Abstract
The evaluation of phthalocyanine labels for the surface-enhanced resonance Raman scattering (SERRS) detection of oligonucleotides is reported. Three phthalocyanine-labelled oligonucleotides were assessed, each containing a different metal centre. Detection limits for each labelled oligonucleotide were determined using two excitation frequencies where possible. Limits of detection as low as 2.8 × 10−11 mol. dm−3 were obtained which are comparable to standard fluorescently labelled probes used in previous SERRS studies. The identification of two phthalocyanine-labelled oligonucleotides without separation was also demonstrated indicating their suitability for multiplexing. This study extends the range of labels suitable for quantitative surface-enhanced resonance Raman scattering with silver nanoparticles and offers more flexibility and choice when considering SERRS for quantitative DNA detection.
INTRODUCTION
Surface-enhanced resonance Raman scattering (SERRS) has been shown to be a more sensitive and specific detection method for the direct analysis of DNA compared to the more commonly used method of fluorescence (1). SERRS produces fingerprint spectra making it a more suitable technique for multiplexed identification of the components of a mixture without separation (2). While multiplexing is possible to a certain degree with fluorescence detection, the technique is limited due to the broad and often overlapping spectra obtained.
While Raman spectroscopy produces relatively weak spectra, enhancements of 1014 have been seen using SERRS (3). In order for this to occur, the analyte must be able to absorb onto a suitable metal surface (providing the surface enhancement) and contain a chromophore approximately coincident with the excitation frequency (leading to resonance enhancement). This combination results in a highly sensitive technique for which single molecule detection has been demonstrated (4,5). While DNA does not meet these requirements, many commonly used fluorescent labels do and as the metal surface commonly used quenches fluorescence, background interference is minimized (6).
Previously, DNA labelled with six commercially available dyes (Cy3, TAMRA, Texas Red, Cy3.5, Cy5 and Rhodamine 6G) has been detected by SERRS using gold nanoparticles with a silver coating singly and as multiplexes (7). In a separate study, silver nanoparticles were used as the metal substrate in the quantitative study of eight oligonucleotides labelled with the commonly used dyes FAM, TET, HEX, TAMRA, R6G, ROX, Cy3 and Cy5 (8). Linear calibration graphs and limits of detection (in some cases as low as 0.5 fmol) were obtained for each of the dyes analysed. That study was then extended to include BODIPY TR-X, Cy3.5, Cy5.5 and Yakima yellow-labelled oligonucleotides (9). Labelled probes have also been used in DNA assays based on SERRS. Vo Dinh et al. used a cresyl fast violet-labelled primer which was successfully incorporated into PCR for subsequent SERRS detection (10). Graham et al. have also demonstrated SERRS detection in multiplex genotyping (11). In this study, the presence or absence of three different genotypes was determined without separation.
While DNA detection by SERRS has been clearly demonstrated to be a powerful technique, there remains a need to expand the range of labels that can be used. Moreover, the range of families of compounds that can be used should be widened as many of the labels used so far in this technique are structurally related and therefore give similar, but distinguishable spectra. This article therefore presents the assessment of three phthalocyanine-labelled oligonucleotides as SERRS probes. Similar dyes have been reported as near-IR fluorescence labels (12) and have also been used to investigate sequence-specific modification of DNA (13). However, their properties as SERRS labels have not been reported until now.
MATERIALS AND METHODS
Labelled oligonucleotides
The labelled oligonucleotides were prepared using a previously reported method (14). The oligonucleotides were synthesized by standard phosphoramidite chemistry and contained a—O-(CH2)3NH2 linker at the 5′-end. Succinimidyl esters of phthalocyanines were then coupled to the oligonucleotides while still attached to the control pore glass (CPG). The conjugates were then cleaved from the CPG with concentrated ammonium hydroxide before purification by preparative reverse phase HPLC on Nucleosil 100-7 C18 column (4.6 × 250 mm, Macherey-Nagel, Düren, Germany). The structures of the phthalocyanines, phthalocyanine-oligonucleotide conjugate absorption maxima and the oligonucleotide sequences are shown in Figure 1. Free oligonucleotide and the conjugates had different Rf values in HPLC as shown in Figure 2. The conjugate UV–Vis spectrum contained both oligonucleotide and phthalocyanine bands. After conjugation, the shape of the spectrum at 600–70 nm is significantly changed in comparison with the absorption spectrum of free phthalocyanine as shown in Figure 3.
