Abstract
Here we report a PCR-based DNA engineering technique for seamless assembly of recombinant molecules from multiple components. We create cloning vector and target molecules flanked with compatible single-stranded (ss) extensions. The vector contains a cassette with two inversely oriented nicking endonuclease sites separated by restriction endonuclease site(s). The spacer sequences between the nicking and restriction sites are tailored to create ss extensions of custom sequence. The vector is then linearized by digestion with nicking and restriction endonucleases. To generate target molecules, a single deoxyuridine (dU) residue is placed 6–10 nt away from the 5′-end of each PCR primer. 5′ of dU the primer sequence is compatible either with an ss extension on the vector or with the ss extension of the next-in-line PCR product. After amplification, the dU is excised from the PCR products with the USER enzyme leaving PCR products flanked by 3′ ss extensions. When mixed together, the linearized vector and PCR products directionally assemble into a recombinant molecule through complementary ss extensions. By varying the design of the PCR primers, the protocol is easily adapted to perform one or more simultaneous DNA manipulations such as directional cloning, site-specific mutagenesis, sequence insertion or deletion and sequence assembly.
INTRODUCTION
The cloning and engineering of genes are widely used techniques to study DNA and protein function. The majority of strategies to engineer genes by site-directed mutagenesis are based on the in vitro extension of a template DNA strand from a synthetic oligonucleotide encoding the desired mutation(s) (1–6). The percentage of mutant clones recovered using these methods of site-directed mutagenesis can vary from 0.1 to >60% depending on the efficiency of the parental (unmutated) DNA elimination step. However, more intricate DNA engineering tasks such as a domain swap between several DNA targets or sequence assembly from multiple fragments largely depend on DNA cleavage by restriction endonucleases and rejoining by DNA ligase. Several strategies for creating and joining two independent DNAs without the use of restriction endonucleases have been developed. One such strategy uses overlap extension employing two PCR steps (7–10). Two intermediate PCR fragments with overlapping end sequences are first amplified, then re-annealed across the overlapping sequences, extended and subsequently amplified with a flanking primer set (7). Though the idea is attractive, the success of overlap PCR depends on efficient cross-annealing of intermediate PCR products and effective removal of unused internal primers to direct the second amplification of the full-length product.
Another known approach for PCR product engineering and cloning is ligase-free UDG cloning (11–13). The method employs PCR amplifications of vector and insert with overlapping PCR primers containing multiple dU residues within the overlap region. Treatment of overlapping PCR products with Uracil DNA Glycosylase (UDG, also known as UNG) generates multiple abasic sites that promote strand separation due to destabilized base pairing. The 3′ single-stranded tails then cross-anneal to form a recombinant product. This strategy has several limitations that have prevented this method from becoming widely used for gene engineering. First, multiple deoxyuridines (dU) must be incorporated into the 5′ end of the primer (at least one-third of the 5′ overlapping tails should consist of dU residues in order to achieve efficient strand separation). In the coding sequences that have low A/T content, it might be difficult to select the overlapping regions. Second, UDG does not cleave the phosphodiester backbone; it only removes the base from the sugar. The assembled recombinant product is left with the protruding single-stranded flaps, which have to be removed in vivo by the repair machinery of the bacterial host thus increasing the possibility of DNA rearrangements in vivo, or by in vitro methods requiring extra manipulations and usually effecting yields (15–17). Finally, compatible dU tails have to be introduced on plasmid vectors by PCR increasing the possibility of incorporating unwanted mutations into the plasmid backbone.
Here we present an improved method based on ligase-free UDG-mediated cloning, referred to as USER™ (Uracil-Specific Excision Reagent) friendly DNA Engineering. The USER™ friendly DNA engineering method allows for multiple changes of the target DNA with a minimum of manipulations by the experimenter. The vector's insertion site is flanked by ss extensions of designed length and nucleotide composition, allowing for ligase-free directional cloning of PCR products. The PCR reactions must be performed with a polymerase which can incorporate a deoxyadenine opposite a dU. Either Taq DNA polymerase or the proofreading DNA polymerase, PfuTurbo Cx Hotstart DNA polymerase (Stratagene) can be used to perform PCR amplification with dU-containing primers. The USER™ enzyme removes the dU residues to generate 3′ ss extensions on PCR-amplified DNA fragments. While the described approach is simple, it provides several advantages: (a) single-stranded extensions may be longer than those produced by restriction endonuclease cleavage; (b) single-stranded extensions are not self-complementary, nor are they complementary to each other, so the vector molecule termini do not re-anneal to form transformable circular DNA; and (c) each single-stranded extension carries a unique nucleotide sequence, thereby permitting the directional assembly of multiple PCR products into the vector.
MATERIALS AND METHODS
Source of bacterial strains, plasmids and enzymes
Eschericia coli NEB5-α competent cells, plasmids pUC193, pGPS2.1 and pNEBR-R1 are from New England Biolabs (Ipswich, MA). Plasmid pBind-Gal4-EcR-053 was a gift from Rheogene (Norristown, PA). Eschericia coli ER2267 cells were made competent using standard CaCl2 procedure. All enzymes used in this study are products of New England Biolabs, except for PfuTurbo Cx Hotstart DNA Polymerase which was from Stratagene (La Jolla, CA). Oligonucleotides and primers were synthesized by the DNA synthesis facility of New England Biolabs and are listed in Table 1.
Table 1.
