Abstract
U12-type introns exist, albeit rarely, in a variety of multicellular organisms. Splicing of U12 intron-containing precursor mRNAs takes place in the U12-type spliceosome that is distinct from the major U2-type spliceosome. Due to incompatibility of these two spliceosomes, alternative splicing involving a U12-type intron may give rise to a relatively complicated impact on gene expression. We studied alternative U12-type intron splicing in an attempt to gain more mechanistic insights. First, we characterized mutually exclusive exon selection of the human JNK2 gene, which involves an unusual intron possessing the U12-type 5′ splice site and the U2-type 3′ splice site. We demonstrated that the long and evolutionary conserved polypyrimidine tract of this hybrid intron provides important signals for inclusion of its downstream alternative exon. In addition, we examined the effects of single nucleotide polymorphisms in the human WDFY1 U12-type intron on pre-mRNA splicing. These results provide mechanistic implications on splice-site selection of U12-type intron splicing. We finally discuss the potential effects of splicing of a U12-type intron with genetic defects or within a set of genes encoding RNA processing factors on global gene expression.
INTRODUCTION
In higher eukaryotes, the majority of genes are interrupted by multiple introns that are excised from precursor mRNA (pre-mRNA) during gene expression. Two distinct types of introns, namely U2 and U12, are found in the genomes of multicellular organisms [reviewed in (1,2)]. U2-type introns predominate, whereas U12-type introns occur with a much lower frequency and are absent in some species such as Caenorhabditis elegans (3). The two intron types are distinguishable by their splice site and branch-site sequences. Almost all pre-mRNA introns have GT and AG dinucleotides at their 5′ and 3′ boundaries, respectively, except for a subgroup with the AT–AC terminal residues (3). The U2- and U12-type introns are spliced by their respective spliceosomes. The two types of the spliceosome share one small nuclear ribonucleoprotein (snRNP), U5, but each has four other specific snRNPs: U1, U2 and U4/U6 in the U2-type spliceosome, and their low-abundance functional analogs, namely U11, U12 and U4atac/U6atac in the U12-type spliceosome [reviewed in (1,2)]. Each snRNA differs from its analogs in the primary sequence but they share a remarkable similarity in the secondary structure (1). Moreover, the two spliceosomes contain a large common set of protein components, and the intricate network of the RNA–RNA interactions is strikingly similar in each spliceosome [reviewed in (1,2,4,5)]. Nevertheless, the individual spliceosomes can only catalyze the removal of their cognate introns.
The U2- and U12-dependent splicing systems might have evolved independently in separate lineages and then merged in a eukaryote progenitor upon lineage fusion (6). These two splicing machineries may have also converged evolutionarily to share common protein factors (4). Another model suggests that the two types of spliceosomal introns may have arisen from two different self-splicing group II introns [reviewed in (7)]. Nevertheless, the scarcity of the U12-type introns in modern organisms may result from their less accurate and slower splicing as compared to the U2-type introns [reviewed in (5)]. Thus, the U12-type introns might have a tendency to convert their sequence to loosely defined U2-type splice site/branch sites via mutational changes during evolution (6).
Phylogenetic analysis reveals that U12-type introns are present in homologous genes encoded by different species that have diverged over 600 million years ago [reviewed in (5)]. The presence of homologous introns in gene family members is largely due to gene amplification throughout evolution (6,8). Interestingly, certain U12-type introns prevail in sets of genes involved in specific cellular processes, e.g. the Ras–Raf signaling pathway (8). Perhaps the splicing of these U12-type introns could regulate the expression of their host genes [reviewed in (5)]. Moreover, the persistence of U12-type introns throughout evolution highlights their indispensable roles in cellular functions [reviewed in (5)].
Here, a bioinformatics scan has identified a dozen new cases of alternative splicing involving U12-type introns, although some might result from aberrant use of splice sites (8). Experimental results indicate that mutations at either terminal nucleotide of U12-type introns could activate alternative splicing via cryptic splice-site utilization (9,10). Preferential activation of cryptic 3′ splice sites (hereafter abbreviated to SS) suggests that the U12-type spliceosome has lower stringency in recognition of the 3′SS, as compared to the 5′SS (9–11). Notably, a mutation in the U12-type intron 5′SS of the tumor suppressor gene LKB1/Serine/threonine–protein kinase 11 (STK11) is associated with the autosomal dominant disorder Peutz–Jeghers syndrome (PJS), underscoring the relevance of nucleotide polymorphisms of U12-type introns in human diseases involving alternative splicing (10). An intriguing case is identified in c-Jun N-terminal kinase (JNK)/SAPK genes, in which the intron between two alternative exons contains the U12-type 5′SS and the U2-type branch site and 3′SS (12–14). It is predicted that this hybrid intron could drive mutually exclusive selection of its flanking exons (13,14). However, the detailed splicing mechanism has not yet been deciphered.
This study was aimed at understanding the mechanisms of alternative splicing of U12-type introns. We experimentally examined the role of cis-elements within the U2–U12 hybrid intron of human JNK2 (c-Jun N-terminal kinase 2) on alternative exon selection and the effect of U12-type intron polymorphisms on splicing of the human WDFY1 pre-mRNA. JNKs function as important cellular signaling kinases in a variety of physiological processes [(15) and reference]. WDFY1 containing WD40 repeats and a phosphatidylinositol 3-phosphate (PI3)-binding FYVE-type zinc finger (16,17) may act as a signaling factor in the PI3 kinase signaling pathways. Therefore, it is interesting to understand whether U12-type intron splicing would impact on their gene expression. Finally, we discuss the presence of U12-type introns in genes encoding pre-mRNA splicing factors and its possible impact on gene expression.
