Abstract
Human immunodeficiency virus type 1 (HIV-1) transcriptional transactivator (Tat) recruits the positive transcription elongation factor b (P-TEFb) to the viral promoter. Consisting of cyclin dependent kinase 9 (Cdk9) and cyclin T1, P-TEFb phosphorylates RNA polymerase II and the negative transcription elongation factor to stimulate the elongation of HIV-1 genes. A major fraction of nuclear P-TEFb is sequestered into a transcriptionally inactive 7SK small nuclear ribonucleoprotein (snRNP) by the coordinated actions of the 7SK small nuclear RNA (snRNA) and hexamethylene bisacetamide (HMBA) induced protein 1 (HEXIM1). In this study, we demonstrate that Tat prevents the formation of and also releases P-TEFb from the 7SK snRNP in vitro and in vivo. This ability of Tat depends on the integrity of its N-terminal activation domain and stems from the high affinity interaction between Tat and cyclin T1, which allows Tat to directly displace HEXIM1 from cyclin T1. Furthermore, we find that in contrast to the Tat-independent activation of the HIV-1 promoter, Tat-dependent HIV-1 transcription is largely insensitive to the inhibition by HEXIM1. Finally, primary blood lymphocytes display a reduced amount of the endogenous 7SK snRNP upon HIV-1 infection. All these data are consistent with the model that Tat not only recruits but also increases the active pool of P-TEFb for efficient HIV-1 transcription.
INTRODUCTION
Transcription of the HIV-1 proviral DNA by RNA polymerase II (RNAPII) is controlled primarily at the level of elongation by the viral Tat protein (1). In its absence, RNAPII clears the HIV-1 promoter but soon arrests due to actions of the negative transcription elongation factor (N-TEF), yielding predominantly short viral transcripts that contain the transactivation response element (TAR) elements at their 5′ ends. To stimulate elongation, Tat binds to TAR via its arginine-rich motif (ARM). Moreover, the activation domain of Tat, consisting of the N-terminal core and cysteine-rich regions, binds cyclin T1 (CycT1), which interacts with Cdk9 to form the positive transcription elongation factor b (P-TEFb) (2). The highly cooperative interactions between Tat, TAR and P-TEFb recruit P–TEFb to the paused transcription complex, where it phosphorylates the C-terminal domain (CTD) of the largest subunit of RNAPII and the components of N-TEF (negative transcription elongation factor). These events convert N–TEF into a positive elongation factor and recruit pre-mRNA splicing and 3′ polyadenylation machineries to the phosphorylated CTD, resulting in efficient elongation and co-transcriptional processing of nascent pre-mRNA (1).
Recent reports indicate that P–TEFb exist in two mutually exclusive complexes that differ in their kinase activities. In HeLa cells, about half of P–TEFb are sequestered in a catalytically inactive complex together with the 7SK snRNA (7SK) and the HEXIM1 protein (3,4). Whereas HEXIM1 binds P–TEFb by interacting directly with CycT1 via its C–terminal region, 7SK serves as a molecular scaffold to facilitate the HEXIM1-P-TEFb binding (5–9). Besides the 7SK-HEXIM1–P-TEFb complex (referred herein as 7SK snRNP), another complex in which a major fraction of nuclear P-TEFb resides is the Brd4–P-TEFb complex (10,11). The Brd4-bound P–TEFb is transcriptionally active and recruited to transcriptional templates possibly due to the ability of Brd4, a bromodomain protein, to bind acetylated histones and the mediator. Importantly, the partitioning of P-TEFb into the inactive and active complexes is dynamic, and several stress-inducing agents are known to disrupt the 7SK snRNP and stimulate the formation of the Brd4–P-TEFb complex (11). Finally, recent findings obtained in heart, breast epithelial, neuroblastoma and murine erythroleukemia cells suggest that a shift in the equilibrium between the two P–TEFb fractions may alter the cellular developmental commitments, leading to either unrestrained growth or terminal differentiation (12–15).
Since most of the P-TEFb are sequestered in the catalytically inactive and active complexes in cells, Tat could in principle modulate their configurations to increase the pool of P-TEFb for efficient HIV-1 transcription. Notably, Tat recruits P-TEFb to the HIV-1 LTR independently of Brd4 (11). However, it is unclear whether Tat affects the inactive 7SK snRNP. In this study, the apparent similarities in the molecular RNA:protein configurations between the HIV-1 TAR-Tat-P-TEFb and the inactive 7SK snRNP prompted us to investigate whether Tat releases P-TEFb from the 7SK snRNP. Indeed, we find that Tat disrupts 7SK snRNP in cells and releases P-TEFb from it via its activation domain. This disruption could be attributed to a direct competition between Tat and HEXIM1 for binding to CycT1. Thus, it appears that HIV-1 has evolved an efficient mechanism that alleviates the negative regulation of P-TEFb by hijacking it from the inactive 7SK snRNP to activate HIV-1 transcription.
