Abstract
High throughput EST and full-length cDNA sequencing have revealed extensive variations at the 3′ ends of mammalian transcripts. Whether all of these changes are biologically meaningful has been the subject of controversy, as such, results may reflect in part transcription or polyadenylation leakage. We selected here a set of tandem poly(A) sites predicted from EST/cDNA sequence analysis that (i) are conserved between human and mouse, (ii) produce alternative 3′ isoforms with unusual size features and (iii) are not documented in current genome databases, and we submitted these sites to experimental validation in mouse tissues. Out of 86 tested poly(A) sites from 44 genes, 84 were individually confirmed using a specially devised RT-PCR strategy. We then focused on validating the exon structure between distant tandem poly(A) sites separated by over 3 kb, and between stop codons and alternative poly(A) sites located at 4.5 kb or more, using a long-distance RT-PCR strategy. In most cases, long transcripts spanning the whole poly(A)–poly(A) or stop-poly(A) distance were detected, confirming that tandem sites were part of the same transcription unit. Given the apparent conservation of these long alternative 3′ ends, different regulatory functions can be foreseen, depending on the location where transcription starts.
INTRODUCTION
During mRNA maturation, mRNA precursors are processed at their 3′ ends in a two-step reaction (1). The primary transcript is endonucleolytically cleaved downstream of a stop codon, at a polyadenylation—poly(A)—site, and adenylate residues are added to the 3′ end to form a poly(A) tail. The 3′ untranslated region (3′ UTR), that extends from the stop codon to the poly(A) site provides binding sites for regulatory binding proteins and plays a key role in mRNA localization, mRNA stability and translation efficiency (2).
It is now established that most transcription units in mammalian genes have multiple poly(A) sites (3,4). Based on available SAGE, EST and cDNA data, the average number of poly(A) sites per gene is 2.1–2.2 for human genes and 1.5–3.3 for mouse genes (4–6). Two main types of alternative polyadenylation profiles have been described based on poly(A) site location on a transcript. Alternative poly(A) sites may be located in the same 3′ exon/3′ UTR and referred to as tandem poly(A) sites, or they may be located in alternative 3′ exons (7,8). The presence of multiple mRNA isoforms that differ only at their 3′ ends in a non-coding region is often related to different stabilities and translation rates. In these cases, the use of tandem poly(A) sites can positively or negatively impact the amount of protein produced per unit of precursor mRNA. The 3′ most poly(A) site usually is the strongest site among all sites, which correlates with the longest mRNA isoform being usually the predominant one (4,9). It has been hypothesized that alternative polyadenylation acts through shortening of mRNA to regulate RNA localization, translation and stability (4).
The structures of 3′ UTRs have been analyzed in different species using different approaches. In the human genome, the average length of the 3′ UTR is 1027 nt for a maximum length of 8555 nt (10). Although several very long-range polyadenylation sites have been described in humans (10,11), such sites are probably under-reported since technical problems are encountered to amplify and clone the full-length sequence of long mRNAs. In a recent study of EST/cDNA-supported poly(A) sites in the human genome, we observed a significant incidence of poly(A) sites up to 10–15 kb past the stop codon and proposed that as many as 5000 human genes may have unreported 3′ extensions (12). Although we introduced quality criteria to evaluate individual poly(A) sites predicted from cDNA data, there has been some reluctance to accept as biologically meaningful results from high throughput screens, since they include cryptic or antisense transcripts as well as significant levels of erroneous sequences (13,14). In order to identify possibly functional instances of alternative polyadenylation, we also studied phylogenetically conserved poly(A) sites, defined as poly(A) sites that are located at the same position in an alignment of orthologous human and mouse 3′ UTRs and supported by cDNAs or ESTs in both species. We identified ∼500 genes with tandem conserved poly(A) sites (15).
In this work, we select a test set of conserved, tandem poly(A) sites producing alternative 3′ isoforms with unusual length features and submit them to individual RT-PCR validation in mouse. Most of the cDNA/EST-based predictions turn out to correspond to specific and reproducible transcripts, providing evidence for a high incidence of alternative transcripts with very long 3′ UTRs in the mouse transcriptome, for which corresponding poly(A) sites are also conserved in humans.