MATERIALS AND METHODS
Computational analysis of U12-type introns
To expand the dataset of human U12-type introns provided by Levine and Durbin (8), we performed a text search based largely on their results. Both 5′ and 3′ splice-site sequences of 404 U12-type intron entries were extracted from the Supplementary Table 1 of Levine and Durbin (8) and subjected to a search in NCBI (http://www.ncbi.nlm.nih.gov/BLAST/) and UCSC (http://genome.ucsc.edu/cgi-bin/hgBlat) for additional information including gene structures and splice-site sequences. We manually annotated each putative U12-type intron and confirmed ∼390 members. Next, we searched for new U12-type introns primarily within their host gene families using the NCBI Gene and Ensembl Gene (http://www.ensembl.org/Homosapiens/index.html/) databases. This search identified ∼110 previously undocumented human U12-type introns, yielding a collection of ∼500 members. Note that a comprehensive splice-site analysis was recently performed in five species and identified 671 U12-type introns in the human genome (18).
Table 1.
U12-type intron-containing genes involved in RNA metabolism
| Gene symbol | Note |
|---|---|
| MORC3 | RNA-binding protein, nuclear matrix-binding (NXP2) |
| NCBP1 | Cap-binding protein |
| NCBP2 | Cap-binding protein |
| CRNKL1 | Splicing factor |
| FUSIP | Splicing factor SRp38 |
| HNRPL | hnRNP |
| HNRPLL | hnRNP |
| HNRPM | hnRNP |
| SNRPE | SmE protein |
| LSM5 | Sm-like protein |
| LSM8 | Sm-like protein |
| LSM12 | Sm-like protein |
| ZC3H8 | Potential role in cleavage and polyadenylation |
| PAN3 | Poly(A)-specific ribonuclease subunit |
| RNPC3 | U11/U12 snRNP 65kDa protein |
| THOC2 | TREX (transcription/export) component |
| EIF3S2 | Translation initiation factor 3 subunit |
| EIF3S12 | Translation initiation factor 3 subunit |
| DCP2 | Pre-mRNA decapping enzyme |
| EXO1 | 5′ to 3′ exonuclease (RNase H) |
| EXOSC1 | Exosome component CSL4 |
| EXOSC2 | Exosome component RRP4 |
| EXOSC5 | Exosome component RRP46 |
| UPF1 | NMD factor |
| MARCH6 | Potential role in mRNA turnover |
| DDX54 | RNA helicase |
| DEADC1 | Deaminase |
| GARS | tRNA synthetase, class II |
| HARS | tRNA synthetase, class II |
| HARSL | tRNA synthetase, class II |
| BXDC5 | Ribosome maturation factor RPF1 |
| NOL1 | Nucleolar protein |
| NOL8 | Nucleolar protein |
| NOL11 | Nucleolar protein |
| NOLA1 | Nucleolar protein |
| RCL1 | RNA 3′-phosphate cyclase |
We obtained the sequences of human JNK2 and its orthologs from Ensembl (GeneView), and annotated the hybrid intron and its flanking exons by using ExonView and GeneSeqView (Ensembl). The JNK family genes of other species were obtained from the orthologs hyperlinks, and their exon/intron boundaries were confirmed by using the UCSC Genome Bioinformatics website. Phylogenetic conservation of the hybrid introns was analyzed by using the AlignX program of VectorNTI (Invitrogen) with a display window size 23. Moreover, to search for single nucleotide polymorphisms (SNPs) of the U12-type introns, 50 U12-type intron-containing genes were analyzed by using SNPper (http://snpper.chip.org/) and Ensemble GeneSeqView.
Plasmid construction
The JNK2 minigene reporter was constructed as follows. Four PCR fragments of the human JNK2 gene were amplified from HEK 293-cell genomic DNA with restriction site-tagged primers. Three fragments contained either a single exon (5, 7 and 8) or two alternative exons (6a and 6b) with the hybrid intron in the middle. Each fragment also extended upstream and downstream from the intronic sequences by ∼120–200 bp. The PCR fragments were ligated and cloned into pcDNA3.1+ (Invitrogen), and the DNA sequence of the resulting minigene was confirmed by sequencing. To swap the two alternative exons with their respective flanking intron sequences (Swap-E + I), the PCR fragment containing exon 6b with a part of the introns was ligated upstream to the fragment containing exon 6a. To swap the exon region only, we designed two 108-bp primers, forward exon 6a and reverse exon 6b. These two primers covered the entire exon 6a and 6b sequences, respectively, and contained the mutations at their 5′ end to create an Nco II site for cloning. PCR was performed with these two primers using the JNK2 minigene as the template. The product was used as a vector backbone to accommodate the hybrid intron through the engineered Nco II sites; the resulting minigene was termed Swap-Eonly. A PCR-based method (QuikChange; Stratagene) was used to generate polypyrimidine (Py)-truncated reporters; the resulting ΔPy1, ΔPy2 and ΔPy reporters lacked nucleotides 46–62, 9–40 and 9–62, upstream of exon 6b, respectively. The AdPy reporter was obtained by insertion of the Py tract sequence (5′-tcatacttatcctgtcccttttttttcc) derived from the first intron of the adenovirus major late gene into the ΔPy reporter.