MATERIALS AND METHODS
Cell culture
The HeLa-based HL3T1 cell line that contains an integrated HIV LTR-luciferase was a gift of Dr Romero (San Francisco State University). The HeLa-based F1C2 cells stably expressing Cdk9-F were described (11). The cells were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum, 100 mM l–glutamine and 50 μg each of penicilin and streptomycin per ml.
Plasmid DNAs
Plasmids coding for GST-T1, Tat and CycT1-binding domain of HEXIM1 (HEXIM1-TBD) were described (7). Plasmid reporter pSLIIBCAT and a plasmid coding for the Rev-CycT1 chimera were described (16). Plasmids coding for Tat-F, Tat-HA, the various mutant Tat proteins and F-HEXIM1 were described (3,6).
Recombinant proteins, RNA and immunological reagents
The anti–CycT1 and anti-Cdk9 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The anti–FLAG M2 antibody and the anti–FLAG M2 beads were purchased from Sigma–Aldrich Corp (St. Louis, MO, USA). Recombinant CycT1, Tat and HEXIM1 proteins, either alone or as GST-chimeras were expressed and purified from E. coli as described (7). A 29–mer HIV-1 TAR RNA was transcribed in vitro from an antisense single-stranded oligonucleotide using the T7 promotor and polymerase system.
GST-pull down assays
For direct competition assays between HEXIM1-TBD, GST-T1 and Tat, ∼2 µg GST or GST-fusion proteins were immobilized on glutathione Sepharose-beads (Amersham) and incubated with 5 µg of HEXIM1-TBD and increasing amounts of Tat (1–20 µg) and TAR. Bindings were performed in 300 µl of 20 mM Tris/HCl (pH 7.4), 100 mM NaCl, 20 µM ZnSO4, 0.5% NP-40, 5 mM β-mercaptoethanol buffer solution at room temperature (RT) for 0.5–1 h. Beads were washed 2–3 times in the same buffer and bound proteins were analyzed by SDS-PAGE and Coomassie staining.
Fluorescence spectroscopy measurements and data analysis
Conditions for the protein binding and replacement assays as well as the subsequent fluorescence measurements have been described previously (7). Binding curves, titration and dissociation experiments were analyzed with Origin 7.0 (OriginLab Corporation, MA, USA). Kinetic dissociation measurements were done using a SX.18MV stopped flow apparatus (Applied Photophysics) by rapidly mixing of a pre-equilibrated solution of 5 µM CycT1 and 5 µM dimeric HEXIM1-TBD* with 10 or 20 µM excess of Tat or Tat/TAR, respectively. The dansyl-based fluorophore was excited at 337 nm, and the fluorescence emission was recorded through a 420 nm cutoff filter. The competition measurements were evaluated with the software Scientist v2.01 (MicroMath Scientific Software, UT, USA).
Affinity purification of FLAG and HA epitope-tagged proteins and their associated factors
For western and northern analyses, FLAG- or HA-tagged HEXIM proteins were expressed in HeLa cells transfected with the Lipofectamine Plus reagent (Invitrogen). Nuclear extracts (NEs) were prepared from the transfected cells 48 h later and immunopurifications were done as described (17). Northern blotting employed the full-length 7SK antisense RNA as a probe as described previously (6). The 7SK snRNP in vitro reconstitution assay was performed as described (18).
Transient transfection and CAT reporter gene assay
HeLa cells were seeded into 6-well plates or 100-mm-diameter petri dishes ∼12 h prior to transfection and transfected with FuGENE6 reagent (Roche Applied Science, Indianapolis, IN, USA). CAT enzymatic assays were performed as described (16).
Luciferase reporter gene assay
Eight hours post-transfection, HL3T1 cells were treated or untreated with PMA (100 ng/ml). After an additional 24 h, cells were washed twice with PBS and lysed by three freeze–thaw cycles. Luciferase activity was measured in a microplate reader (LB96V MicroLumat Plus) using 50 µl of each sample. To normalize the results, protein concentrations were measured using Bradford reagent (Biorad).
