Abstract
Successful PCR starts with proper priming between an oligonucleotide primer and the template DNA. However, the inevitable risk of mismatched priming cannot be avoided in the currently used primer system, even though considerable time and effort are devoted to primer design and optimization of reaction conditions. Here, we report a novel dual priming oligonucleotide (DPO) which contains two separate priming regions joined by a polydeoxyinosine linker. The linker assumes a bubble-like structure which itself is not involved in priming, but rather delineates the boundary between the two parts of the primer. This structure results in two primer segments with distinct annealing properties: a longer 5′-segment that initiates stable priming, and a short 3′-segment that determines target-specific extension. This DPO-based system is a fundamental tool for blocking extension of non-specifically primed templates, and thereby generates consistently high PCR specificity even under less than optimal PCR conditions. The strength and utility of the DPO system are demonstrated here using multiplex PCR and SNP genotyping PCR.
INTRODUCTION
Since the development of the polymerase chain reaction (PCR), a variety of modifications in primer design and reaction conditions have been proposed to enhance and optimize specificity (1–3), but a fundamental solution for eliminating non-specific priming still remains a challenge and limits the versatility of PCR in nucleic-acid-based tests (NATs). Achieving a high level of specificity in priming currently requires very rigid primer search parameters and optimization of PCR conditions. Even when these requirements are satisfied, a high risk of non-specific priming is inevitable.
In order to fundamentally block non-specific priming, we have developed a novel dual priming oligonucleotide (DPO) system that is structurally and functionally different from the primer system currently in widespread use. The conventional priming system is based on a single priming event between the primer and template, in which mismatches in priming can lead to extension of non-specific products. In contrast, the DPO system has two separate primer segments, one of which is longer than the other, joined by a polydeoxyinosine [poly(I)] linker (Figure 1A). Deoxyinosine (I) is known to have a relatively low melting temperature compared to the natural bases, due to weaker hydrogen bonding (4). Thus, we hypothesized that a poly(I) linker inserted between two stretches of natural bases would form a bubble-like structure and separate a single primer into two functional regions at a certain annealing temperature: a 5′-segment 18–25 nt in length and a 3′-segment 6–12 nt in length. This unequal distribution of nucleotides leads to different annealing preferences for each segment. The longer 5′-segment preferentially binds to the template DNA and initiates stable annealing, whereas the short 3′-segment selectively binds to its target site and blocks non-specific annealing. Therefore, only target-specific extension will result from the successive priming of the 5′-segment and the 3′-segment of the DPO.
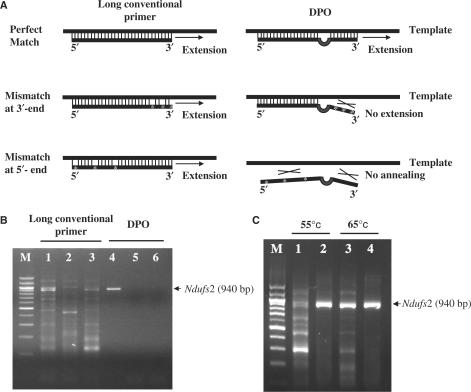
Figure 1.
Strategy and validation of DPO-based PCR. (A) Schematic representation of long conventional primer-based and DPO-based PCR strategies. (B) Comparison of 3′-RACE products of Ndufs2 obtained using long conventional primers (left) and DPO primers (right), containing different internal mismatched nucleotides at an annealing temperature of 60°C. Perfect match (lanes 1 and 4), 3 bp mismatches in the 3′-segment (lanes 2 and 5), and 3 bp mismatches in the 5′-segment (lanes 3 and 6). (C) Comparison of 3′-RACE products of Ndufs2 obtained using the perfectly matched long conventional primers (lanes 1 and 3) and DPO primers (lanes 2 and 4) at a low (55°C) and a high (65°C) annealing temperature. M, the 100-bp-size marker (Seegene). The primers are described in Table 1.
In this article, we describe and demonstrate how effectively DPO eliminates extension of non-specifically primed templates and generates high PCR specificity under a range of sub-optimal or stringent reaction conditions. In particular, we show that DPO is successfully applied in one example of complex PCR manipulations such as multiplex PCR of the respiratory viruses and one example of single nucleotide polymorphism (SNP) genotyping PCR in CYP2C19 gene, which have not been perfected to date.