To construct the WDFY1 minigene reporter, HeLa cell genomic DNA was used as template for PCR amplification of four separate fragments of WDFY1; each fragment encompassed a single exon (4 to 7) with its flanking intron sequence of ∼200 bp. The PCR products were appropriately ligated and cloned into vector pcDNA3.1+, and their DNA sequence was confirmed. Exogenous U12 snRNA was expressed from the pU12-DIR2 vector under the control of the native promoter and terminator of the U12 gene (19); expression level of the exogenous U12 snRNA was ∼4-fold higher than that of the endogenous one (19). Site-directed mutagenesis (QuikChange) was performed to introduce the desired mutations into the WDFY1 minigenes and pU12-DIR2. The mutant WDFY1 minigenes used in this study were 5′ + 6CU, B-4CU, B-4CA, B-4CG and 5′Bdm (see Results section for details). The U12 snRNA mutant contained changes at nucleotides 5 (C to A) and 22 (G to U).
In vivo splicing assay
HeLa, HEK 293 and N2A cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin/glutamine (Invitrogen). P19 cells were maintained in Alpha medium (Invitrogene) containing 7.5% bovine calf serum and 2.5% FBS. For the in vivo splicing assay, ∼6 × 105 HEK 293 cells were transiently transfected with 1.6 μg of reported minigene using Lipofectamine 2000 (Invitrogen) for 24 h. To examine the compensatory effect of exogenous U12 snRNA, 1 µg of the reporter and 3 µg of the U12 snRNA expression vector were co-transfected. To analyze the splicing products, total RNA was extracted using the TRIzol reagent (Invitrogen) and converted to first-strand cDNA using SuperScript III reverse transcriptase with either oligo dT (12–18 mers; Invitrogen) or minigene-specific BGH (5′-tagaaggcacagtcgagg; Invitrogen) as primer. To detect WDFY1 transcripts, RT was performed using the BGH primer followed by PCR amplification with primers 5′-gctagcatggaatttcacgtttctg (exon 4; forward) and 5′-ctcgagtcaatgatggccctgaag (exon 7; reverse). The reverse primer was 5′ end-labeled with 32P using T4-polynucleotide kinase (New England BioLabs) and 10 000 cpm (∼108 cpm per µg) of labeled primer was used for each PCR reaction. The PCR products were analyzed by electrophoresis on a denaturing 6% polyacrylamide gel. To detect JNK2 minigene transcripts, PCR was performed using forward exon 5 primer (5′-gatttgaagcctagcaacattg) and reverse BHG primer, and the products were digested with EcoRV before electrophoresis on 2% agarose gels. Endogenous JNK2 transcripts were examined analogously except that exon 7 primer (5′-cagttggctgaagtttcttc) instead of BHG was used for PCR.
RESULTS
A previous report identified ∼400 U12-type introns within the human genome (8). Since database entries continuously increase, we attempted to search for additional human U12-type introns. By using a less comprehensive text search (see Materials and Methods section), our collection of human U12-type introns was tentatively expanded to ∼500. The new additions were primarily the members of the known U12-type intron-containing gene families. In this study, we took advantage of this pool of U12-type introns to search for alternative splicing and nucleotide polymorphisms of U12-type introns. We also categorized U12-type intron host genes according to their cellular functions and herein discuss our findings.
Alternative splicing involving a U2–U12 hybrid intron
The JNK family of genes in vertebrates contains a U2–U12 hybrid intron that possesses the U12-type 5′SS and U2-type 3′ end sequence (Figure 1A). This intron cannot be properly spliced, but, in conjunction with its upstream U2-type or downstream U12-type introns, it may play a role in mutually exclusive selection of exon 6 (12,13). This alternative splicing control generates mRNA isoforms, some of which exhibit tissue specificity. For example, the exon 6a-containing JNK2/SAPKα/MAKP9 isoforms are preferentially expressed in neurons, whereas ubiquitous transcripts contain exon 6b (15). This observation indicates the biological importance of the U2–U12 intron-mediated splicing control.
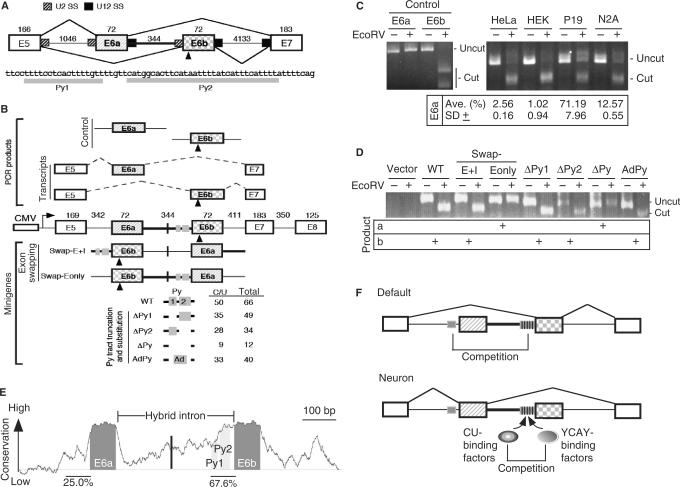
Figure 1.