Figure 1.
Structure of phthalocyanine conjugates.
Figure 2.
Reverse-phase HPLC data for preparation of oligonucleotide conjugates. A mixture of 20% acetonitrile (A) and water (B), both containing 0.05 M triethylamine-acetate buffer (pH = 7.0) was used as the mobile phase. The following gradient elution was run: 0 to 50 min: 0 to 100% A. The flow rate was 2.0 ml/min, and the detection wavelength was 260 nm. 1 – starting oligonucleotide; 2 – conjugate with PtcCo.
Figure 3.
Absorption spectra of free phthalocyanine (- - - - -) and its oligonucleotide conjugate (——) for (A) Ptc Co, (B) Ptc Al and (C) Ptc Zn.
Silver nanoparticle preparation
Citrate-reduced silver nanoparticles were prepared in a colloidal suspension using a modified Lee and Meisel procedure (15).
Instrumentation
The labelled oligonucleotides were analysed using three excitation wavelengths. A Renishaw inVIA microscope system with a 514.5 nm argon ion laser, using a ×20/0.4 long working distance objective to focus the laser beam into a microtitre plate well containing the sample was used. A Renishaw Ramascope system 2000 was used with a 632.8 nm helium-neon laser or a 785 nm Renishaw diode laser. A ×20 objective was used to focus the laser beam using both these wavelengths into a microtitre plate containing the sample.
Sample preparation
For the determination of the limits of detection, the dye-labelled oligonucleotides were diluted to various concentrations using sterile water (18.2 MΩ. cm). Samples were then prepared for SERRS analysis by adding 7 μL of dye-labelled oligonucleotide and 10 μL of 0.1 mol dm−3 spermine, followed by 175 μL of water and finally 175 μL of silver nanoparticles. The samples were analysed within 1 min of the addition of the silver nanoparticles and each concentration was analysed five times. The aggregation process of the nanoparticles is dynamic and in order to provide reproducibility the measurements were carried out within the same time frame each time. The spectra were obtained with the spectrometer grating centred at 1400 cm−1 and with a 10 s accumulation time. The spectra were baseline corrected using the GRAMS/32 software, and the average peak height of the strongest peak in the spectra was plotted against the concentration after being normalized to the silicon standard peak.
For the assessment of the suitability of the dye-labelled oligonucleotides for multiplexing, a mixture of two dye-labelled oligonucleotides was added, to a total volume of 30 μL or less, to 10 μL of 0.1 mol dm−3 spermine, followed by 175 μL of water and finally 175 μL of silver nanoparticles.
RESULTS AND DISCUSSION
Phthalocyanines were chosen as labels for this study as they are chemically and photochemically stable and have large extinction coefficients (>105 cm−1 M−1) (12). In addition, they can be made either fluorescent or non-fluorescent depending on the choice of metal centre (12). They are highly versatile as they can be modified via their metal centre or substituent side chains giving rise to distinct Raman spectra. The three phthalocyanine labels used in this study and the oligonucleotide sequences are shown in Figure 1. Modification of the oligonucleotide sequence with propargylamine as in previous studies was not required for SERRS detection. Each of the phthalocyanine labels used had a different metal centre—aluminium (PtcAl), zinc (PtcZn) and cobalt (PtcCo)—and each had different side chains—SO2NH(CH2)5COOH, SCH2COOH and COOH. The absorption maximum varies for each dye label as this is determined by the metal centre present and the substituents around the ring. The absorption maxima are as follows: PtcAl—640 nm, PtcZn—680 nm and PtcCo—625 nm. This indicates that the labels are more in resonance with the 632.8 nm laser excitation source than the 514.5 or 785 nm excitation.
A 0.1 mol. dm−3 solution of spermine, a naturally occurring tetramine, was used in this study as an aggregating agent as this had previously been shown to give maximum SERRS signal enhancement in the analysis of dye-labelled oligonucleotides (8). The spermine neutralizes the negatively charged phosphate backbone of the oligonucleotide, which otherwise may inhibit attachment onto the negatively charged citrate-coated silver surface but will also aid hybridization (16). It also aggregates the silver nanoparticles into discrete clusters producing the roughened metal surface required for the SERRS effect.