Oligonucleotide sequences
| Oligonucleotides for vector pNEB206A constructiona |
| 5′-taaGCTGAGGGAAAGTCTAGAGGATCCTCTAGATGTCTCCTCAGCgttt |
| 5′-aaacGCTGAGGAGACATCTAGAGGATCCTCTAGACTTTCCCTCAGCttaat |
| Oligonucleotides for USER enzyme activity assay |
| 5′-GATTTCATTTTTTATTUATAACTTTATATTGb |
| 5′-CAATATAAAGTTATAAATAAAAAATGAAATC |
| PCR primers for Cat gene engineering and cloningc |
| Left: GGAGACAUCGGATCCATACCTGTGACGGAAG-3′ |
| Right: GGGAAAGUGGATCCAGGCGTTTAAGGGCACC-3′ |
| P1: ATATGGGAUAGTGTTCACCCTTGTTACACC-3′ |
| P2: ATCCCATAUGACCAGCTCACCGTCTTTCATTGCCATAC-3′ |
| P3: AGGTTCAUGCTGCCGTTTGTGATGGCTTCCATGTCG-3′ |
| P4: ATGAACCUGAATCGCCAGCGGCATC-3′ |
| PCR primers for RheoActivator gene engineering and cloningd |
| R47: GGGAAAGUAACATGGTGATGCGGTTTTGGCAGTAC |
| R48: ATGGCAGUTGTTACGACATTTTGGAAAG |
| R49: ACTGCCAUCGCCCGCCCCGTTGACGCA |
| R50: ACTGGAGCUTAAGTTCGAGACTGTTGTGTC |
| R51: AGCTCCAGUGACTCTCTTAAGGTAGCCT |
| R52: AGGTCCCAUGTTGGCACCGGTCCTAGGCTGTGGAGAGAAAGGCAAAGTGG |
| R53: ATGGGACCUAAAAAGAAGCGTAAAGTCGCC |
| R54: ATCCCCGUCCCCCAACATATCCAGATCGAAATCGTCTAGCGCGT |
| R55: ACGGGGAUTCCCCAGGGCCGGGATTTACCCCCCA |
| R56: ACAGGCAUCTCGAACTCCCCACCGTACTCGTCAATTCC |
| R57: ATGCCTGTGGACAGGATTCTGGAGGCAGAGCTTGCTGTGG |
| R58: AGGAAAAGUGTGGAATCCTCTTCGCCCACTCAACAAG |
| R59: ACTTTTCCUCCTTGCCTCTGGATGACCAGGTCATATTGCTGCGGGCAGGC |
| R60: GGAGACAUATCTCCAATAAGGCGAAAGAAAAAC |
aSequence shown in uppercase defines cassette boundaries. Sequence shown in lowercase represent PacI and PmeI compatible ends that allow re-creating PacI and PmeI sites in pNEB206A vector.
bOligonucleotide has fluorescein label on 5′ and 3′ ends.
cThe 5′ ends of primer sequences shown in boldface can be cleaved-off from the corresponding PCR product with the USER enzyme thus creating PCR fragment flanked with 3′-single-stranded extensions.
dpNEB206A-compatible sequences in primers R47 and R60 are shown in boldface. The insertion sequence in primer R52 is shown in italic. The point mismatches in the primers compared to the template sequence are underlined.
Construction and linearization of pNEB206A plasmid
To construct the pNEB206A vector, a double-stranded cassette was designed to have PacI and PmeI compatible ends (Table 1). The cassette was then subcloned into pUC193 that was linearized with PacI and PmeI restriction endonucleases. After insertion of the cassette, the multiple cloning site of pNEB206A is still in frame with the lacZα gene, allowing screening for recombinants using α-complementation. To prepare linear pNEB206A, 10 μg of plasmid DNA was digested with 40 units of XbaI in 100 μl of NEBuffer 4 for 18 h at 37°C and then nicked with 20 units of Nt.BbvCI for 1 h at 37°C. The cleaved DNA was purified by phenol–chloroform extraction and ethanol precipitation, dissolved in 100 μl of TE buffer (10 mM Tris-HCl, pH 8.0, 0.1 mM EDTA) and stored at −20°C. The concentration of the linearized vector pNEB206A was determined spectroscopically at A260 and was adjusted to 50 ng/μl.
USER enzyme activity
USER enzyme is a mixture of E. coli uracil DNA glycosylase and endonuclease VIII (18,19). Together these two enzymes create a nick in a 34-mer oligonucleotide duplex containing a single dU paired with a deoxyadenine. To establish the optimal ratio of the enzymes in the mixture, various amounts (ng) of EndoVIII were pre-mixed with 0.2 units of UDG (New England Biolabs) and the resulting mixtures were assayed for complete nicking of 10 pmol of substrate in 15 min at 37°C in a 10 μl reaction buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 10 mM DTT, 1 mM ATP, 20 μg/ml BSA). The reactions were quenched by the addition of 10 μl of 95% formamide, 0.1% xylene cyanol, 0.1% bromphenol blue, 10 mM EDTA, pH 11, and the reaction products were analyzed on a 15% TBE-Urea denaturing gel (Invitrogen, Carlsbad, Ca).
PCR amplification
PCR amplifications were performed using either Taq DNA polymerase or PfuTurbo Cx Hotstart DNA polymerase (PfuCx) following manufacturer's recommendations. PfuCx is a genetically engineered derivative of Pfu DNA polymerase carrying a point mutation V93Q that abolishes PCR inhibition by dU (20,21). Each 50 μl PCR reaction contained 20 ng of template DNA, 0.2 mM dNTPs, 0.2 mM of each primer and 0.5 μl of DNA polymerase. The template was amplified for 25 cycles using cycling protocols as follows. For Taq DNA polymerase: the initial denaturation is 5 min at 94°C; denaturation for 30 s at 94°C, annealing for 1 min at 55°C, polymerization is from 20 s to 1 min at 72°C; final polymerization is for 5 min at 72°C. For PfuCx DNA polymerase: initial denaturation is 5 min at 95°C; denaturation for 30 s at 95°C, annealing for 1 min at 55°C, polymerization is from 20 s to 1 min at 72°C; final polymerization is 5 min, at 72°C. The amount of each PCR product in 5 μl of PCR mixture was evaluated by agarose-gel electrophoresis. The PCR product concentration should be at least 0.02 pmol/μl or higher for efficient PCR fragment assembly.
Assembly and transformation
For chloramphenicol acetyltransferase (Cat) gene engineering, four assembly reactions were performed using PCR fragments that had been amplified with either Taq DNA polymerase or PfuCx DNA polymerase. Each assembly reaction (20 μl) contained 1 μl (50 ng) of linear pNEB206A vector and 1 μl (1 unit) of USER enzyme. Five microlitre aliquots of the completed PCR reactions were added directly to the assembly reactions as indicated: assembly reaction 1—PCR reaction Cat926; assembly reaction 2—PCR reactions Cat142 and Cat792; assembly reaction 3—PCR reactions Cat461 and Cat474; assembly reaction 4—PCR reactions Cat142, Cat327 and Cat474. The final volume of each reaction was adjusted to 20 μl by adding the appropriate volume of 1X PfuCx polymerase reaction buffer. The assembly reactions were incubated for 15 min at 37°C to cleave at dU residues and for an additional 15 min at room temperature to allow annealing of the complementary extensions. Fifty microlitres of the competent cells were transformed with 5 μl of each assembly reaction. E. coli ER2267 strain was used for transformations with Taq DNA polymerase-derived assembly reactions, whereas E. coli NEB 5-α strain was used for PfuCx polymerase-derived assembly reactions. Transformants were selected by plating 50 μl of transformation reaction (from 1 ml of total outgrowth volume) on 3 LB plates supplemented with Amp (0.1 mg/ml), IPTG (0.2 mM) and X-gal (0.04 mg/ml). The white (recombinant) and blue (vector background) colonies were counted after 18 h incubation at 37°C. The percentage of recombinant colonies was determined by dividing the number of white colonies by the total number of transformants. Assembly efficiency was determined by comparing the number of transformants recovered from each assembly reaction to the number of transformants resulting from transformation of 50 ng of uncut plasmid pNEB206A.
