Abstract
Recent studies have identified large sets of genes in embryonic stem and embryonal carcinoma cells that are associated with the transcription factors Sox2 and Oct-3/4. Other studies have shown that Sox2 and Oct-3/4 work together cooperatively to stimulate the transcription of their own genes as well as a network of genes required for embryogenesis. Moreover, small changes in the levels of Sox2:Oct-3/4 target genes alter the fate of stem cells. Although positive feedforward and feedback loops have been proposed to explain the activation of these genes, little is known about the mechanisms that prevent their overexpression. Here, we demonstrate that elevating Sox2 levels inhibits the endogenous expression of five Sox2:Oct-3/4 target genes. In addition, we show that Sox2 repression is dependent on the binding sites for Sox2 and Oct-3/4. We also demonstrate that inhibition is dependent on the C-terminus of Sox2, which contains its transactivation domain. Finally, our studies argue that overexpression of neither Oct-3/4 nor Nanog broadly inhibits Sox2:Oct-3/4 target genes. Collectively, these studies provide new insights into the diversity of mechanisms that control Sox2:Oct-3/4 target genes and argue that Sox2 functions as a molecular rheostat for the control of a key transcriptional regulatory network.
INTRODUCTION
New technologies are providing the foundation for deciphering transcriptional regulatory networks that orchestrate mammalian embryogenesis (1,2). Transcriptional regulatory networks consist of functional interactions between regulatory genes and a much larger set of downstream target genes. Downstream target genes possess cis-regulatory regions (e.g. enhancers) able to respond to and integrate multiple signals from regulatory genes, many of which code for transcription factors (TFs). Enhancers within the regulatory genes themselves allow for more complex and precise control of the entire network. Decoding and understanding transcriptional regulatory networks will not only require an extensive cataloging of TFs and the genes to which they bind, but will also require a detailed understanding of how these gene interactions are modulated by chromatin structure and the expression levels of key TFs. Moreover, understanding interactions between key regulatory genes, which influence the temporal and spatial expression of one another by feedforward and feedback regulatory loops, will provide novel insights into the cross talk between different transcriptional regulatory networks.
Recent efforts using ChIP-Chip (1) and ChIP-PET studies (2) have begun to identify large groups of genes in embryonic stem (ES) cells that are associated with three TFs, Nanog, Sox2 and Oct-3/4 (also known as Oct-3 and Oct-4). Interest in this trio of TFs has grown considerably with the recognition that each is essential for proper mammalian embryogenesis, as well as the self-renewal and pluripotency of ES cells (3–6). Intriguingly, hundreds of genes identified in these genome-wide studies were found to be co-occupied by two and, in many cases, by all three TFs (1,2). Thus far, the cis-regulatory elements present in the vast majority of these genes, which are responsible for binding the three TFs, remain to be determined. However, it is likely that the regulatory regions of many of these genes possess remarkably similar DNA sequences, which are responsible for the concerted binding of two or all three of these TFs. In this regard, more conventional approaches have identified a group of at least seven genes that are regulated by closely spaced HMG and POU motifs (referred to as an HMG/POU cassette), which bind Sox2 and Oct-3/4, respectively. Although small sequence differences exist between HMG/POU cassettes (7), the consensus sequence generated has been useful in the identification of other genes regulated by the combined action of Sox2 and Oct-3/4 (8,9). The importance of HMG/POU cassettes is highlighted by the finding that their disruption virtually eliminates enhancer function and subsequent promoter activation of each gene (8–17). As such, the HMG/POU cassette of each gene forms the core of an enhancer that regulates the transcription of these genes. In this study, we refer to genes that contain functional HMG/POU cassettes as Sox2:Oct-3/4 target genes.
The network of Sox2:Oct-3/4 target genes includes Sox2, Oct-3/4, Nanog, FGF-4, UTF1, Fbx15, Lefty1 and many more are expected to be identified in the near future. Each of these genes is expressed in ES cells and their tumor-derived counterparts, embryonal carcinoma (EC) cells. Furthermore, with the possible exception of Lefty1, each of the genes is substantially down-regulated upon differentiation of both ES and EC cells, due to the down-regulation of Sox2 and Oct-3/4 (5,11,12,14,16,18–20). The finding that Sox2 and Oct-3/4 work together cooperatively to regulate the transcription of their own genes (15,16) argues that Sox2 and Oct-3/4 proteins function as central nodes in a critical transcriptional regulatory network that controls the expression of other essential genes. As in the cases of Sox2, Oct-3/4 and Nanog, FGF-4 is essential for embryogenesis (3–6,21). Less is known about the remaining three identified Sox2:Oct-3/4 target genes, Lefty1, UTF1 and Fbx15. Although Lefty1 is not essential for the earliest stages of mammalian development, disruption of this gene causes serious defects in left–right patterning of visceral organs (22). UTF1 may be essential for development given that UTF1 null ES cells proliferate more slowly when compared to their wild-type counterparts (23). Thus far, Fbx15 is the only known Sox2:Oct-3/4 target gene that has not yet been shown to play a prominent role in normal development (14).
Given the importance of Sox2:Oct-3/4 target genes during development, it is no surprise that their expression is tightly controlled. Small changes in Oct-3/4 levels drastically alter cell fate. A 2-fold increase in Oct-3/4 levels causes ES cells to differentiate into endoderm and mesoderm, whereas knockdown of Oct-3/4 causes ES cells to differentiate into trophectoderm-like cells (24). Modifying Nanog levels also alters the behavior of ES cells. Overexpression of Nanog in mouse ES cells negates their need for LIF (6). In contrast, knockdown of Nanog results in the differentiation of ES cells to cells with the properties of endoderm (6,25). Interestingly, Nanog has been reported recently to interact with Sall4 to activate target gene transcription, including the autoregulation of the Nanog and Sall4 genes themselves, in a fashion similar to Sox2 and Oct-3/4 (26). Nanog has also been shown to function as part of a feedforward/feedback loop in conjunction with Oct-3/4 and FoxD3 (27). Clearly, the consequences of altering the levels of Oct-3/4 and Nanog have provided useful insights into the developmental potential of ES cells and EC cells. So far, much less attention has been placed on understanding how altering the levels of Sox2 influences cell fate. Although knockdown of Sox2 causes ES cells to differentiate (28), there are no reports describing how elevating Sox2 levels influences the regulation of Sox2:Oct-3/4 target genes.
Current evidence indicates that Sox2 and Oct-3/4 work cooperatively to stimulate the transcription of a number of critical genes (8–17). Given the need to tightly control the expression of these genes, one would expect that negative feedback loops exist to ensure that the levels of these genes are controlled properly. Earlier work suggested that this may be the case for FGF-4. Although it is well known that Sox2 and Oct-3/4 work in concert to stimulate FGF-4 transcription (10,11,29,30), overexpression of Sox2 has also been shown to inhibit the function of the FGF-4 enhancer in promoter/reporter gene constructs (31). In this study, we have examined the possibility that overexpression of Sox2 exerts a much broader effect over Sox2:Oct-3/4 target genes. We determined that overexpression of Sox2 inhibits the activities of Sox2, FGF-4, Nanog, UTF1 and Oct-3/4 promoter/reporter gene constructs in both EC cells and ES cells. Importantly, we determined that overexpression of Sox2 in F9 EC cells inhibits the endogenous expression of five Sox2:Oct-3/4 target genes, Sox2, FGF-4, Nanog, UTF1 and Oct-3/4. Fbx15 and Lefty1 were not examined in this study. We also determined that this effect is mediated through the HMG/POU cassettes in the enhancers of these genes. Moreover, we demonstrate that the effect of Sox2 is mediated by its C-terminal region, which contains its transactivation domain (TAD). Finally, we show that although overexpression of Oct-3/4 and Nanog each inhibits its own promoter, their overexpression does not appear to broadly inhibit the promoters of other Sox2:Oct-3/4 target genes.
MATERIALS AND METHODS
Cell culture conditions
Stock cultures of mouse F9 EC cells and P19 EC cells were maintained in Dulbecco's modified Eagle's medium (Life Technologies, Inc.) supplemented with 10% fetal bovine serum (HyClone, Logan UT) as reported previously (32). D3 ES cells were cultured in the absence of a feeder layer in medium supplemented with leukemia-inhibiting factor as described previously (29). Stock cultures and all experimental cultures were maintained at 37°C in a moist atmosphere of 95% air and 5% CO2.