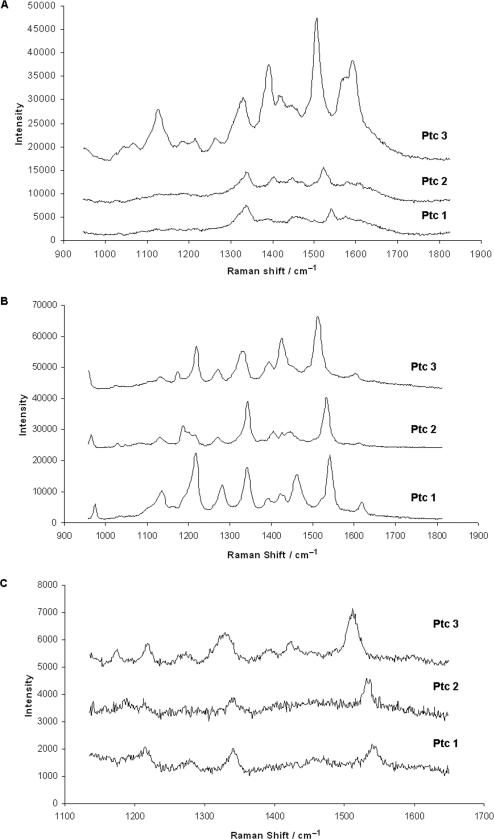
The SERRS spectra of the three-labelled oligonucleotides using the three excitation wavelengths are shown in Figure 4. Only weak SERRS was obtained using 785 nm excitation and only at the highest concentration. The oligonucleotides labelled with PtcCo and PtcAl also only gave weak SERRS with the highest concentration at 514.5 nm excitation. PtcZn, however, gave markedly stronger signals with a different pattern of relative intensities from that recorded at 632.8 nm. The signals come from the phthalocyanine and are not affected by the oligonucleotide sequence. This is due to the resonance contribution from the label dominating the spectra.
Figure 4.
SERRS Spectra from the three phthalocyanine-labelled oligonucleotides using a 10 s scan and excitation at (A) 514.5 nm, (B) 632.8 nm and (C) 785 nm. The final concentration of sample for (A) and (C) was 1.9 × 10−8 mol dm−3 and for (B) was 3.9 × 10−9 mol dm−3. All spectra have been base line corrected.
The electronic spectrum in PtcZn has an extended shoulder from the B band up to about 520 nm which is not present in the spectra of PtcCo and PtcAl (13). This may permit a more effective resonance contribution to the surface enhancement by electronic coupling with the B band. The increased relative intensity of the bands at about 1600 cm−1 is also observed in the resonance Raman spectra of phthalocyanines excited in the B band region.
The most effective SERRS spectra were obtained using 632.8 nm excitation which is in resonance with the Q band of the dyes for which λmax ranged from 625 to 675 nm. Within the band envelop of the plasmon resonance for some small clusters lies 632.8 nm, which is also a frequency and therefore gives effective surface enhancement from the aggregated colloid. The most intense band in each spectrum was between 1510 and 1545 cm−1 and its position is directly related to the metal ion present in the complex (18). The major feature of this vibration is the large displacement on the C-N-C bridges between the benzopyrrole groups. The frequency shift is therefore dependent on both the strength of the bridge bonds and the effect that the metal ion has on the shape of the ring. For example, whereas CoPtc has a cavity diameter of 3.82 Å in its equilibrium state ZnPtc has a cavity diameter of around 3.96 Å. As well as affecting the frequency of the intense band at about 1510 and 1545 cm−1, the loss of effective D4h symmetry causes changes in the electronic spectra as discussed above for ZnPtc and differences between molecules in the relative intensity of some of the weaker peaks in the SERRS spectra.
