presence of stalled mutant (mt) RNAP βIA117 KA118 EA119 elongation complex (open triangles) or in the presence of a wild-type transcription initiation complex (filled squares). The rate of TFO displacement by the stalled wild-type elongation complex in the absence of Mfd is also shown (open diamonds).
To determine whether the effect of stalled elongation complexes on Mfd required direct protein–protein contact between the two, or was the result of a particular DNA conformation arising from the transcription complex, we repeated the experiments using RNAP βIA117 KA118 EA119. Stalled transcription complexes formed using this RNAP derivative did not stimulate the TFO displacement activity of Mfd, indicating that the contact between the Mfd RID and the β subunit of RNAP is required for this effect.
These experiments show that incorporation of full-length Mfd into an RNAP–DNA–Mfd complex has the same effect on its TFO displacement activity as deletion of D7. This suggests that interaction of Mfd with a stalled transcription elongation complex overcomes the autoinhibitory effect of D7.
DISCUSSION
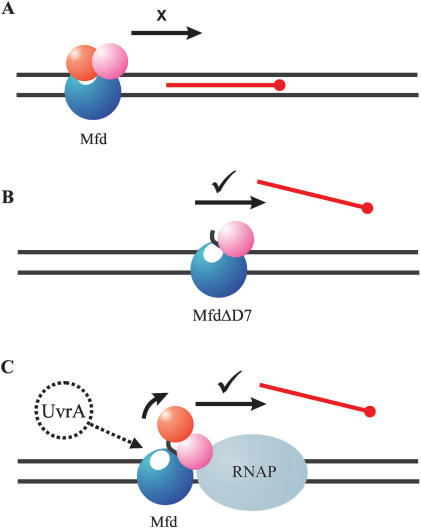
Like many members of the helicase superfamilies Mfd does not function in isolation, but acts in concert with other proteins as part of a macromolecular complex. During transcription-coupled repair its DNA translocation activity forms just one link in a chain of events that leads to RNAP displacement and the recruitment of DNA repair enzymes. Successful completion of the pathway is likely to require coordinated control of the various steps. In this work, we have found that DNA translocation activity of Mfd is regulated by the action of an autoinhibitory domain. In the isolated protein domain D7 inhibits the translocation activity. This inhibitory effect is relieved when the Mfd protein interacts with a stalled RNAP to form the initial complex on the transcription-coupled DNA repair pathway, suggesting that the interaction with RNAP results in repositioning of the autoinhibitory domain (Figure 7).
Figure 7.
Model for the control of Mfd activity by autoinhibitory domain D7. (A) In wild-type Mfd, the translocation activity is repressed by the action of D7 (red). (B) The autoinhibitory effect can be relieved by deletion of D7, and the truncated protein can displace a TFO in the absence of other factors. (C) The autoinhibitory effect is also relieved by the interaction of wild-type Mfd with RNAP. Mfd binds to RNAP via the RID (magenta), and D7 is repositioned. The repositioning of D7 activates the DNA translocation activity of Mfd, and may also reveal a binding site for UvrA (white ellipse: (12)). TFO displacement may result either from RNAP being pushed through the TFO-binding site (as shown), or by Mfd continuing to translocate DNA after displacing RNAP.
How might D7 inhibit DNA translocation activity? Deletion of D7 increased the ATPase activity of the protein even in the absence of DNA. This indicates that the inhibitory effect of the domain occurs at least in part by preventing nucleotide binding, hydrolysis and/or release in a manner that is independent of the interaction of Mfd with its DNA substrate. In the crystal structure of Mfd, D7 is not close to the predicted path of DNA across the protein and does not interact directly with domains D5 and D6, which contain the motifs important for ATP hydrolysis and DNA translocation (although it is important to note that the crystal structure was obtained in the absence of nucleotides, and significant structural rearrangements are predicted to occur on ATP binding). However, D7 is structurally linked to the translocase domains via the hook helices, which lead on from the TRG motif and interact with the relay helix linking the translocase domains to the RID (Figure 1). It is therefore feasible that deletion or movement of D7 could affect ATPase and translocation activity via transmission of conformational changes through the structure. In any such allosteric effects of D7 the TRG motif is likely to play an important role.