Transient transfection assays
F9 EC cells and P19 EC cells were seeded at 500 000 cells per 100-mm dish and transfected in duplicate the following day using the calcium phosphate precipitation method as described previously (29). In addition to 12 µg of a promoter/reporter construct, the cells were co-transfected with 1 µg of the internal standard pCMV-β-gal (Clontech, Palo Alto, CA, USA) to normalize for any differences in transfection efficiency. Where indicated, either 1 or 3 µg of a CMV-driven expression vector was co-transfected with the promoter/reporter construct. D3 ES cells were seeded at 150 000 cells per 60-mm dish and transfected in duplicate the following day using Lipofectamine™ 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA), as described previously (33), using 4.5 µg of promoter/reporter construct and 1 µg of pCMV-β-gal. Where indicated, 0.3 or 1.0 µg of the Sox2 expression vector was co-transfected. For all transfections, DNA from the empty vector, CMV5, was added where needed to ensure that all cells were transfected with the same amount of DNA. Calcium phosphate transfected cells were washed twice 24 h after transfection with serum-free medium and refed with serum-containing medium. With all transfections, chloramphenicol acetyltransferase (CAT) and β-galactosidase activities were determined 48 h after transfection as described previously (29).
Plasmid constructs
The plasmid construct FGF-4 + E (previously named 427T + E) contains 427 bp of the mouse FGF-4 promoter and 316 bp of the mouse FGF-4 enhancer separated by a CAT reporter gene (29,34,35). The construct FGF-4 + E(HPmut), previously named 427T + EnGSp, is similar to FGF-4 + E except that the HMG/POU cassette has been replaced with a binding site for the DNA-binding domain (DBD) of the yeast TF GAL4 (13).
The construct Nanog + E contains the mouse Nanog promoter and enhancer cloned directly upstream of the CAT reporter gene. The region from −812 to +171 of the Nanog transcription start site was PCR amplified from mouse genomic DNA using the following primers: 5′-cgtgatGTCGACAATTTCTTCTTCCATTGCTTAGACGG-3′ and 5′-tgcAGATCTAAGGGATTTCTGAAAAGGTTTTAGGC-3′. The SalI and BglII restriction sites shown in italics and a small piece of extra DNA shown in lowercase allow for restriction enzyme digestion and cloning of the fragment containing the Nanog promoter and enhancer into the previously described pBLCAT7 reporter plasmid (35).
The construct UTF1 + E was described previously (7) and contains 321 bp of the mouse UTF1 promoter (−202 to +119) (12,36) cloned upstream of the CAT reporter gene as well as 139 bp of the mouse UTF1 enhancer (12) cloned downstream of the CAT reporter gene. The construct UTF1 + E(HPmut) was created from UTF1 + E by replacing the HMG/POU cassette with a DNA sequence able to bind the DBD of the yeast TF GAL4. This change was made by the cassette exchange method (7), using the following primers: 5′-tggaAGATCTCGGAGGACAGTCCACCGACGGCTCATCCTGAGGCTC-3′ and 5′-tggaAGATCTCCGGCAGCAGCTTCCTTTCC-3′ where the GAL4 site is underlined, the BglII sites are in italics, and a small piece of extra DNA is in lowercase (to facilitate restriction enzyme digestion).
The Oct-3/4 promoter and enhancer elements were also amplified from mouse genomic DNA and cloned into pBLCAT7 upstream of the CAT reporter gene to create Oct-3/4 + E using the following primers: 5′-ttctGTCGACTCTAGGCACGCTTAGGGC-3′ and 5′-ttctAGATCTCCGAGCCGGGGGCCTGGTGG-3′ (SalI and BglII sites in italics and extra bases in lowercase). This amplified a fragment spanning −2295 to +33 of the Oct-3/4 gene, which includes the promoter and both the proximal and distal enhancer (16,37). The construct Sox2 + US/DSE contains the Sox2 promoter and CAT reporter gene as well an upstream Sox2 enhancer element and a downstream Sox2 enhancer element (containing the HMG/POU cassette), all placed relative to their natural positions. The mouse Sox2 promoter was described previously (38) and consists of the −528 to +238 region (relative to the transcription start site). The upstream Sox2 enhancer was cloned from genomic mouse DNA with the following primers: 5′-cctcCATATGTCAAATAGGG CCCTTTTCAG-3′ and 5′-cctcCATATGAAGCCAACT GACAATGTTGTGG-3′ (NdeI sites in italics and extra bases in lowercase), which amplify a 392 bp fragment ∼4 kb upstream of the Sox2 transcription start site previously described as the ‘0.4a’ Sox2 enhancer region (39). The downstream Sox2 enhancer (containing the HMG/POU cassette) was cloned with the following primers: 5′-catGAGCTCGGTTCCCCTCTAATTAATG CAGAGAC-3′ and 5′-catGAGCTCATACTGTCCATT GGCTGGAGTTCC-3′ (SacI sites in italics and extra bases in lowercase), which amplify a 211 bp region ∼2 kb downstream of the Sox2 gene. The construct Sox2 + US/DSE(HPmut) contains the GAL4-binding site in place of the HMG/POU cassette. It was generated by the cassette exchange method, using the following primers: 5′-ggaAGATCTCGGAGGACAGTCCACCGAGGATTATTCACGTGGTAATGAGC-3′ and 5′-tggaAGATCTTGCCCGAGCCCGGGAAATTCTTTTAGAG-3′ where the GAL4 site is underlined, the BglII sites are in italics, and a small piece of extra DNA is in lowercase. Site-directed mutagenesis was used to destroy the POU motif of the Sox2 + US/DSE construct, creating Sox2 + US/DSE(Pmut), using the following primers: 5′-CCCGGGCTCGGGCAGCCATTGTGgccggcATAGGATTATTCACG-3′ and 5′-CGTGAATAATCCTATgccggcCACAATGGCTGCCCGAGCCCGGG-3′ where a new NaeI site (italics) was introduced in place of the POU motif (underlined). Changes from wild-type sequence are shown in lowercase.
The contruct TK contains the herpes simplex virus (HSV) thymidine kinase (TK) promoter cloned upstream of the CAT reporter gene. TK + E contains the HSV TK promoter plus the FGF-4 enhancer (as in FGF-4 + E) downstream of the CAT reporter gene. Both constructs were described previously (13).
All expression vectors are driven by the CMV promoter. CMV-Sox2 contains a cDNA for the entire coding region of mouse Sox2, as described previously (30). Likewise, CMV-Oct-3/4 contains the coding region of mouse Oct-3/4, as described previously (30). The expression vector CMV-Sox11 and the chimeras CMV-Sox2-2-11 (Sox2 HMG/DBD domain with Sox11 TAD) and CMV-Sox11-11-2 (Sox11 HMG/DBD domain and Sox2 TAD) were also described previously (40). The empty expression vector GFP is also known as pEGFP-C1 (Clontech). GFP-Sox2 is a fusion of GFP and the full-length Sox2 cDNA and was described previously (30). The CMV-Nanog expression vector contains the full-length mouse Nanog cDNA (1.0 kb) amplified from F9 EC cell RNA with the following primers: 5′-cctcAAGCTTTTCAGAAATCCCTTCCCTCGCC-3′ and 5′-ctccTCTAGAAGGAAGGAACCTGGCTTTG CCC-3′ where HindIII and XbaI restriction sites are shown in italics (extra DNA for restriction enzyme digestion in lowercase) allowing for cloning into the CMV expression vector.
GFP transfection and cell sorting
F9 EC cells were seeded at 500 000 cells per 100-mm dish and transfected using Lipofectamine 2000 Reagent™, as described previously (33). In short, after 24 h, each plate was transfected with either 3 µg empty GFP vector or 3 µg GFP-Sox2. After an additional 24 h, cells were prepared for sorting as follows. Each plate was washed two times with 5 ml of PBS. Cells were then trypsinized, collected in serum-containing medium and transferred to 1.5 ml Eppendorf tubes before being spun down at 4°C and 3000 r.p.m. for 5 min. The resulting cell pellet was resuspended in 1 ml of serum-containing medium per plate and GFP positive cells were sorted and collected in the UNMC Cell Analysis Facility using a BD FACSAria™ Flow Cytometer (San Jose, CA, USA). Data was acquired and analyzed with DiVa 5.0.1 software.