RESULTS
Generating plasmid vectors with unique single-stranded extensions
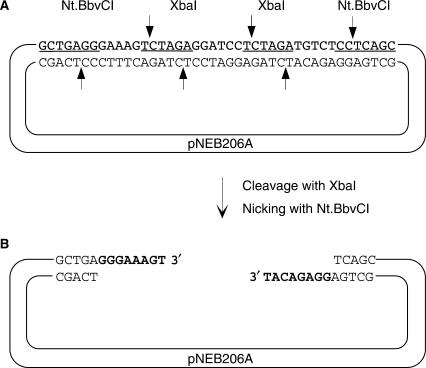
Recently, a variety of site-specific nicking endonucleases were derived by genetically engineering their corresponding restriction endonucleases (22–24). These enzymes nick only one strand in double-stranded DNA and the orientation of the recognition sequence defines whether the top or bottom strand is nicked. When two inversely oriented nicking sites are placed within a short distance, a double-strand break may be achieved due to the strand dissociation across the spacer sequence. However, the generated single-stranded extensions are complementary to each other as they solely derive from the spacer sequence. By incorporating one or more restriction endonuclease sites into the spacer sequence it is possible to divide it into two halves, each having a unique sequence, so the single-stranded extensions generated after the cleavage with restriction and nicking endonucleases are not complementary to each other. The spacer sequence between two nicking sites may be easily customized. This flexibility is available by changing the order, orientation and spacing of the nicking sites with respect to the adjacent restriction site(s) in the cassette.
A double-stranded oligonucleotide cassette containing two inversely oriented nicking endonuclease Nt.BbvCI sites (24) separated by two XbaI restriction endonuclease sites were synthesized by annealing two complementary single-stranded oligonucleotides (Table 1). Within the cassette, each XbaI site was separated from the adjacent Nt.BbvCI site by unique, 5-nucleotide long spacer sequence as shown in Figure 1A. This cassette was then subcloned into a polylinker of pNEB193 vector to make a derivative referred to as pNEB206A. pNEB206A plasmid was double-digested with XbaI and Nt.BbvCI as described in Materials and Methods yielding linearized vector flanked by unique 8-nucleotide long 3′ ss extensions (Figure 1B). The nicking reaction should be performed exactly as described in Materials and Methods because Nt.BbvCI possesses a residual double-strand cleavage activity, which is observed when vector DNA is over-digested. The linearized vector was tested for complete digestion by a transformation assay. Transformation results yielded 4.0 × 103 blue colonies per microgram of the linearized vector compared to 5.0 × 106 blue colonies per microgram of uncut vector, indicating that only 0.08% of the vector remains uncut.
Figure 1.
Schematic representation of the pNEB206A cloning vector. (A) pNEB206A vector was constructed by ligating a synthetic double-stranded cassette into the PacI and PmeI sites of pNEB193. Within the cassette, XbaI and Nt.BbvCI recognition sequences are underlined; cleavage sites are shown by arrows. (B) For USER friendly cloning, pNEB206A is double-digested with XbaI and Nt.BbvCI as described in the Materials and methods section to produce linearized vector flanked by 3′ single-stranded extensions on both ends.
USER friendly DNA engineering method
An overview
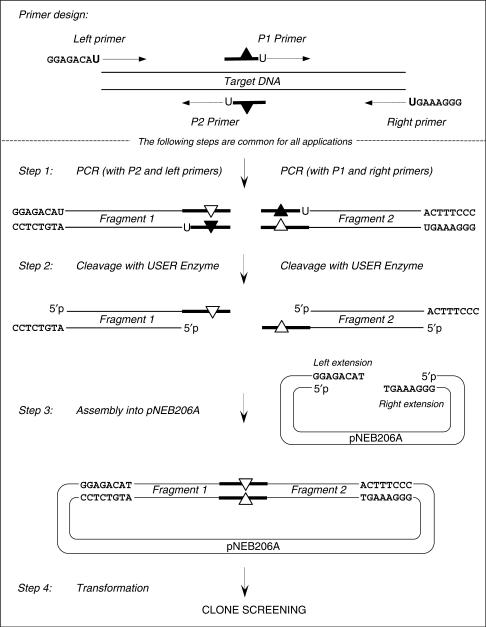
A general overview of this technique is presented in Figure 2. The target DNA is amplified as two overlapping intermediate fragments in separate amplification reactions. Accordingly, two pairs of primers, (P1 + Right) and (P2 + Left) are required for the two amplification reactions. The overlapping primers, P1 and P2, are selected at the location of nucleotide sequence manipulation, so that the desired sequence changes can be engineered into the primer sequences. The P1 and P2 primers prime opposite strands of the target DNA and overlap each other by 6–10 nucleotides at their 5′ ends. Each overlapping primer contains one dU residue that flanks the overlap region on the 3′ side. To provide vector-compatible extensions on PCR-amplified fragments, the Left and Right primers are designed with 8 additional nucleotides at their 5′ ends. These 8 nucleotides are identical to single-stranded extensions on the linearized vector pNEB206A (Figure 1B), except for a 3′ deoxythymine, which in the primer sequences is replaced by a dU residue. Downstream of these 8 nucleotides, the primer sequences are complementary to target DNA specific sequences. The two amplification reactions are then performed with primer pairs (Left primer + P2) and (P1 + Right primer). In the PCR reaction, the polymerases incorporate an adenine opposite the template-strand dU. Next, the dU residues are excised from the amplified fragments using USER enzyme in the assembly reaction as described in the Methods section. After phosphodiester bond breakage, the terminal 5′ single-stranded oligonucleotides dissociate, leaving the PCR fragments flanked by a 3′ single-stranded extension on each end. The linearized vector pNEB206A and the USER-treated PCR fragments directionally assemble into the desired recombinant molecule, as the inside 3′ single-stranded extensions of PCR fragments are complementary to each other, while the outside 3′ single-stranded extensions of PCR fragments are complementary to the single-stranded extensions on the linearized vector (Figure 2). Because the extensions are 6–10 bases in length, ligation is not required. The assembly reaction is extremely efficient and guarantees high yield of desired recombinants, which only depends on the competence of the cell host. The construct is now ready for transformation of chemically competent E. coli cells.
Figure 2.
Schematic description of the USER friendly DNA Engineering method. Lines with the arrowheads symbolize PCR primers. Boldface lines within the P1 and P2 primers and PCR products symbolize complementary overlapping sequences. Black triangles refer to the point mismatches in the primers and white triangles refer to the resulting mutations in the PCR products. dU is indicated by ‘U’. The specific sequences GGAGACAU in the left primer and GGGAAAGU in the right primer are designed for their compatibility with the vector pNEB206A sequence.