Glycerol gradient sedimentation analysis
NEs, prepared from HeLa cells either untransfected or transfected with the WT Tat–F–expressing vector at 48 h post-transfection, were subjected to ultracentrifugation in a Sorvall SW41 Ti rotor at 38 000 r.p.m. for 21 h at 4°C in a 10 ml glycerol gradient solution (10–30%) containing 20 mM HEPES, pH 7.9, 0.3 M KCl, 0.2 mM EDTA and 0.1% NP40. Fractions were collected and analyzed as described (17).
Infection of primary cells
Primary blood lymphocytes (PBLs; 5 × 107 cells) from healthy donors were stimulated with PHA (3 µg/ml) and IL-2 (100 U/ml) for 3 days prior to infection with two different amounts of the HIV-1 Lai strain (0.5 and 5 ng of p24). The cells were exposed to the virus for 3 h in 2% FCS medium and were maintained in the RPMI medium containing 10% FCS, supplemented with PHA and IL-2. Five days later, the efficiency of infection was verified by measuring the amounts of p24 in the supernatants, and the cells were harvested for glycerol gradient sedimentation analysis.
RESULTS
Tat displaces HEXIM1 from the native 7SK snRNP in vitro
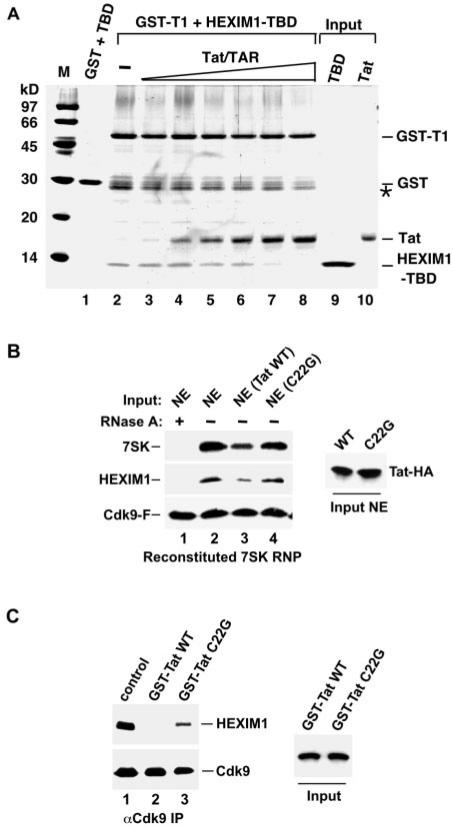
Previous studies demonstrated that HEXIM1 and Tat interact with CycT1 in a region that is immediately C-terminal to the cyclin box (4,7,19), suggesting that the interactions of Tat and HEXIM1 with CycT1 may be mutually exclusive. To test this possibility, we performed a binding assay with recombinant proteins to study the competition between Tat and the CycT1–binding domain of HEXIM1 (HEXIM1-TBD; aa 255–359) (7) for binding to the chimeric GST-CycT1 (GST-T1; aa 1–292). Data in Figure 1A indicate that HEXIM1-TBD bound to GST–T1 but not GST alone in the absence of Tat (compare lanes 1 and 2). However, the addition of increasing amounts of HIV-1 Tat protein and TAR RNA (kept at an 1:1 molar ratio) displaced HEXIM1-TBD from GST-T1 in a dose-dependent manner (lanes 3–8). Notably, the absence of TAR slightly decreased the ability of Tat to compete with HEXIM1-TBD for binding to CycT1 in vitro (data not shown). This issue will be addressed in more detail in Figure 2.
Figure 1.
Tat disrupts the HEXIM1-P-TEFb interaction in vitro. (A) Competition between HEXIM1-TBD and Tat for binding to GST-T1. GST alone (lane 1) or GST-T1 (lanes 2–8) was incubated with HEXIM1-TBD. Increasing amounts of Tat and TAR (kept at an 1:1 molar ratio) were added to the reactions in lanes 3–8. The bound proteins were detected in a Coomassie–stained SDS-gel. Lanes 9 and 10 represent 40% of the input proteins. The bands denoted with a star (*) represent degradation products of GST-T1. (B) Tat suppresses the in vitro reconstitution of 7SK snRNP. To the immobilized P-TEFb (through Cdk9-F), RNase A-treated or untreated HeLa NE (lanes 1 and 2) or NE containing the wild-type or mutant Tat-HA proteins (lanes 3 and 4) was added as indicated. The amounts of 7SK and HEXIM1 bound to P-TEFb (left panel) and the levels of Tat-HA proteins in NE (right panel) are determined by northern and western blotting. (C) Tat disrupts the 7SK snRNP in vitro. 7SK snRNP immobilized on anti-Cdk9 beads were incubated with the wild-type or mutant C22G GST-Tat chimeras as indicated. The levels of Cdk9-bound HEXIM1 (left panel) and the input GST-Tat chimeras (right panel) are determined by western blotting.
