Abstract
The global regulator Mlc is a repressor of several genes and operons that are involved in sugar uptake and metabolism. A Salmonella enterica serovar Typhimurium mlc mutant showed reduced levels of invasion and cytotoxicity compared to the wild-type, and exhibited reduced expression levels of hilD, hilA and invF, which are regulatory genes in the Salmonella pathogenicity island 1 (SPI1). However, the effects of Mlc on hilD expression and bacterial invasiveness were not seen in the hilE mutant, and hilE expression was increased in the mlc mutant, which suggests that Mlc exerts positive effects on the expression of SPI1 genes by reducing the expression of HilE, which is known to down-regulate the expression of SPI1 genes through direct interaction with HilD. We found that the two known promoters of hilE were not modulated by Mlc, and we identified a third promoter, designated P3, which was repressed by Mlc. The gel mobility shift assay and footprinting analysis revealed that Mlc repressed hilE in a direct manner by binding to two distinct sites in the hilE P3 promoter region. The specific down-regulation of hilD observed in the presence of Mlc regulon-inducible sugars, such as glucose and mannose, could not be detected in the mlc mutant. Based on these results, we propose that Mlc functions to sense the availability of sugars and is linked to virulence gene regulation by its ability to control hilE expression in Salmonella.
INTRODUCTION
Salmonella enterica serovar Typhimurium is a facultative intracellular pathogen that initiates disease, which is normally limited to gastroenteritis in humans. However, this bacterium causes systemic disease in mice and has been used as an animal model of typhoid fever. Since Salmonella is acquired usually by oral ingestion of contaminated materials, a key step in the infection process is passage across the intestinal epithelium by invasion of M cells in Peyer's patches (1). Many of the genes required for intestinal penetration and invasion of host cells are carried on the 40-kb region at centisome 63, which is called Salmonella pathogenicity island 1 (SPI1) (2). SPI1 contains at least 37 genes, which encode various components of the type III secretion systems (T3SSs), its regulators and its secreted effectors (3). The effector proteins mediate actin cytoskeleton rearrangement, in the form of large membrane ruffles, which engulf the bacteria into the host cell (4,5).
The expression of SPI1 is controlled in response to a specific combination of environmental signals, including pH, oxygen tension, medium osmolarity, bile, Mg2+ concentration, short-chain fatty acids (SCFAS), and growth stage of the bacteria (3,6). Although many factors and a complex mechanism related to environmental stimuli are involved in the modulation of SPI1 genes, they are thought to converge on the activation of several transcriptional regulators encoded within SPI1, such as HilA, HilC, HilD and InvF (7–9). HilA plays a crucial role in the expression of genes that encode the SPI1-T3SS apparatus, the prg and inv/spa operons, by binding upstream of prgH and invF (10). The expression of invF leads to the induction of several effector genes encoded both within SPI1 (sic/sip operon) and outside SPI1 (sigD/sopB and sopE), with SicA as a co-factor (11). HilC and HilD, which are members of the AraC/XylS family, have been postulated to act as a derepressor of hilA expression by counteracting the action of negative regulatory elements at the hilA promoter (12,13). However, it has been shown that HilD provides an essential activating function for hilA in the absence of negative regulators (14). HilC and HilD also directly activate the alternative promoter of the invF operon, which is independent of HilA (15,16).
Similar to HilC and HilD, RtsA activates the expression of SPI1 genes by binding upstream of the master regulatory gene, hilA, to induce its expression (9,17). Recently, Baxter and Jones (18) have shown that HilE is an important Salmonella-specific regulator of hilA expression. A null mutation in hilE causes an increase in hilA expression and invasion. Using two-hybrid analysis, it has been shown that HilE interacts with HilD, which suggests that HilE represses hilA expression by inhibiting the activity of HilD through a protein–protein interaction (19,20). Several other genes have been identified that exert positive or negative effects on these direct regulators of SPI1 in response to the changes in environmental conditions. These genes include those that encode several two-component regulatory systems (PhoP/Q, PhoBR, OmpR/EnvZ and SirA/BarA), post-transcriptional systems (CsrAB, RNase E, PNPase and Lon), nucleoid proteins (HU, FIS, H-NS and Hha), signaling molecules (ppGpp and NpnN), and other regulatory proteins (FliZ, FadD and CpxA) (6,7,19,21,22).
The phosphoenolpyruvate: sugar phosphotransferase system (PTS) is the major sugar transport system in many Gram-positive and Gram-negative bacterial species. In the animal model, attenuation of virulence has been noted for Salmonella strains that carry mutations in the pts, crr, cya or crp genes, which encode the general energy-coupling enzymes of the PTS, enzyme IIAGlc of the PTS, adenylate cyclase and cyclic AMP receptor protein, respectively (23,24). Mlc is a global regulator of carbohydrate metabolism and controls several genes involved in sugar utilization (25–27). Therefore, it seemed possible that Mlc also affects the virulence of Salmonella. In the present study, a Salmonella Typhimurium mlc mutant was constructed to investigate the contribution of Mlc to the virulence phenotype. We have found that Mlc activates SPI1 gene expression by repressing hilE expression.
MATERIALS AND METHODS
Bacterial strains and growth condition
The strains used in this study are listed in Table 1. Bacteria were routinely grown in LB broth (1% bactotryptone, 0.5% bacto yeast extract, 1% NaCl) at 37°C. When necessary, the medium was supplemented with ampicillin (100 μg/ml) or kanamycin (100 μg/ml). Tryptone broth (TB) (1% bactotryptone, 0.8% NaCl) buffered with 0.1 M MOPS (pH 7.0) was used to investigate the effect of various carbohydrate sources on the expression of hilE and hilD. As needed, the following supplements were added to the TB: 0.2% (w/v) glucose, mannose, arabinose or glycerol. Low oxygen bacterial growth condition (SPI-inducing condition) was used to induce invasion gene transcription (8,14). Briefly, a stationary-phase culture that had been grown overnight with shaking was used as the stock culture. The stock culture was inoculated into fresh LB broth at a 1:100 dilution, and grown in static culture to the late exponential phase for 4 h (OD600 ∼ 0.6). The isogenic hilE/mlc double-mutant strain SR1305 was obtained through P22 HT-mediated transduction to SR1304 of the mutant allele (hilE::cam) from the hilE mutant strain BJ2462 (18).
Table 1.