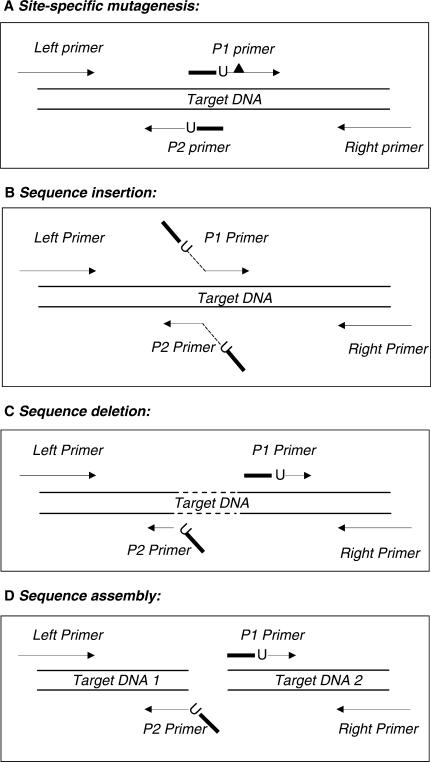
The proposed method is easily adapted to perform different DNA manipulations because the only variable is the design of the PCR primers, whereas steps 1 to 4 are identical for virtually any DNA manipulation (Figure 2). For example, directional cloning of any PCR product into pNEB206A, may be achieved by using the Left and Right primers shown in Figure 2. Two choices of overlapping primer P1 and P2 design, suitable for site-specific mutagenesis are depicted in Figures 2 and 3A. To perform sequence insertion, the design of overlapping primer design should be adapted as shown in Figure 3B. To perform sequence deletion or sequence assembly, the design of overlapping primers should be adapted as shown in Figure 3C or D, respectively.
Figure 3.
PCR primer design for various DNA manipulations. Only the primer design aspects of the indicated DNA manipulations are shown. The indicated DNA manipulations are completed by performing steps 1–4 shown in Figure 2. Lines with the arrowheads symbolize priming sequences in PCR primers. Boldface lines within primers symbolize overlapping sequences. dU is indicated by ‘U’ and flank the overlapping sequences on their 3′ side. (A) Site-specific mutagenesis. The desired sequence change, shown as a black triangle, is introduced downstream of dU into either of the overlapping primers P1 or P2. An alternative is shown in Figure 2 where sequence changes are introduced into the overlap sequence of both primers. (B) Nucleotide sequence insertion. Overlapping primers P1 and P2 precisely prime the template adjacent to the insertion site. The desired non-priming sequences are added to the 5′ ends of both primers (shown as angled lines). The overlapping sequence is created from the 5′ terminal 6–10 nucleotides of the insertion sequences (shown as the boldface angled lines), while the rest of the insertion sequence (shown as a dotted angled line) may be of any desired length. (C) Nucleotide sequence deletion. The overlapping primers P1 and P2 prime distant locations precisely adjacent to the targeted deletion region (shown as a dotted line). To create the overlapping sequence, the 5′ end of one primer (P2) is supplemented by a non-priming sequence complementary to the 5′ end sequence of the other primer (P1). (D) Sequence assembly. The overlapping primers, P1 and P2, prime different DNA templates. A complementary copy of the 5′ terminal sequence of one primer (P1) is added to the 5′ terminus of the other primer (P2) to create the overlapping sequence necessary for joining two independent DNAs.
Preparation of the USER enzyme specific for dU
dU residues can be incorporated into DNA by chemically synthesizing a primer containing dU and extending the dU primer with the PCR reaction, thus creating a unique target for uracil DNA glycosylase that specifically acts on deoxyuracil (18). While UDG is capable of excision of a dU base, creating an abasic (AP) site, it does not cleave the phosphodiester backbone. To create a ‘dU-nicking’ tool, UDG reactions must be supplemented with an additional enzymatic activity capable of cleaving phosphodiester bonds specifically at AP sites. The technique detailed here, requires a ‘dU-nicking’ tool that leaves a 5-phosphate which is suitable for ligation in vivo.
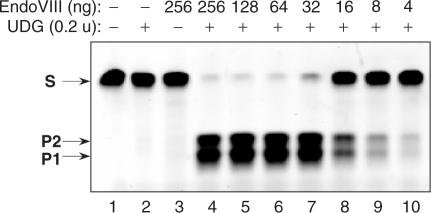
Upon screening various AP lyases, we found E. coli endonuclease VIII (EndoVIII DNA glycosylase/AP lyase) possesses the most appropriate AP site cleavage characteristics. To determine the optimal ratio of enzymes, a series of UDG and EndoVIII enzyme mixes were prepared. In the mixes, the concentration of EndoVIII varied from 4 to 256 ng, while the concentration of UDG was kept constant at 0.2 activity units, which is enough to release 12 pmol of uracil per minute (New England Biolabs). The resulting mixtures were assayed for complete nicking of 10 pmol ds oligonucleotide substrate containing a single dU residue as described in Materials and Methods. The results of the assay (Figure 4) show that complete digestion of substrate occurs with the mixture that has 32 ng of EndoVIII. This enzyme mix, USER enzyme, removes the dU nucleotide in a double-stranded DNA molecule, leaving a single nucleotide gap bordered by 5′ and 3′ phosphates.
Figure 4.
USER enzyme activity assay. A 34-mer oligonucleotide duplex (10 pmol) containing a single dU paired with a deoxyadenine (Table 1) was incubated with a series of EndoVIII and UDG enzyme mixtures for 15 min at 37°C in a 10 μl of T4 DNA ligase reaction buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 10 mM DTT, 1 mM ATP, 20 μg/ml BSA). The reactions were quenched by the addition of 10 μl of 95% formamide, 0.1% xylene cyanol, 0.1% bromophenol blue, 10 mM EDTA, pH 11, and the reaction products were analyzedon a 15% TBE-Urea denaturing gel. S, 34-nt oligonucleotide substrate. P1 and P2, 15 nt and 18 nt cleavage products, respectively. Two product bands of differing size result from the fact that the uracil is not in the centre of the substrate. Lane 1—no enzyme added. Lane 2—reaction contains 0.2 units of UDG. Lane 3—reaction contains 256 ng of EndoVIII. Lanes 4–10, reaction contains 0.2 units of UDG and the amount of EndoVIII shown above the respective lanes.
Assembly and directional cloning of PCR products into pNEB206A
For method development, the chloramphenicol acetyltransferase (Cat) gene of the pGPS2.1 plasmid (New England Biolabs) was chosen as a target on which to perform sequence manipulations. At position 248 nt of the Cat gene, the silent mutation G to C was introduced to create an NdeI restriction site (sequence changes are shown by asterisks in Figure 5A). At position 575 nt, the codon CAT was replaced by GCT to introduce a His193Ala substitution, which creates the catalytically inactive variant of chloramphenicol acetyltransferase (25).