MATERIALS AND METHODS
mRNA preparation and controls
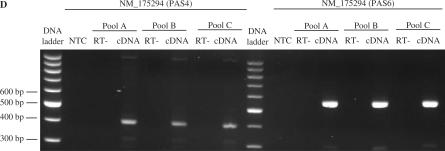
Embryonic fibroblast NIH 3T3 cells and embryonic fibroblasts from Balb/c wild-type mouse (MEF) wt were kindly provided by Joel Tardivel (INSERM U624, Marseille) and cultivated in DMEM (Invitrogen) and 10% (v/v) Fetal Calf Serum (Invitrogen). RNAs were prepared from 70% confluent cells using TRIzol reagent (Invitrogen). mRNAs from thymic epithelial 427.1, thymic stromal 1308.1, thymic medullary epithelial MTE, lymphoma WEHI, mastocytoma P815 and fibroblast LMTK cells were kindly provided by Dr Denis Puthier (INSERM ERM206, Marseille). mRNAs were also obtained from murine tissue (Balb/c mouse) using the TRIzol reagent. Prior to RT-PCR, mRNAs were treated with a RNAse free DNAse (Promega), according to the manufacturer's recommendations. Quality was assessed by analytical gel and PCR amplification with primers for beta actin 5′-GACTCCGGTGACGGGGTCACC-3′ and 5′-CACGATGGAGGGGCCGGACTC-3′. We confirmed the absence of amplification from primers located downstream of poly(A) sites for six candidates having no other described poly(A) site downstream of the tested one. Primers used were 5′-AATCCTTGGGCAACTTGATG-3′ and 5′-TGGCCAGTTTTCCTTTTGAG-3′ for Gmfb (439 bp), 5′-CCCCATCCTCAGCAGATAAA-3′ and 5′-ATGGATGGATGGGTACATGG-3′ for Cdyl2 (375 bp), 5′-TTGTTGCTGTTTGAGGCTTG-3′ and 5′-AGGTGTTGCAGACACAGCAG-3′ for Hdh (494 bp), 5′-TTTGCACCCTTCACACTGTC-3′ and 5′-GCAGTTTTTAGCTGCCCAAC-3′ for Cpeb2 (360 bp), 5′-GCCTTGGAATGGTTCACTGT-3′ and 5′-CCCCTTTTCCCTCATCAAAT-3′ for Klf7 (366 bp), 5′-TGTTGTGTGCCTCTCTCAGG-3′ and 5′-ACCTGTGTGTGCACCTGTGT-3′ for Atp9a (434 bp). Results are shown in Figure 1C for two candidates, Cpeb2 and Atp9a.
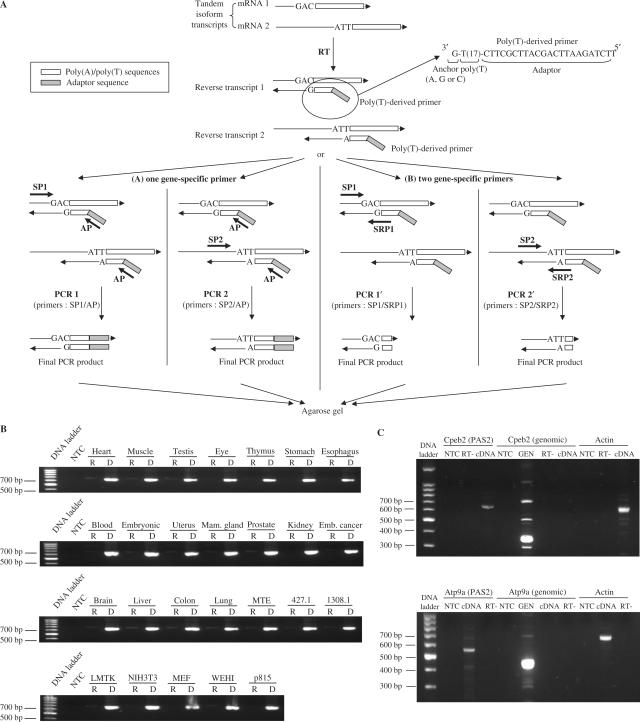
Figure 1.
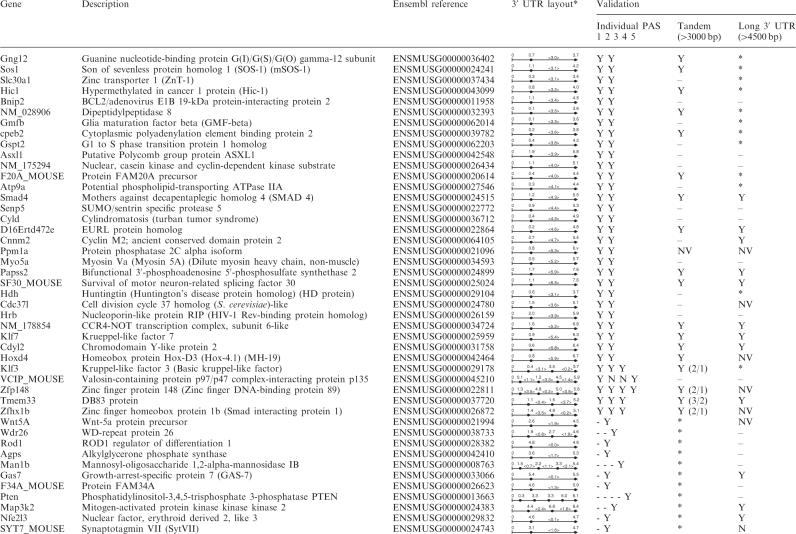
Individual poly(A) site validation. (A) Outline of experiment. mRNAs are reverse-transcribed using a pool of poly(T)-derived primers containing a sequence of 17 T, with an adaptor sequence in its 5′ end, and a one-nucleotide anchor in its 3′ end (A, G or C). (A) The PCR amplification is done with a forward gene-specific primer for each tested poly(A) site (SP1 or SP2) and the adaptor primer (AP) as a reverse primer and the PCR product are resolved on agarose gel. (B) When no satisfying result is obtained, a reverse gene-specific primer is also designed (SRP1 or SRP2) that overlaps part of the poly(A) tail and the sequence just upstream the poly(A) site. B: Controls. Quality of samples was assessed by PCR amplification with primers for beta actin. mRNAs were treated with a RNAse-free DNAse and cDNAs were prepared with reverse transcriptase (D for cDNA) or without (R for RT-). NTC stands for ‘no template control.’ Expected size: 651 bp. (C) Absence of amplification using primers downstream of poly(A) site. A PCR amplification was performed using the primers designed to validate the last poly(A) site, (Cpeb2, PAS2 [649 bp expected] and Atp9a, PAS2 [516 bp expected]) or primers downstream of this last poly(A) site, i.e. located in intergenic DNA (Cpeb2, genomic [360 bp expected] and Atp9a, genomic [434 bp expected]). Templates were cDNAs prepared with reverse transcriptase (cDNA) or without (RT-), or genomic DNA (GEN). NTC stands for ‘no template control.’ (D) Example. RT-PCR is performed as described from the three pools of cDNAs (A, B or C) for the two NM_175294 tested isoforms corresponding to use of poly(A) sites 4 (399 bp expected) and 6 (573 bp expected). Templates were cDNAs prepared with reverse transcriptase (cDNA) or without (RT-). NTC stands for ‘no template control’.