The bacterial strains and plasmids used in this study
| Strains or plasmids | Genotype | References |
|---|---|---|
| S. Typhimurium | ||
| SL1344 | Wild type, rpsL hisG | 16 |
| MGS-7 | SL5283, r−m+ galE recDΔmutS Camr | 28 |
| BJ2462 | SL1344, hilE::cam, Camr | 20 |
| SR1304 | SL1344, mlc::kan, Kanr | This study |
| SR1305 | SR1304, hilE::cam, Camr | This study |
| E. coli | ||
| SR5055 | MC4100 mlc::kan, Kanr | Laboratory collection |
| BL21(DE3) | F− ompT hsdSB with a λ prophage carrying the T7 RNA polymerase | Novagen |
| Plasmid | ||
| pUC19 | Ampr | Laboratory collection |
| pJB3 | hilD expression from lac promoter in pZC320 | 14 |
| pJB5 | hilD::lacZ reporter vector, Ampr | 14 |
| pMAB69 | hilE::lacZ reporter vector, Tetr | 18 |
| pKB | pUC19 containing the mlc promoter and structure region | This study |
| pET-15b | Ampr, N-terminal His-tag vector | Novagen |
| pET-Mlc | pET-15b containing the mlc structure gene | This study |
Construction of S. Typhimurium mlc mutant
The MGS-7 strain (Table 1) was used for the transfer by transduction of chromosomal DNA from E. coli to S. Typhimurium and for efficient recombination into the genomic DNA of the recipient, as strains that carry the galE mutation are particularly suitable as hosts for phages P1 and P22 (28). When Salmonella galE mutants are grown in the presence of high concentrations (∼1%) of galactose and glucose, the LPS produced is the smooth form (P22-sensitive phenotype), whereas when they are grown in the absence of galactose, the LPS produced is rough (P1-sensitive phenotype). Escherichia coli phage P1 transductions were performed to transfer the mlc::kan region of E. coli to MGS-7 (S. Typhimurium galE mutant), and phage P22 transductions were performed from the mlc-mutated MGS-7 to the wild-type S. Typhimurium SL1344. Mutation of the mlc gene was confirmed by PCR.
Plasmid construction
pKB, which contains the mlc promoter and structural region, was constructed by inserting the DNA fragment that spans −473 to +1281 relative to the mlc translation start codon into the PstI site of plasmid pUC19. An 1750-bp PCR fragment of the mlc region was amplified from the chromosomal DNA using the forward primer 5′-GTCTGACAGAACTGCAGGAAGAACCTTTCG-3′ and the reverse primer 5′-GATATGGCAAGGGCCTGCAGCTTGAGTTAG-3′ (PstI site underlined). The pET-Mlc plasmid used for purification of the Salmonella Mlc protein was generated by cloning the DNA segment that spans positions −13 to +1306 relative to the translation start codon into the NdeI and BamHI sites downstream of the His-tag element in plasmid pET-15b (Novagen). The PCR fragment of the mlc structural gene was amplified using the forward primer 5′-AAAGGGAGTGACATATGGTTGCTGATAGTC-3′ and the reverse primer 5′-AAATAATACAGTGGATCCAGTCTAAGATAT-3′, which introduced NdeI and BamHI sites (underlined), respectively. The clone was verified by DNA sequencing.
Invasion and replication assays
The HeLa and RAW264.7 cells were cultured in DMEM and the HEp-2 cells were grown at 37°C under 5% CO2 in RPMI 1640 medium that was supplemented with 10% fetal bovine serum (FBS), penicillin (50 U/ml) and streptomycin (50 U/ml). Confluent monolayers for infection with bacteria were prepared in 24-well tissue culture plates, and Salmonella (∼2 × 106 CFU/well), which were washed with PBS and suspended in pre-warmed medium, were then added to the cell monolayers (∼2 × 105 cells/well) at a multiplicity of infection (MOI) of 10. Invasion assays were conducted with bacteria that were grown to exponential phase in static cultures, using previously described protocols (29). Survival of the opsonized S. Typhimurium strains in RAW 264.7 cells was determined as previously described (30,31). Briefly, aerated cultures grown to stationary phase were opsonized for 15 min in DMEM that contained 10% FBS and 10% normal mouse serum, and the bacteria were then added to the RAW 264.7 cells.
LDH assay for cytotoxicity
Lactate dehydrogenase (LDH) is a cytoplasmic enzyme, the presence of which in the culture medium reflects the loss of plasma membrane integrity. LDH activity in the culture supernatants was measured using the colorimetric Cytotox 96 kit (Promega). The RAW 264.7 murine macrophage-like cell line was seeded into a 96-well plate at a density of 5 × 104 cells/well and incubated for 24 h at 37°C. Before infection with the bacteria, the medium was replaced with serum-free DMEM medium. The cells were infected with Salmonella bacteria, which were grown to exponential phase without shaking, at an MOI of 10. After 1 h of incubation, gentamicin (100 μg/ml) was added, to kill the extracellular bacteria. At 4 h post-infection, the culture supernatants were collected for analysis. Cytotoxicity was quantified colorimetrically with the CytoTox 96 kit and the percentage of cytotoxicity was calculated according to the formula: 100 × [(experimental release − spontaneous release)/(total release − spontaneous release)], in which spontaneous release is the amount of LDH activity in the supernatant of uninfected cells, and total release is the LDH activity in macrophage lysates.
β-Galactosidase assay
β-Galactosidase assays were performed according to the standard method of Miller (32).
Primer extension assay
Total RNA was isolated from the bacteria using the Trizol reagent (Life Technologies). To study SPI1 gene expression, the 32P-labeled primers (50 000 c.p.m.) (Table 2) were co-precipitated with 30 µg of total RNA. Primer extension reactions were performed as described by Lim et al. (29).
Table 2.
Oligonucleotide primers used for primer extension analysis
| Gene | Nucleotide sequences (5′ → 3′) | Complementary regionc |
|---|---|---|
| hilA | TAATAATATTGTTATAACTAACTGTGATTA | ∼−216 to −245 |
| invFA | CATTGTGTCGGCTTTCAGAAAATGACATAT | ∼−1 to +28 |
| invFD | GGAGTTAATATGAAAAAATTTTATAGCTGT | ∼−447 to −476 |
| hilC | GGAAATTTGTTCGGCTGTTGAAGGTGATTA | ∼+45 to +74 |
| hilD | TTTAATTTGCTGCCGGGTATTTGTCAAAAG | ∼+73 to +102 |
| hilE | CAATGAAAGAACGTTCCATTTTCCAGCCA | ∼+2 to +30a |
| hilE | ATATCAATATCATTTCTTATTTTTATCCGA | ∼−188 to −217b |
| prgH | CTGTCAGCAATGGAAACTCACAGCCGTTCA | ∼+71 to +100 |
| sigD | AGGTTTTTTGTAGGCTTTTAAAAGCCTCCT | ∼+44 to +73 |
ahilE3 primer used for the analysis of P1 and P2 transcript.
bhilE5 primer used for the analysis of P3 transcript.
cNumbers of nucleotides were determined relative to translational start site respectively.
Real-time PCR analysis
Bacterial cells were grown to exponential phase in LB medium without shaking, and then total RNA was isolated using RNeasy Mini Kit (Qiagen). After DNase treatment (Ambion), cDNA was synthesized from 1 μg of RNA using Omniscript RT kit (Qiagen) and random hexamers (Invitrogen) according to the manufacturer's instruction. Quantification of cDNA was carried out using IQ SYBR Green PCR Supermix (Bio-Rad), and real-time amplification of PCR product was analyzed using iCycler IQ™ (Bio-Rad). The amplification program consisted of one cycle at 95°C for 5 min, followed by 40 cycles at 95°C for 30 s, 52°C for 30 s, and 72°C for 30 s. The relative amount of cDNA was calculated using a standard curve obtained from PCR on serially diluted genomic DNA as templates. mRNA expression levels of target genes were normalized to 16S rRNA expression level. The sequences of the primers used are presented in Table 3.