MATERIALS AND METHODS
DPO design
The DPO is composed of three regions, a longer 5′-segment a shorter 3′-segment and a poly(I) linker that bridges these two segments. In designing the DPO (Figure 1A), the position of the 3′-segment was determined first, at a site where 6–12 bases had a 40–80% GC content and the Tm was not considered. Five deoxyinosines were designated for the poly(I) linker since they had generated the best result when 3–8 deoxyinosines were tested to determine the optimum length of the linker. The 5′-segment of the DPO was automatically determined by the sequence of the bases upstream of the 3′-segment and extended 18–25 bases, until the Tm was >65°C. The secondary structure and dimers were not considered in the design of the DPO because the 3′-segment alone, which is physically separated by the linker from the 5′-segment, is too short to form such structures stably. Viral genes are highly variable; therefore, in order to generate virus-specific DPOs, the length of the 5′-segment of the DPO was increased so that it had an even higher Tm (70–90°C). The long conventional primers comprise sequences identical to the DPO primers except the poly(I) linker regions. The sequences of the primers, as well as their annealing temperatures, are given in Table 1. For correct prediction of the Tm values of primers which contains deoxyinosine, we used HYTHER™ (http://ozone3.chem.wayne.edu/) which adopts the nearest neighbor model with optimized thermodynamic parameters for deoxyinosine pairs in DNA duplex (5).
Table 1.
Details of conventional primers and DPO primers used for this study
| Primer namea | Direction | Sequence (5′ – 3′)b | Tm(°C)c |
|---|---|---|---|
| Ndufs2 | |||
| Ndufs2/Df-00d | Forward | CTTCTCTCTTCGGATTGATGAGIIIIIGGAGATGC | 83 (77) |
| Ndufs2/Lcf-00d | Forward | CTTCTCTCTTCGGATTGATGAGGTGGAGGAGATGC | 88 |
| Ndufs2/Df-33d | Forward | CTTCTCTCTTCGGATTGATGAGIIIIIGCAGTTGG | 84 (77) |
| Ndufs2/Lcf-33d | Forward | CTTCTCTCTTCGGATTGATGAGGTGGAGCAGTTGG | 88 |
| Ndufs2/Df-53d | Forward | CTACTCTCTTGGGATTGAAGAGIIIIIGGAGATGC | 83 (77) |
| Ndufs2/Lcf-53d | Forward | CTACTCTCTTGGGATTGAAGAGGTGGAGGAGATGC | 88 |
| dT18-ACP1 | Reverse | CTGTGAATGCTGCGACTACGATIIIIITTTTTTTTTTTTTTTTTT | 82 (82) |
| dT15-ACP2 | Reverse | CTGTGAATGCTGCGACTACGATIIIIITTTTTTTTTTTTTTT | 82 (82) |
| RV detection | |||
| INFa/Df | Forward | AGGCCCCCTCAAAGCCGAGAIIIIICAGAGACTTG | 92 (90) |
| INFa/Lcf | Forward | AGGCCCCCTCAAAGCCGAGATCGCGCAGAGACTTG | 96 |
| INFa/Dr | Reverse | ATAGCCTTAGCTGTAGTGCTGGCIIIIICCATTCTGTT | 91 (84) |
| INFa/Lcr | Reverse | ATAGCCTTAGCTGTAGTGCTGGCTAAAACCATTCTGTT | 87 |
| INFb/Df | Forward | TTGGCTATGACIGAAAGIATAACCIIIIICAGCCCAA | 87 (75) |
| INFB/Lcf | Forward | TTGGCTATGACTGAAAGAATAACCAGAGACAGCCCAA | 88 |
| INFb/Dr | Reverse | TTACATGTTCGGTAAAAITCGTTTAIIIIITCCATACATG | 80 (75) |
| INFB/Lcf | Reverse | TTACATGTTCGGTAAAAGTCGTTTATTCCTTCCATACATG | 83 |
| OC43/Df | Forward | TATGTTAGGCCGATAATTGAGGACTIIIIIACTCTGACGG | 86 (80) |
| OC43/Lcf | Forward | TATGTTAGGCCGATAATTGAGGACTACTTTACTCTGACGG | 86 |
| OC43/Dr | Reverse | GTAATTACCGACTTTGGACTTAACATIIIIIGCAAAACCAC | 83 (78) |