The role of the U2–U12 hybrid intron in alternative splicing of human JNK2 pre-mRNA. (A) Schematic diagram shows exons 5 to 7 of JNK2. Between alternative exon 6a and 6b is the U2–U12 hybrid intron (heavy line); mutually exclusive selection of either exon generates mRNA isoforms. The numbers indicate the exon or intron length in nucleotides. U2 and U12 splice sites are indicated by hatched and filled boxes, respectively. The triangle indicates an EcoRV restriction site in exon 6b. Below the diagram is the sequence of the Py tract of the hybrid intron. (B) The minigene contains exons 5 to 8 of human JNK2, in which all introns except for the hybrid one are internally truncated (length indicated by the numbers). Transcription of the minigenes is driven by the CMV promoter. Translation start codon was in frame introduced in exon 5. Within the hybrid intron, the vertical line and grey boxes indicate the site for exon 6a/b swap and CU-rich sequences, respectively. The E6a and E6b control PCR products were amplified from the minigene, whereas the E6a and E6b transcripts represent the RT-PCR products that were amplified from the JNK transcripts. EcoRV digestion of the exon 6b-containing DNA fragment (472 bp) yielded two bands (294 and 178 bp). The modified minigenes are also depicted; the numbers of total and C/U residues of their polypyrimidine (Py) tract are listed. (C) Analysis of the JNK transcripts of four cell lines. The first four lanes show the E6a and E6b control fragments with or without EcoRV digestion. RT-PCR DNA fragments amplified from the endogenous JNK2 transcripts were of 316 bp; EcoRV digestion of the exon 6b-containing product generates two nearly comigrating fragments (cut) of 171 and 145 bp. Percentage of E6a-containing transcripts [cut/(cut + uncut) × 100%] in four cell lines was measured from three independent experiments; average with standard deviation is indicated. (D) HEK 293 cells were transfected with a minigene reporter as indicted. Total RNA was collected 24-h post-transfection and analyzed as in panel B. Shown on the gel are uncut PCR products (634 bp) and the larger digested fragment (463 bp). (E) The graph represents absolute sequence complexity (Y-axis) of a JNK2 genomic segment containing the alternative exon 6a/b and the hybrid intron (X-axis) from eight vertebrates (human, chimp, dog, cow, rat, mouse, opossum and chicken). Conservation of the Py tracts is indicated as percentage. (F) Model shows that mutually exclusive exon (hatched and squared boxes) selection of the JNK2 gene is driven by the U2–U12 intron (heavy line). The Py tract (grey box) of the hybrid intron dominates over that upstream of exon 6a, thus leading to exon 6b inclusion as the default pathway, particularly in non-neuronal cells. In neuronal cells, specific splicing activators, such as Nova, induce the use of exon 6a (21,22). On the other hand, since several YCAY elements (vertical lines) exist in the Py tract of the hybrid intron, YCAY-binding factors (such as Nova) may antagonize the activity of the ubiquitous CU-rich element-binding factors to reduce the utilization of exon 6b.
Previous reports have shown that neuronal expression of exon 6a-containing JNK2 isoforms is likely to be driven by neuron-specific Nova proteins (20,21). However, to date, the questions as to why non-neuronal JNK2 transcripts preferentially include exon 6b and what the role of the hybrid intron plays in alternative exon selection have not been experimentally investigated. We therefore set out to address these questions using a minigene system. First, we examined alternative splicing of JNK2 in several mammalian cell lines. RT-PCR was performed using primers specific to common exons 5 and 7. Since an EcoRV restriction site resides within exon 6b but not in 6a (Figure 1A), the amplified PCR fragments containing either alternative exon can be distinguished by EcoRV digestion (Figure 1C, control). EcoRV-resistant products were observed in neuronal cells (Figure 1C, P19 and N2A) but barely detected in non-neuronal cells (HeLa and HEK 293). This result was expected and it indicated that exon 6a is utilized in neuronal JNK2 transcripts. Moreover, predominant expression of the exon 6b-containing isoforms in HeLa and HEK 293 cells suggests that exon 6b selection is a default splicing pathway in cells lacking specific splicing regulators such as Nova proteins (20,21).
Next, we established a JNK2 minigene to examine whether the hybrid intron determines exon 6 selection. The minigene contained a genomic fragment of human JNK2 from exon 5 to exon 8, within which all introns except for the hybrid one were internally truncated (Figure 1B). Expression of the JNK2 minigene in HEK 293 cells was examined by RT-PCR-EcoRV digestion. As observed with endogenous JNK2 (Figure 1C), the minigene transcripts exclusively contained exon 6b but not 6a in HEK 293 cells (Figure 1D, WT). We then swapped exon 6a and 6b, each together with a part of its adjacent introns, in the minigene to examine whether exon 6a is thus activated in HEK 293 cells (Figure 1B, Swap-E + I). Note that the intron between the swapped exons still remained as a hybrid, i.e. the U12-type 5′SS and U2-type 3′SS. The result showed that exon 6b still dominated over 6a in the JNK2 transcripts (Figure 1D), indicating that utilization of exon 6 in HEK 293 cells is not determined by its position but perhaps by its sequence. To distinguish whether exon 6b or its adjacent intron sequence provides putative signals for exon choice, another swapping minigene was constructed, in which only the alternative exon parts were exchanged (Figure 1B, Swap-Eonly). Splicing of this minigene transcript showed exclusive use of exon 6a (Figure 1D), indicating that intron instead of exon sequence contributes to the alternative exon selection at least in a non-neuronal cell line. We noted that the polypyrimidine (Py) tract of the hybrid intron is unusually long and it shows a higher degree of conservation across several vertebrate species (Figure 1E; 67.6% conservation vs 15–25% conservation of several other JNK2 introns). Moreover, as compared to the intron upstream of exon 6a, the hybrid intron has a higher C/U content in the Py tract (Figure 1B, 26 C/Us in the 3′ 66 nucleotides of intron 5 vs 50 in the hybrid intron). To examine whether the Py tract of the hybrid intron contributes to default utilization of exon 6b, we made several deletion mutants of the minigene (Figure 1B). Figure 1D shows that a partial Py sequence still retained the full activity for exon 6b inclusion (ΔPy1 and ΔPy2), whereas deletion of an entire Py tract drove a complete switch from exon 6b to 6a (ΔPy). Nevertheless, a heterologous Py tract, derived from an intron of the adenovirus major late transcript, completely restored the use of exon 6b (Figure 1D, AdPy). Therefore, our data suggested that a Py tract, perhaps with a substantially high density of C/U residues, is sufficient to activate exon 6b utilization in JNK2.