Table 3.
Primers for real-time PCR analysis
| Gene | Primer sequence (5′ to 3′) | ||
|---|---|---|---|
| Forward | Reverse | Size (bp) | |
| hilA | GTCCGGTCGTAGTGGTGTCT | CGGCAGTTCTTCGTAATGGT | 182 |
| invF | TGTCGCACCAGTATCAGGAG | AAATAGCGCGAAACTCAGGA | 155 |
| hilE | AAAGCCGGATCAAAGGTTTT | CTTTCACCGTTTTCCCGTTA | 180 |
| rrs | CGGGGAGGAAGGTGTTGTG | CAGCCCGGGGATTTCACATC | 178 |
Purification of the Mlc protein
Cultures of E. coli strain BL21(DE3) that carried pET-Mlc were grown at 37°C in LB that was supplemented with ampicillin (100 μg/ml). Mlc under the control of the T7 promoter was induced with 0.01 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in the early log phase of growth. After induction, the cells were allowed to grow for 5 h, and were then harvested by centrifugation. The N-terminal-His6-tagged Mlc protein was purified from cell extracts by Ni2+ affinity chromatography, according to the Novagen standard protocol.
Gel mobility shift assay
The 340-bp hilE promoter DNA fragment (−581 to −241) was amplified by PCR using the hilE-F2 (5′-GTAGCGTTGGATCGTTTCGTGTTC-3′) and hilE-R2 (5′-TCCACCGAATCGGAATATAGACAATTC-3′) primers. The PCR product was purified from an agarose gel using a gel extraction kit (Qiagen), and labeled with [γ-32P] ATP. The binding buffer for this assay contained 20 mM Tris-acetate (pH 8.0), 3 mM magnesium acetate, 200 mM potassium glutamate, 100 μg/ml BSA, 1 mM dithiothreitol and 1% sucrose. For the Mlc–DNA binding interactions, 2 nM of labeled DNA fragments were mixed with the Mlc protein and 100 ng of poly dI-dC (Amersham Pharmacia) as the DNA competitor in 20 μl of buffer. For competition assays, an excess of unlabeled probe was added. The binding mixture was incubated at room temperature for 15 min and analyzed by electrophoresis on a 6% polyacrylamide gel.
DNase I footprinting
A DNA fragment that contained the hilE promoter was amplified by PCR using [γ-32P]ATP-labeled hilE-F2 primer and unlabeled hilE-R2 primer. The PCR product was purified from an agarose gel using a gel extraction kit (Qiagen). Mlc–DNA binding was performed in 40 μl of binding buffer under the conditions used in the gel mobility shift assay. DNase I solution (5 μl; 10 ng DNase I per reaction) was added to the binding mixture, which was then placed at room temperature for 1 min. DNase I activity was terminated by the addition of 200 μl of stop solution that contained 0.4 M sodium acetate, 10 mM EDTA and 100 μg/ml yeast tRNA. After phenol extraction and ethanol precipitation, the pellet was dissolved in sequencing dye, and resolved on a 6% polyacrylamide gel that contained 8 M urea.
RESULTS
Mutation of the mlc gene reduces Salmonella invasiveness
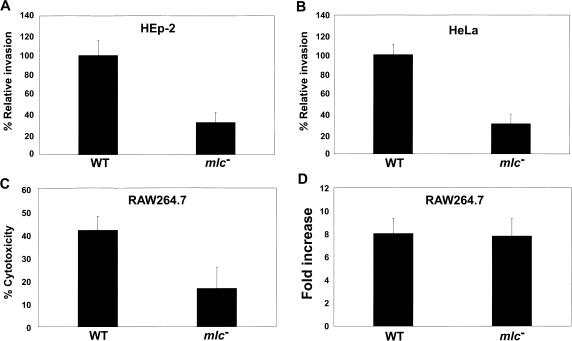
The process of invasion into non-phagocytic epithelial cells, which is known to be mediated by proteins secreted by the SPI1-encoded type III machinery (3), is an important initial step in the pathogenesis of S. Typhimurium. We examined the ability of an mlc mutant, SR1304, to invade cultured HEp-2 and HeLa epithelial cells. SL1344 was used as the control strain, and the invasiveness of this strain was arbitrarily set at 100%. When the Salmonella bacteria were grown in static cultures to the exponential phase (SPI1-inducing condition), the invasiveness of SR1304 for both epithelial cell lines was reduced by about 3-fold, compared to that of SL1344 (Figure 1A and B).
Figure 1.
Analysis of the virulence of the Salmonella mlc mutant strain. All of the cell infection experiments were carried out with wild-type (SL1344) and mlc− mutant (SR1304) strains. HEp-2 (A) and HeLa cells (B) were infected with Salmonella grown to exponential phase without shaking. Infected cells were lysed 2 h after infection, and dilutions of the suspension were plated onto LB agar medium, to enumerate colony-forming units (CFUs). The data are presented as percentages of the CFU of the wild-type strain. (C) For the cytotoxicity assay, RAW 264.7 macrophage cells were infected with Salmonella grown to exponential phase without shaking, and then assayed for LDH release. (D) The intracellular replication assay was carried out with opsonized bacteria grown to stationary phase under aerobic conditions. After 2 and 18 h of infection, the eukaryotic cells were lysed and the viable intracellular (gentamicin-protected) bacteria were counted. The values shown represent fold-increases, which were calculated as the ratios of intracellular bacteria 2 and 18 h after bacterial entry. These assays were performed at least twice in triplicate, to allow calculation of means and standard deviations.
S. Typhimurium induces the apoptosis of infected macrophages. This process is rapid, specific and depends on the T3SS encoded within SPI1 (33,34). To discover whether the mlc mutant grown under the SPI1-inducing condition affects the capacity to induce apoptosis in macrophages, the release of the cytoplasmic enzyme LDH was determined in a cytotoxicity assay (33). Figure 1C shows that SL1344 killed 42% of the macrophages, whereas SR1304 killed only 16% of the macrophages 4 h after infection, which demonstrates that the mlc gene plays an important role in the expression of genes involved in cytotoxicity. We also tested the effect of the mlc mutation on the ability to replicate within the RAW264.7 macrophage cell line. After cultivation to the stationary phase with aeration, the bacteria were opsonized with 10% normal mouse serum and added to the cell monolayer (30). When the number of intracellular bacteria were measured at 2 and 18 h post-infection, the degree of replication was similar for SL1344 and SR1304 (Figure 1D). Collectively, these results suggest that the main target of Mlc action in Salmonella virulence is SPI1, which is known to encode various genes that are required for Salmonella invasion of host cells.