| OC43/Lcr | Reverse | GTAATTACCGACTTTGGACTTAACATAAACAGCAAAACCAC | 84 |
| RSVa/Df | Forward | AGAATTTTATCAATCAACATGCAGTGIIIIIAGCAAAGGCT | 85 (77) |
| RSVa/Lcf | Forward | AGAATTTTATCAATCAACATGCAGTGCAGTCAGCAAAGGCT | 88 |
| RSVa/Dr | Reverse | ATTGTTGAGTGTATAATTCATAAACCTTGGIIIIICTCTTCTGGC | 85 (78) |
| RSVa/Lcr | Reverse | ATTGTTGAGTGTATAATTCATAAACCTTGGTAGTTCTCTTCTGGC | 85 |
| RSVb/Df | Forward | TCAGTCTATCAAATGGGGTCAGTGIIIIIACCAGCAAAG | 87 (81) |
| RSVb/Lcf | Forward | TCAGTCTATCAAATGGGGTCAGTGTTTTAACCAGCAAAG | 86 |
| RSVb/Dr | Reverse | ATTACACCATAGATAGGTAGCTGTACAACIIIIICAAGGACTTC | 86 (80) |
| RSVb/Lcr | Reverse | ATTACACCATAGATAGGTAGCTGTACAACATATGCAAGGACTTC | 85 |
| SNP detection | |||
| CYP2C19/Df | Forward | GCTTTTATACTATCAAAAGCAGGIIIIIGTCTAGGAAATG | 80 (75) |
| CYP2C19/Lcf | Forward | GCTTTTATACTATCAAAAGCAGGTATAAGTCTAGGAAATG | 81 |
| CYP2C19/Dr-a1e | Reverse | CAAGGTTTTTAAGTAATTTGTTIIIIITTCC GGG GGG |
81 (71) |
| CYP2C19/Lcr-a1e | Reverse | CAAGGTTTTTAAGTAATTTGTTATGGGTTCC GGG GGG |
84 |
| CYP2C19/Df-a2e | Forward | TAATTTTCCCACTATCATTGAIIIIITCCC GGA GGA |
82 (72) |
| CYP2C19/Lcf-a2e | Forward | TAATTTTCCCACTATCATTGATTATTTCCC GGA GGA |
81 |
| CYP2C19/Dr | Reverse | AAACTAGTCAATGAATCACAAATIIIIIAGCAGTCAC | 81 (74) |
| CYP2C19/Lcr | Reverse | AAACTAGTCAATGAATCACAAATACGCAAGCAGTCAC | 84 |
aLcf, Lcr, Df and Dr represent long conventional forward primer, long conventional reverse primer, DPO forward primer and DPO reverse primer, respectively.
bThe positions of deoxyinosine (I) are highlighted in bold. The positions of mismatched bases in Ndufs2-specific primers are underlined. The position of a single base variation in CYP2C19-specific primers is highlighted in gray.
cThe Tm value of the entire primer sequence. The Tm value of the 5′-end portion of the DPO is also indicated in parentheses. The Tm values of the primers were predicted using HYTHER™ which adopts the nearest neighbor model with optimized thermodynamic parameters for deoxyinosine pairs in DNA duplex (http://ozone3.chem.wayne.edu/).
d00, 33, 53 represent a perfectly matched primer, a 3 base mismatch in the 3′-end of the primer, a 3 base mismatch in the 5′-end of the primer, respectively.
ea1 and a2 represent allele 1-specific primer and allele 2-specific primer, respectively.
3′-RACE of Ndufs2
Total RNA extracted from an embryonic day 14.5 (E14.5) mouse embryo was reverse-transcribed using the primer dT18-ACP1 (Seegene, Korea) and M-MLV reverse transcriptase (Promega), according to the manufacturer's instructions. 3′-RACE fragments containing partial sequences from the Ndufs2 gene (940 bp) were amplified using 2× Mastermix (Solgent, Korea) with an Ndufs2-specific 5′ primer and dT15-ACP2 (Seegene). After preheating at 94°C for 5 min, 30 amplification cycles were carried out in a thermal cycler (GeneAmp PCR System 9700; Applied Biosystems) under the following conditions: denaturation at 94°C for 40 s, annealing at 55–65°C for 40 s and extension at 72°C for 40 s. Amplification was completed with a final extension step at 72°C for 5 min.