In summary, the Py tract of the hybrid intron provides a signal for the default selection of exon 6b in non-neuronal cells, perhaps by competing with the use of the 3′ splice site prior to exon 6a (Figure 1F).
Effects of SNPs in alternative splicing of a U12-type intron
Meanwhile we also investigated genetic variations of U12-type introns that might activate alternative splicing. We selected ∼50 U12-type intron-containing genes from the ∼500 member pool to search for their SNPs within or near the splice-site or branch-site sequences of the U12-type introns. In addition to LKB1 (10), our analysis identified SNPs in the U12-type intron of another gene, WDFY1. The WDFY1 gene encodes a previously uncharacterized protein containing WD40 repeats and a zinc finger for PI3 binding (16,17). The two variable nucleotides detected in the WDFY1 U12-type intron are located within the 5′SS consensus and upstream of the branch site. Nucleotide polymorphisms in the intron consensus sequences could directly affect pre-mRNA splicing, resulting in differential levels of isoforms between individuals (22). Since U12-type introns exist in a number of genes encoding WD repeat-proteins and PI3 signaling factors (8), we therefore went on to characterize the effects of the detected U12-type intron SNPs on splicing of the WDFY1 pre-mRNA.
Cryptic 5′SS utilization is induced by a mutation in the 5′SS consensus sequence of the U12-type intron
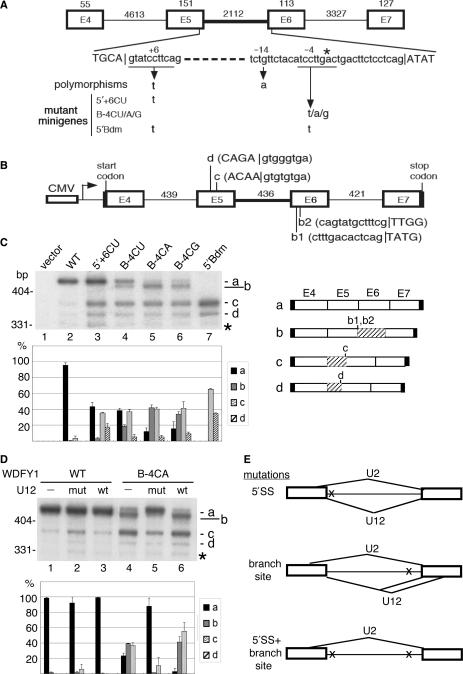
Apparently, the detected polymorphisms in the WDFY1 U12-type intron include nucleotide 6 of the 5′SS (refSNP:rs604990) and nucleotide 14 upstream of the branch site (refSNP:rs530065) (Figure 2A; hereafter termed 5′ + 6 and B-14, respectively). A minigene construct was created containing WDFY1 exons 4 to 7 with internally truncated introns between these exons (Figure 2B). The wild-type minigene was transiently transfected into HEK 293 cells, and the RNA products were analyzed by RT-PCR using 32P-labeled primers. A major cDNA product (band a) corresponded to the correctly spliced mRNA, as confirmed by sequencing (Figure 2C, lane 2). Nucleotide 6 of intron 5 was then changed from C to U to match the detected SNP at the 5′SS (5′ + 6CU). With this mutant minigene, two shorter cDNA fragments became more visible (Figure 2C, lane 3, bands c and d) and were then characterized as the products generated by using cryptic 5′SSs in exon 5 (Figure 2B, c and d sites). Because these cryptic 5′SSs conform to the U2-type consensus, the U12 5′ + 6CU mutation might activate the U2-type spliceosome to use cryptic 5′SSs and splice them to the 3′SS of the U12-type intron, which harbors the terminal dinucleotide AG. Our results thus indicate that the 5′SS mutation at the +6 position of a U12-type intron impairs its recognition by the U11 snRNP and allows the activation of cryptic 5′SSs by the U2-type spliceosome. This is somewhat different from what was observed with the first nucleotide mutation in the LKB1 U12-type intron, which could generate aberrant splicing products by using various combinations of splice sites by both types of the spliceosome (10).
Figure 2.
Effects of single nucleotide polymorphisms of the human WDFY1 U12-type intron. (A) Schematic diagram shows exons 4 to 7 of the human WDFY1 gene. Nucleotide variations were found at residue 6 (+6) of the 5′SS consensus sequence and residue 14 (−14) upstream of the branch site. The minigenes used for the splicing assay contain a single mutation at either the 5′ + 6 or the B-4 position or dual mutations (5′Bdm) at both sites. The numbers indicate the exon or intron length in nucleotides. (B) The minigene is composed of exons 4 to 7 with internally truncated introns (length indicated by the numbers). Translation start and stop codons are in frame at the 5′ end of exon 4 and 3′ end of exon 7, respectively. The use of the cryptic splice sites b1, b2, d, c yielded aberrant transcripts (see panel C). (C) HEK 293 cells were transfected with a minigene reporter as indicted. Total RNA was collected 24-h post-transfection and subjected to RT-PCR analysis using 32P-labeled primers. The identity of the cDNA products is shown at the right; hatched boxes represent truncated exons. The bar graph shows the relative abundance of the major splicing products as a percentage; the data were obtained from two to three independent experiments. The ∼330-bp band (asterisk) included a variety of aberrantly spliced JNK products, none of which expressed dominantly, and therefore was tentatively ignored in the graph. (D) A U12 snRNA expression vector was co-transfected with the wild-type or B-4CA WDFY1 minigene into HEK 293 cells. Splicing of the WDFY1 transcripts was examined as in panel C. The bar graph is as in panel C. (E) Model shows alternative splicing of the WDFY1 U12-type intron induced by genetic mutations. A 5′SS mutation activates cryptic U2-type 5′SSs in exon 5 (upper panel). A branch-site consensus mutation promotes the use of cryptic 5′SSs in exon 5 by the U2-type spliceosome or cryptic 3′SSs in exon 6 by the U2-type spliceosome (middle panel). Bottom: A double mutant is spliced only by the U2-type spliceosome, yielding aberrant mRNA products (lower panel).