RT-PCR
cDNAs were prepared from 1 μg total RNAs with an equimolar pool of three modified poly(T) primers: 5′-TTCTAGAATTCAGCATTCGCTTCTTTTTTTTTTTTTTTTTA-3′, 5′-TTCTAGAATTCAGCATTCGCTTCTTTTTTTTTTTTTTTTTG-3′, 5′-TTCTAGAATTCAGCATTCGCTTCTTTTTTTTTTTTTTTTTC-3′ using the superscript II reverse transcriptase (Invitrogen). The reverse transcription lasted 50 min for individual poly(A) site validation and 2 h for other validations. For individual poly(A) site validation, PCR was performed with the Phusion high fidelity PCR mix (Finnzymes) with an annealing temperature of 60°C and a 30 s extension at 72°C for all experiments. Most of the PCRs were done with the reverse adaptor primer (AP): 5′-TTCTAGAATTCAGCATTCGCTTC-3′ or a gene-specific reverse primer and a gene-specific forward primer (list in Supplementary Table 3). To validate transcripts with long distance between two poly(A) sites or long 3′ UTR, the Platinium Taq DNA polymerase (Invitrogen) was used and an annealing temperature between 55 and 62°C was chosen depending on the Tm of the reverse primers. Extension was performed at 68°C, for 1 min/kb. PCR products were resolved on 1–2% agarose gel, depending on the expected size.
PCR product purification and digestion
PCR products were purified with the Qiaquick PCR purification kit (Qiagen) and submitted to digestions with restriction enzymes at 37°C for 2 h. Digestion products were resolved on a 1% agarose gel.
Real-time quantitative RT-PCR
Real-time RT-PCR amplification was performed using primer sets for GAPDH (5′-GGGTGTGAACCAGGAGAAAT-3′ and 5′-TTCCACAATGCCAAAGTTGT-3′, 118 bp), and Atp9A genes (5′-CTACATTGCCTCCCTGGTGT-3′ and 5′-ACAGCTGACCAAGGTGATGA-3′ [103 bp] to amplify both isoforms, 5′-CATGCAAACAGACCCATCTC-3′ and 5′-GGATGCAAGTGCTGAAAAGA-3′ [105 bp] for the largest isoform). Synthesis of the first-strand cDNA was carried out as previously described with 5 μg RNAs. The RT product was then amplified for 40 cycles using the Power SYBR Green PCR Master Mix according to the manufacturer's recommendations (Applied Biosystems), on ABI 7000 sequence detector (2 min at 50°C, 10 min at 95°C, 15 s at 95°C, 30 s at 60°C, 30 s at 72°C). Serial dilutions of cDNA (5 logs) generated from the Wehi and p815 cell lines were used to generate a standard curve for each tested isoform and GAPDH, to confirm that they were all amplified with a comparable and high efficiency. Then, relative expression of both Atp9a isoforms was evaluated with the comparative CT method. Relative expression values for each isoform were expressed as a ratio of isoform expression level to GAPDH expression level in the same tissue. Each assay was repeated three times (triplicate), a negative control without template or with RT- RNAs was always conducted with every amplification. The PCR products were resolved on 1% agarose gels.
RESULTS
Alternative poly(A) site database construction
Initial poly(A) site prediction was performed through mapping of ESTs and full-length cDNAs onto the human and mouse genomes as described in Ara et al. (2006) (15). We collected as putative tandem sites all sets of two or more predicted poly(A) sites occurring in the 10 kb region downstream of the 3′-most stop codon of an annotated gene. This comprised sites lying within annotated 3′ UTRs and also a significant number of sites lying in apparent intergenic regions. In order to enrich this collection in biologically significant tandem sites, we aligned the 10 kb 3′ regions of orthologous human and mouse genes and retained only those sites that were exactly superimposed in the pairwise alignment and had EST/cDNA support in both human and mouse. This short list of 1096 phylogenetically conserved tandem poly(A) sites from about 500 genes is available in Supplementary Table 1 of Ara et al. (2006) (15). These data were further enriched through incorporation of SAGE tags uniquely supporting transcript isoforms and expression information extracted from cDNA, EST and SAGE library data. Tissue-specific expression of 3′ isoforms was assessed based on EST and SAGE library counts followed by statistical tests of the overrepresentation of libraries. Results are available as part of the AltPAS database (http://tagc.univ-mrs.fr/pub). A special user interface was devised to facilitate examination of the ∼1000 poly(A) sites and assist the following selection procedure.