Figure 5.
Outline of the chloramphenicol (Cat) gene mutagenesis and assembly experiment. (A) PCR primer design strategy. Lines with the arrowheads symbolize priming sequences in the PCR primers. Left and Right cloning primers contain 5′ extensions compatible with the single-stranded extensions on the linearized pNEB206A vector. The overlapping primers P1/P2 and P3/P4 are selected in the vicinity of the targeted mutations. Asterisks designate the point mismatches in the primers compared to the template sequence. Overlapping primers are complementary to each other within the boxed sequences. The 3′ dT of the boxed sequence in the primer sequences is replaced by dU. (B) Schematic representation of six PCR products amplified using the indicated primers. Cycling conditions are described in Materials and Methods. The label, Cat926 etc. shows the PCR fragment size in bp. The ends of the PCR products are flanked by the compatible overlapping sequences with a single dU residue at the junction. (C) Four assembly reactions were carried out using the indicated PCR fragments to assemble a full-length Cat gene carrying the indicated phenotype into pNEB206A.
PCR primers used for Cat gene engineering and cloning are listed in Table 1. Primer locations and their directions within the Cat gene are shown in Figure 5A. Six PCR fragments from the Cat gene were amplified either with Taq or PfuCx polymerases using PCR primers shown in Figure 5B. Two sets of assembly reactions, a set for each DNA polymerase derived PCR products, were set up as described in Materials and Methods and shown in Figure 5C. Assembly reactions were incubated for 15 min at 37°C to cleave at dU residues and then incubated for an additional 15 min at room temperature to allow annealing of the complementary extensions. Five microlitres of each assembly reaction was used for transformation and blue-white selection was applied to distinguish transformants carrying empty vector, which should form blue colonies. Since the template pGPS 2.1 does not replicate in ordinary lab strains of E. coli (26,27) all white colonies recovered from transformation were counted as recombinants.
The Cat gene assembly data is summarized in Table 2. The percentage of recombinant colonies, which was determined by dividing the number of white colonies by the total number of transformants, was calculated for each assembly reaction. As can be seen from the presented data, the PfuCx-derived assembly reactions have a slightly higher percentage of recombinants compared to Taq. The lower percentage of recombinants for Taq-derived assembly reactions may be explained by the variability of the 3′ ends of PCR products. Due to the absence of 3′ to 5′ proofreading activity, Taq polymerase tends to add an extra base on the ends of PCR fragments. PCR fragments carrying extra bases interfere with efficient assembly. This is easily noticeable when comparing three-fragment assembly reactions: the percentage of Taq-derived recombinants is 84% compared to 95% obtained from PfuCx-derived assembly reactions.
Table 2.
Transformation efficiency of assembled PCR fragments generated either by Taq DNA polymerase or PfuCx DNA polymerase
| Number of PCR fragments assembled (fragment size in bp) | Number of white colonies (recombinants)a c.f.u./50 ng vector | Percentage of recombinant coloniesb (%) | Assembly efficiencyc (%) | |||
|---|---|---|---|---|---|---|
| Taq DNA polymerase | PfuCx DNA polymerase | Taq DNA polymerase | PfuCx DNA polymerase | Taq DNA polymerase | PfuCx DNA polymerase | |
| One (926) | 1.12 × 104 | 1.52 × 105 | 98 | 99 | 9.7 | 11.0 |
| Two (142,792) | 0.44 × 104 | 0.85 × 105 | 91 | 98 | 3.8 | 6.2 |
| Two (461, 474) | 0.73 × 104 | 0.86 × 105 | 96 | 98 | 6.3 | 6.2 |
| Three (142, 327,474) | 0.26 × 104 | 0.36 × 105 | 84 | 95 | 2.3 | 2.6 |
aBlue–white selection was performed to distinguish the recombinant colonies (white) from the colonies carrying vector background (blue).
bPercentage of recombinants was determined by dividing the number of white colonies by the total number of transformants.
cAssembly efficiency shows what fraction of transformants is recovered from each assembly reaction compared to the total cell competency. Cell competency of ER2267competent cells used for transformation of Taq-generated PCR fragments was 1.15 × 105 c.f.u. per 50 ng of uncut vector pNEB206A. Cell competency of NEB5-α competent cells used for transformation of PfuCx-generated PCR fragments was 1.38 × 106 c.f.u. per 50 ng of uncut vector pNEB206A.
Next, we compared the fraction of recombinants recovered from the assembly of one, two or three PCR fragments. To determine assembly efficiency, the number of recombinants obtained from each assembly reaction was divided by the number of transformants obtained from transformation of 50 ng of uncut pNEB206A plasmid, which was set to 100% (Table 2). The fraction of recombinants obtained from one-fragment assembly was similar for both tested DNA polymerases falling in the range of 10% of the total cell competency. When compared to one-fragment assembly, the fraction of recombinants obtained from two- and three-fragment assemblies was approximately 2-fold (3.8–6.3%) and 4-fold (2.3–2.6%) lower, respectively.
To verify recombinant plasmids for the expected sequence changes, as well as for the presence of errors introduced by PCR, plasmid DNA was purified from 48 randomly selected white colonies (6 colonies from each assembly reaction) and sequenced across the Cat gene. Sequencing data showed that all constructs recovered from assembly reactions 2 and 4 (Figure 5C) had a newly introduced NdeI site, and in all constructs from assembly reactions 3 and 4 the codon CAT was replaced by codon GCT indicating that mutagenesis efficiency was 100%. No sequence rearrangements were detected at the vector–fragment or fragment–fragment junctions (in total 72 junctions in 48 constructs), indicating exceptionally high precision of assembly reactions. However, Taq- and PfuCx-polymerase-derived constructs differ in the number of PCR errors. While no PCR errors were detected in PfuCx-polymerase-derived constructs, Taq DNA polymerase introduced 13 PCR errors per 24 constructs, which equals to ∼0.6 errors per each kilobase.
As noted above, the pGPS2.1 template does not replicate in ordinary E. coli strains (26,27), therefore the template background was not an issue in experiments detailed here. However, elimination of plasmid template is crucial in applications where PCR fragments are amplified from ordinary plasmids carrying the ampicillin-resistant gene. We have tested different template elimination protocols, and the most efficient and the least labor-intensive protocol is treatment of PCR products with DpnI, which cleaves methylated DNA (i.e. template DNA), but does not cleave PCR products. Since DpnI works in the majority of PCR buffers, digestion may be performed directly in the PCR fragment mix. No additional purification step is required after digestion, but before proceeding to the assembly step, DpnI should be heat-inactivated by incubation at 80°C for 20 min to prevent digestion of the pNEB206A vector.