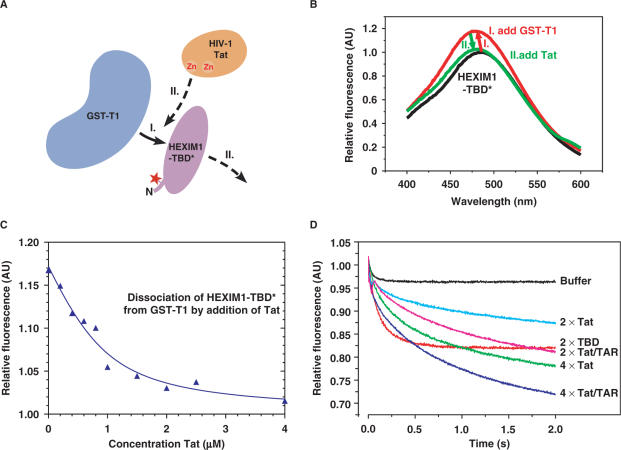
Figure 2.
Tat displaces HEXIM1-TBD from CycT1 due to its higher affinity for CycT1. (A) A schematic diagram of the main steps of the fluorescence competition assay. GST-T1 was first added to HEXIM1-TBD* (step I), followed by the addition of Tat (step II). (B) Fluorescence emission spectra of the two-step titrations as diagrammed in A. To 1.0 µM HEXIM1-TBD* (black curve), 1.0 µM of GST-T1 was added (red), followed by an excess of 5.0 µM Tat (green). (C) Dissociation of the HEXIM1-TBD* from GST-T1 by increasing concentrations of Tat. The displacement of HEXIM1-TBD* from GST-T1 by Tat was observed by equilibrium fluorescence titration. To a preformed complex of 2.0 µM HEXIM-TBD* and 1.0 µM GST-T1, Tat was added at concentrations from 0.2–4 µM. (D) Kinetics of the Tat- or Tat/TAR-mediated displacement of HEXIM1-TBD* from GST-T1 as measured by stopped flow techniques. To a pre-equilibrated solution of 5 µM GST-T1 and 5 µM dimeric HEXIM1-TBD*, Tat or Tat/TAR was injected at a concentration of 10 (2×) or 20 µM (4×) to displace HEXIM1-TBD* over the indicated time periods (in seconds). The displacement caused by buffer alone or 10 µM (2×) unlabeled HEXIM1-TBD (labeled as TBD) is also presented as a comparison.
The above experiments used recombinant HEXIM1 and CycT1 protein fragments. To extend these findings, we asked whether Tat could displace HEXIM1 from CycT1 in the context of the native 7SK snRNP in vitro. First, to examine the effect of Tat on the formation of the 7SK snRNP in HeLa nuclear extracts (NE), we employed a previously described in vitro reconstitution assay (18). In this system, the HeLa-based F1C2 cell line expressing the FLAG–tagged Cdk9 protein (Cdk9-F) was used to immobilize P-TEFb on anti-FLAG beads. Whereas high salt washed off 7SK and HEXIM1 from the immobilized P-TEFb, the 7SK snRNP could be reconstituted with the addition of HeLa NE.
Indeed, NE prepared from untransfected HeLa cells reconstituted the interactions of HEXIM1 and 7SK with P-TEFb and the presence of RNase A prevented these interactions (Figure 1B, compare lanes 1 and 2). Importantly, the addition of NE prepared from HeLa cells expressing the transfected, HA-tagged wild-type Tat protein (Tat-HA) reduced the bindings of HEXIM1 and 7SK to P-TEFb. In contrast, the incubation with NE containing the mutant Tat protein (C22G), which does not bind CycT1 due to a point mutation in its activation domain (19), failed to reduce these bindings (Figure 1B, compare lanes 3 and 4). Thus, Tat was able to compete with the full-length HEXIM1 for binding to CycT1 even in the context of the native 7SK snRNP.