Candidate selection for experimental validation
We focused on the most intriguing candidate poly(A) sites which were the ones located at long distances from each other or from the stop codon, and which were not yet annotated in the Ensembl human and mouse genome databases (16). We performed two distinct candidate selections. First, tandem alternative poly(A) sites were sorted according to the length separating two consecutive sites, and sites separated by a distance of 3000 nt or more were chosen for experimental validation. This group was composed of 34 candidates representing 75 poly(A) sites, some candidates having more than two conserved tandem poly(A) sites. Second, the 3′ most poly(A) site was sorted for each gene of our gene list, and poly(A) sites located over 4500 nt from the 3′-most stop codon were selected. This group was composed of 34 candidates as well. Twenty-two candidates were common to the two groups that totaled 86 distinct poly(A) sites and transcripts. This final list contained all conserved tandem poly(A) sites from the computational pipeline (15) satisfying the topological criteria, only excluding genes annotated as unnamed or incomplete.
The gene list and topology of tandem poly(A) sites are presented in Table 1. Precise cleavage site location and numbers of EST/cDNA supporting poly(A) sites in the mouse genes and their human counterparts are shown in Supplementary Table 1. A more detailed description of the mouse sites is given in Supplementary Table 2, including location and sequence of the poly(A) signal, presence of SAGE or full-length cDNA supporting the transcript, evaluation of the cleavage site homogeneity and presence of AU-Rich Elements (ARE) and Upstream Sequence Elements (USE) in surrounding sequences. Among the 86 selected poly(A) sites, 53 were associated with a canonical AAUAAA poly(A) signal (61.1%), in agreement with overall poly(A) signal frequencies (17). Comparison of the signal sequence at the 3′-most and the 5′-most poly(A) signals of candidates having only two tandem poly(A) sites did not show a significant difference in the representation of the AAUAAA signal.
Table 1.
Gene list and topology of tandem poly(A) sites
Selection of the cleavage site is determined mainly by the distance between the upstream polyadenylation signal and the downstream elements composed of U/GU-rich elements (18), but seems to be quite imprecise (3,4,19,20). We define cleavage site homogeneity as the ratio between the number of EST-cDNA ending exactly at a given cleavage site and the total number of EST-cDNA supporting the poly(A) site. In our selection, only 17 transcripts had a ratio over 0.7, which reflects a high rate of cleavage site heterogeneity. Whether this was due to biological reality or technical artifacts such as misalignments of low quality EST sequences remains to be established.
We sought known regulatory sequences in the 3′ UTR (ARE) and in the vicinity of the poly(A) signal (USE) (Supplementary Table 2). AREs are found in the 3′ UTR of many mRNAs and represent a common determinant for mRNA instability. In the case of tandem poly(A) sites, the use of the proximal poly(A) signal could result in exclusion of a part of the 3′ UTR harboring AREs (21). Three classes of AREs have been described, two AUUUA-containing classes and one U-rich region-containing class (18). We scanned the region preceding the poly(A) site of each transcript to search for the AUUUA motif. For 5′-proximal sites, the scanned region was composed of the sequence between the stop codon and the poly(A) site. For distal sites, the scanned region ranged from the previous conserved poly(A) site to the distal site. ARE densities were not significantly different between proximal and distal sites (data not shown).
USEs are U-rich elements upstream of the poly(A) signal that can positively enhance polyadenylation efficiency in a spacing-dependent way (22). We scanned the 100-nt-sequence upstream of each poly(A) signal for the consensus USE sequence AU(2–5)GURA (22) or putative-derived USE sequences AU(2–5)G/CU/ARA/U/C (21,23). Most transcripts (67.4%) had at least one USE sequence but no significant difference could be established between USE frequencies near proximal and distal poly(A) signals (data not shown).
Validation of individual poly(A) sites
We experimentally validated the 3′ extremities of all individual candidates using RT-PCR. First, total mRNAs were extracted from different cell lines, treated with a RNAse-free DNAse to remove genomic DNA contamination, and reverse-transcribed using a pool of poly(T)-derived primers containing a sequence of 17 T, a 5′ adaptor sequence and a one-nucleotide anchor at the 3′ end (mixed A, G or C). These primers target the beginning of the poly(A) tail of transcripts, since the anchor must hybridize with the nucleotide just upstream of the first A of the poly(A) tail to allow extension (Figure 1A). The benefits of this method are (1) to circumvent the problem of cleavage site heterogeneity, and (2) to append the same adaptor sequence to all reverse–transcribed cDNAs (Figure 1A). Sample quality was assessed by PCR amplification with primers for beta actin (Figure 1B).
To ensure that long 3′ isoform products were not the result of genomic contamination, we also designed probes targeting genomic DNA downstream of distal poly(A) sites. This was done for six genes where no further poly(A) site was predicted downstream of the long-distance site tested. As expected, DNA amplification was obtained with the genomic primers using genomic DNA, but in no case could we amplify any product using the genomic primers and cDNA. Detailed controls are shown in Figure 1C for two genes, Cpeb2 and Atp9a.