RNA isolation and cDNA synthesis
GFP positive cells were obtained, spun down and resuspended in 800 µl TRIzol® Reagent (Invitrogen). RNA was isolated according to the manufacturer's protocol with the exception of increasing the precipitation step to overnight at −20°C. RNA pellets were resuspended in 20 µl RNase/DNase-free H2O. The concentration of RNA was determined by UV spectrophotometry. RNA was treated with amplification grade DNase I (Invitrogen). Next, cDNA was synthesized using the SuperScript® III First-Strand Synthesis SuperMix (Invitrogen).
Quantitative PCR
The cDNA generated from GFP and GFP-Sox2 transfected cell isolates was subjected to SYBR green qPCR on the Cepheid SmartCycler® using software Version 2.0c (Sunnyvale, CA, USA). Gene expression was assayed using RT2 Real-Time™ SYBR Green PCR master mix from SuperArray BioScience Corporation (Frederick, MD, USA) according to the manufacturer's protocol using the following gene-specific primers. Primer sets specific for mouse GAPDH (PPM02946A), FGF-4 (PPM03044A), UTF1 (PPM04713A), Oct-3/4 (also known as Pou5f1, PPM04726A) and Sox2 (PPM04762A) were obtained from SuperArray, with the catalog numbers shown in parentheses. The Sox2 primer was used to detect overexpression of GFP-Sox2. Nanog-specific primer sequences were obtained from PrimerBank, Nanog pair 3 (PrimerBank ID 31338864a3): upper 5′-CCTGATTCTT CTACCAGTCCCA-3′ and lower 5′-GGCCTGAGAGA ACACAGTCC-3′; which create a 123 bp amplicon (41). Endogenous Sox2 gene expression was detected using a primer set specific for a portion of the Sox2 5′ untranslated region (not present in the expression plasmid). The primers, upper 5′-AAGGAGAGAAGTTTGGAGCC-3′ and lower 5′-TCTGGCGGAGAATAGTTGG-3′ create a 153 bp amplicon. Relative gene expression for each transfection (GFP and GFP-Sox2) was calculated according to the SuperArray protocol and normalized to GAPDH. Gene expression for GFP-Sox2 transfected cells was reported as a fraction of the corresponding expression in control (GFP transfected) cells.
Western blot analysis
F9 EC cells were transfected with GFP-Sox2 and subjected to sorting as described above. Nuclear extracts were prepared from GFP-positive cells using the NE-PER™ nuclear and cytoplasmic extraction kit (Pierce, Rockford, IL, USA). Nuclear extracts were prepared in the presence of protease inhibitors: aprotinin (2.5 kallikrein-inactivating U/ml), PMSF (0.2 mM), soybean trypsin inhibitor (20 µg/ml), pepstatin A (1 µg/ml) and leupeptin (1 µg/ml) (42). Nuclear extracts also contained the protein phosphatase inhibitor NaF (5 mM) and NaPPi (30 mM). The resultant nuclear extract was separated on a 4–20% SDS PAGE gel and then transferred to an Immobilon-P PDVF membrane (Millipore, Bedford, MA, USA). Western blot analysis was performed using the Chemicon International, Inc. (Temecula, CA, USA) anti-Sox2 antibody (AB5603) at a 1 μg/ml concentration and an alkaline phosphatase conjugated goat α-rabbit secondary antibody (1:1000). Sox2 migrated as a ∼35 kDa protein [as described previously, (7)] and GFP-Sox2 migrated at ∼60 kDa (due to the ∼25 kDa GFP epitope). Proteins were detected using an enhanced chemiflourescence kit (Amersham Biosciences) and scanned on a Storm PhosphorImager and quantified using ImageQuant analysis software (Molecular Dynamics, Sunnyvale, CA).
RESULTS
Overexpression of Sox2 results in inhibition of Sox2 promoter activity
Recent work has shown that Sox2 is critical for the expression of a network of Sox2:Oct-3/4 target genes. However, since Sox2 contributes to the activation of its own transcription, it was unclear whether mechanisms exist to prevent the overexpression of Sox2 and other Sox2:Oct-3/4 target genes. To test this possibility, we examined the effects of Sox2 overexpression in F9 EC cells. F9 EC cells were chosen because they have been used extensively to study the expression of Sox2:Oct-3/4 target genes (43). These cells not only express the same set of Sox2:Oct-3/4 target genes as ES cells, the expression of these genes turns off at the transcriptional level when ES cells and F9 EC cells differentiate (5,11,12,14,16,18–20). In our initial studies, F9 EC cells were transiently transfected with an expression vector for Sox2 plus a Sox2 promoter/reporter gene construct. The Sox2 promoter/reporter gene construct used for these studies contains the two enhancers known to be functional in EC and ES cells (15,39). One enhancer is located ∼4 kb upstream of the Sox2 transcription start site and the second, which contains the HMG/POU cassette, is located ∼2 kb downstream of the single exon of Sox2. In our promoter/reporter gene construct, the placement of the two Sox2 enhancers reflects their position in the endogenous Sox2 gene (Figure 1A). We determined that Sox2 overexpression inhibited the activity of our Sox2 promoter/enhancer gene construct in a dose-dependent manner (Figure 1A). In contrast, another Sox family member, Sox11, which has a DBD that exhibits nearly 70% sequence identity with Sox2 (40), had no effect on promoter/reporter activity when overexpressed in F9 EC cells (Figure 1B). Moreover, overexpression of neither Oct-3/4 (Figure 1A and B) nor Nanog (see below) inhibited the activity of the Sox2 promoter/enhancer gene construct.
Figure 1.
Effect of Sox2 Oct-3/4 and Sox11 overexpression on Sox2 promoter activity. Schematic of the Sox2 promoter/reporter construct is shown. The gray boxes indicate critical enhancers, the basal Sox2 promoter is represented by a black box adjacent to the CAT reporter gene, and the relative HMG/POU cassette is indicated with an asterisk. F9 EC cells were transiently transfected and assayed as described in the Materials and methods section. The cells were transfected with 12 µg of the Sox2 promoter/reporter gene construct (Sox2 + US/DSE) plus 1 µg of the CMV-β-gal expression vector. In (A), 1 or 3 µg of either the Sox2 or Oct-3/4 expression vector was co-transfected. In (B), 3 µg of the indicated expression vector (Sox2, Oct-3/4 or Sox11) was co-transfected. Reporter activity was normalized to that of β-galactosidase and the activity of Sox2 + US/DSE (with no overexpression) was set to 1. The data shown represents the mean and standard deviation of duplicate samples from a representative experiment. Each experiment was repeated twice and similar results were obtained.
Sox2 overexpression inhibits the promoter of multiple Sox2:Oct-3/4 target genes
Given the effect of Sox2 overexpression on its own promoter/reporter gene construct, we tested whether Sox2 exerted similar effects on other Sox2:Oct-3/4 target genes. Using a panel of promoter/reporter constructs, we determined that Sox2 overexpression inhibited the activity of FGF-4, Oct-3/4, Nanog and UTF1 promoter/reporter gene constructs in a dose-dependent fashion (Figure 2). Each of the promoter/reporter gene constructs contained the appropriate promoter and enhancer regions, including a functional HMG/POU cassette within their respective enhancers. Furthermore, the placement of the enhancers within the promoter/reporter gene constructs reflects their positions, relative to the transcription start site, in their endogenous genes (see Materials and methods section for details). Remarkably, each promoter/enhancer construct exhibited a similar dose-dependent inhibition in response to Sox2 overexpression, which suggests that a common mechanism may be responsible for the inhibition of each promoter/reporter gene construct. Interestingly, the residual activity observed in response to Sox2 overexpression was greater than the activity of equivalent constructs in which the HMG/POU cassette was disrupted, which is typically >90% lower than the activity of the wild-type promoter/reporter gene construct (8,11–13,15–17,44).
Figure 2.
Effect of Sox2 overexpression on multiple Sox2:Oct-3/4 target gene promoters. Schematics of each of the promoter/reporter constructs are shown. The gray boxes indicate critical enhancers, the basal promoter is represented by a black box adjacent to the CAT reporter gene, and the relative HMG/POU cassette is indicated with an asterisk. F9 EC cells were transfected with 12 µg of the indicated promoter/reporter gene construct plus 1 µg of the CMV-β-gal expression vector. Where indicated, 1 or 3 µg of the Sox2 expression vector was co-transfected. Reporter activity was normalized to that of β-galactosidase and the activity of each individual promoter/reporter construct (with no Sox2 overexpression) was set to 1. Overexpression values are shown relative to each particular gene construct. The data shown represents the mean and standard deviation of duplicate samples from a representative experiment. This experiment was repeated twice and similar results were obtained.