A branch-site mutation of the U12-type intron causes aberrant selection of both 5′ and 3′SSs
The SNP at the B-14 site (a G to A mutation) was initially investigated by using a minigene construct, but it had no detectable effect on splicing (data not shown). To understand more about the impact of branch-site consensus mutations on splicing, we generated mutations at the −4 position, which slightly varies among U12-type introns [reviewed in (5)]. The C to U change (B-4CU) activated bands c and d, as observed with the 5′ + 6CU mutation, and also generated a minor product, band b (Figure 2C, lane 4). Sequence determination revealed that band b contained two cDNAs generated by the use of cryptic 3′SSs in exon 6 (Figure 2B, b1 and b2 sites). When the B-4C residue was substituted by a purine, the level of these aberrant splicing products was particularly elevated with a concomitant decrease of the correctly spliced RNA (Figure 2C, lanes 5 and 6 for B-4CA and B-4CG, respectively). The cryptic 3′SSs detected in exon 6 were probably activated by the U12-type spliceosome because the 5′SS used for splicing was unchanged. Nevertheless, the B-4 mutations in WDFY1 could also activate cryptic 5′SSs through the use of the U2-type spliceosome (Figure 2C, lanes 4–6, bands c and d and Figure 2E).
A 5′SS and branch-site double mutation suppresses U12-type spliceosome-mediated splicing
Next, we examined the effect of a double mutation on U12-type intron splicing. The 5′ + 6CU mutation, in conjunction with a mild branch-site mutation, B-4CU, was introduced into the WDFY1 minigene. With this double mutant, bands a and b were no longer detected (Figure 2C, lane 7). Of note, both correctly spliced mRNA (band a) and aberrant splicing products detected in band b were generated by the U12-type spliceosome, albeit the latter through the use of cryptic 3′SSs. Therefore, our result indicated that the activity of the U12-type spliceosome was completely abrogated when both the 5′SS and branch site were mutated. Moreover, bands c and d were enhanced with the double mutant 5′Bdm (Figure 2C, lane 7), suggesting that the U2-type spliceosome overwhelmed for splicing in this case due to complete inactivation of the U12 splicing.
Aberrant splicing caused by a branch-site mutation can be corrected by a compensatory U12 snRNA mutant
The above data showed that alternative splice-site utilization is activated by nucleotide alterations in the 5′SS or branch-site consensus sequences in WDFY1. We next examined whether such splicing events can be reversed by coexpression of compensatory snRNA mutants (23). Since the 5′SS sequence is sequentially recognized by the U11 and U6atac snRNAs during splicing [reviewed in (5)], to simplify the experiments, we investigated a branch-site mutation. The B-4CA mutant, which altered the utilization of both 5′ and 3′SSs, was subjected to experimental study. To rescue its splicing defects, nucleotide 22 of the U12 snRNA was changed to restore its base-pairing interaction with the B-4CA branch site and a concomitant change at nucleotide 5 was introduced to maintain the helix I stem (19). The B-4CA minigene was co-transfected with the expression vector encoding U12 snRNA. Expression of exogenous U12 snRNA was driven by the U12 promoter and detected by RT-PCR (data not shown). Wild-type U12 snRNA had no significant effect on splicing of either wild-type or B-4AC reporter (Figure 2D, lanes 3 and 6). Overexpression of the mutant U12 snRNA slightly increased the level of band c produced from the wild-type reporter (Figure 2D, lane 2); this was probably due to competition between the mutant and endogenous wild-type U12 snRNAs. Nevertheless, the splicing defect of B-4CA was almost completely rescued by this mutant U12 snRNA, except for a residual amount of band c (Figure 2D, lane 5). Thus, U12 snRNA primarily suppressed cryptic U12 3′SS utilization and acted in an allele-specific manner, suggesting that the base-pairing interaction between U12 snRNA and the branch site was restored.
Taken together, our data indicated that nucleotide alternations within the U12-type intron consensus elements could induce cryptic-site utilization by either type of the spliceosome (Figure 2E), and this result is essentially consistent with the previous reports (9–11).
U12-type introns in mRNA processing factors
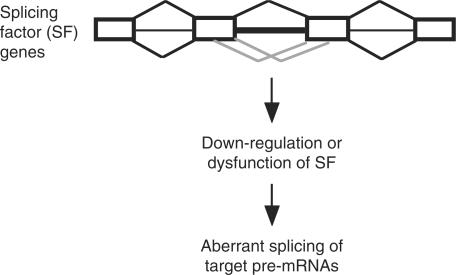
It has been reported that U12-type introns are present in several large gene families primarily due to gene amplification (8). Conceivably, those U12-type intron-containing paralogs have similar cellular functions; examples include voltage-dependent calcium channel subunits, mitogen-activated protein kinases (MAPK), RAP guanine nucleotide exchange factors (RAPGEF) and importin-β family nuclear transporters (8 and Supplementary Table 1). A previous report has indicated that U12-type introns are over-represented in genes implicated in Ras–Raf signaling pathways (8). We also observed that U12-type introns prevail in some functionally but not genetically related genes, such as those involved in chromatin remodeling and transcriptional control (Supplementary Table 2) and those involved in mRNA metabolism (Table 1). Some of these mRNA processing factors may directly participate in pre-mRNA splicing or even in alternative splicing regulation. Moreover, factors such as transportin and SR protein kinases involved in cellular metabolism of splicing regulatory factors also possess U12-type introns (8). Given that splicing of U12-type introns is a rate-determining step (24), expression of these genes might be controlled at the level of pre-mRNA splicing, which could subsequently lead to global regulation of alternative splicing (Figure 3). If this is the case, splicing of RNPC3 that encodes an essential splicing factor associated with the U11/U12 snRNP (25) may have a feedback control on U12-type intron splicing; this hypothesis remains to be investigated.