In order to speed up validation, cDNAs were pooled in three samples: cDNAs from thymic origin, named group A (cDNAs from 427.1, 1308.1 and MTE cells), cDNAs from embryonic and/or fibroblast origin, named group B (LMTK, NIH 3T3 and MEF cells) and cDNAs from oncogenic origin, named group C (WEHI and p815 cells). PCR amplification was then performed using a forward gene-specific primer for each isoform and the adaptor sequence as a common reverse primer (Figure 1A) and PCR products were resolved on agarose gels. When successful, this strategy typically produced a single band for each targeted 3′ variant. Examples of such PCR amplifications are shown in Figure 1D. In some cases, the expected polyadenylated transcripts were fished out together with other transcripts corresponding to distinct neighboring poly(A) sites, or partial cDNA sequences caused by internal priming at adenine stretches, thus producing multiple bands on the gel. When such unsatisfactory results were obtained (no band or multiple bands), a specific reverse primer was designed that overlapped part of the poly(A) tail and the sequence just upstream the poly(A) site (Figure 1A) and was used to perform the PCR amplification together with the forward-specific primer.
Complete results are presented in Table 1 (column ‘individual PAS’) and with further detail in Supplementary Table 3. When a single band was obtained at the expected size, the poly(A) site was considered as validated for the tested cDNA pool (note ‘Y’ in Supplementary Table 3). When two bands were observed, one at the expected size and one that could be explained (because of its size) by the presence of a neighboring poly(A) site or internal priming site, the site was also considered as validated. When two or three bands were observed, one at the expected size and at least one that could not be explained, the poly(A) site was considered as validated with reservation (noted ‘y’ in Supplementary Table 3). Finally, when no band or no specific band was obtained, the experiment was repeated to confirm that the negative amplification was not due to a technical problem and the site was not validated in this cDNA pool (noted ‘N’ in Supplementary Table 3). Of the 86 transcripts, only two were not validated in any of the three pools of tested cDNAs: the two transcript isoforms of the VCIP_MOUSE gene (poly(A) sites 2 and 8). As these poly(A) sites were supported by less than 3 ESTs, it is possible that these transcripts were either spurious or expressed at a very low level and not detectable with our method in our pools of cell lines. Seventy-five transcript ends (87.2%) were validated in all three cDNA pools.
Poly(A) sites with predicted tissue expression biases were further tested using total mRNAs from 23 different murine tissues. Results are shown in Supplementary Table 3. For all 31 transcripts with a predicted tissue bias, RT-PCR confirmed expression in the corresponding tissues. Although many transcripts showed expression in brain, this bias is not significant when considering that brain tissues have higher EST coverage and most of the predicted expression biases were in brain.
Validation of tandem poly(A) sites
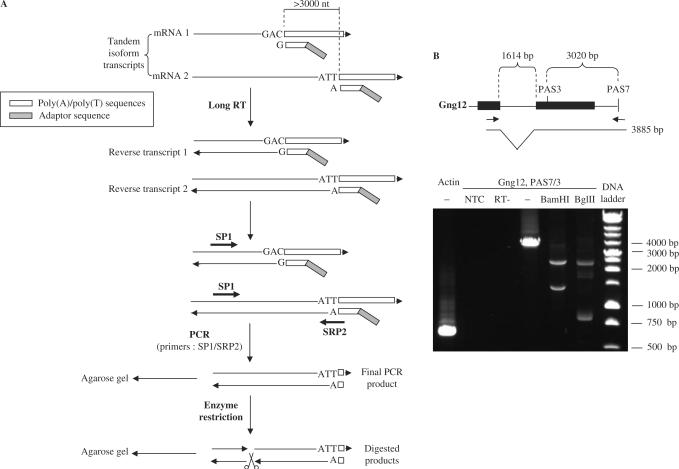
The previous experiment did not distinguish tandem poly(A) sites, i.e. sites located in the same 3′ exon, from poly(A) sites resulting from alternative 3′ exons. To validate the tandem topology, one must check that no intron lies between the two confirmed poly(A) sites. This validation was performed on 19 randomly selected candidates from the first validation group, using cDNAs from NIH 3T3 and MEF cells. Reverse transcription was performed with the same modified poly(T) primers described previously. Then, PCR amplification was performed using the specific forward primer used to check the validity of the proximal poly(A) site and a specific reverse primer for the distal poly(A) site that overlapped part of the poly(A) tail (Figure 2A). This strategy should reveal intervening introns as these would lead to shorter PCR products than expected. Further, bands obtained in agarose gels were purified and digested with restriction enzymes, and digestion profiles were compared to those expected from transcript sequences. Two positive digestion profiles from two different enzymes were required to confirm each transcript. Results obtained for the distal tandem poly(A) sites of Gng12, a two-exon gene, are shown in Figure 2B. The distance between the two poly(A) sites is 3020 nt. The forward primer targeted the first exon while the reverse primer targeted the transcript termination along with part of the poly(A) tail. The PCR product was digested with BamHI or BglII. Selected primers and results are listed in Supplementary Table 4, and results are summarized in Table 1 (column ‘tandem’). Eighteen out of 19 tandems were fully confirmed as intron-less while one (Ppm1a) could not be confirmed by restriction enzyme digestion, although band position was compatible with a continuous transcript spanning the two consecutive poly(A) sites. Only two of the 19 confirmed tandems (Gng12 and cpeb2) had previous support from a FANTOM3 full-length cDNAs spanning the two consecutive poly(A) sites.
Figure 2.