We considered the possibility that high levels of Sox2 lead to a reduction in the levels of Oct-3/4, and, as a result, cause a reduction in the expression of all Sox2:Oct-3/4 target genes. We tested this possibility by examining whether the inhibition by Sox2 could be reversed by overexpressing Oct-3/4. However, in combination with Sox2, Oct-3/4 overexpression did not reverse the inhibition observed with Sox2 overexpression (Figure 3). Interestingly, overexpression of Oct-3/4 on its own increased the activities of both the Sox2 and the FGF-4 promoter/reporter gene constructs (Figure 3). Moreover, as shown below, overexpression of Oct-3/4 in the absence of Sox2 substantially inhibits the activity of the Oct-3/4 promoter/reporter gene construct. Hence, down-regulation of Oct-3/4 does not appear to be primarily responsible for the effects of Sox2 overexpression.
Figure 3.
Effect of Sox2 plus Oct-3/4 overexpression on the Sox2 and FGF-4 promoters. Schematics of each of the promoter/reporter constructs are shown. The gray boxes indicate critical enhancers, the basal promoter is represented by a black box adjacent to the CAT reporter gene, and the relative HMG/POU cassette is indicated with an asterisk. F9 EC cells were transfected with 12 µg of either (A) Sox2 + US/DSE or (B) FGF-4 + E plus 1 µg of the CMV-β-gal expression vector. Where indicated, 1, 2 or 3 µg of the Sox2 and/or the Oct-3/4 expression vector was co-transfected. Reporter activity was normalized to that of β-galactosidase and the activity of the reporter construct (with no overexpression) was set to 1. Overexpression values are shown relative to the given promoter/reporter construct. The data shown represents the mean and standard deviation of duplicate samples from representative experiments. Each experiment was repeated twice and similar results were obtained.
Sox2 overexpression inhibits the expression of endogenous Sox2:Oct-3/4 target genes
Given the broad effects of Sox2 overexpression observed above, we examined the effects of Sox2 overexpression on the endogenous expression of Sox2:Oct-3/4 target genes. For this purpose, we used a chimeric protein containing GFP fused to full-length Sox2 (GFP-Sox2), which like Sox2, inhibits the activities of constructs containing promoter/enhancer regions of Sox2:Oct-3/4 target genes (data not shown). More specifically, F9 EC cells were transiently transfected with either an expression vector for GFP (control) or an expression vector for GFP-Sox2. One day later, GFP positive cells for each condition were collected by cell sorting and their RNA isolated and analyzed by real-time qPCR using GAPDH expression for normalization (Figure 4A). Consistent with the results described above, we determined that mRNA for FGF-4, Oct-3/4, UTF1 and Nanog were each reduced by ∼50% in cells transfected with GFP-Sox2. Using a primer set that detects a region of Sox2 mRNA not present in the GFP-Sox2 expression vector, we detected a reduction (averaging 30%) of endogenous Sox2 gene expression. We suspect there are several reasons why the reduction in endogenous gene expression (∼50%) was less than that seen in the promoter/reporter gene assays (∼80%). The most likely explanation is the difference in time courses used in the two assays, 24 and 48 h, respectively. In addition, different endpoints were assayed—mRNA as opposed to CAT enzymatic activity. To examine the level of GFP-Sox2 protein in the transfected cells, GFP-Sox2 positive cells were isolated 24 h later. Western blot analysis was performed on nuclear extracts prepared from the isolated cells using a Sox2 antibody. This antibody detects both endogenously expressed Sox2 and the larger GFP-Sox2 fusion protein and the relative intensities of Sox2 and GFP-Sox2 were measured as described in the Materials and methods section. We determined that GFP-Sox2 levels were ∼2-fold higher than endogenously produced Sox2 (Figure 4B). If the levels of endogenous Sox2 have not changed significantly, and may have actually decreased in response to GFP-Sox2, then the combined level of Sox2 and GFP-Sox2 in the transfected cells was at most 3-fold higher than the normal levels of Sox2. In related studies, we determined that the viability of the F9 EC cells was not compromised by overexpression of Sox2. When replated after cell sorting, the plating efficiency and morphology of the GFP and the GFP-Sox2 cells were very similar to one another and to that of untreated F9 EC cells (data not shown). Taken together, our findings argue that Sox2 overexpression not only inhibits the activity of promoter/reporter constructs of Sox2:Oct-3/4 target genes, but also inhibits the expression of their endogenous counterparts.
Figure 4.
Effect of Sox2 overexpression on endogenous Sox2:Oct-3/4 target gene expression. (A) F9 EC cells were transfected with either GFP or GFP-Sox2, and GFP positive cells were collected and processed as described in the Materials and methods section. Between these two cell populations, the levels of expression of five genes were compared after normalizing the values to the expression of GAPDH. Target gene expression in GFP-Sox2-transfected cells is reported as a fraction of the corresponding expression in control cells (GFP-transfected). The values shown represent the mean and standard deviation after averaging values from two separate isolates of GFP positive cells and at least two qPCR assays of each gene. (B) Western blot analysis of Sox2 protein in F9 EC cells transfected with the GFP-Sox2 expression plasmid and collected by flow cytometry. GFP-Sox2 and endogenous Sox2 are indicated.
Sox2 inhibition is mediated through a functional HMG/POU cassette
Next, we examined the underlying mechanism(s) responsible for the inhibitory effects of Sox2. The finding that each of the five promoter/reporter constructs was inhibited to a similar extent by Sox2 overexpression raised the possibility that the inhibitory effects of Sox2 are mediated by a common mechanism. A common denominator among each of the five genes is the presence of an HMG/POU cassette in a critical enhancer present in each of the genes. The involvement of HMG/POU cassettes is consistent with our previous finding that Sox2 overexpression does not inhibit the FGF-4 promoter in constructs lacking the FGF-4 enhancer nor does it inhibit an FGF-4 promoter/reporter gene construct in which the HMG/POU cassette of the FGF-4 enhancer is disrupted (31). Similarly, Sox2 overexpression does not inhibit the activity of a promoter/reporter construct containing the Sox2 promoter, but lacking the Sox2 enhancers (data not shown).
To examine the role of the HMG/POU cassettes of these genes, we initially tested the effects of Sox2 overexpression on the activity of a heterologous promoter/reporter construct pCATSO3, which contains six copies of the FGF-4 HMG/POU cassette upstream of a minimal SV40 promoter (30). As in the case of the five promoter/reporter gene constructs tested above (Figures 1 and 2), Sox2 overexpression caused a dose-dependent decrease in the activity of pCATSO3 in F9 EC cells (data not shown). Next, we compared the effects of Sox2 overexpression on a panel of promoter/reporter constructs for the Sox2, UTF1 and FGF-4 genes in which the HMG/POU cassette present in each of their enhancers had been disrupted by site-directed mutagenesis. As discussed above, disruption of the HMG/POU cassettes of these constructs reduces their activity ∼90%. In contrast to the effects of Sox2 on the wild-type constructs, Sox2 overexpression did not inhibit the activity of constructs with ablated HMG/POU cassettes (Figure 5A). Thus, it appears that the inhibitory effects of Sox2 are mediated through the HMG/POU cassette of Sox2:Oct-3/4 target genes. Surprisingly, Sox2 overexpression actually increased the activities of the modified promoter/reporter gene constructs and this was also true for a Nanog promoter/reporter gene construct in which its HMG/POU cassette had been disrupted (data not shown). Currently, it is unclear why Sox2 stimulates the mutant promoter/reporter genes constructs. Possible explanations are addressed in the Discussion section.
Figure 5.