Next, we asked whether Tat could disrupt a preformed 7SK snRNP, which was immunopurified from HeLa NE with antibodies directed against the endogenous Cdk9. While the addition of the wild-type GST-Tat chimera completely disrupted the HEXIM1-P-TEFb interaction, the mutant GST-Tat C22G had a significantly reduced effect (Figure 1C, lanes 2 and 3). The residual activity displayed by this mutant could be due to its weak, but still detectable binding to P-TEFb (data not shown). Taken together, these results suggest that Tat not only decreases the formation of but also disrupts a preformed 7SK snRNP in vitro. Moreover, this ability of Tat likely stems from its direct displacement of HEXIM1 from CycT1.
Tat displaces HEXIM1 from CycT1 in vitro due to its higher affinity for CycT1
To directly compare the CycT1-binding abilities of HEXIM1 and Tat, we performed a series of fluorescence emission assays, which detect protein complex formation based on the general principle that a solvent exposed fluorescence label emits a signal of lower intensity than a label in a tight molecular environment. Following a two-step titration strategy outlined in Figure 2A, an equal concentration of GST-T1 was added to a solution containing 1 µM of fluorescently labeled HEXIM1-TBD (HEXIM1-TBD*). This was followed by the addition of 5 µM of recombinant Tat protein to the reaction mixture. The equilibrium titration of this reaction scheme was monitored by fluorescence emission spectroscopy (Figure 2B). Starting from the spectra of unbound HEXIM1-TBD* (black line), we first observed an increase in fluorescence emission upon the HEXIM1-TBD*–GST-T1 complex formation (red line). However, upon the addition of Tat, the spectra returned to the ground state (green line), implicating a release of HEXIM1-TBD* by the binding of Tat to GST-T1.
Next, we followed the dissociation of HEXIM1-TBD* from GST-T1 by the addition of Tat in a fluorescence competition experiment involving 2.0 µM of HEXIM1-TBD* and 1.0 µM GST-T1 proteins (Figure 2C). Increasing concentrations of Tat from 0.2 to 4 µM led to a decrease in the fluorescence signal, indicating the ability of Tat to displace HEXIM1-TBD* from GST–T1. A fit of the steady-state curve yielded an ∼10-fold higher affinity of GST–T1 for Tat than for HEXIM1-TBD*.
Finally, to analyze the impact of the HIV-1 TAR RNA on the ability Tat to displace HEXIM1-TBD* from GST-T1 in a quantitative manner, we performed fast stopped flow kinetic measurements (Figure 2D). To a pre-equilibrated solution containing 5 µM GST-T1 and 5 µM dimeric HEXIM1–TBD*, Tat was injected at a concentration of 10 (2×) or 20 µM (4×). Compared to buffer alone, a dose-dependent decrease in the fluorescence signal and thus release of HEXIM–TBD* from GST-T1 was observed. Addition of equal concentrations of TAR to Tat further enhanced this displacement (Figure 2D). From a fit of the competition displacement curves, we determined the kinetic parameters of the GST-T1–Tat interaction. The presence of TAR apparently increased the kon rate by a factor of 1.5–2 and decreased koff by a factor of 2–3. Overall, the affinity of Tat/TAR to GST-T1 compared to Tat alone was increased by a factor of 5, leading to an increased displacement of HEXIM1-TBD* from GST-T1. Taken together, these results suggest that Tat displays a higher affinity for CycT1 than does HEXIM1 and is thus able to directly displace HEXIM1 from CycT1. Moreover, the presence of TAR enhances the ability of recombinant Tat to disrupt the HEXIM1–CycT1 interaction possibly due to stabilization of the Tat conformation and/or its solubility.
Tat prevents the binding of HEXIM1 to P-TEFb in vivo
The in vitro observations presented so far prompted us to examine whether Tat could block the binding of HEXIM1 to P-TEFb in vivo. To test this hypothesis, we examined the abilities of wild-type and mutant Tat proteins to compete with co-expressed HEXIM1 for binding to the endogenous P-TEFb in HeLa cells. As indicated above, this ability of Tat is expected to depend on the integrity of the Tat N-terminal activation domain that is involved in the Tat–CycT1 interaction. Additionally, the arginine-rich motif (ARM) of Tat, which is critical for TAR RNA binding and nuclear localization, could contribute to this effect as well.