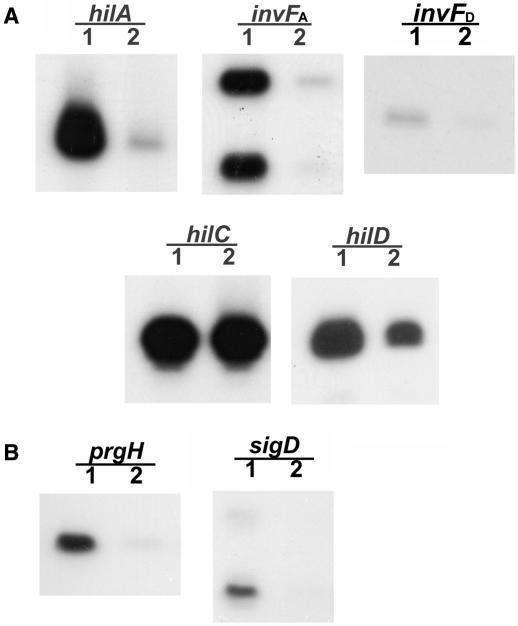
The expression levels of hilA, invF and hilD, but not of hilC, are decreased in the mlc mutant
To identify the target gene in SPI1 that is regulated by Mlc, the expression patterns of the SPI1-regulatory genes, which include hilA, invF, hilC and hilD, were examined by primer extension analysis of total RNA from SL1344 and SR1304 grown statically to the exponential phase. The transcriptional start sites of these genes have been reported previously (12,16,35,36). SR1304 exhibited a >10-fold reduction in hilA and HilA-dependent invF (invFA) expression, and about a 3-fold reduction in hilD and HilC/D-dependent invF (invFD) expression, compared to SL1344. However, hilC expression was unchanged (Figure 2A). We also observed reduced expression of the prgH and sigD genes, which are known to be regulated by HilA and InvF (10,11,29,35), in SR1304 (Figure 2B). The expression levels of hilA and invF were previously shown to be affected by both HilC and HilD (8,9,16) but since hilC levels are unaffected by the mlc mutation (Figure 2A) we speculated that Mlc was regulating hilA and invF expression indirectly probably by modulating the expression of hilD or the higher regulator of hilD.
Figure 2.
Effects of mlc mutation on the expression of SPI1-regulatory genes (A) and apparatus/effector genes (B) in S. Typhimurium, as analyzed by primer extension analysis. Total RNA was prepared from SL1344 (lane 1; wild type) and SR1304 (lane 2; mlc− mutant), which were grown to exponential growth phase in LB medium without shaking. Aliquots of 30 μg of total RNA were co-precipitated and annealed with end-labeled primers. Reactions were performed as described in the Materials and Methods section. The products were resolved on a 6% sequencing gel.
Mlc modulates the expression of SPI1 genes by repressing hilE
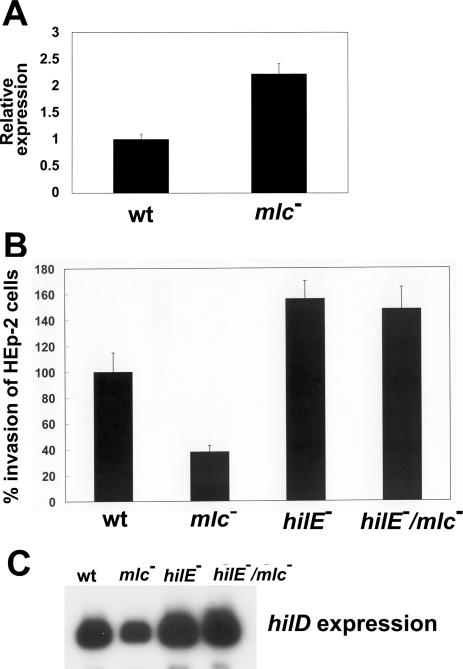
Subsequently, we studied whether hilE expression was affected by Mlc. Baxter et al. (20) have found that HilE plays an important regulatory role in the expression of the Salmonella-invasive phenotype by affecting hilA transcription through direct interaction with HilD. To study the effect of Mlc on hilE expression, we measured the expression of hilE-lacZY on the low-copy plasmid pMAB69, which contains the hilE promoter region from −886 to +121, relative to the translation start site. When these cells were grown in static cultures to exponential phase, SL1344-pMAB69 expressed 28.9 ± 3.5 Miller units of β-galactosidase, whereas SR1304-pMAB69 expressed 49.5 ± 5.3 Miller units of β-galactosidase; the level of hilE expression in the mlc mutant was almost 1.8-fold higher than that in the wild-type strain. The hilE mRNA level measured by real-time PCR also revealed that hilE expression was increased in SR1304 by about 2-fold (Figure 3A). These results suggest that Mlc can act as a negative regulator of hilE.
Figure 3.
Effects of the hilE/mlc/double mutation on Salmonella invasiveness and hilD expression. (A) mRNA level of hilE gene determined by real-time PCR analysis. SL1344 (wt) and SR1304 (mlc−) were grown in LB medium without shaking. Total RNA was isolated from aliquots of cells obtained at the exponential phase. Expression levels of the target genes were normalized to that of 16S rRNA gene. Real-time PCR assay was performed three times in duplicate. (B) HEp-2 cells were infected with SL1344 (wild-type), SR1304 (mlc− mutant), BJ2462 (hilE− mutant) and SR1305 (hilE−/mlc− double mutant), which were grown to exponential phase without shaking. The CFU of intracellular bacteria were assessed 2 h after infection. The values are presented as percentages of the wild-type invasion, which was set at 100%. This assay was performed at least twice in triplicate, to allow calculation of means and standard deviations. (C) Thirty micrograms of total RNA from the indicated strains, extracted at the exponential growth phase in LB medium, were subjected to primer extension analysis.
The role of HilE in the regulation of SPI1 gene expression by Mlc was further studied by comparing the invasive abilities of the hilE and hilE/mlc mutant strains of Salmonella. Invasiveness for HEp-2 cells was reduced to 39.1% of the wild-type level by mlc mutation, whereas it was increased by 1.57-fold by hilE mutation (Figure 3B). However, the hilE/mlc double mutant showed an almost similar invasion ability to that of the hilE mutant (Figure 3B), which suggests that the effect of the mlc mutation on the invasive phenotype is mainly dependent upon HilE function. The requirement of hilE for Mlc function was reflected in the levels of hilD transcription. While hilD expression was reduced in the mlc mutant, it was slightly higher than the wild-type level in both the hilE and hilE/mlc mutant strains (Figure 3C). Additionally, the over-expression of HilD from the plasmid pJB3 containing hilD gene under the control of a lac promoter caused the induction of hilA by ∼2-fold and invF by ∼6-fold irrespective of hilE or mlc mutation when real-time PCR was used to compare the expression levels (data not shown). Since HilD activates the transcription of hilA (14), which in turn can activate HilA-dependent invFA expression (10), and directly activates HilC/D-dependent invFD expression (16), these results establish that the mlc mutation exerts a negative effect on SPI1 gene expression, mainly by increasing the level of hilE expression. The low level of hilD expression in the mlc mutant can be explained by the fact that the increased level of HilE in the mlc mutant can repress the activity of HilD, which is known to act as an activator of its own expression (9,36).