Effect of Sox2 overexpression on Sox2:Oct-3/4 target gene constructs containing mutant HMG/POU cassettes. Schematics of each of the promoter/reporter constructs are shown. The gray boxes indicate critical enhancers, the basal promoter is represented by a black box adjacent to the CAT reporter gene, and the relative HMG/POU cassette (or mutant) is indicated with an asterisk. (A) In each promoter/reporter construct, the HMG/POU cassette was destroyed. (B) Where indicated, the POU motif in the HMG/POU cassette was mutated. F9 EC cells were transfected with 12 µg of the indicated promoter/reporter gene construct plus 1 µg of the CMV-β-gal expression vector. Where indicated, 1 or 3 µg of the Sox2 expression vector was co-transfected. Reporter activity was normalized to that of β-galactosidase and the activity of each individual promoter/reporter construct (with no overexpression) was set to 1. Overexpression values are shown relative to each particular gene construct. The data shown represents the mean and standard deviation of duplicate samples from representative experiments. This experiment was repeated twice and similar results were obtained.
The synergism between Sox2 and Oct-3/4, when bound to the HMG/POU cassettes, is well established (7,8,10–12,14–17,28,30). Hence, we considered the possibility that only the HMG motif, which binds Sox2, is necessary for inhibition by Sox2 overexpression. This possibility was tested by transfecting increasing amounts of Sox2 in conjunction with a Sox2 promoter/reporter construct containing a mutated POU motif, but a functional HMG motif [Sox2 + US/DSE(Pmut)]. However, Sox2 overexpression had no inhibitory effect, and actually stimulated promoter activity, similar to that seen when the entire HMG/POU cassette was disrupted (Figure 5B). Hence, it appears that the HMG portion of these cassettes, on its own, is not sufficient for repression via elevated levels of Sox2, and that an intact HMG/POU cassette is required.
The requirement for a functional HMG/POU cassette suggested that Sox2 overexpression does not exert a general effect on transcription. This is consistent with a lack of an inhibitory effect of Sox2 on other promoters. In addition to not observing any inhibitory effects of Sox2 overexpression on the FGF-4 and Sox2 promoters (in the absence of their respective enhancers), we did not observe any effects of Sox2 overexpression on the CMV and SV40 promoters (data not shown). Similarly, Sox2 overexpression had no effect on the HSV TK promoter (Figure 6). However, when the HSV TK promoter was coupled with the FGF-4 enhancer, which contains an HMG/POU cassette, promoter/reporter activity was significantly repressed by the elevated levels of Sox2 (Figure 6). Taken together, our findings argue that Sox2 overexpression does not exert a general effect on transcription, but rather influences the function of strong enhancers that contain HMG/POU cassettes.
Figure 6.
Effect of Sox2 overexpression on the HSV TK promoter. Schematics of each of the promoter/reporter constructs are shown. The gray box indicates the FGF-4 enhancer (HMG/POU cassette is indicated with an asterisk) and the basal TK promoter is represented by a black box adjacent to the CAT reporter gene. F9 EC cells were transfected with 12 µg of the indicated promoter/reporter gene construct plus 1 µg of the CMV-β-gal expression vector. Where indicated, 3 µg of the Sox2 expression vector was co-transfected. Reporter activity was normalized to that of β-galactosidase and the activity of the TK promoter/reporter construct (with no enhancer and no overexpression) was set to 1. All other values are shown relative to that particular gene construct (TK). The data shown represents the mean and standard deviation of duplicate samples from a representative experiment. This experiment was repeated twice and similar results were obtained.
Sox2 inhibition is mediated by its TAD
We also examined which domain of Sox2 is responsible for mediating repression. For these studies, we used chimeric proteins containing different domains of Sox2 and Sox11 (40). Sox2 and Sox11 each have an N-terminal DBD (HMG domain), which share nearly 70% sequence homology. In contrast, their C-terminal domains, which contain their TADs, exhibit no significant homology. Although Sox11 inhibits the FGF-4 promoter/reporter gene construct (31), it does not inhibit the activity of the Sox2 promoter/reporter gene construct (Figure 1B) or the activity of the Nanog promoter/reporter gene construct (see below). This enabled us to test whether the HMG domain and/or the C-terminal domain of Sox2 is responsible for its inhibitory effects. Specifically, we tested two chimeric constructs. One contained the N-terminal region as well as the DBD of Sox2 coupled to the Sox11 C-terminal domain (Sox2-2-11). The other contained the N-terminal domain and the DBD of Sox11 coupled to the Sox2 C-terminal domain (Sox11-11-2) (Figure 7A). When overexpressed in conjunction with either a Sox2 or a Nanog promoter/reporter construct in F9 EC cells, Sox11-11-2 and Sox2 were equally inhibitory, whereas Sox2-2-11 stimulated both promoters (Figures 7B and C). These results argue strongly that the C-terminal domain of Sox2 contained in Sox11-11-2 (Sox2 amino acids 131–319), is sufficient for full inhibition. Given that the Sox2 TAD is spread over amino acids 152–189 and 255–319 (30,45), our findings further suggest that the TAD of Sox2 is responsible for its inhibitory effect. Consistent with this possibility, overexpression of a truncated form of Sox2, which lacks amino acids 292–319, did not inhibit promoter activity (data not shown). In this regard, we previously demonstrated that the ability of this truncated protein to transactivate was abrogated (30). Surprisingly, Sox2-2-11 stimulated both the Sox2 and the Nanog promoter/reporter gene constructs. We suspect that this is due to the fact that the Sox11 TAD in Sox2-2-11 is far stronger than the TAD of Sox2 (40) and that Sox2 and Sox11 are likely to mediate their effects on transcription through the action of different co-activators.
Figure 7.
Effect of chimeric Sox2/11 overexpression on Sox2 and Nanog promoter activity. (A) Schematics of chimeric Sox constructs. Sox2-2-11 contains the Sox2 DBD coupled with the Sox11 TAD whereas Sox11-11-2 contains the Sox11 DBD coupled with the Sox2 TAD. (B) Effect of chimeric Sox overexpression on Sox2 promoter activity. (C) Effect of chimeric Sox overexpression on Nanog promoter activity. In both (B) and (C), a schematic of the promoter/reporter construct is shown—the gray boxes indicate critical enhancers, the basal promoter is represented by a black box adjacent to the CAT reporter gene, and the relative HMG/POU cassette is indicated with an asterisk. F9 EC cells were transfected with 12 µg of the indicated promoter/reporter gene construct plus 1 µg of the CMV-β-gal expression vector. Where indicated, 3 µg of either the Sox2, Sox2-2-11 or Sox11-11-2 expression vector was co-transfected. Reporter activity was normalized to that of β-galactosidase and the activity of each individual promoter/reporter construct (with no overexpression) was set to 1. Overexpression values are shown relative to each particular gene construct. The data shown represents the mean and standard deviation of duplicate samples from a representative experiment. Each experiment was repeated twice and similar results were obtained.
Sox2 overexpression inhibits Sox2:Oct-3/4 target genes in P19 EC cells and D3 ES cells
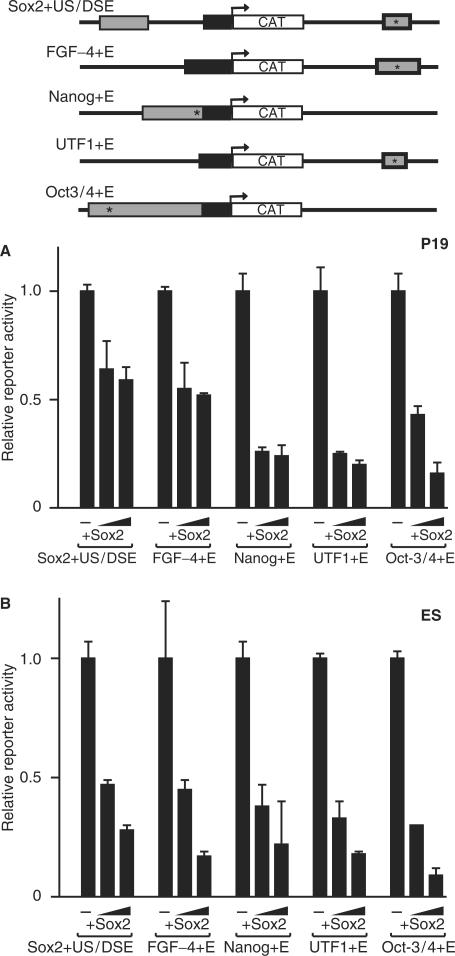
To extend the findings with F9 EC cells, the effects of Sox2 overexpression were examined in mouse P19 EC cells. Like F9 EC cells, P19 EC cells are frequently used in gene expression studies. For these studies, P19 EC cells were transiently transfected with a panel of Sox2:Oct-3/4 target gene promoter/reporter constructs and increasing amounts of a Sox2 expression vector. As in the case of F9 EC cells, Sox2 overexpression significantly inhibited the activities of the Sox2, FGF-4, Nanog, UTF1 and Oct-3/4 promoter/reporter gene constructs (Figure 8A). The levels of inhibition of the Nanog, UTF1 and Oct-3/4 promoter/reporter gene constructs were similar to those observed in F9 EC cells. However, the activity of the FGF-4 promoter/reporter construct was not inhibited as strongly as that observed in F9 EC cells. This is likely to be due to the FGF-4 enhancer, which contains an HMG/POU cassette, having much lower overall activity in P19 EC cells (7). To extend these findings further, mouse D3 ES cells were transiently transfected with a Sox2 expression vector and promoter/reporter gene constructs for the Sox2, FGF-4, Nanog, UTF1 and Oct-3/4. Similar to F9 and P19 EC cells, overexpression of Sox2 caused significant inhibition of the Sox2, FGF-4, Nanog, UTF1 and Oct-3/4 promoter/reporter gene constructs in ES cells (Figure 8B). Hence, the findings with P19 EC cells and D3 ES cells argue that Sox2 inhibition of Sox2:Oct-3/4 target genes is not cell-line specific.