Tandem poly(A) site validation. (A) Outline of experiment. RNAs are reverse-transcribed into cDNAs using a pool of poly(T)-derived primers as described in Methods. The PCR amplification is done with the forward gene-specific primer used to validate the proximal poly(A) (SP1) and a reverse gene-specific primer that targets the distal poly(A) site and overlaps part of the distal poly(A) tail (SRP2). Restrictions with restriction enzymes are performed to assess the size. (B) Example. RT-PCR is performed as described from NIH3T3 and MEF mRNAs to test the poly(A) site 7 of Gng12 from cDNAs prepared with reverse transcriptase or without (RT-) or without template (NTC). The forward primer, that was used to validate the poly(A) site 3 of the gene, targets the first exon while the reverse primer targeted end of the transcript with part of the poly(A) tail (scheme). The PCR product is digested with BamHI (expected products: 1438 bp, 2447 bp) or BglII (expected products: 2250 bp, 798 bp, 837 bp) or left undigested (−).
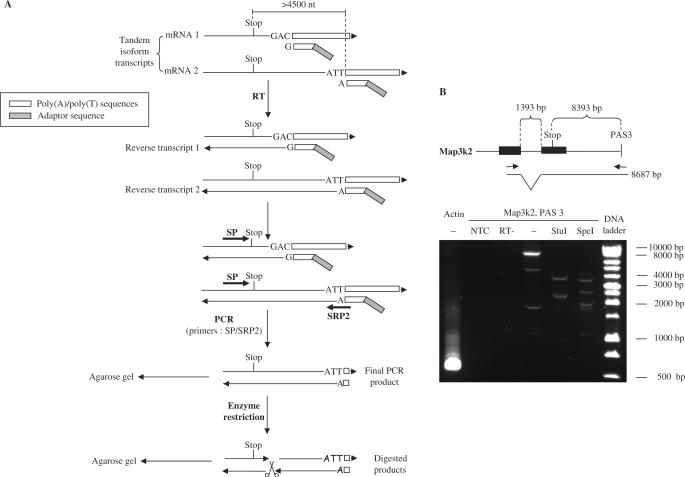
Validation of very long 3′ UTR extensions
The 3′ UTR of a mature mRNA extends from the stop codon to the poly(A) site. For validation of exceptionally long 3′ UTRs (>4.5 kb), the forward primer was selected so as to target a sequence upstream of the stop codon, in most cases in the exon preceding the last exon (Figure 3A). This validation was performed on 19 randomly selected distal isoforms from the second group, using cDNAs from NIH 3T3 and MEF cells. Two expected digestion profiles were required to validate each transcript. As an example, validation of the most distal human/mouse-conserved poly(A) site of Map3k2 is shown in Figure 3B. The distance between the stop codon and poly(A) site was 8393 nt. The forward primer targeted exon 18 while the reverse primer targeted a sequence upstream the poly(A) site and part of the poly(A) tail in exon 19 (Figure 3B). The PCR product was digested with StuI or SpeI. Results of this validation for 19 distal isoforms are shown together with primers in Supplementary Table 5 and summarized in Table 1 (column ‘long 3′ UTR’). Twelve distal poly(A) sites were fully validated by RT-PCR and restriction digestion (noted ‘Y’), while six more yield PCR products of expected sizes but could not be resolved by restriction digestion (noted ‘NV’). Only one out of 19 long UTRs tested (Syt7) was not confirmed at all.
Figure 3.
Validation of transcripts with very long 3′ UTR. (A) Outline of experiment. RNAs are reverse-transcribed into cDNAs using a pool of poly(T)-derived primers. The PCR amplification is done with a forward gene-specific primer that targets a sequence upstream the stop codon (SP) and a reverse gene-specific primer that targets the distal poly(A) site and overlaps part of the distal poly(A) tail (SRP2). Digestions with restriction enzymes are performed to assess fragment sizes. (B) Example. RT-PCR is performed as described from NIH3T3 and MEF mRNAs to test the poly(A) site 3 of Map3k2 from cDNAs prepared with reverse transcriptase or without (RT-) or without template (NTC). The forward primer targets the exon 18 while the reverse primer targeted end of the transcript on exon 19 with part of the poly(A) tail (scheme). The PCR product is digested with StuI (expected products : 2393 bp, 2280 bp, 3859 bp, 155 bp) or SpeI (2664 bp, 3698 bp, 347 bp, 1978 bp) or left undigested (–).
DISCUSSION
In mammalian cells, the cleavage/polyadenylation specificity factor (CPSF) recognizes and binds the poly(A) signal while the cleavage stimulation factor (CstF) binds the U/GU-rich downstream element. Cooperative interactions and assembly of CPSF and CstF with other proteins like poly(A) polymerase (PAP) are important for cleavage of the pre-mRNA and addition of ∼250 adenylate residues at the 3′ end (1). Choice of a poly(A) signal in a competitive environment as it is the case for tandem poly(A) sites seems to be a multifactorial process that involves steric constraints (23), presence of enhancer sequences like USE (21) and availability of cleavage/polyadenylation factors and their modulators (7).