The frequency dependence of the main peak on molecular structure provides multiplexing potential for this class of dyes. The most intense peaks are distinct enough to enable a mixture of two phthalocyanine-labelled oligonucleotides to be easily identified in a mixture as shown in Figure 5. The oligonucleotides labelled with PtcCo and PtcZn were analysed in two mixtures of differing ratios. Despite being in the same region, the major peak for each label could easily be identified and the peak height was seen to vary with the quantity of sample present. The instrument was set to provide a broad spectral range but by concentrating only on the 1500–1550 cm−1 region, greater numbers of phthalocyanines could be identified in a mixture.
Figure 5.
Spectra of mixtures of PtcCo and PtcZn with the following ratios: A–1:2 and B–2:1. Spectra obtained using 632.8 nm laser excitation with a 10s accumulation.
Limits of detection were determined for each of the labelled oligonucleotides using 632.8 nm excitation and for PtcZn, using 514.5 nm excitation. The concentration graphs obtained are shown in Figure 6. These show the linear concentration dependence of the labelled oligonucleotides using both excitation wavelengths. The error bars represent ±1 SD and the R.S.D. for the data set varied between 3 and 17%. Again, as shown in other DNA detection studies with SERRS, the technique has been shown to deliver quantitative and reproducible results. The limits of detection obtained are shown in Table 1. Similar detection limits were seen for the three labels using 632.8 nm excitation. As noted earlier, strong signals were obtained for PtcZn at 514.5 nm allowing limits of detection to be determined. The limits of detection for the phthalocyanine-labelled oligonucleotides are comparable to those previously obtained from commonly available fluorescently labelled oligonucleotides. The limit of detection does vary with the nature of the label and in previous studies (8), this has ranged from 56 (TET) to 1 (Rhodamine 6G) pmoldm−3 which are several orders of magnitude lower in limit of detection than achieved by fluorescence using routinely available instrumentation (1). An important feature of SERRS is that the detection limits in a multiplex are the same as for a labelled oligonucleotide on its own. This is highly significant as similar techniques such as fluorescence can experience a deterioration in sensitivity when multiplexed analyses are attempted.
Figure 6.
Calibration graph obtained for (A) PtcZn using 514.5 nm laser excitation and (B) Ptc Co, Ptc Al and Ptc Zn using 632.8 nm excitation. The error bars shown are ±1 S.D. and each point is the average of five repeat samples.
Table 1.
The detection limits for phthalocyanine labelled oligonucleotides at each excitation frequency
| Dye label | Detection limit (mol dm−3) | |
|---|---|---|
| 514.5 nm | 632.8 nm | |
| PtcCo | – | 3.2 × 10−11 |
| PtcAl | – | 2.8 × 10−11 |
| PtcZn | 1.4 × 10−10 | 3.2 × 10−11 |
CONCLUSIONS
Phthalocyanine dyes have been shown for the first time to be suitable labels for the detection of DNA by SERRS and were analysed at three wavelengths. Linear concentration-dependent graphs of SERRS intensity of three phthalocyanine labels using 632.8 nm excitation were obtained from which limits of detection were determined. These were comparable to the detection limits reported using 514.5 nm excitation by Faulds et al. (8) and are at biologically relevant concentrations. The addition of phthalocyanines as a new group of SERRS-DNA labels will allow increased multiplexing abilities using this technique. This study has also demonstrated that a mixture of two phthalocyanine labelled oligonucleotides can easily be identified without separation. Larger multiplexes should also be possible using only phthalocyanine labels due to the significant change in main peak position depending on the metal centre present. This also demonstrates multiplexing using 632.8 nm excitation rather than 514.5 nm excitation used in the previous study by Graham et al. (11). Thus, phthalocyanines have been assessed as labels for the SERRS detection of DNA at three wavelengths and their potential to enhance the multiplexing capacity of this technique has been demonstrated.