Figure 8.
Effect of Sox2 overexpression on Sox2:Oct-3/4 target genes in P19 EC cells and D3 ES cells. (A) Effects of Sox2 overexpression on Sox2, FGF-4, Nanog, UTF1 and Oct-3/4 promoter/reporter gene constructs in P19 EC cells. (B) Effects of Sox2 overexpression on Sox2, FGF-4, Nanog and Oct-3/4 promoter/reporter gene constructs in D3 ES cells. Schematics of each of the promoter/reporter constructs are shown. The gray boxes indicate critical enhancers, the basal promoter is represented by a black box adjacent to the CAT reporter gene, and the relative HMG/POU cassette is indicated with an asterisk. In (A), P19 EC cells were transfected with 12 µg of the indicated promoter/reporter gene construct plus 1 µg of the CMV-β-gal expression vector. Where indicated, the cells were also transfected with 1 or 3 µg of the Sox2 expression vector. In (B), D3 ES cells were transfected with 4.5 µg of the indicated promoter/reporter gene construct plus 1 µg of the CMV-β-gal expression vector. Where indicated, the cells were also transfected with 0.3 or 1 µg of the Sox2 expression vector. In both (A) and (B), reporter activity was normalized to that of β-galactosidase and the activity of each individual promoter/reporter construct (with no Sox2 overexpression) was set to 1. Overexpression values are shown relative to each particular gene construct. The data shown represents the mean and standard deviation of duplicate samples from a representative experiment. Each experiment was repeated and similar results were obtained.
Neither Oct-3/4 nor Nanog broadly inhibit the promoters of Sox2:Oct-3/4 target genes
Recent studies employing ChIP-Chip and ChIP-PET technologies have shown that Sox2, Oct-3/4 and Nanog are each associated with the regulatory regions of a large number of genes (1,2). In many instances, two or all three of these TFs are associated in close proximity within the same region of the gene. Hence, it has been argued that Sox2, Oct-3/4 and Nanog function as master regulators that exert a strong influence over a vast array of target genes. Given these findings, we examined whether overexpression of Oct-3/4 or Nanog also exert broad effects over the expression of Sox2:Oct-3/4 target genes. As discussed above, Oct-3/4 did not inhibit the activity of the Sox2 or the FGF-4 promoter/reporter gene constructs (Figures 3A and B). Thus, we tested the effect of overexpressing Oct-3/4 on an Oct-3/4 promoter/reporter construct. Increasing levels of Oct-3/4 inhibited its own promoter in a fashion similar to Sox2 overexpression (Figure 9A). Moreover, overexpression of Oct-3/4 also inhibited the activity of the Nanog promoter/reporter gene construct (data not shown), as reported by others working with ES cells (27). We also determined that overexpression of Nanog inhibited the activity of the Nanog promoter/reporter gene construct. However, Nanog overexpression exerted little or no effect over the Oct-3/4 promoter/reporter gene construct (Figure 9A) or the Sox2 promoter/reporter gene construct (Figure 9B). Collectively, our results argue that Sox2 overexpression exerts an overarching effect over Sox2:Oct-3/4 target genes; whereas, overexpression of Oct-3/4 and Nanog exert more restricted effects over Sox2:Oct-3/4 target genes.
Figure 9.
Effect of Oct-3/4 and Nanog overexpression on promoter activity. (A) Effect of Oct-3/4 overexpression on Oct-3/4 promoter activity. (B) Effect of Nanog overexpression on Nanog and Sox2 promoter activity. Schematics of each of the promoter/reporter constructs are shown. The gray boxes indicate critical enhancers, the basal promoter is represented by a black box adjacent to the CAT reporter gene, and the relative HMG/POU cassette is indicated with an asterisk. F9 EC cells were transfected as indicated with 12 µg of either the Oct-3/4, Nanog or Sox2 promoter/reporter gene constructs (Oct-3/4 + E, Nanog + E or Sox2 + US/DSE) plus 1 µg of the CMV-β-gal expression vector. Where indicated, 1 or 3 µg of the Oct-3/4, Sox2 or Nanog expression vector was co-transfected. Reporter activity was normalized to that of β-galactosidase and the activity of each individual promoter/reporter construct (with no overexpression) was set to 1. Overexpression values are shown relative to each particular gene construct. The data shown represents the mean and standard deviation of duplicate samples from a representative experiment. Each experiment was repeated twice and similar results were obtained.
DISCUSSION
Recent studies argue that Sox2 and Oct-3/4 work in concert to control a transcriptional regulatory network that helps orchestrate mammalian embryogenesis as well as the self-renewal and pluripotency of ES cells. Although the full spectrum of genes regulated by Sox2 and Oct-3/4 is only beginning to emerge, Sox2 and Oct-3/4 are known to work cooperatively to stimulate the transcription of their own genes and the transcription of a growing number of downstream targets, many of which are required for normal development. Previous studies have shed significant light into the mechanisms by which Sox2:Oct-3/4 target genes are activated (8–17). However, much less is known about negative feedback controls that serve to prevent the overexpression of these genes. The studies described in this manuscript provide new insights into the diversity of mechanisms used to tightly control transcription of Sox2:Oct-3/4 target genes. Sox2 is a central player in this control having a biphasic effect on this network of genes. Importantly, we demonstrate that overexpression of Sox2 not only inhibits the activity of promoter/reporter gene constructs for five different Sox2:Oct-3/4 target genes, but the endogenous expression of these genes is inhibited within one day when Sox2 levels are elevated ∼3-fold or less. In addition, our studies argue that the inhibitory effects of Sox2 are mediated through the HMG/POU cassettes present in the enhancers of these genes. Our studies further demonstrate that the region containing the TAD of Sox2, and not the DBD, is necessary for this inhibition. Lastly, the studies presented here show that, unlike Sox2, overexpression of neither Oct-3/4 nor Nanog exerts broad control over Sox2:Oct-3/4 target genes.
Although overexpression of Sox2 exerts broad control over Sox2:Oct-3/4 target genes, multiple lines of evidence argue that the effects of Sox2 overexpression are specific. Moreover, Oct-3/4 and Nanog do not exert the same broad-based inhibitory effects observed with Sox2. Importantly, our studies argue that Sox2 overexpression does not exert a widespread repression of general transcription. Sox2 overexpression did not inhibit several viral promoters, including the CMV, SV40 or HSV TK promoters. Similarly, Sox2 overexpression did not inhibit the activities of Sox2 or FGF-4 promoter/reporter constructs that lack their enhancers. In addition, promoter/reporter gene constructs of these genes, in which the HMG/POU cassettes were disrupted, were not inhibited. In fact, they were stimulated by Sox2 overexpression. Currently, it is unclear why Sox2 stimulated the mutant promoter/reporter gene constructs. It is possible that the stimulatory effects of Sox2 are not specific. Overexpression of Sox2 increases the basal activity of the promoter-less parent construct several fold (data not shown). However, the basal activity of the parent construct is very low, and much lower than the activities of our promoter/reporter gene constructs in which the HMG/POU cassettes were disrupted. Moreover, previous findings have shown that the cryptic activity within the parent vector used in our studies is silenced when promoters and strong enhancers, such as those used in our studies, are inserted into the vector (35). Alternatively, the regulatory regions inserted in our promoter/reporter gene constructs may contain additional Sox2 response elements. Previous studies have shown that the enhancer present in our FGF-4 promoter/reporter gene construct possesses a second functional HMG motif located ∼20 bp upstream of the HMG motif in the FGF-4 HMG/POU cassette (46). However, disruption of this HMG motif in conjunction with disruption of the HMG/POU cassette did not ablate the stimulatory effect observed with Sox2 overexpression. Further study will be needed to resolve this question.