The majority of documented long-range, alternative polyadenylation sites result from the analysis of high throughput data such as EST sequences (4,11,12,15). A recent EST and cDNA mapping on the human and mouse genome has predicted on the order of 5000 poly(A) sites lying in the 5–10 kb region following annotated transcription ends (12). However, as experimental confirmations of such long-range poly(A) sites are limited and their biological significance is still uncertain, gene-by-gene validation studies such as the present one are still needed. To perform individual RT-PCR validation we needed a reasonably sized subset of long-range sites, hence further selection criteria were required. We opted to focus only on those ∼1000 tandem poly(A) sites that were conserved between orthologous 3′ regions in human and mouse. Combining this with distance requirements (>3 kb between sites, or >4.5 kb from stop codon) we ended up with a short list of 86 individual poly(A) sites from 44 different genes. Our experimental validations showed that (1) all but two tested poly(A) sites were real; (2) at least 18 out of 19 tested tandem sites were actually located in the same exon and did not result from alternative splicing; and (3) between 11 and 18 out of 19 tested long 3′ UTR were actually transcribed as a continuous block and were not the product of independent transcription units.
The qualitative PCR strategy we used to validate individual poly(A) sites does not preclude amplification of low abundance transcripts that may be of lesser functional significance. EST counts do not support a lower expression of longer isoforms: the average numbers of EST/isoform in our gene set are 21.5 and 21.4 for shorter and longer isoforms, respectively. However, certain long isoforms exhibit modest experimental support. For instance the distal site of Atp9a is supported by 3 ESTs versus 143 for the proximal site (Supplementary Table 1), and is only confirmed in the Wehi and p815 cell lines (Supplementary Table 3). We further evaluated the expression of both Atp9a isoforms with real-time quantitative RT-PCR (Supplementary Figure 1). Expression of the longest isoform was very low indeed in the Wehi and p815 cell lines but drastically increased in kidney (72X increase) and testis (53X increase). This indicates that even though a transcript may be detected at low abundance in cell lines, a higher expression in specific tissues or under specific conditions is possible and should not be overlooked, as context-specific expression confers a real biological interest to transcript isoforms.
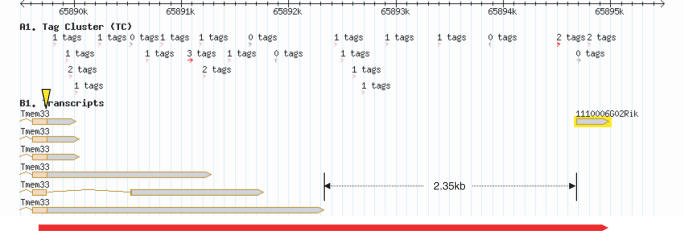
Among 22 validated 3′ UTR extensions (either through amplification of entire UTRs or inter-poly(A) site regions), 12 had support from FANTOM3 full-length cDNAs. Interestingly, in seven cases, the full-length cDNA from which a distal poly(A) site was predicted was completely disjointed from the annotated transcription unit. An example is shown in Figure 4 for gene Tmem33 coding for the DB83 membrane protein (24). Although the FANTOM3 cDNA supporting the distal site is located at about 2.5 kb from the end of the transcription unit, we showed by RT-PCR that another transcript exists (bottom red strip) that bridges this gap while encompassing the complete 5.2 kb UTR. Considering that we observed seven such instances of gap-spanning out of 12 poly(A) sites with full-length cDNA support (Sos1, NM_028906, Hoxd4, Tmem33, Gas7, F34A, Map3k2), we may expect extended 3′ UTRs of these sort to occur quite frequently. It is not clear however, how much of these transcripts span the entire coding region. Only one of the 12 full-length cDNAs corresponding to distal poly(A) sites starts at exon one of the annotated gene. In all other cases, cDNAs have their transcription start sites (TSS) in internal exons or in the last exon. As multiple TSSs arise in the 3′ regions of transcription units (13,14), we expect that a fraction of our long-range poly(A) sites correspond to such ‘late’ TSSs. However, examples of ‘gap spanning’ such as Tmem33 show us that for a given TSS in the 3′ UTR, other TSSs that are closer to the gene 5′ end may also exist that are not present in current full-length cDNA collections. The majority of the long 3′ UTRs we tested had a TSS in the coding region or upstream, as forward PCR primers were always chosen upstream of the stop codon, therefore they contained at least a part of the coding region. Depending on the integrity of this coding region, the functions of these extended transcripts may involve protein expression control as in the COX-2 example above or more indirect mechanisms. For instance, a long 3′ UTR extension could titrate binding proteins away from the shorter isoforms, or be involved in sense–antisense regulation of a flanking gene (25).
Figure 4.
Example of an extended UTR bridging the gap to a distant full-length cDNA. Screenshot from the Riken genomic element viewer (http://fantom3.gsc.riken.jp/) displaying both full-length cDNAs and CAGE tags (transcription starts) for gene Tmem33. The stop codon position is indicated with a yellow triangle. The bottom red strip represents the RT-PCR validated transcript (5′ end is not determined). We located a putative poly(A) site for gene Tmem33 at around 5.2 kb from the stop codon, supported by a single full-length cDNA (highlighted). The 2.35 kb gap between this unique cDNA and the longest Tmem33 transcript caused Riken to predict two independent transcription units (TUs) for these entities.