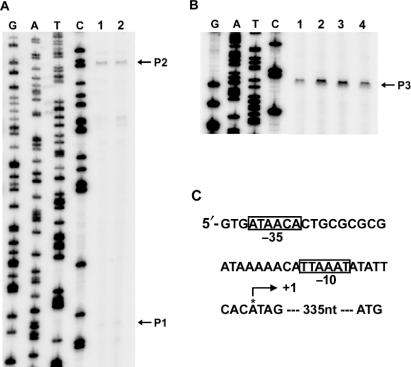
The hilE P3 promoter is repressed by Mlc
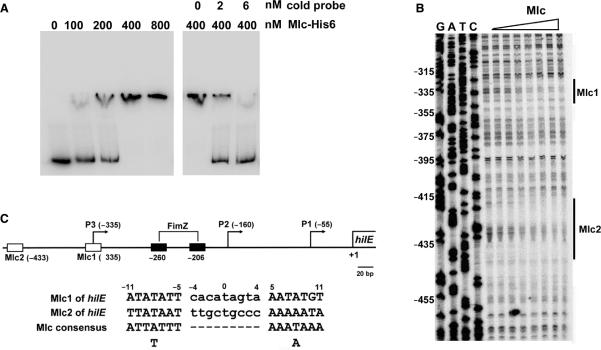
Baxter and Jones (18) have identified two independent hilE promoters, P1 and P2. We performed a primer extension assay of total RNA isolated from SL1344 grown to exponential phase in static culture, using the primer described by Baxter and Jones (18), to discover which promoter is regulated by Mlc. The transcriptional start site of one transcript, designated P1, agreed with the previous report, although the promoter activity was very low under the conditions used in our experiment (Figure 4A). We detected a stronger transcript, designated P2, upstream of P1, although the transcription start site of P2 was mapped 12 nucleotides upstream of that reported by Baxter and Jones (18) (Figures 4A and 5C). This difference could be attributed to differences in the experimental model conditions; in our study, total RNA from a Salmonella strain was used in the primer extension assay, whereas Baxter and Jones (18) used total RNA prepared from E. coli harboring the hilE reporter plasmid pMAB69. However, the activities of the two promoters were not changed by mlc mutation. Using a primer extension assay with the hilE5 primer (Table 2), the promoter of hilE, which is designated as P3, was newly identified; this promoter initiates 335 nucleotides upstream of the translation start site of hilE (Figure 4B, lane 1). We observed putative −10 (TTAAAT) and −35 (ATAACA) motifs for hilE P3 (Figure 4C) (37). Transcription from the P3 promoter was increased substantially by mlc mutation (Figure 4B, lane 2). The specificity of the Mlc effect was verified by complementation, as the plasmid pKB, which expresses mlc, restored the transcriptional level of hilE in SR1304 almost to that of the wild-type strain (Figure 4B, lane 4).
Figure 4.
Identification of an Mlc-regulated hilE promoter. Primer extension analysis was performed using total RNA from SL1344 (lane 1: wild type), SR1304 (lane 2: mlc− mutant), SR1304 harboring pUC19 (lane 3) and SR1304 harboring pKB (lane 4; Mlc over-expression), all of which were grown to exponential phase in LB medium without shaking. Aliquots of the RNA (30 μg) were subjected to primer extension analysis, and a sequence ladder was generated using the same end-labeled primer that was used for the primer extension analysis. (A) The effects of the mlc mutation on the P1 and P2 promoters were examined using the hilE3 primer (see Table 2). (B) The transcriptional start site of the new promoter (P3), which is modulated by the mlc mutation, was identified using the hilE5 primer (see Table 2). (C) The start site (+1) is indicated by an arrow, and the promoter elements that resemble the consensus −10 and −35 sites are boxed.
Figure 5.
The Mlc protein binds to the hilE promoter. (A) Gel mobility shift assay of hilE promoter DNA with the purified Mlc-His6 protein. Labeled hilE promoter DNA (2 nM) was incubated with 100 ng of poly dI-dC DNA competitor and various amounts of Mlc-His6, as indicated at the top of the left panel. Labeled DNA (2 nM) was mixed with 400 nM of Mlc-His6 and various concentrations of unlabeled hilE promoter DNA (cold probe), as indicated at the top of the right panel. The DNA–protein complexes were resolved by electrophoresis in a 6% polyacrylamide gel. (B) DNase I footprinting analysis of the hilE promoter DNA was performed with a probe for the non-coding strand. The hilE promoter DNA was incubated with purified Mlc-His6, which was diluted through a 2-fold series of dilutions in 1× binding buffer to the desired concentration (lanes 1–8: 0, 18.75, 37.5, 75, 150, 300, 600 and 1200 nM, respectively). The protected regions of the two Mlc sites are indicated with solid vertical lines and marked as Mlc1 and Mlc2. The numbering on the left is based on the translational start site for hilE. (C) Schematic representation of the locations of the promoters and putative protein-binding sites in the hilE promoter. The numbering is relative to the translational start site for hilE. The transcriptional start sites of P1 (−55), P2 (−160) and P3 (−335) are shown with arrows. The binding sites for Mlc and FimZ are shown by open and black boxes, respectively, and the numbers under the boxes indicate the centers of the binding sites. The sequence of each Mlc-binding site in the hilE promoter region is numbered relative to the center of the binding site and is indicated along with the consensus sequence for the Mlc-binding site. The locations of the P1 and P2 promoters and FimZ-binding sites have been reported by Baxter and Jones (18). The map is drawn to scale.
Mlc directly represses hilE by binding to the P3 promoter
A gel mobility shift assay was performed with purified Mlc, to test whether Mlc directly represses the hilE P3 promoter. The Mlc-His6 protein was produced in E. coli and purified to >90% homogeneity by Ni2+ affinity chromatography. The activity of the purified Mlc-His6 protein was verified with the control gel mobility shift experiment employing the Salmonella ptsG DNA fragment that contains the known Mlc-binding site (data not shown). When the labeled hilE promoter DNA was incubated with 2-fold dilutions of the purified Mlc-His6 protein (100–800 nM) in the presence of the non-specific DNA competitor poly dI-dC, the concentration-dependent formation of protein–DNA complexes was observed (Figure 5A). The addition of cold probe released the labeled probe from the retarded complex (Figure 5A, right panel), which indicates specific binding of Mlc to the hilE P3 promoter DNA.
The precise locations of the Mlc-binding sites were determined by DNase I footprinting with His-tagged Mlc. Two Mlc-binding sites, one at position −10 to +12 (Mlc 1) and the other at position −86 to −108 (Mlc 2) with respect to the transcriptional start site of hilE P3, were identified (Figure 5B). Inspection of the DNA sequences at these sites showed high-level homology with the known consensus Mlc-binding sequence, which has the conserved TT-9 bp-AA motif and an AT-rich region at positions ±7 to ±11, showing imperfect dyad symmetry (38) (Figure 5C). These results clearly demonstrate that Mlc can regulate directly the hilE P3 promoter by binding to the promoter.