Our studies argue that the TAD of Sox2 is likely to be responsible for its inhibitory action. Sox2 overexpression may lead to the expression of a repressor that acts on Sox2:Oct-3/4 target genes. Alternatively, Sox2 overexpression may interfere with the function of one or more essential coactivators. Given that TADs activate transcription by interacting with coactivators and are likely to do so by interacting with more than one coactivator (47–49), Sox2 overexpression may function by a squelching mechanism (50), whereby incomplete Sox2:cofactor complexes are generated that are unable to activate Sox2:Oct-3/4 target genes. This model is similar to one proposed to explain the inhibitory effects observed when overexpressing scaffold proteins (51). In the case of Sox2, this seems likely given that it also physically interacts with Oct-3/4 when bound to HMG/POU cassettes (11,44,52). Currently very little is known about the coactivators involved in the transcription of Sox2:Oct-3/4 target genes. The one exception is p300 (13,30). Previous studies have shown that p300 can mediate the effects of the GAL4-Sox2 fusion protein as well as the combined effects of Sox2 and Oct-3/4 on promoter/reporter gene constructs (30). Moreover, ChIP studies have shown that p300 is recruited to the FGF-4 enhancer in F9 EC cells (13), and our unpublished studies indicate that this is also true for the enhancer regions of the Nanog gene and the Oct-3/4 gene where their HMG/POU cassettes are located (Mallanna and Rizzino, unpublished data). This prompted us to test whether overexpression of p300 could reverse the inhibitory effects of Sox2. However, overexpression of p300 did not reverse the inhibitory effects of Sox2 (data not shown). Hence, Sox2 overexpression may influence the proper function of one or more other coactivators.
An important outcome from the work described in this study is the difference in the responses to overexpression of Sox2, Oct-3/4 and Nanog (Figure 10). The response to Sox2 is biphasic; whereas, the responses of most Sox2:Oct-3/4 target genes to Oct-3/4 and Nanog are not. At levels found in EC cells, Sox2 and Oct-3/4 activate Sox2:Oct-3/4 target genes (Figure 10A). However, when Sox2 levels rise, they begin to inhibit the expression of Sox2:Oct-3/4 target genes (Figure 10B). Although Sox2 exerts a general effect over Sox2:Oct-3/4 target genes, overexpression of Oct-3/4 or Nanog does not (Figure 10C). Overexpression of Oct-3/4 inhibits the activities of Oct-3/4 and Nanog promoter/reporter gene constructs in EC cells, as it does in ES cells (27). However, overexpression of Oct-3/4 slightly stimulates the activities of Sox2 and FGF-4 promoter/reporter gene constructs, rather than inhibiting them. Given the known role of Oct-3/4 in the activation of the Sox2 and FGF-4 genes, this is not overly surprising. As is the case of Oct-3/4, overexpression of Nanog leads to inhibition of its own promoter (Figure 10C). Nonetheless, the failure of Nanog to inhibit the Sox2 or the Oct-3/4 promoter/reporter gene construct argues that overexpression of Nanog does not exert a general restraining effect over Sox2:Oct-3/4 target genes.
Figure 10.
Model of Sox2, Oct-3/4 and Nanog feedforward and feedback gene regulation. Sox2 is shown as a central node in a network of genes critical to embryogenesis as well as stem cell self-renewal and pluripotency. (A) At normal levels, Sox2 and Oct-3/4 activate transcription of all known Sox2:Oct-3/4 target genes. (B) Increasing levels of Sox2 leads to inhibition of target gene expression. (C) Increasing levels of Oct-3/4 inhibits expression of only Oct-3/4 and Nanog, and high levels of Nanog lead to inhibition of Nanog gene expression, but not other Sox2:Oct-3/4 target genes. Presumably, there are additional Sox2:Oct-3/4 target genes that are regulated in a similar fashion.
In summary, the findings described in this study provide new insights into the diversity of mechanisms used to tightly control transcription of Sox2:Oct-3/4 target genes. Importantly, our studies in conjunction with the large number of earlier studies (8–17), which demonstrated the role of Sox2 in the activation of Sox2:Oct-3/4 target genes, argue that Sox2 functions as a molecular rheostat that carefully controls a key transcriptional regulatory network required for proper embryogenesis, as well as self-renewal and pluripotency of ES and EC cells. Given our expanded understanding of how Sox2 influences Sox2:Oct-3/4 target genes, it will be important to examine more closely how overexpression of Sox2 influences the growth and differentiation of stem cells. In this regard, we did not observe any phenotypic effects of transient overexpression of Sox2 in F9 EC cells. Interestingly, Sox2 overexpression (2- to 20-fold increase in mRNA) has been shown to promote the differentiation of ES cells into neuroectoderm, but it did not alter their ability to self-renew (53).
ACKNOWLEDGEMENTS
Charles Kuszynski and Linda Wilkie of the Cell Analysis Core Facility are thanked for their technical advice and assistance. This work was supported by a grant from the National Institute of General Medical Sciences (GM80751). Core facilities used in connection with this work was supported in part by a Cancer Center Support Grant (CA36727). B.B. and J.K. were supported in part by a Cancer Biology Training Grant (CA09476). Funding to pay the Open Access publication charge was provided by the grant information: GM80751.
Conflict of interest statement. None declared.
REFERENCES
- 1.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 3.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 4.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 6.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 7.Boer B, Bernadt CT, Desler M, Wilder PJ, Kopp JL, Rizzino A. Differential activity of the FGF-4 enhancer in F9 and P19 embryonal carcinoma cells. J. Cell. Physiol. 2006;208:97–108. doi: 10.1002/jcp.20635. [DOI] [PubMed] [Google Scholar]
- 8.Kuroda T, Tada M, Kubota H, Kimura H, Hatano SY, Suemori H, Nakatsuji N, Tada T. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 2005;25:2475–2485. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakatake Y, Fukui N, Iwamatsu Y, Masui S, Takahashi K, Yagi R, Yagi K, Miyazaki J, Matoba R, et al. Klf4 cooperates with oct3/4 and sox2 to activate the lefty1 core promoter in embryonic stem cells. Mol. Cell. Biol. 2006;26:7772–7782. doi: 10.1128/MCB.00468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dailey L, Yuan H, Basilico C. Interaction between a novel F9-specific factor and octamer-binding proteins is required for cell-type-restricted activity of the fibroblast growth factor 4 enhancer. Mol. Cell. Biol. 1994;14:7758–7769. doi: 10.1128/mcb.14.12.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 12.Nishimoto M, Fukushima A, Okuda A, Muramatsu M. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 1999;19:5453–5465. doi: 10.1128/mcb.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nowling T, Bernadt C, Johnson L, Desler M, Rizzino A. The co-activator p300 associates physically with and can mediate the action of the distal enhancer of the FGF-4 gene. J. Biol. Chem. 2003;278:13696–13705. doi: 10.1074/jbc.M207567200. [DOI] [PubMed] [Google Scholar]
- 14.Tokuzawa Y, Kaiho E, Maruyama M, Takahashi K, Mitsui K, Maeda M, Niwa H, Yamanaka S. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol. Cell. Biol. 2003;23:2699–2708. doi: 10.1128/MCB.23.8.2699-2708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 2002;30:3202–3213. doi: 10.1093/nar/gkf435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J. Biol. Chem. 2005;280:5307–5317. doi: 10.1074/jbc.M410015200. [DOI] [PubMed] [Google Scholar]