Figure 3.
U12-type introns present in splicing factor genes may impact on gene expression. When the level or activity of a splicing factor encoded by a U12-type intron-containing gene is altered due to defective U12-type intron splicing (grey lines), aberrant splicing of its target pre-mRNAs may thus be observed.
DISCUSSION
To better understand alternative splicing of U12-type introns, we experimentally examined alternative splicing events that were either mediated by a U2–U12 hybrid intron or induced by genetic mutations within the U12-type intron consensus elements. Moreover, the presence of U12-type introns in a set of splicing factor genes leads us to hypothesize that their splicing, particularly removal of the U12-type intron(s), might have some impact(s) on global gene expression.
Our analysis towards the role of a U2–U12 hybrid intron in alternative exon selection first reveals that the Py tract of the human JNK2 hybrid intron is exceptionally long and conserved and contains a high C/U content. The experiments of exon swapping and intron truncation and substitution provided evidence suggesting that exon 6b inclusion, a prevalent pathway in non-neuronal cells, primarily attributes to the Py tract of the hybrid intron. In particular, the exon-only swapping result could almost exclude the possibility that exonic sequences have any significant effect on exon selection at least in non-neuronal cells. Moreover, we hypothesize that ubiquitous splicing activators could bind to C/U-rich elements within the Py tract of the hybrid intron, leading to the default splicing (Figure 1F). On the other hand, overexpression of neuronal Nova proteins could induce exon 6a inclusion via binding to multiple YCAY elements (21). Note that the hybrid intron also harbors YCAY elements, and five of them are particularly embedded in the Py tract (Figure 1F). Thus, it is also possible that Nova proteins antagonize the activity of the putative CU-element binding activators via binding to these YCAY sites, leading to suppression of exon 6b utilization in neuronal cells (Figure 1F). Interestingly, the Py tract prior to exon 6a, although poor in its CU content, does not contain the Nova binding sites. Perhaps, without competition between the Py and YCAY elements, exon 6a can be activated in neuronal cells. Note that some of the JNK homologs in pufferfish (Fugu rubripes) and zebrafish (Danio rerio) contain only one exon 6 (or equivalent), of which the upstream and downstream introns are U2- and U12-type, respectively. Perhaps, the hybrid intron emerged by duplication of an ancestral exon 6 along with a part of its adjacent introns, and its presence inevitably drove the exclusive use of duplicated exon 6.
Nucleotide alterations at the splice-site consensus elements of U12-type introns can activate cryptic splice-site utilization (9–11). Our results show that a U12 5′SS mutation in WDFY1 preferentially induced aberrant 5′SSs through the action of the U2-type spliceosome (Figure 2E). Therefore, consistent with previous reports (9–11), when the U12-type spliceosome fails to recognize altered 5′SS sequence of a U12-type intron, the abundant U2-type spliceosome may take over the 3′SS of this intron to pair it with a cryptic U2-type 5′SS. Aberrant splicing was also observed with a branch-site mutation; however, utilization of aberrant 3′SSs by the U12-type spliceosome was particularly activated under this condition (Figure 2E). Therefore, inefficient use of a variant branch-site sequence may drive the U12-type spliceosome to search for an aberrant branch site/3′SS for splicing. However, the U12-type spliceosome functions only when the 5′SS can be well recognized. Perhaps the authentic 5′SS sequence provides a site for U11 snRNP binding, which allows its U12 partner to locate an appropriate, albeit cryptic, sequence for splicing. When a U12-type intron has mutations at both the 5′SS and branch site, the U2-type spliceosome could fully occupy this intron and function to yield aberrant splicing products (Figure 2E). Together, our data support previous observations that the U12-type spliceosome has a more stringent sequence requirement for the 5′SS than the branch site/3′SS; in other words, U12-type intron splicing can take place at variable 3′SS sequences.
U12-type introns, though more conserved than U2-type introns, have nucleotide variations in the splice-site elements [reviewed in (5)]. In general, U12-type intron splicing has a more stringent requirement for intronic elements and is likely to be rate limiting in the reaction [reviewed in (5)]. Therefore, nucleotide variations and polymorphisms in U12-type introns might profoundly affect splicing and any combination of nucleotide changes could exacerbate splicing defects, as evidenced by our data (Figure 2). Moreover, we speculate that the spectrum of splicing defects resulting from U12 intron polymorphisms somewhat differs from that of U2-type introns because aberrant U12-type intron splicing often generates products by use of cryptic splice sites rather than through exon skipping (Figure 2E). As compared with exon skipping, aberrant splice-site utilization has a higher probability of generating products with premature translation stop codons, which in turn results in down-regulation of gene expression (26). We thus conclude that the splicing of U12-type introns has a substantial impact on expression of specific genes.