Sugars that induce the Mlc regulon repress hilD expression by activating hilE
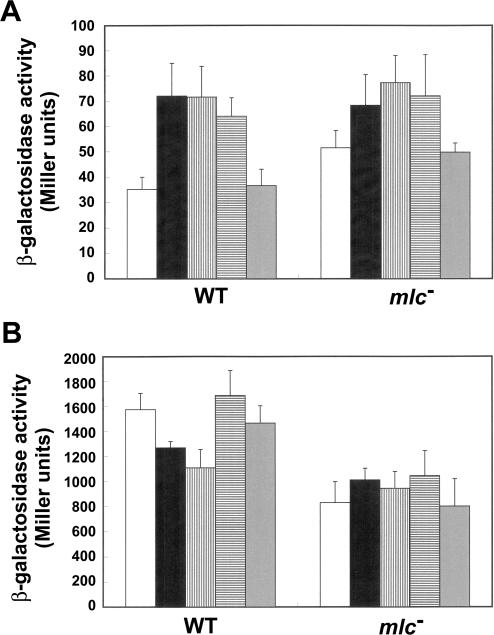
Sequestration of Mlc by the unphosphorylated form of glucose permease, enzyme IICBGlc, is known to displace Mlc from its DNA-binding sites, thereby allowing transcription of its target genes (25–27). The Mlc regulon can be induced by glucose and, to a lesser extent, by mannose (39). The effects of various carbon sources on the expression of hilE and hilD were studied with two Mlc regulon-inducing sugars, glucose and mannose, and two Mlc regulon-non-inducing carbohydrates, arabinose and glycerol. SL1344 and SR1304 carrying either pMAB69 (hilE-lacZY) or pJB5 (hilD-lacZY) were cultivated in TB that was buffered to pH 7.0 with 0.1 M MOPS, to minimize pH effects arising from growth in the presence of the various carbon sources. In the wild-type strain, glucose and mannose increased hilE expression 2-fold and hilE activation reduced hilD expression (Figure 6). The expression levels of hilE and hilD remained almost unaffected in the presence of glycerol. Interestingly, arabinose also activated hilE, albeit to a lesser degree than either glucose or mannose. Nevertheless, this caused a minute increase in hilD expression. In the absence of sugars, the mlc mutation increased hilE expression 1.5-fold, with a concomitant reduction in hilD expression. On the other hand, the expression in SR1304 of both hilE and hilD was increased in the presence of glucose, mannose or arabinose. These results imply that multiple regulatory pathways are involved in the regulation of hilE and hilD by carbohydrates.
Figure 6.
Effect of various carbohydrates on hilE and hilD expression levels. The β-galactosidase activities were measured for (A) the hilE-lacZY reporter plasmid pMAB69, and (B) the hilD-lacZY reporter plasmid pJB5, in both SL1344 (wild-type) and SR1304 (mlc− mutant). Cultures were grown to exponential phase in MOPS-buffered TB medium in static cultures without carbohydrate (white bars) or with 0.2% glucose (black bars), mannose (vertical hatched bars), arabinose (horizontal hatched bars) or glycerol (gray bars). The β-galactosidase assays were performed at least three times.
DISCUSSION
Analysis of the Salmonella mlc mutant revealed that invasiveness for epithelial cells was impaired by the mlc mutation (Figure 1). It is well known that SPI1 is required for the invasion of host cells and induction of macrophage apoptosis (2,3,40). The genetic evidence to date is consistent with a regulatory cascade of transcriptional activation, in which HilD, HilA and InvF act sequentially to activate SPI1 expression. In brief, HilD binds directly to sites upstream of hilA and invFD and acts as an activator (14,16,36). HilA activates the transcription of invFA and SPI1-T3SS apparatus genes required for the secretion of effector proteins, whereas the expression of effector proteins is regulated by InvF (10,11). This regulatory cascade implies that the reduced invasive phenotype of mlc mutant is the result of hilD repression, which affects both hilA and invF expression (Figures 1 and 2). The 2–3-fold decrease in hilD expression caused by the mlc mutation is sufficient to account for the nearly 10-fold decrease in hilA and invFA transcription, owing to the nature of the feed-forward regulatory loop, in that moderate effects on HilD production are amplified (9). Although the Salmonella mlc mutant was less invasive for epithelial cells than the wild-type strain, the extent of the reduction in invasiveness and cytotoxicity of the mlc mutant was not as great as we had expected, considering the observed reduction in SPI1 gene expression (Figure 2). This may be partly due to the expression of hilC, a de-repressor of hilA, which was not reduced in the mlc mutant (Figure 2A). The hilC mutation had little effect on SPI1 gene expression or the invasive phenotype, whereas the over-expression of hilC suppressed the hilD mutation, thereby promoting high levels of hilA and invF expression, which suggests that HilC can partially compensate for the loss of function of HilD and the invasive phenotype (8,9,15). We have reported previously that there may be an independent hilC regulatory pathway that is not applicable to hilA or invFA regulation when bacteria respond to changes in osmolarity (29).
Our data clearly demonstrate that Mlc directly regulates hilE expression by binding to the hilE P3 promoter (Figure 5). The two Mlc-binding sites in hilE P3 identified by the gel mobility shift assay and DNase I footprinting analysis show high-level homology with the known consensus Mlc-binding sequences; additionally, the two Mlc-binding sites are separated by 98 nt, as is the case for other promoters that have two Mlc-binding sites (41). The transcriptional start site of the hilE P3 promoter lies 335 nt upstream of the ATG start codon of hilE (Figures 4B and 5C). It is unusual that the 5′-untranslated region (UTR) of mRNA is more than 300 nt in bacteria (42). However, this length of 5′-UTR has been observed in other SPI1 regulators. The HilC/D-dependent transcriptional start site of the invFD promoter is 631 bp upstream of the invF open reading frame (16). The UTR of the hilA gene is also up to 350 nt in length, and is suggested to be involved in the complex regulation of hilA in response to environmental signals (8,12). It has been demonstrated that the region 190–270-bp upstream of the hilE promoter is required for the activation of hilE P2 expression by FimYZ, which is a response regulator that is involved in the expression of type 1 fimbriae and motility genes (18, Figure 5C). Therefore, we cannot rule out the possibility that additional cis- or trans-acting regulatory elements, which have not yet been characterized, are involved in hilE expression.
Glucose and mannose induce the Mlc regulon (39) by dephosphorylating EIICBGlc, which then sequesters Mlc from its binding sites (27,43). It has been suggested that glucose plays a negative role in the expression of invasion-associated genes in Salmonella. The addition of excess glucose to the culture medium results in the reduction of cell association by S. Typhimurium (44). The glucose present in DLB (LB broth diluted 1:5) decreased hilA expression 2.5-fold, as compared to DLB without glucose, while lactose and arabinose had little effect on hilA expression (45). The results presented in this study support the notion that hilE expression is activated as a result of Mlc sequestration by unphosphorylated EIICBGlc in the presence of glucose. We observed a 2-fold activation of hilE and concomitant repression of hilD in the presence of glucose or mannose, which are known inducers of the Mlc regulon, when TB was used (Figure 6). However, the effect of the sugar was not seen when LB was used (data not shown). We speculate that the nutrient-rich LB broth may mask the effect of the sugar on hilE expression because the effect of sugar addition on hilA expression was more distinct in DLB than LB (45). In addition, hilE expression was also increased in the presence of arabinose, which suggests an additional regulatory mechanism mediated by arabinose.