- 17.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida T, Muramatsu H, Muramatsu T, Sakamoto H, Katoh O, Sugimura T, Terada M. Differential expression of two homologous and clustered oncogenes, Hst1 and Int-2, during differentiation of F9 cells. Biochem. Biophys. Res. Commun. 1988;157:618–625. doi: 10.1016/s0006-291x(88)80295-x. [DOI] [PubMed] [Google Scholar]
- 19.Peters G, Brookes S, Smith R, Placzek M, Dickson C. The mouse homolog of the hst/k-FGF gene is adjacent to int-2 and is activated by proviral insertion in some virally induced mammary tumors. Proc. Natl. Acad. Sci. U.S.A. 1989;86:5678–5682. doi: 10.1073/pnas.86.15.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velcich A, Delli-Bovi P, Mansukhani A, Ziff EB, Basilico C. Expression of the K-fgf protooncogene is repressed during differentiation of F9 cells. Oncogene Res. 1989;5:31–37. [PubMed] [Google Scholar]
- 21.Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- 22.Meno C, Shimono A, Saijoh Y, Yashiro K, Mochida K, Ohishi S, Noji S, Kondoh H, Hamada H. Lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell. 1998;94:287–297. doi: 10.1016/s0092-8674(00)81472-5. [DOI] [PubMed] [Google Scholar]
- 23.Nishimoto M, Miyagi S, Yamagishi T, Sakaguchi T, Niwa H, Muramatsu M, Okuda A. Oct-3/4 maintains the proliferative embryonic stem cell state via specific binding to a variant octamer sequence in the regulatory region of the UTF1 locus. Mol. Cell. Biol. 2005;25:5084–5094. doi: 10.1128/MCB.25.12.5084-5094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 25.Hough SR, Clements I, Welch PJ, Wiederholt KA. Differentiation of mouse embryonic stem cells after RNA interference-mediated silencing of OCT4 and Nanog. Stem Cells. 2006;24:1467–1475. doi: 10.1634/stemcells.2005-0475. [DOI] [PubMed] [Google Scholar]
- 26.Wu Q, Chen X, Zhang J, Loh YH, Low TY, Zhang W, Zhang W, Sze SK, Lim B, et al. Sall4 interacts with nanog and co-occupies Nanog genomic sites in embryonic stem cells. J. Biol. Chem. 2006;281:24090–24094. doi: 10.1074/jbc.C600122200. [DOI] [PubMed] [Google Scholar]
- 27.Pan G, Li J, Zhou Y, Zheng H, Pei D. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. FASEB J. 2006;20:1730–1732. doi: 10.1096/fj.05-5543fje. [DOI] [PubMed] [Google Scholar]
- 28.Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma YG, Rosfjord E, Huebert C, Wilder P, Tiesman J, Kelly D, Rizzino A. Transcriptional regulation of the murine k-FGF gene in embryonic cell lines. Dev. Biol. 1992;154:45–54. doi: 10.1016/0012-1606(92)90046-j. [DOI] [PubMed] [Google Scholar]
- 30.Nowling TK, Johnson LR, Wiebe MS, Rizzino A. Identification of the transactivation domain of the transcription factor Sox-2 and an associated co-activator. J. Biol. Chem. 2000;275:3810–3818. doi: 10.1074/jbc.275.6.3810. [DOI] [PubMed] [Google Scholar]
- 31.Bernadt CT, Nowling T, Rizzino A. Transcription factor Sox-2 inhibits co-activator stimulated transcription. Mol. Reprod. Dev. 2004;69:260–267. doi: 10.1002/mrd.20168. [DOI] [PubMed] [Google Scholar]
- 32.Rizzino A, Orme LS, De Larco JE. Embryonal carcinoma cell growth and differentiation. Production of and response to molecules with transforming growth factor activity. Exp. Cell Res. 1983;143:143–152. doi: 10.1016/0014-4827(83)90116-7. [DOI] [PubMed] [Google Scholar]
- 33.Nowling T, Desler M, Kuszynski C, Rizzino A. Transfection of embryonal carcinoma cells at high efficiency using liposome-mediated transfection. Mol. Reprod. Dev. 2002;63:309–317. doi: 10.1002/mrd.90014. [DOI] [PubMed] [Google Scholar]
- 34.Johnson LR, Johnson TK, Desler M, Luster TA, Nowling T, Lewis RE, Rizzino A. Effects of B-Myb on gene transcription: phosphorylation-dependent activity and acetylation by p300. J. Biol. Chem. 2002;277:4088–4097. doi: 10.1074/jbc.M105112200. [DOI] [PubMed] [Google Scholar]
- 35.Rosfjord E, Lamb K, Rizzino A. Cryptic promoter activity within the backbone of a plasmid commonly used to prepare promoter/reporter gene constructs. In Vitro Cell. Dev. Biol. Anim. 1994;30A:477–481. doi: 10.1007/BF02631317. [DOI] [PubMed] [Google Scholar]
- 36.Nishimoto M, Fukushima A, Miyagi S, Suzuki Y, Sugano S, Matsuda Y, Hori T, Muramatsu M, Okuda A. Structural analyses of the UTF1 gene encoding a transcriptional coactivator expressed in pluripotent embryonic stem cells. Biochem. Biophys. Res. Commun. 2001;285:945–953. doi: 10.1006/bbrc.2001.5265. [DOI] [PubMed] [Google Scholar]
- 37.Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- 38.Wiebe MS, Wilder PJ, Kelly D, Rizzino A. Isolation, characterization, and differential expression of the murine Sox-2 promoter. Gene. 2000;246:383–393. doi: 10.1016/s0378-1119(00)00086-x. [DOI] [PubMed] [Google Scholar]
- 39.Catena R, Tiveron C, Ronchi A, Porta S, Ferri A, Tatangelo L, Cavallaro M, Favaro R, Ottolenghi S, et al. Conserved POU binding DNA sites in the Sox2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J. Biol. Chem. 2004;279:41846–41857. doi: 10.1074/jbc.M405514200. [DOI] [PubMed] [Google Scholar]
- 40.Wiebe MS, Nowling TK, Rizzino A. Identification of novel domains within Sox-2 and Sox-11 involved in autoinhibition of DNA binding and partnership specificity. J. Biol. Chem. 2003;278:17901–17911. doi: 10.1074/jbc.M212211200. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scholtz B, Lamb K, Rosfjord E, Kingsley M, Rizzino A. Appearance of nuclear protease activity after embryonal carcinoma cells undergo differentiation. Dev. Biol. 1996;173:420–427. doi: 10.1006/dbio.1996.0037. [DOI] [PubMed] [Google Scholar]
- 43.Rizzino A. Embryonic stem cells provide a powerful and versatile model system. In: Litwack G, editor. Vitamins and Hormones. Philadelphia: Academic Press, Philadelphia; 2002. pp. 1–42. [DOI] [PubMed] [Google Scholar]
- 44.Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ambrosetti DC, Scholer HR, Dailey L, Basilico C. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J. Biol. Chem. 2000;275:23387–23397. doi: 10.1074/jbc.M000932200. [DOI] [PubMed] [Google Scholar]
- 46.Luster TA, Rizzino A. Regulation of the FGF-4 gene by a complex distal enhancer that functions in part as an enhanceosome. Gene. 2003;323:163–172. doi: 10.1016/j.gene.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 47.Chen D, Huang SM, Stallcup MR. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 2000;275:40810–40816. doi: 10.1074/jbc.M005459200. [DOI] [PubMed] [Google Scholar]
- 48.Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 49.Zhang C, Baudino TA, Dowd DR, Tokumaru H, Wang W, MacDonald PN. Ternary complexes and cooperative interplay between NCoA-62/Ski-interacting protein and steroid receptor coactivators in vitamin D receptor-mediated transcription. J. Biol. Chem. 2001;276:40614–40620. doi: 10.1074/jbc.M106263200. [DOI] [PubMed] [Google Scholar]
- 50.Cahill MA, Ernst WH, Janknecht R, Nordheim A. Regulatory squelching. FEBS Lett. 1994;344:105–108. doi: 10.1016/0014-5793(94)00320-3. [DOI] [PubMed] [Google Scholar]
- 51.Kortum RL, Lewis RE. The molecular scaffold KSR1 regulates the proliferative and oncogenic potential of cells. Mol. Cell. Biol. 2004;24:4407–4416. doi: 10.1128/MCB.24.10.4407-4416.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17:2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao S, Nichols J, Smith AG, Li M. SoxB transcription factors specify neuroectodermal lineage choice in ES cells. Mol. Cell Neurosci. 2004;27:332–342. doi: 10.1016/j.mcn.2004.08.002. [DOI] [PubMed] [Google Scholar]