Rheoactivator gene engineering
The pNEBR-R1 plasmid (Receptor plasmid for RheoSwitch® Mammalian Inducible Expression System) expresses an engineered nuclear receptor heterodimer consisting of two proteins, RheoReceptor-1 (Choristoneura furniferana ecdysone receptor, CfEcR) and RheoActivator (Hybrid Human/LocusT retinoid X receptor) (33,34). The two proteins constitute the holoreceptor and regulate transcription of genes cloned into the expression vector, pNEBR-X1 (New England Biolabs). The RheoActivator gene in pNEBR-R1 is expressed from the constitutive ubiquitin UbC promoter. We employed the USER friendly DNA engineering method to modify the nucleotide sequence of RheoActivator gene. A single-format experiment was designed to achieve multiple goals such as: (i) to position the RheoActivator gene under the control of the CMV (early cytomegalovirus) promoter; (ii) to delete NcoI, PvuI and PstI restriction sites from the CMV promoter; (iii) to delete ApaI, SmaI, EcoRI, BclI and two BamHI restriction sites from the Rheoactivator gene and (iv) to insert two new restriction sites (AvrII and AgeI) between the CMV promoter and the Rheoactivator gene. To achieve this, a directional assembly of seven intermediate PCR fragments into pNEB206A vector was performed. The 3′terminal portion (540 bp) of the CMV promoter was amplified as three intermediate fragments, 153 bp, 184 bp and 218 bp in length. The junction sites within the CMV promoter sequence were chosen in the vicinity of the NcoI, PvuI and PstI sites targeted for elimination so that the desired sequence changes could be engineered into the primer sequences. Similarly, the Rheoactivator gene sequence was split into four intermediate fragments, 144 bp, 142 bp, 190 bp and 470 bp in length, in the vicinity of ApaI, SmaI, EcoRI + BamHI and BamHI + BclI sites targeted for elimination. Fourteen PCR primers carrying the intended sequence changes were designed to generate seven intermediate PCR fragments (Table 1). The 5′ ends of the flanking primers (R47 and R60) were extended by 8-nucleotide sequences suitable for cloning into pNEB206A vector. The six pairs of the overlapping primers (R48 and R49, R50 and R51, R52 and R53, R54 and R55, R56 and R57, R58 and R59) were designed to generate 8–9-nucleotide long overlaps at the respective junctions of the PCR fragments. All PCR primers contained a single dU residue flanking the 3′ side of the overlap sequence. Three overlapping fragments of the CMV promoter were amplified from pBind-Gal4-cf-EcR plasmid template using the PfuCx DNA Polymerase and primer pairs R47 + R48, R49 + R50 and R51 + R52. The four overlapping RheoActivator gene fragments were amplified from pNEBR-R1 plasmid template using primer pairs R53 + R54, R55 + R56, R57 + R58 and R59 + R60. Agarose gel electrophoresis of the PCR products showed the expected PCR-amplified DNA fragments (Figure 6A). The proper assembly of the intermediate fragments was verified by performing a 15-min control ligation of the USER-treated PCR products. Ligation is necessary to avoid dissociation of the assembled products during agarose gel electrophoresis. Agarose gel pattern of a 10 μl sample from the ligation reaction is shown in Figure 6A, lane 9. As expected, a variety of intermediate ligation products were formed, with the largest product being 1500 bp (indicated by white arrow). This is the size of the intended CMV promoter and Rheoactivator gene fusion, indicating a full-length assembly product was formed during directional assembly of seven intermediate USER-treated PCR fragments.
Figure 6.
Rheoactivator gene engineering. (A) Electrophoretic analysis of the PCR products amplified using PfuCx DNA Polymerase. 5 μl of each PCR reaction corresponding either to the CMV promoter amplification products (Lanes 1–3) or to the Rheoactivator gene amplification products (Lanes 4–7) were loaded on a 1% agarose gel. Lane 1—PCR with primers R47 + R48 (product size 153 bp). Lane 2—PCR with primers R49 + R50 (product size 184 bp). Lane 3—PCR with primers R51 + R52 (product size 218 bp). Lane 4—PCR with primers R53 + R54 (product size 144 bp). Lane 5—PCR with primers R55 + R56 (product size 142 bp). Lane 6—PCR with primers R57 + R58 (product size 190 bp). Lane 7—PCR with primers R59 + R60 (product size 470 bp). Lane 8—50 μg of the linearized pNEB206A vector. Lane 9—control ligation of the USER-treated PCR fragments shown in Lanes 1–7. White arrow indicates the 1500-bp ligation product that represents the full-length CMVpromoter and Rheoactivator gene fusion. Lane M corresponds to the 2-log DNA ladder. (B) Clone screening by PCR amplification directly from 10 white colonies with primers R51 and R56 that will only amplify clones carrying a CMV promoter and Rheoactivator gene fusion. (C), Plasmid DNA from clones 1, 2, 4, 5, 7 and 8 were analyzed by restriction digestion with BbvCI restriction endonuclease.
For assembly into the pNEB206A vector, 5 μl aliquots from each PCR reaction were combined and digested with 2 μl (40 units) of DpnI restriction endonuclease for 1 h at 37°C to destroy template plasmids. After completion of digestion, DpnI was inactivated by incubation for 20 min at 80°C. The PCR fragment mix was supplemented with 2 μl (100 ng) of the linearized pNEB206A and 2 μl (2 units) of the USER enzyme. The assembly reaction was then incubated for 15 min at 37°C and an additional 30 min at room temperature. Escherichia coli NEB5-α competent cells were transformed with 5 μl of assembly reaction as described in the Materials and methods section. The transformants were selected on ampicillin plates by performing blue/white screening. In total, 28 white (recombinant) and 98 blue (vector background) colonies were recovered from a 100 μl aliquot of transformation reaction. Next, 10 individual white colonies were screened by colony PCR using primers R51 and R56 that will only amplify clones carrying a fusion of the CMV promoter and the Rheoactivator gene. The expected 500-bp PCR fragment was produced in 8 out of 10 clones, suggesting these clones carry the properly merged chimeric fusion (Figure 6B). The restriction digestion analysis of plasmid DNA purified from six positive clones revealed an expected 1500-bp size insert that corresponded to the full-length fusion of the CMV promoter and Rheoactivator gene (Figure 6C). To confirm nucleotide sequence, all six plasmids were sequenced across the insert. Nucleotide sequence analysis showed that all clones carried the full set of intended sequence changes, indicating 100% mutagenesis efficiency. No sequence divergence at the PCR fragment junctions was observed. However, three out of six clones showed either PCR-based or primer-based point mutations.