Multiplex RT-PCR for the detection of respiratory viruses
Total RNA extracted from nasal aspirate samples from five patients was reverse-transcribed using random hexamer primers (Fermentas) and M-MLV reverse transcriptase (Promega). Multiplex RT-PCR was performed on the five cDNA samples to detect five different virus-specific genes using 2× Mastermix (Solgent). The following gene segments were amplified: the segment 7 gene of the influenza A virus (513 bp), the segment 1 gene of the influenza B virus (455 bp), the F gene of the respiratory syncytial virus B (391 bp), the F gene of the respiratory syncytial virus A (273 bp) and the M gene of coronavirus OC43 (231 bp). As a negative control, sterile deionized water was used as the template instead of nucleic acid. As a positive control, plasmids containing amplicons of the same length were used. After preheating at 94°C for 15 min, 45 amplification cycles were carried out in the thermal cycler (same as above) under the following conditions: denaturation at 94°C for 30 s, annealing at 60°C for 90 s and extension at 72°C for 90 s. Amplification was completed with a final extension step at 72°C for 10 min.
Multiplex PCR for detection of an SNP in CYP2C19
Genomic DNA was extracted from nine human blood samples, which consisted of three samples for each of three different genotypes at bp 681 in exon 5 of CYP2C19 (allele1/allele1, allele1/allele2, allele2/allele2). The allele-specific DPO primers were designed to have an SNP in the middle of the 3′-segment because such a position maximizes disturbance of the 3′-segment annealing. Multiplex PCR analysis of the genomic DNA was performed to detect allele 1 (321 bp) and allele 2 (232 bp), together with a general primer to detect CYP2C19 (492 bp) using 2× Mastermix (Solgent). After a preheating step at 94°C for 5 min, 35 amplification cycles were carried out in the thermal cycler (same as above) under the following conditions: denaturation at 94°C for 30 s, annealing at 63°C for 30 s and extension at 72°C for 30 s. Amplification was completed with a final extension step at 72°C for 5 min.
RESULTS
Effect of DPO on PCR specificity
The difference in overall strategy of the conventional and the DPO-based PCR approaches is illustrated in Figure 1A. Conventional primers have a single priming region and extension may proceed even in the presence of mismatches between a primer and a template. In contrast, DPO has two priming regions and extension proceeds only when the two priming segments are perfectly matched with the sequence of a template (Figure 1A, perfect match). Even though the 5′-segment of the DPO binds to the template, extension will not proceed if there is any mismatch in the 3′-segment of the DPO (Figure 1A, mismatch at 3′-end). If the 5′-segment does not bind to the template due to mismatches, the 3′-segment alone, which is 6–12 nt in length, has a Tm too low (below 30°C) to bind its template at the generally used annealing temperatures of 55–65°C (Figure 1A, mismatch at 5′-end).
To demonstrate our hypothesis, we conducted 3′-RACE of Ndufs2 with DPO and long conventional primers carrying three mismatched bases in 5′- or 3′-ends (Figure 1B). The conventional primers generated non-specific products under all conditions (lanes 1, 2 and 3), whereas the perfectly matched DPO primer generated only one target product (lane 4), and did not amplify any PCR product when three mismatched bases were incorporated in either the 3′-segment (lane 5) or the 5′-segment (lane 6).
We also hypothesized that the DPO primer will maintain a high level of specificity over a wide range of annealing temperatures due to its structural feature. The 3′-RACE of Ndufs2 was further conducted at a low (55°C) and a high (65°C) annealing temperature. As shown in Figure 1C, the long conventional primers reduced the non-specific products by increasing the annealing temperature to 65°C (lanes 1 and 3), while the DPO primer produced only one target product over a wide range of annealing temperatures (55 and 65°C) (lanes 2 and 4). These results clearly support our hypothesis that the unique dual-specificity annealing properties of the DPO primer block non-specific priming events even under less than optimal PCR conditions.