Some 3′ UTR segments can be more conserved than coding exons (26), reflecting an unexpected selective pressure in this region. As multiple targets for regulatory proteins or RNAs are hosted in 3′ UTRs (27), one can hypothesize that conservation of specific alternative 3′ ends together with specific 3′ UTR elements might reflect novel regulatory mechanisms. This is observed for instance in the alternative polyadenylation of cyclooxygenase 2 (COX-2). The proximal and the distal poly(A) sites of human COX-2 are located at 0.6 and 2.5 kb from the stop codon respectively, a pattern conserved in mouse and rat. Two ARE-rich regions located on both sides of the first poly(A) site and bound by two proteins with opposite effects have been described. The RNA stability factor HuR binds the proximal AREs, enhancing the stability of both isoforms (28) while tristetraprolin binds only the most distal AREs and destabilizes the longer isoform (29). The two poly(A) sites are regulated in a tissue-specific manner and the proximal site contains three USEs that are crucial for its usage (21). This example shows that usage of distal poly(A) sites can impact protein synthesis in response to environmental constraints such as cell cycle progress or growth factor stimulation. All the tandem sites confirmed here are conserved in humans and therefore are candidates for such regulatory mechanisms involving specific target sites in the longer isoform. As these long transcripts are relatively difficult to clone, they may be under-represented in high throughput cDNA collections. Additional efforts are required to assess the full extent of these transcripts and their relevance to the overall transcriptional landscape.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
The authors acknowledge the European Commission for the grant (LSHG-CT-2003-503329) that supported this work and paid the Open Access publication charges.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decker CJ, Parker R. Diversity of cytoplasmic functions for the 3′ untranslated region of eukaryotic transcripts. Curr. Opin. Cell Biol. 1995;7:386–392. doi: 10.1016/0955-0674(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 3.Beaudoing E, Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11:1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Texier V, Riethoven JJ, Kumanduri V, Gopalakrishnan C, Lopez F, Gautheret D, Thanaraj TA. AltTrans: transcript pattern variants annotated for both alternative splicing and alternative polyadenylation. BMC Bioinformatics. 2006;23:169. doi: 10.1186/1471-2105-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siddiqui AS, Khattra J, Delaney AD, Zhao Y, Astell C, Asano J, Babakaiff R, Barber S, Beland J, et al. A mouse atlas of gene expression: large-scale digital gene-expression profiles from precisely defined developing C57BL/6J mouse tissues and cells. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18485–18490. doi: 10.1073/pnas.0509455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan J, Marr TG. Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Res. 2005;15:369–375. doi: 10.1101/gr.3109605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Legendre M, Gautheret D. Sequence determinants in human polyadenylation site selection. BMC Genomics. 2003;4:7. doi: 10.1186/1471-2164-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iseli C, Stevenson BJ, de Souza SJ, Samaia HB, Camargo AA, Buetow KH, Strausberg RL, Simpson AJ, Bucher P, et al. Long-range heterogeneity at the 3′ ends of human mRNAs. Genome Res. 2002;12:1068–1074. doi: 10.1101/gr.62002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez F, Granjeaud S, Ara T, Ghattas B, Gautheret D. The disparate nature of ‘intergenic’ polyadenylation sites. RNA. 2006;10:1794–1801. doi: 10.1261/rna.136206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carninci P. Tagging mammalian transcription complexity. Trends Genet. 2006;9:501–510. doi: 10.1016/j.tig.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 15.Ara T, Lopez F, Ritchie W, Benech P, Gautheret D. Conservation of alternative polyadenylation patterns in mammalian genes. BMC Genomics. 2006;7:189. doi: 10.1186/1471-2164-7-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birney E, Andrews TD, Bevan P, Caccamo M, Chen Y, Clarke L, Coates G, Cuff J, Curwen V, et al. An overview of Ensembl. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 19.Pauws E, van Kampen AH, van de Graaf SA, de Vijlder JJ, Ris-Stalpers C. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res. 2001;29:1690–1694. doi: 10.1093/nar/29.8.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajarnavis A, Korf I, Durbin R. A probabilistic model of 3′ end formation in Caenorhabditis elegans. Nucleic Acids Res. 2004;32:3392–3399. doi: 10.1093/nar/gkh656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall-Pogar T, Zhang H, Tian B, Lutz CS. Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res. 2005;33:2565–2579. doi: 10.1093/nar/gki544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schek N, Cooke C, Alwine JC. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol. Cell. Biol. 1992;12:5386–5393. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Natalizio BJ, Muniz LC, Arhin GK, Wilusz J, Lutz CS. Upstream elements present in the 3′-untranslated region of collagen genes influence the processing efficiency of overlapping polyadenylation signals. J. Biol. Chem. 2002;277:42733–42740. doi: 10.1074/jbc.M208070200. [DOI] [PubMed] [Google Scholar]
- 24.Nakadai T, Kishimoto T, Kokura K, Ohkawa N, Makino Y, Muramatsu M, Tamura T. Cloning of a novel rat gene, DB83, that encodes a putative membrane protein. DNA Res. 1998;5:315–317. doi: 10.1093/dnares/5.5.315. [DOI] [PubMed] [Google Scholar]
- 25.Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fritz DT, Liu D, Xu J, Jiang S, Rogers MB. Conservation of Bmp2 post-transcriptional regulatory mechanisms. J. Biol. Chem. 2004;279:48950–48958. doi: 10.1074/jbc.M409620200. [DOI] [PubMed] [Google Scholar]
- 27.Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawaoka H, Dixon DA, Oates JA, Boutaud O. Tristetraprolin binds to the 3′-untranslated region of cyclooxygenase-2 mRNA. A polyadenylation variant in a cancer cell line lacks the binding site. J. Biol. Chem. 2003;278:13928–13935. doi: 10.1074/jbc.M300016200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.