DISCUSSIONS
Two new techniques were employed to develop a new approach to customize DNA sequences. The first technique uses a new group of nicking endonucleases, which cleave only one strand of ds DNA in a site-specific manner (22–24). This site-specific nicking is used to create insertion sites in vectors with unique non-palindromic 3′ ss extensions. Ideally, the vector backbone should not have additional nicking sites besides those in the cassette. Thus, to create pNEB206A vector we used the nicking endonuclease Nt.BbvCI (24) which recognizes the seven base pair sequence, CC↓TGAGG, and which is not present (or infrequently present) in the sequences of commonly used cloning vectors. We have now determined that a complete absence of additional nicking sites is not essential as long as nicked sites on opposite strands are separated by at least 100 bp. Currently, a variety of site-specific nicking endonucleases recognizing either six or seven base pair sequences are commercially available from New England Biolabs, therefore the design of single-stranded extensions on the vectors may be custom modified by replacing the nicking/restriction sites in the cassette. Recently, a comprehensive set of the USER compatible vectors has been constructed by other researchers for constitutive expression in a variety of host organisms (35).
The other technique employed to customize DNA sequences is an improvement to a ‘ligase-free UDG cloning’ method (11). DNA amplification using primers with a single dU placed near their 5′ end followed by incubation with the USER enzyme creates PCR fragments with unique non-palindromic 3′ ss extensions. This allows for accurate, multi-fragment assemblies among the multiple PCR fragments, as well as insertion into the vector, all in one step.
The concept and experiments presented here are flexible, in that, many different nicking enzyme specificities could be used. Furthermore, different modified bases along with their cognate ‘repair’ enzyme can be used to create breaks near the 5′ ends of amplified fragments. Several E. coli DNA glycosylases/AP lyases are capable of breaking phosphodiester bonds at the AP-sites (19,28,29). For example, formamidopyrimidine-DNA glycosylase, also known as FPG, can excise a variety of oxidized purines (29), whereas either DNA endonucleasae III or endonuclease VIII both can excise a variety of oxidized pyrimidines (19,28). Due to the AP-lyase activity, which can act independently of associated glycosylase activity, these enzymes are also capable of breaking phosphodiester bonds at AP sites, but Endo III breaks the first phosphodiester bond 3′ to the AP site (28), whereas FPG and EndoVIII are capable of breaking phosphodiester bonds on both 3′ and 5′ sides of the AP site (19,29). In general, the combination of DNA glycosylases in the mixture depends on (i) the type of modified nucleotide to be excised and (ii) the type of termini desired at the nick location. For example, by combining the uracil DNA glycosylase hSMUG1 (30) with EndoVIII we have created a nicking tool, which is specific for hydroxymethyluracil (data not shown). A combination of three enzymes, UDG, EndoIV and EndoVIII, was used to nick at dU and generate a 5′ phosphate and 3′ hydroxyl at DNA termini (data not shown).
We created an efficient and fast method for assembly of desired DNA molecules from multiple PCR fragments, referred to as the USER friendly DNA engineering method. The most distinctive feature of the technique is its universality, as it combines nucleotide sequence manipulation, PCR fragment assembly and directional cloning in a single experimental format. By modifications in the primers, which, during DNA amplification are incorporated into the ends of the PCR products, virtually any envisioned DNA manipulation may be performed at any location of a target DNA. The junction of PCR fragments across single-stranded extensions generated by the USER enzyme is extremely precise. Since the assembly reaction is ligase free, the extensions with mismatched sequences do not yield stable recombinant molecules and are lost during transformation. In addition, when proofreading polymerase PfuCx is used for DNA amplification, the PCR errors are rare. PfuCx DNA polymerase is a genetically engineered derivative of Pfu polymerase (21). No wild-type proofreading DNA polymerases, such as Vent, Deep-Vent or Pfu, can yield PCR product from primers carrying dU (14,20). Archaeal DNA polymerases possess a uracil-binding pocket which blocks extension past a uridine (20,21). In addition to PfuCx, the other DNA polymerase compatible with the USER friendly DNA engineering method is Taq DNA polymerase. Despite the fact that Taq polymerase has a higher error rate, we have successfully used it in numerous site-specific mutagenesis and gene fusion experiments where a two or three PCR fragment assembly was required.
To date, the highest number of PCR fragments successfully assembled in one reaction was seven. It should be pointed out that the efficiency of directional assembly of PCR fragments to a great degree depends on the uniqueness of nucleotide sequences at the junctions. However, in some cases, the choices for junction sequence selection are very limited due to unfavorable A/T content of the target DNA. Thus, when more than five PCR fragments are assembled in the same reaction, there is a greater possibility that junctions will have similar sequences and the fragments may tend to assemble incorrectly, which significantly reduces the number of recombinants. For example, in one of our experiments the goal was to delete seven restriction sites from the LexA gene, thus a six-fragment assembly was performed to introduce seven silent mutations simultaneously. Initially, the percentage of recombinants was very poor (14%), but by shifting the position of one faulty junction we were able to eliminate the mis-annealing of two intermediate fragments and the yield of recombinants was increased to 60%.
In addition to altering the sequence of a target DNA, the USER enzyme could be used in conjunction with PfuCx DNA polymerase to amplify the cloning vector and in the same reaction add sequence junctions compatible for assembly of targeted PCR fragments. This approach was successfully employed to generate precise protein fusions for rapid protein purification using either the IMPACT™ (Intein Mediated Purification with an Affinity Chitin-binding Tag) system or the pMAL™ Protein Fusion and Purification system (31,32). In conclusion, the rapidity, precision and efficacy of the USER friendly DNA engineering method outweigh the higher costs of dU in PCR primers, as well as a limited choice of DNA polymerases.