The TRG motif contains a pair of helices that interact with helicase motif VI, providing a link to the ATPase catalytic site (12,16). In the RecG–ADP–DNA structure, these helices form a hairpin with two closely juxtaposed arginine residues at the base, whereas in the Mfd structure, which contains no nucleotide, these helices have moved apart (12,16). It seems that this structure forms a ‘spring-loaded’ switch that changes conformation during the nucleotide hydrolysis cycle: nucleotide binding stabilizes the closed conformation, and charge repulsion between the two arginines favours the open conformation. It has previously been proposed that opening and closing of the TRG helical hairpin may be transmitted to the RID and D7 via the hook helices, and so the state of the TRG motif could control the position of these domains (12). Our findings suggest the relationship may be reciprocal, and D7 may regulate ATPase and DNA translocation activities by controlling the position of the hook helices and TRG motif. In the isolated full-length protein, D7 may occupy a conformation that constrains the opening and closing of the TRG motif and hence inhibits ATPase activity and DNA translocation; if D7 is deleted this constraint is removed. However, the TRG motif does not function solely as a relay in the autoinhibitory mechanism as it is clear that the TRG motif is essential for Mfd function even in the absence of D7. The RA953 substitution, which removes one of the two key arginines within the TRG motif, abolishes the DNA translocation activity of MfdΔD7. Furthermore, although disruption of the TRG motif has no effect on the ATPase activity of MfdΔD7 in the absence of DNA it reduces the ability of DNA to stimulate the ATPase activity.
How might interaction with RNAP relieve the inhibitory effect of D7? The simplest explanation is that interaction between Mfd and RNAP causes D7 to be repositioned. In the Mfd crystal structure, D7 packs between D2 and D4 (the RID). As discussed above the juxtaposition of D4, D7 and the TRG motif is also potentially linked by changes in the interaction between the hook and relay helices. Interaction of RNAP with the RID may therefore directly affect the position of D7. Alternatively, D7 may make a separate interaction with RNAP that results in its repositioning. If this is the case this secondary interaction is presumably weaker than the characterized RID–β subunit interaction, as a C-terminal fragment of Mfd containing D7 did not bind to RNAP in pull-down assays that detected the interaction made by fragments that included the RID (14).
The RID contact site lies within the mobile β1 domain of the RNAP β subunit, movement of which is one of the conformational changes associated with the opening and closing of the main channel of RNAP (29). In this work, we found that the interaction between Mfd and the β subunit of RNAP was essential for RNAP displacement by MfdΔD7, despite the fact that the truncated protein was able to rapidly displace a TFO from DNA. This result shows that DNA translocation activity alone is not sufficient to enable MfdΔD7 to displace a stalled transcription complex from DNA: it must also bind to RNAP. Why must the MfdΔD7 motor be tethered to RNAP in order to displace it? We have suggested previously that translocation by Mfd tethered to β1 might drive a remodelling process that opens the RNAP main channel (21), in addition to rewinding the transcription bubble (18), and this would be consistent with the findings reported here. An alternative possibility is that the Mfd–RNAP interaction may allow RNAP to act as a ‘processivity factor’ for Mfd, ensuring that Mfd continues to push against the transcription complex until it is displaced. Stalled transcription complexes are stabilized by multiple interactions between proteins, RNA and DNA, and are likely to present more of an obstacle to a translocating motor protein than a triplex does. We do not know how many ATPase cycles are required for Mfd to displace a transcription complex, and unlike a TFO the transcription complex is dynamic and may slide backwards to its starting position if Mfd dissociates before displacement is complete (30,31). When associated with a transcription complex Mfd is tethered to the DNA both directly, via its translocase domains, and indirectly, via the contact of the RID with the transcription complex. This means that translocation will not necessarily terminate if the translocase domains transiently dissociate from the DNA (in contrast to the situation that would arise with free MfdΔD7). In this context, the transcription complex may therefore play a similar role to the wedge domain of RecG, which increases the processivity of that motor by acting as a sliding clamp on the DNA (19).
Autoinhibition and context-specific activation may prove to be a widespread strategy for the control of helicase proteins that function within macromolecular assemblies. It is important that the activity of such enzymes is strictly regulated, as their activities are costly to the cell in terms of ATP hydrolysis and likely to be deleterious if performed inappropriately. The helicase activity of the E. coli Rep protein (a superfamily 1 helicase that functions in DNA replication), and the ATPase and DNA-binding activities of UvrB (a superfamily 2 helicase that detects DNA damage in complex with UvrA) have both been shown recently to be regulated by autoinhibitory domains (32,33). Rep autoinhibition is thought to be relieved by dimerization, and it has been suggested that autoinhibition of UvrB may be relieved by interaction with UvrA. The regulatory mechanism uncovered in our work ensures that DNA translocation by Mfd occurs only in the correct context of an Mfd–RNAP complex. The fact that the control process involves domain D7, which is also implicated in regulating the interaction of Mfd with the repair protein UvrA, suggests that the transcription-coupled repair process as a whole is highly coordinated.
SUPPLEMENTARY DATA
Supplementary Data available at NAR Online.