Interestingly, carbohydrate regulation of hilE and hilD expression in the mlc mutant created a different story. All of the carbohydrates tested in this study, with the exception of glycerol, increased hilD expression by ∼1.1–1.2-fold, even though hilE expression in the mlc mutant was increased by ∼30% in the presence of the sugars tested (Figure 6). These results suggest that the presence of complex regulatory mechanisms for hilE is required for optimal regulation of the SPI1 genes, as can be expected from the complex regulatory networks identified in Salmonella (46). It has been suggested that the two-component regulatory system PhoR-PhoB leads to increased hilE P2 expression and subsequent repression of hilA and invasion genes (7,18). The PhoR sensor kinase phosphorylates PhoB when extracellular Pi levels are low, and the phosphorylated PhoB then binds and activates the promoters in the Pho regulon. However, even in the absence of PhoR, many carbon sources are known to activate the Pho regulon via CreC, which is a PhoR homolog (47). Thus, carbon metabolism sensed by PhoB/R may also affect hilE expression (22), and this may be one of the reasons for the elevated hilE expression in the presence of carbohydrates in the mlc mutant (Figure 6A).
Recently, Teplitski et al. (48) have reported that the expression of sirA, which is a response regulator for BarA (49), is decreased in the presence of 50 mM glucose. The repressive effect of glucose was also observed for the downstream members of the SirA regulon, such as csrB, csrC and hilA (48,49). Collectively, these results show that SPI1 is not activated when Salmonella is grown in the presence of glucose (Figure 6; 44,45,48). Since the environmental conditions faced by Salmonella change constantly during passage through the intestine of the animal host, this bacterium should use multiple signals to modulate the virulence genes needed for survival. It is known that S. Typhimurium uses bile or SCFAs including acetate, propionate and butyrate, as environmental signals to modulate its invasion of the gastrointestinal tract (50–52). We propose that Salmonella can use the glucose concentration of the mammalian intestinal tract as one of many signals for the regulation of invasion genes. Salmonella may use Mlc to sense the availability of sugars, thereby allowing decisions as to when and where to initiate the expression of genes involved in invasion. S. Typhimurium typically invades the distal small intestine (ileum) of the mammalian upper gastrointestinal tract (53). Simple sugars, such as glucose, are rarely encountered in the distal ileum, as most are absorbed in the proximal portion of the small intestine (54). The relatively high glucose concentration in the proximal small intestine may repress SPI1 gene expression through Mlc, perhaps together with PhoR-PhoB and/or SirA, whereas upon transit to the distal ileum, the glucose concentration becomes low enough to activate the invasion genes of SPI1. The effects of various sugars on the differential regulation of SPI1 genes require further investigation to understand their roles in Salmonella pathogenesis.
ACKNOWLEDGEMENTS
This work was supported by a grant of the Korea Health 21 R&D project, Ministry of Health & Welfare, Republic of Korea (01-PJ10-PG6-01GM02-0002 and 00-PJ1-PG3-22000-0008). J.Y. and C.P were the recipients of a graduate fellowship provided by the Ministry of Education through the Brain Korea 21 Project. Funding to pay the Open Access publication charge was provided by the Brain Korea 21 Project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jones BD, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mills DB, Bajaj V, Lee CA. A 40 kilobase chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 genome. Mol. Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 3.Lostroh CP, Lee CA. The Salmonella pathogenicity island-1 type III secretion system. Microb. Infect. 2001;3:1281–1291. doi: 10.1016/s1286-4579(01)01488-5. [DOI] [PubMed] [Google Scholar]
- 4.Galán JE. Salmonella interactions with host cells: Type III secretion at work. Annu. Rev. Cell. Dev. Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 5.Guiney DG, Lesnick M. Targeting of the actin cytoskeleton during infection by Salmonella strains. Clin. Immunol. 2005;114:248–255. doi: 10.1016/j.clim.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Altier C. Genetic and environmental control of Salmonella invasion. J. Microbiol. 2005;43:85–92. [PubMed] [Google Scholar]
- 7.Lucas RL, Lostroh P, DiRusso CC, Spector MP, Wanner BL, Lee CA. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2000;182:1872–1882. doi: 10.1128/jb.182.7.1872-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas RL, Lee CA. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2001;183:2733–2745. doi: 10.1128/JB.183.9.2733-2745.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellermeier CD, Ellermeier JR, Slauch JM. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2005;57:691–705. doi: 10.1111/j.1365-2958.2005.04737.x. [DOI] [PubMed] [Google Scholar]
- 10.Lostroh CP, Lee CA. The HilA box and sequences outside it determines the magnitude of HilA-dependent activation of PprgH form Salmonella pathogenicity island 1. J. Bacteriol. 2001;183:4876–4885. doi: 10.1128/JB.183.16.4876-4885.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darwin KH, Miller VL. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 2001;20:1850–1862. doi: 10.1093/emboj/20.8.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schechter LM, Damrauer SM, Lee CA. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 1999;32:629–642. doi: 10.1046/j.1365-2958.1999.01381.x. [DOI] [PubMed] [Google Scholar]
- 13.Schechter LM, Lee CA. AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 2001;40:1289–1299. doi: 10.1046/j.1365-2958.2001.02462.x. [DOI] [PubMed] [Google Scholar]
- 14.Boddicker JD, Knosp BM, Jones BD. Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators. J. Bacteriol. 2003;185:525–533. doi: 10.1128/JB.185.2.525-533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rakeman JL, Bonifield HR, Miller SI. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 1999;181:3096–3104. doi: 10.1128/jb.181.10.3096-3104.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akabar S, Schechter LM, Lostroh CP, Lee CA. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol. Microbiol. 2003;47:715–728. doi: 10.1046/j.1365-2958.2003.03322.x. [DOI] [PubMed] [Google Scholar]
- 17.Ellermeier CD, Slauch JM. RtsA coordinately regulates DsbA and the Salmonella pathogenicity island I type III secretion system. J. Bacteriol. 2004;186:68–79. doi: 10.1128/JB.186.1.68-79.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baxter MA, Jones BD. The fimYZ genes regulate Salmonella enterica serovar Typhimurium invasion in addition to type I fimbrial expression and bacterial motility. Infect. Immun. 2005;73:1377–1385. doi: 10.1128/IAI.73.3.1377-1385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fahlen TF, Mathur N, Jones BD. Identification and characterization of mutants with increased expression of hilA, the invasion gene transcriptional activator of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 2000;28:25–35. doi: 10.1111/j.1574-695X.2000.tb01453.x. [DOI] [PubMed] [Google Scholar]
- 20.Baxter MA, Fahlen TF, Wilson RL, Jones BD. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 2003;71:1295–1305. doi: 10.1128/IAI.71.3.1295-1305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song M, Kim HJ, Kim EY, Shin M, Lee HC, Hong Y, Rhee JH, Yoon H, Ryu S, et al. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J. Biol. Chem. 2004;279:34183–34190. doi: 10.1074/jbc.M313491200. [DOI] [PubMed] [Google Scholar]