ACKNOWLEDGEMENT
The authors would like to thank the British Council for funding and forging the collaboration that led to this paper.Funding to pay for the open access publication charges for this article was provided by the Royal Society of Chemistry's Analytical Trust Fund through the award of the Analytical Grand Prix to DG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Faulds K, Barbagallo RP, Keer JT, Smith WE, Graham D. SERRS as a more sensitive technique for the detection of labelled oligonucleotides compared to fluorescence. Analyst. 2004;129:567–568. doi: 10.1039/b406423b. [DOI] [PubMed] [Google Scholar]
- 2.Munro CH, Smith WE, White PC. Qualitative and semi-quantitative trace analysis of acidic monoazo dyes by surface-enhance resonance Raman. Analyst. 1995;120:993–1003. [Google Scholar]
- 3.Stacy AM, Van Duyne RP. Surface enhanced Raman and resonance Raman spectroscopy in a nonaqueous electrochemical environment: Tris(2,2′-bipyridine)ruthenium(II) adsorbed on silver from acetonitrile. Phys. Chem. Lett. 1983;102:365–370. [Google Scholar]
- 4.Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari RR, Feld MS. Single molecule detection using surface-enhanced Raman scattering (SERS) Phys. Rev. Lett. 1997;78:1667–1670. [Google Scholar]
- 5.Nie S, Emory S. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 6.Rodger C, Smith WE, Dent D, Edmondsun J. Surface-enhanced resonance-Raman scattering: An informative probe of surfaces. J. Chem. Soc. Dalton Trans. 1996;5:791–799. [Google Scholar]
- 7.Cao YC, Jin R, Mirkin CA. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 8.Faulds K, Smith WE, Graham D. Evaluation of Surface-enhanced resonance Raman scattering for quantitative DNA analysis. Anal. Chem. 2004;76:412–417. doi: 10.1021/ac035060c. [DOI] [PubMed] [Google Scholar]
- 9.Faulds K, Stewart L, Smith WE, Graham D. Quantitative detection of dye labelled DNA using surface enhanced resonance Raman scattering (SERRS) from silver nanoparticles. Talanta. 2005;67:667–671. doi: 10.1016/j.talanta.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Isola NR, Stokes DL, Vo-Dinh T. Surface enhanced Raman gene probe for HIV detection. Anal. Chem. 1998;70:1352–1356. doi: 10.1021/ac970901z. [DOI] [PubMed] [Google Scholar]
- 11.Graham D, Mallinder DJ, Whitcombe D, Watson ND, Smith WE. Single multiplex genotyping by surface-enhanced resonance Raman scattering. Anal. Chem. 2002;74:1069–1074. doi: 10.1021/ac0155456. [DOI] [PubMed] [Google Scholar]
- 12.Hammer RP, Owens CV, Hwang SH, Sayes CM, Soper SA. asymmetrical, water-soluble Phthalocyanine dyes for covalent labeling of oligonucleotides. Bioconjugate Chem. 2002;13:1244–1252. doi: 10.1021/bc0155869. [DOI] [PubMed] [Google Scholar]
- 13.Chernonosov AA, Koval VV, Knorre DG, Chernenko AA, Derkacheva VM, Lukyanets EA, Fedorova OS. Conjugates of phthalocyanines with oligonucleotides as reagents for sensitized or catalytic DNA modification. Bioinorg. Chem. Appl. 2006;2006:1–8. doi: 10.1155/BCA/2006/63703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koval VV, Chernonosov AA, Abramova TV, Ivanova TM, Fedorova OS, Derkacheva VM, Lukyanets EA. Photosenstized and catalytical oxidation of DNA by metallophthalocyanine-oligonucleotide conjugates. Nucleosides, Nucleotides and Nucleic Acids. 2001;20:1259–1262. doi: 10.1081/NCN-100002531. [DOI] [PubMed] [Google Scholar]
- 15.Lee PC, Meisel D. Adsorption and surface-enhanced Raman of silver and gold sols. J. Phys. Chem. 1982;86:3391–3395. [Google Scholar]
- 16.Basu HS, Marton LJ. The interaction of spermine and pentamines with DNA. Biochem. J. 1987;244:243–246. doi: 10.1042/bj2440243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koval VV, Chernonosov AA, Abramova TV, Ivanova TM, Fedorova OS, Knorre DG. The Synthesis of a Cobalt(II) Tetracarboxyphthalocyanine–deoxyribooligonucleotide conjugate as a reagent for the directed DNA modification. Russ. J. Bioorg. Chem. 2000;26:104–110. [PubMed] [Google Scholar]
- 18.Tackley DR, Dent G, Smith WE. Phthalocyanines: structure and vibrations. Phys. Chem. Chem. Phys. 2001;3:1419–1426. [Google Scholar]