Application 1: multiplex PCR
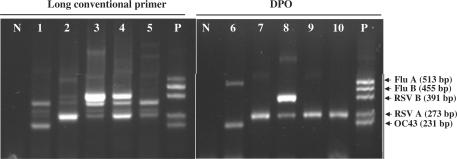
We evaluated the DPO-based system in a multiplex PCR application for the detection of five different human respiratory viruses. Most respiratory tract infections are fairly mild, but they are highly infectious, and can sometimes result in severe symptoms requiring hospitalization, or even lead to death. Traditional detection tools such as cell culture and antigenic detection are usually slow and may be inaccurate as well. Various multiplex PCR-based detection tools have been developed and introduced to direct appropriate therapy and to avoid the use of unnecessary antibiotics (6,7). However, current multiplex PCR-based assays require further validation, such as nested PCR or a probe hybridization assay, due to their high rate of false positives (8,9). We developed a DPO-based multiplex PCR assay for the detection of five respiratory viruses: influenza A, influenza B, respiratory syncytial virus A, respiratory syncytial virus B and coronavirus OC43 (Figure 2). As expected, long conventional primers generated many non-specific bands, most likely due to non-specific annealing or primer competition. In contrast, the DPO primer generated target-specific viral fragments, and no false positives. In addition, DPO primers detected the influenza A virus in patient 1 (lane 6), which was not detected using the conventional primer system (lane 1). The presence of respiratory viruses in each patient was further validated by sequencing the fragment. These results indicate that a DPO-based multiplex PCR approach is a reliable tool for detecting multiple pathogens.
Figure 2.
Detection of multiple respiratory viruses. Comparison of PCR products generated using DPO-based multiplex RT-PCR (right) and long conventional primer-based multiplex RT-PCR (left) from five patient samples. N, negative control; P, positive control; Flu A, influenza A virus; Flu B, influenza B virus; RSV B, respiratory syncytial virus B; RSV A, respiratory syncytial virus A; OC43, coronavirus OC43. The primers are described in Table 1.
Application 2: SNP genotyping PCR
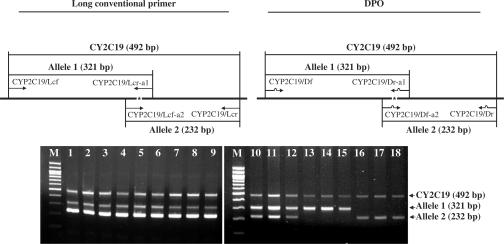
We further evaluated the DPO-based multiplex PCR system for the detection of a single nucleotide polymorphism (SNP) in CYP2C19. This polymorphism is one of the most thoroughly characterized SNPs, in which a single base pair substitution (G → A) at position 681 in exon 5 of CYP2C19 results in a non-functional protein and affects the metabolism of a number of commonly used drugs (10). SNPs are the most common type of DNA sequence variation in the human genome, and are an important genetic factor in the origin and development of complex genetic traits in humans (11,12). Appropriate methods for SNP genotyping are important in many fields of biological science, and several different methods for detecting SNPs in the human genome have been introduced (13). However, an ideal PCR-based SNP detection method, one that is simple and provides an accurate genotype without additional verification steps such as sequencing, does not currently exist. We carried out multiplex PCR analysis of nine human genomic DNA samples with known genotypes at the CYP2C19 locus (Figure 3, lanes 1–3 and 10–12, allele 1/allele 2; lanes 4–6 and 13–15, allele 1/allele 1; lanes 7–9 and 16–18, allele 2/allele 2) using two different DPO primer sets to detect the three different allelic combinations (Figure 3). DPO-based multiplex PCR clearly distinguished between the different alleles of CYP2C19, while long conventional primer-based multiplex PCR did not.
Figure 3.
Detection of an SNP in CYP2C19. Schematic representation of primer positions, and comparison of PCR products generated using DPO-based multiplex RT-PCR (right) and long conventional primer-based multiplex RT-PCR (left) from nine human blood samples. The asterisk indicates the position of the SNP in CY2C19. M, the 100-bp-size marker (Seegene). The primers are described in Table 1.