ACKNOWLEDGEMENTS
The authors are grateful to Richard J. Roberts and William Jack for reviewing the manuscript. We also thank Elisabeth Raleigh and Fiona Stewart for scientific and technical assistance, Laurie Mazzola for DNA sequencing and John Buswell for oligonucleotide synthesis. We are especially thankful for the environment and suppor provided by Don Comb. Funding to pay the Open Access publication charge was provided by New England Biolabs.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. U.S.A. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hofer B, Kuhlein B. A high efficiency method for site-directed mutagenesis with any plasmid. Gene. 1989;84:153–157. doi: 10.1016/0378-1119(89)90149-2. [DOI] [PubMed] [Google Scholar]
- 3.Taylor JW, Ott J, Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985;13:8764–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashimoto-Gotoh T, Mizuno T, Ogasahara Y, Nakagawa M. An oligodeoxyribonucleotide-directed dual amber method for site-directed mutagenesis. Gene. 1995;152:271–275. doi: 10.1016/0378-1119(94)00750-m. [DOI] [PubMed] [Google Scholar]
- 5.Deng WP, Nickoloff JA. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 6.Andrews CA, Lesley SA. Selection strategy for site-directed mutagenesis based on altered beta-lactamase specificity. Biotechniques. 1998;24:972–974. doi: 10.2144/98246st03. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interaction. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho SF, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 9.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 10.Ge L, Rudolph P. Simultaneous introduction of multiple mutations using overlap extension PCR. Biotechniques. 1997;22:28–30. doi: 10.2144/97221bm03. [DOI] [PubMed] [Google Scholar]
- 11.Nisson PE, Rashtchian A, Watkins PC. Rapid and efficient cloning of Alu-PCR products using uracil DNA glycosylase. PCR Methods Appl. 1991;1:120–123. doi: 10.1101/gr.1.2.120. [DOI] [PubMed] [Google Scholar]
- 12.Rashtchian A, Thornton CG, Heidecker G. A novel method for site-directed mutagenesis using PCR and uracil DNA glycosylase. PCR Methods Appl. 1992;2:124–130. doi: 10.1101/gr.2.2.124. [DOI] [PubMed] [Google Scholar]
- 13.Booth PM, Buchman GW, Rashtchian A. Assembly and cloning of coding sequences for neurotrophic factors directly from genomic DNA using polymerase chain reaction and uracil DNA glycosylase. Gene. 1994;146:303–308. doi: 10.1016/0378-1119(94)90310-7. [DOI] [PubMed] [Google Scholar]
- 14.Lasken RS, Schuster DM, Rashtchian A. Archaebacterial DNA polymerases tightly bind uracil-containing DNA. J. Biol. Chem. 1996;271:17692–17696. doi: 10.1074/jbc.271.30.17692. [DOI] [PubMed] [Google Scholar]
- 15.Rashtchian A. Novel methods for cloning and engineering genes using the polymerase chain reaction. Curr. Opin. Biotechnol. 1995;6:30–36. doi: 10.1016/0958-1669(95)80006-9. [DOI] [PubMed] [Google Scholar]
- 16.Smith C, Day PJR, Walker MR. Generation of cohesive ends on PCR products by UDG-mediated excision of dU, and amplification for cloning into restriction digest-linearized vectors. PCR Methods Appl. 1993;2:328–332. doi: 10.1101/gr.2.4.328. [DOI] [PubMed] [Google Scholar]
- 17.Watson DE, Bennett GN. Cloning and assembly of PCR products using modified primers and DNA repair enzymes. Biotechniques. 1997;23:858–864. doi: 10.2144/97235st01. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl T, Ljungquist S, Siegert W, Nyberg B, Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 1977;252:3286–3294. [PubMed] [Google Scholar]
- 19.Jiang D, Hatahet Z, Melamede RJ, Kow YW, Wallace SS. Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem. 1997;272:32230–32239. doi: 10.1074/jbc.272.51.32230. [DOI] [PubMed] [Google Scholar]
- 20.Greagg MA, Fogg MJ, Panayotou G, Evans SJ, Connolly BA, Pearl LH. A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9045–9050. doi: 10.1073/pnas.96.16.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fogg MJ, Pearl LH, Connolly BA. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 2002;9:922–927. doi: 10.1038/nsb867. [DOI] [PubMed] [Google Scholar]
- 22.Besnier CE, Kong H. Converting MlyI endonuclease into a nicking enzyme by changing its oligomerization state. EMBO Rep. 2001;9:782–786. doi: 10.1093/embo-reports/kve175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Z, Samuelson JC, Zhou J, Dore A, Xu SY. Engineering strand-specific DNA nicking enzymes from the type IIS restriction endonucleases BsaI, BsmBI, and BsmAI. J. Mol. Biol. 2004;337:573–583. doi: 10.1016/j.jmb.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Heiter DF, Lunnen KD, Wilson GG. Site-specific DNA-nicking mutants of the heterodimeric restriction endonuclease R.BbvCI. J. Mol. Biol. 2005;348:631–640. doi: 10.1016/j.jmb.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 25.Leslie AG, Moody PC, Shaw WV. Structure of chloramphenicol acetyltransferase at 1.75-A resolution. Proc. Natl. Acad. Sci. U.S.A. 1988;85:4133–4137. doi: 10.1073/pnas.85.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolter R, Inuzuka M, Helinski DR. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 27.Metcalf WW, Jiang W, Wanner BL. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene. 1994;138:1–7. doi: 10.1016/0378-1119(94)90776-5. [DOI] [PubMed] [Google Scholar]
- 28.Dizdaroglu M, Laval J, Boiteux S. Substrate specificity of the Escherichia coli endonuclease III: excision of thymine- and cytosine-derived lesions in DNA produced by radiation-generated free radicals. Biochemistry. 1993;32:12105–12111. doi: 10.1021/bi00096a022. [DOI] [PubMed] [Google Scholar]
- 29.Graves RJ, Felzenszwalb I, Laval J, O'Connor TR. Excision of 5′-terminal deoxyribose phosphate from damaged DNA is catalyzed by the Fpg protein of Escherichia coli. J. Biol. Chem. 1992;267:14429–14435. [PubMed] [Google Scholar]
- 30.Masaoka A, Matsubara M, Hasegawa R, Tanaka T, Kurisu S, Terato H, Ohyama Y, Karino N, Matsuda A, et al. Mammalian 5-formyluracil-DNA glycosylase. 2. Role of SMUG1 uracil-DNA glycosylase in repair of 5-formyluracil and other oxidized and deaminated base lesions. Biochemistry. 2003;42:5003–5012. doi: 10.1021/bi0273213. [DOI] [PubMed] [Google Scholar]
- 31.Chong S, Mersha FB, Comb DG, Scott ME, Landry D, Vence LM, Perler FB, Benner J, Kucera RB, et al. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 32.Maina CV, Riggs PD, Grandea AG, Slatko BE, Moran LS, Tagliamonte JA, McReynolds LA, Guan CD. An Escherichia coli vector to express and purify foreign proteins by fusion to and separation from maltose-binding protein. Gene. 1988;74:365–373. doi: 10.1016/0378-1119(88)90170-9. [DOI] [PubMed] [Google Scholar]
- 33.Palli SR, Kapitskaya MZ, Kumar MB, Cress DE. Eur. J. Biochem. 2003;270:1308–1315. doi: 10.1046/j.1432-1033.2003.03501.x. [DOI] [PubMed] [Google Scholar]
- 34.Karzenowski D, Potter DW, Padidam M. Inducible control of transgene expression with ecdysone receptor: gene switches with high sensitivity, robust expression, and reduced size. Biotechniques. 2005;39:191–196. doi: 10.2144/05392ST01. [DOI] [PubMed] [Google Scholar]
- 35.Nour-Eldin HH, Hansen BG, Norholm MH, Jensen JK, Halkier BA. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 2006;34:e122. doi: 10.1093/nar/gkl635. [DOI] [PMC free article] [PubMed] [Google Scholar]