- 22.Jones BD. Salmonella invasion gene regulation: a story of environmental awareness. J. Microbiol. 2005;43:110–117. [PubMed] [Google Scholar]
- 23.Saier MH, Reizer J. The bacterial phosphotransferase system: new frontiers 30 years later. Mol. Microbiol. 1994;13:755–764. doi: 10.1111/j.1365-2958.1994.tb00468.x. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy MJ, Yancey RJ, Jr, Sanchez MS, Rzepkowski RA, Kelly SM, Curtiss R., III Attenuation and immunogenicity of Δcya Δcrp derivatives of Salmonella choleraesuis in pigs. Infect. Immun. 1999;67:4628–4636. doi: 10.1128/iai.67.9.4628-4636.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S-J, Boos W, Bouché J-P, Plumbridge J. Signal transduction between a membrane-bound transporter, PtsG, and a soluble transcription factor, Mlc, of Escherichia coli. EMBO J. 2000;19:5353–5361. doi: 10.1093/emboj/19.20.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka Y, Kimata K, Aiba H. A novel regulatory role of glucose transporter of Escherichia coli: membrane sequestration of a global repressor Mlc. EMBO J. 2000;19:5344–5352. doi: 10.1093/emboj/19.20.5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nam T-W, Cho S-H, Shin D, Kim J-H, Jeong J-Y, Lee J-H, Roe J-H, Peterkofsky A, Kang S-O, et al. The Escherichia coli glucose transporter enzyme IICBGlc recruits the global repressor Mlc. EMBO J. 2001;20:491–498. doi: 10.1093/emboj/20.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melvin GS, Bradford WG, Jefrey JE, Wade AN, Bradley DJ, Michael AA. Mutation of the htrB gene in a virulent Salmonella typhimurium strain by intergeneric transduction: strain construction and phenotypic characterization. J. Bacteriol. 1997;179:5521–5533. doi: 10.1128/jb.179.17.5521-5533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim S, Yong K, Ryu S. Analysis of Salmonella pathogenicity island 1 expression in response to the changes of osmolarity. J. Microbiol. Biotechnol. 2005;15:175–182. [Google Scholar]
- 30.Cirillo DM, Valdivia RH, Monack DM, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 31.Lim S, Kim B, Choi HS, Lee Y, Ryu S. Fis is required for proper regulation of ssaG expression in Salmonella enterica serovar Typhimurium. Microb. Pathog. 2006;41:33–42. doi: 10.1016/j.micpath.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Miller J. Experiments in Molecular Genetics. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 33.Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasion SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos RL, Tsolis RM, Baumler AJ, Smith R, III, Adams LG. Salmonella enterica serovar Typhimurium induces cell death in bovine monocyte-derived macrophages by early sipB-dependent and delayed sipB-independent mechanisms. Infect. Immun. 2001;69:2293–2301. doi: 10.1128/IAI.69.4.2293-2301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lostroh CP, Bajaj V, Lee CA. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol. Microbiol. 2000;37:300–315. doi: 10.1046/j.1365-2958.2000.01991.x. [DOI] [PubMed] [Google Scholar]
- 36.Oleknovich IN, Kadner RJ. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2002;184:4148–4160. doi: 10.1128/JB.184.15.4148-4160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell JE, Zheng D, Busby SJW, Minchin SD. Identification and analysis of ‘extended -10’ promoters in Escherichia coli. Nucleic Acids Res. 2003;31:4689–4695. doi: 10.1093/nar/gkg694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plumbridge J. DNA binding sites for the Mlc and NagC proteins: regulation of nagE encoding the N-acetylglucosamine-specific transporter in Escherichia coli. Nucleic Acids Res. 2001;29:506–514. doi: 10.1093/nar/29.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plumbridge J. Regulation of gene expression in the PTS in Escherichia coli: the role and interactions of Mlc. Curr. Opin. Microbiol. 2002;5:187–193. doi: 10.1016/s1369-5274(02)00296-5. [DOI] [PubMed] [Google Scholar]
- 40.Lundberg U, Vinatzer U, Berdnik D, von Gabain A, Baccarini M. Growth phase-regulated induction of Salmonella-induced macrophage apoptosis correlates with transient expression of SPI-1 genes. J. Bacteriol. 1999;181:3433–3437. doi: 10.1128/jb.181.11.3433-3437.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimata K, Inada T, Tagami A, Aiba H. A global repressor (Mlc) is involved in glucose induction of the ptsG gene encoding major glucose transporter in Escherichia coli. Mol. Microbiol. 1998;29:1509–1519. doi: 10.1046/j.1365-2958.1998.01035.x. [DOI] [PubMed] [Google Scholar]
- 42.Bechhofer D. 5′ mRNA stabilizers. In: Belasco J, Brawerman G, editors. Control of Messenger RNA Stability. San Diego, CA, USA: Academic Press, Inc.; 1993. pp. 31–52. [Google Scholar]
- 43.Tanaka Y, Itoh F, Kimata K, Aiba H. Membrane localization itself but not binding to IICBGlc is directly responsible for the inactivation of the global repressor Mlc in Escherichia coli. Mol. Microbiol. 2004;53:941–951. doi: 10.1111/j.1365-2958.2004.04179.x. [DOI] [PubMed] [Google Scholar]
- 44.Schiemann DA. Association with MDCK epithelial cells by Salmonella typhimurium is reduced during utilization of carbohydrates. Infect. Immun. 1995;63:1462–1467. doi: 10.1128/iai.63.4.1462-1467.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Durant JA, Corrier DE, Stanker LH, Ricke SC. Expression of the hilA Salmonella typhimurium gene in a poultry Salm. enteritidis isolate in response to lactate and nutrients. J. Appl. Microbiol. 2000;89:63–69. doi: 10.1046/j.1365-2672.2000.01089.x. [DOI] [PubMed] [Google Scholar]
- 46.Becker D, Selbach M, Rollenhagen C, Ballmaier M, Meyer TF, Mann M, Bumann D. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature. 2006;440:303–307. doi: 10.1038/nature04616. [DOI] [PubMed] [Google Scholar]
- 47.Waner BL. Signal transduction and cross regulation in the Escherichia coli phosphate regulon by RhoR, CreC, and acetyl phosphate. In: Hoch JA, Silhavy TJ, editors. Two-Component Signal Transduction. American Society for Microbiology Press, Washington, DC, USA; 1995. pp. 203–221. [Google Scholar]
- 48.Teplitski M, Goodier RI, Ahmer BM. Catabolite repression of the SirA regulatory cascade in Salmonella enterica. Int. J. Med. Microbiol. 2006;296:449–466. doi: 10.1016/j.ijmm.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 49.Altier C, Suyemoto M, Ruiz AI, Burnham KD, Maurer R. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 2000;35:635–646. doi: 10.1046/j.1365-2958.2000.01734.x. [DOI] [PubMed] [Google Scholar]
- 50.Prouty AM, Gunn JS. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect. Immun. 2000;68:6763–6769. doi: 10.1128/iai.68.12.6763-6769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawhon SD, Maurer R, Suyemoto M, Altier C. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 2002;46:1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- 52.Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A, Hinton JC, Van Immerseel F. Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl. Environ. Microbiol. 2006;72:946–949. doi: 10.1128/AEM.72.1.946-949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carter PB, Collins FM. The route of enteric infection in normal mice. J. Exp. Med. 1974;139:1189–1203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferraris RP. Dietary and developmental regulation of intestinal sugar transport. Biochem. J. 2001;360:265–276. doi: 10.1042/0264-6021:3600265. [DOI] [PMC free article] [PubMed] [Google Scholar]