Herein we report that U12-type introns prevail in some genes coding for RNA processing factors; of particular interest is a set of splicing factors that participate in constitutive and/or regulated splicing. Given that U12-type intron splicing is rate-limiting (24) and less tolerant to nucleotide variations in the splice-site and branch-site elements (10 and this study), the yield and the quality/activity of those RNA processing factors can thus be controlled by splicing of their U12-type intron(s). Through the functions of these proteins, U12-type intron splicing may have a considerable impact on global gene expression at the level of alternative splicing regulation (Figure 3).
SUPPLEMENTARY DATA
Supplementary data is available at NAR online.
ACKNOWLEDGEMENTS
We thank Trees-Juen Chuang (Taipei), Tetsuro Hirose (Tokyo) and Yi-Tao Yu (Rochester) for comments on the manuscript and Tim C. Taylor for editing the manuscript. This work was supported by Academia Sinica Research Award to W.-Y. Tarn. Funding to pay the Open Access publication charge was provided by Academia Sinica.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tarn WY, Steitz JA. Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends Biochem. Sci. 1997;22:132–137. doi: 10.1016/s0968-0004(97)01018-9. [DOI] [PubMed] [Google Scholar]
- 2.Burge CB, Tuschl T, Sharp PA. Splicing of precursor to mRNA by the spliceosomes. In: Gesteland RF, Cech T, Aktins JF, editors. The RNA World II. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 3.Sharp PA, Burge CB. Classification of intron: U2-type or U12-type. Cell. 1997;91:875–879. doi: 10.1016/s0092-8674(00)80479-1. [DOI] [PubMed] [Google Scholar]
- 4.Will CL, Luhrmann R. Splicing of a rare class of intron by the U12-dependent spliceosome. J. Biol. Chem. 2005;386:713–724. doi: 10.1515/BC.2005.084. [DOI] [PubMed] [Google Scholar]
- 5.Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell. Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 6.Burge CB, Padgett RA, Sharp PA. Evolutionary fates and origins of U12-type introns. Mol. Cell. 1998;2:773–785. doi: 10.1016/s1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- 7.Lynch M, Richardson AO. The evolution of spliceosomes introns. Curr. Opin. Genet. Dev. 2002;12:701–710. doi: 10.1016/s0959-437x(02)00360-x. [DOI] [PubMed] [Google Scholar]
- 8.Levine A, Durbin R. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 2001;29:4006–4013. doi: 10.1093/nar/29.19.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietrich RC, Peris MJ, Seyboldt AS, Padgett RA. Role of the 3′ splice site in U12-dependent intron splicing. Mol. Cell. Biol. 2001;21:1942–1952. doi: 10.1128/MCB.21.6.1942-1952.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hastings ML, Resta N, Traum D, Stella A, Guanti G, Krainer AR. An LKB1 AT-AC intron mutation causes Peutz-Jeghers syndrome via splicing at noncanonical cryptic splice sites. Nat. Struct. Mol. Biol. 2005;12:54–59. doi: 10.1038/nsmb873. [DOI] [PubMed] [Google Scholar]
- 11.Dietrich RC, Fuller JD, Padgett RA. A mutational analysis of U12-dependent splice site dinucleotides. RNA. 2005;11:1430–1440. doi: 10.1261/rna.7206305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letunic I, Copley RR, Bork P. Common exon duplication in animals and its role in alternative splicing. Human Mol. Genet. 2002;13:1561–1567. doi: 10.1093/hmg/11.13.1561. [DOI] [PubMed] [Google Scholar]
- 13.Smith CWJ. Alternative splicing-when two's a crowd. Cell. 2005;123:1–7. doi: 10.1016/j.cell.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Graveley BR. Mutually exclusively splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casanova E, Garate C, Ovalle S, Calvo P, Chinchetru MA. Identification of four splice variants of the mouse stress-activated protein kinase JNK/SAPK a-isoform. NeuroReport. 1996;7:1320–1324. doi: 10.1097/00001756-199605170-00021. [DOI] [PubMed] [Google Scholar]
- 16.Ridley SH, Ktistakis N, Davidson K, Anderson KE, Manifava M, Ellson CD, Lipp P, Bootman M, Coadwell J, et al. FENS-1 and DFCP1 are FYVE domain-containing proteins with distinct functions in the endosomal and golgi compartments. J. Cell Sci. 2001;114:3991–4000. doi: 10.1242/jcs.114.22.3991. [DOI] [PubMed] [Google Scholar]
- 17.Fritzius T, Burkard G, Haas E, Heinrich J, Schweneker M, Bosse M, Zimmermann S, Frey AD, Caelers A, et al. A WD-FYVE protein binds to the kinases Akt and PKCzeta/lambda. Biochem. J. 2006;399:9–20. doi: 10.1042/BJ20060511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarn WY, Yario TA, Steitz JA. U12 snRNA in vertebrates: evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of introns. RNA. 1995;1:644–656. [PMC free article] [PubMed] [Google Scholar]
- 20.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 21.Relogio A, Ben-Dov C, Baum M, Ruggiu M, Gemund C, Benes V, Darnell RB, Valcarcel J. Alternative splicing microarrays reveal functional expression of neuron-specific regulators in Hodgkin Lymphoma cells. J. Biol. Chem. 2005;280:4779–4784. doi: 10.1074/jbc.M411976200. [DOI] [PubMed] [Google Scholar]
- 22.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 23.Hall SL, Padgett RA. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science. 1996;271:1716–1718. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- 24.Patel AA, McCarchy M, Steitz JA. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 2002;21:3804–3815. doi: 10.1093/emboj/cdf297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benecke H, Luhrmann R, Will CL. The U11/U12 snRNP 65K protein acts as a molecular bridge, binding the U12 snRNA and U11-59K protein. EMBO J. 2005;24:3057–3069. doi: 10.1038/sj.emboj.7600765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]