DISCUSSION
We have developed a new primer technology, the DPO system. The DPO system differs structurally and functionally from the conventional primer system by including a poly(I) linker, which is one of the most commonly used universal bases (14), between two segments of primer sequences (Figure 1A). In general, primers >25 bases are rarely used, since the Tms of 25 mers or the longer primers can be over 70°C, which is too high for proper annealing (15). This is a fundamental limitation in current conventional primer design. However, the long DPO primer (34–42 mer) is divided into two distinct target-specific priming segments by the presence of the poly(I) linker, and, thus, does not suffer from the limitations of a high Tm. In addition, the two priming segments of differing lengths have distinct priming functions. For example, at the general annealing temperatures of 55–65°C, stable annealing is initiated only by the longer 5′-segment since it has a high enough Tm (over 50°C) to bind to the template. Target-specific extension is then determined by the shorter 3′-segment, resulting in unparalleled high specificity. In order to demonstrate the usefulness of DPO, we compared the performance of the DPO system to that of the conventional primer system in multiplex PCR and SNP genotyping PCR. Overall our results demonstrate that the DPO system has dramatically improved performance.
DPO primers are easier to design than conventional primers. In designing conventional primers, the sequence should be carefully checked for certain primer design features such as primer length, melting temperature, GC content and secondary structure (16). In contrast, we believe that a DPO can be designed based on almost any sequence of interest since the poly(I) linker prevents formation of secondary structure and effectively eliminates non-specific priming. This is very advantageous, particularly for detection of pathogens such as virus and bacteria since their sequences are highly variable and available primer sites are highly restricted.
We have demonstrated the successful application of DPO primers in one multiplex reaction. Multiplex PCR is a rapid and economical tool (6), but when a large bank of genes is amplified with multi-primer sets, conventional primers often produce false positives due to primer competition, to primer dimers or to the different melting temperatures of the different primers. Therefore, current multiplex PCR-based assays require further validation, such as nested PCR or a probe hybridization assay (7,8). However, DPO allows specific detection of a large number of pathogens without any false result because the bubble-like structure of the poly(I) linker in DPO efficiently prevents primer-dimer and hairpin structure formation. The example presented here (Figure 2) demonstrates the successful use of the DPO-based multiplex PCR for simultaneous detection of multiple respiratory viruses with one PCR step. The high specificity without production of any non-specific bands or false-positive products clearly demonstrates the great potential of DPO-based multiplex PCR approaches to be a reliable, rapid, practical and cost-effective detection method.
We have demonstrated the successful application of DPO primers for one SNP genotyping PCR. In general, conventional primers cannot offer reliable results for SNP genotyping and current SNP genotyping methods require additional steps after amplification of an SNP-containing region such as RFLP (17), sequencing (18) or hybridization (19). These are neither rapid nor easy to manipulate, and they require large initial investments in equipment. In contrast, the example presented here (Figure 3) demonstrates that SNP genotyping can be simply achieved in one PCR step by using the DPO system.
Finally, DPO can be used for any PCR approach which makes use of the ability of primers to specifically hybridize to complementary sequences except for the case of long and accurate PCR (LA-PCR) which is achieved by DNA polymerases having proofreading activity (e.g. Pfu). It was found that such DNA polymerases have a tendency to completely drop off the DNA when they encounter inosine in a PCR primer (20).
In this article, we demonstrated that the DPO system can be successfully used in multiplex PCR and SNP genotyping PCR. We propose that combination of DPO-based multiplex PCR with high-throughput analysis methods such as DNA microarray amplification or quantitative analysis methods such as real-time PCR will further extend the potential of this approach.
ACKNOWLEDGEMENTS
We thank Sun-Hwa Joung for critical reading of the manuscript. Funding to pay the Open Access publication charge was provided by Seegene.
Conflict of interest statement. None declared.