ACKNOWLEDGEMENTS
This work was funded by research grants from the BBSRC (BB/C507053) to N.J.S. and Wellcome Trust (067439) to M.D.S. M.D.S. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Sciences. Funding to pay the Open Access publication charge was provided by a Wellcome Trust “Value in People” grant to the University of Bristol, reference 081949.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gorbalenya AE, Koonin EV. Helicases – amino-acid-sequence comparisons and structure-function-relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 2.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 3.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 4.Tornaletti S, Hanawalt PC. Effect of DNA lesions on transcription elongation. Biochimie. 1999;81:139–146. doi: 10.1016/s0300-9084(99)80046-7. [DOI] [PubMed] [Google Scholar]
- 5.Scicchitano DA, Olesnicky EC, Dimitri A. Transcription and DNA adducts: what happens when the message gets cut off? DNA Repair. 2004;3:1537–1548. doi: 10.1016/j.dnarep.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: transcription-coupled repair or transcriptional mutagenesis? Chem. Rev. 2006;106:474–488. doi: 10.1021/cr040466q. [DOI] [PubMed] [Google Scholar]
- 7.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J. Biol. Chem. 1990;265:21330–21336. [PubMed] [Google Scholar]
- 8.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 9.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 10.Zalieckas JM, Wray LV, ,Jr, Ferson AE, Fisher SH. Transcription-repair coupling factor is involved in carbon catabolite repression of the Bacillus subtilis hut and gnt operons. Mol. Microbiol. 1998;27:1031–1038. doi: 10.1046/j.1365-2958.1998.00751.x. [DOI] [PubMed] [Google Scholar]
- 11.Washburn RS, Wang Y, Gottesman ME. Role of E. coli transcription-repair coupling factor Mfd in Nun-mediated transcription termination. J. Mol. Biol. 2003;329:655–662. doi: 10.1016/s0022-2836(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 12.Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 13.Assenmacher N, Wenig K, Lammens A, Hopfner KP. Structural basis for transcription-coupled repair: the N terminus of Mfd resembles UvrB with degenerate ATPase motifs. J. Mol. Biol. 2006;355:675–683. doi: 10.1016/j.jmb.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 14.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J. Biol. Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 15.Park JS, Marr MT, Roberts JW. E. coli transcription repair coupling factor (Mfd Protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 16.Mahdi AA, Briggs GS, Sharples GJ, Wen Q, Lloyd RG. A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J. 2003;22:724–734. doi: 10.1093/emboj/cdg043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chambers AL, Smith AJ, Savery NJ. A DNA translocation motif in the bacterial transcription-repair coupling factor, Mfd. Nucleic Acids Res. 2003;31:6409–6418. doi: 10.1093/nar/gkg868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park JS, Roberts JW. Role of DNA bubble rewinding in enzymatic transcription termination. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briggs GS, Mahdi AA, Wen Q, Lloyd RG. DNA binding by the substrate specificity (wedge) domain of RecG helicase suggests a role in processivity. J. Biol. Chem. 2005;280:13921–13927. doi: 10.1074/jbc.M412054200. [DOI] [PubMed] [Google Scholar]
- 20.Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 21.Smith AJ, Savery NJ. RNA polymerase mutants defective in the initiation of transcription-coupled DNA repair. Nucleic Acids Res. 2005;33:755–764. doi: 10.1093/nar/gki225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolb A, Kotlarz D, Kusano S, Ishihama A. Selectivity of the Escherichia coli RNA polymerase E sigma 38 for overlapping promoters and ability to support CRP activation. Nucleic Acids Res. 1995;23:819–826. doi: 10.1093/nar/23.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin JR, Krummel B, Chamberlin MJ. Isolation and properties of transcribing ternary complexes of Escherichia coli RNA polymerase positioned at a single template base. J. Mol. Biol. 1987;196:85–100. doi: 10.1016/0022-2836(87)90512-2. [DOI] [PubMed] [Google Scholar]
- 24.Kiianitsa K, Solinger JA, Heyer WD. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal. Biochem. 2003;321:266–271. doi: 10.1016/s0003-2697(03)00461-5. [DOI] [PubMed] [Google Scholar]
- 25.Firman K, Szczelkun MD. Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krummel B, Chamberlin MJ. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Individual complexes halted along different transcription units have distinct and unexpected biochemical properties. J. Mol. Biol. 1992;225:221–237. doi: 10.1016/0022-2836(92)90917-9. [DOI] [PubMed] [Google Scholar]
- 27.Mahdi AA, McGlynn P, Levett SD, Lloyd RG. DNA binding and helicase domains of the Escherichia coli recombination protein RecG. Nucleic Acids Res. 1997;25:3875–3880. doi: 10.1093/nar/25.19.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darst SA. Bacterial RNA polymerase. Curr. Opin. Struct. Biol. 2001;11:155–162. doi: 10.1016/s0959-440x(00)00185-8. [DOI] [PubMed] [Google Scholar]
- 29.Murakami KS, Darst SA. Bacterial RNA polymerases: the whole story. Curr. Opin. Struct. Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 30.Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 31.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 32.Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10076–10081. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, DellaVecchia MJ, Skorvaga M, Croteau DL, Erie DA, Van Houten B. UvrB Domain 4, an autoinhibitory gate for regulation of DNA binding and ATPase activity. J. Biol. Chem. 2006;281:15227–15237. doi: 10.1074/jbc.M601476200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.