REFERENCES
- 1.Birch DE, Kolmodin J, Wong J, Zangenberg GA, Zoccoli MA, McKinney N, Young KKK, Laird WJ. The use of a thermally activated DNA polymerase PCR gives improved specificity, sensitivity and product yield without additives or extra process steps. Nature. 1996;381:445–446. [Google Scholar]
- 2.Brownie J, Shawcross S, Theaker J, Whitcombe D, Ferrie R, Newton C, Little S. The elimination of primer-dimer accumulation in PCR. Nucleic Acids Res. 1997;25:3235–3241. doi: 10.1093/nar/25.16.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ailenberg M, Silverman M. Controlled hot start and improved specificity in carrying out PCR utilizing touch-up and loop incorporated primers (TULIPS) Biotechniques. 2000;29:1018–1020. 1022–1024. doi: 10.2144/00295st03. [DOI] [PubMed] [Google Scholar]
- 4.Bergstrom DE, Zhang P, Johnson WT. Comparison of the base pairing properties of a series of nitroazole nucleobase analogs in the oligodeoxyribonucleotide sequence 5′- d(CGCXAATTYGCG)-3′. Nucleic Acids Res. 1997;25:1935–1942. doi: 10.1093/nar/25.10.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watkins NE, Jr, SantaLucia J., Jr Nearest-neighbor thermodynamics of deoxyinosine pairs in DNA duplexes. Nucleic Acids Res. 2005;33:6258–6267. doi: 10.1093/nar/gki918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. Multiplex PCR: optimization and application in diagnostic virology. Clin. Microbiol. Rev. 2000;13:559–570. doi: 10.1128/cmr.13.4.559-570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grondahl B, Puppe W, Hoppe A, Kuhne I, Weigl JAI, Schmitt H-J. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J. Clin. Microbiol. 1999;37:1–7. doi: 10.1128/jcm.37.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindiyeh M, Levy V, Azar R, Varsano N, Regev L, Shalev Y, Grossman Z, Mendelson E. Evaluation of a multiplex real-time reverse transcriptase PCR assay for detection and differentiation of influenza viruses A and B during the 2001-2002 influenza season in Israel. J. Clin. Microbiol. 2005;43:589–595. doi: 10.1128/JCM.43.2.589-595.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellau-Pujol S, Vabret A, Legrand L, Dina J, Gouarin S, Petitjean-Lecherbonnier J, Pozzetto B, Ginevra C, Freymuth F. Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J. Virol. Methods. 2005;126:53–63. doi: 10.1016/j.jviromet.2005.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Morais SMF, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J. Biol. Chem. 1994;269:15419–15422. [PubMed] [Google Scholar]
- 11.Miller RD, Kwok PY. The birth and death of human single-nucleotide polymorphisms: new experimental evidence and implications for human history and medicine. Hum. Mol. Genet. 2001;10:2195–2198. doi: 10.1093/hmg/10.20.2195. [DOI] [PubMed] [Google Scholar]
- 12.Group TISMW. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 13.Kwok PY. Methods for genotyping single nucleotide polymorphisms. Annu. Rev. Genomics. Hum. Genet. 2001;2:235–258. doi: 10.1146/annurev.genom.2.1.235. [DOI] [PubMed] [Google Scholar]
- 14.Dovas CI, Efthimiou K, Katis NI. Generic detection and differentiation of tobamoviruses by a spot nested RT-PCR-RFLP using dI-containing primers along with homologous dG-containing primers. J. Virol. Methods. 2004;117:137–144. doi: 10.1016/j.jviromet.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Wu DY, Ugozzoli L, Pal BK, Qian J, Wallace RB. The effect of temperature and oligonucleotide primer length on the specificity and efficiency of amplification by the polymerase chain reaction. DNA Cell. Biol. 1991;10:233–238. doi: 10.1089/dna.1991.10.233. [DOI] [PubMed] [Google Scholar]
- 16.Lowe T, Sharefkin J, Yang SQ, Dieffenbach CW. A computer program for selection of oligonucleotide primers for polymerase chain reactions. Nucleic Acids Res. 1990;18:1757–1761. doi: 10.1093/nar/18.7.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alonso-Navarro H, Martinez C, Garcia-Martin E, Benito-Leon J, Garcia-Ferrer I, Vazquez-Torres P, Puertas I, Lopez-Alburquerque T, Agundez JA, et al. CYP2C19 polymorphism and risk for essential tremor. Eur. Neurol. 2006;56:119–123. doi: 10.1159/000095702. [DOI] [PubMed] [Google Scholar]
- 18.Bender K. SNaPshot for pharmacogenetics by minisequencing. Methods Mol. Biol. 2005;297:243–252. doi: 10.1385/1-59259-867-6:243. [DOI] [PubMed] [Google Scholar]
- 19.Matsubara Y, Kure S. Detection of single nucleotide substitution by competitive allele-specific short oligonucleotide hybridization (CASSOH) with immunochromatographic strip. Hum. Mutat. 2003;22:166–172. doi: 10.1002/humu.10247. [DOI] [PubMed] [Google Scholar]
- 20.Knittel T, Picard D. PCR with degenerate primers containing deoxyinosine fails with Pfu DNA Polymerase. PCR Methods Appl. 1993;2:346–347. doi: 10.1101/gr.2.4.346. [DOI] [PubMed] [Google Scholar]