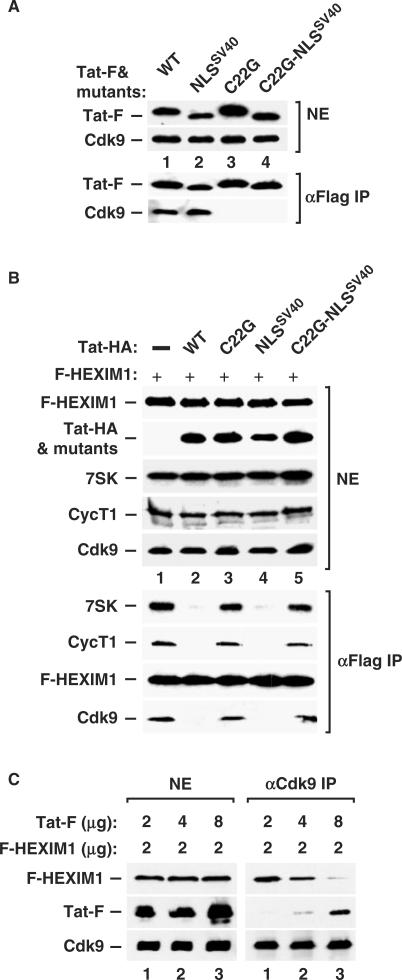
First, we examined the abilities of the various FLAG-tagged Tat proteins (Tat-F) to interact with the Cdk9 subunit of the endogenous P-TEFb (Figure 3A). Western blotting of anti–FLAG immunoprecipitates revealed that wild-type Tat-F interacted with Cdk9 in the absence of the HIV-1 TAR RNA (lane 1). Similarly, a mutant Tat (Tat-NLSSV40), whose ARM was replaced with the nuclear localization signal (NLS) derived from the SV40 T antigen, also bound Cdk9 (lane 2), further implicating a TAR-independent interaction between Tat and P–TEFb. As expected, the C22G mutation within the Tat activation domain in either the wild–type or mutant Tat-NLSSV40 background abolished this interaction (Figure 3A, lanes 3 and 4).
Figure 3.
Tat prevents HEXIM1 from binding to P-TEFb in vivo. (A) Western analyses of the levels of Cdk9 as well as the indicated wild-type or mutant Tat-F proteins in NEs of transfected HeLa cells (upper panels; all plasmids transfected at 2 µg per 15-cm dish) and the levels of Cdk9 associated with Tat-F in the anti-FLAG immunoprecipitates (lower panels). (B) HeLa cells were co-transfected with appropriate empty vectors (−) or vectors coding for F-HEXIM1 (2 µg/15-cm dish) and various Tat-HA proteins (2 µg) as indicated (total 4 µg plasmid DNA per dish). The levels of F-HEXIM1, Tat-HA, 7SK, CycT1 and Cdk9 in NEs (top five panels) and the amounts of 7SK, CycT1 and Cdk9 bound to the immunoprecipitated F-HEXIM1 (bottom four panels) are examined by western and northern blotting. (C) HeLa cells were co-transfected with a constant amount (2 µg/15-cm dish) of the plasmid coding for F-HEXIM1 and increasing amounts (2, 4 and 8 µg) of the plasmid coding for wild-type Tat-F. The levels of F-HEXIM1, Tat-F and Cdk9 proteins in NEs of transfected cells as well as the amounts of F-HEXIM1 and Tat-F proteins bound to the immunoprecipitated Cdk9 are detected by western blotting and shown in the left and right panels, respectively.
Consistent with their abilities to interact with P-TEFb, both wild-type Tat and Tat–NLSSV40 efficiently prevented the interaction of the co-expressed F-HEXIM1 protein with endogenous P-TEFb (Figure 3B, lanes 2 and 4). In contrast, neither Tat C22G nor C22G–NLSSV40 was able to disrupt the interaction between F-HEXIM1 and P-TEFb (lanes 3 and 5). These observations indicate that the ability of Tat to prevent the formation of the 7SK snRNP depended on the interaction of Tat with P-TEFb but not the TAR RNA. Thus, these in vivo results differed from the in vitro fluorescence competition data described above (Figure 2D), which revealed a TAR RNA-mediated enhancement of the displacement of HEXIM1-TBD from GST-T1 by Tat. As discussed below, this discrepancy could result from an improved, TAR-dependent folding and structural integrity of bacterially produced recombinant Tat proteins.
To demonstrate the direct competition between Tat and HEXIM1 for binding to P-TEFb in vivo, we performed a titration experiment in which levels of F-HEXIM1 were kept constant while amounts of the co-expressed Tat-F were increased in 2-fold increments (Figure 3C). The interactions of these two FLAG-tagged proteins with the endogenous P–TEFb were determined by anti-Cdk9 immunoprecipitation followed by western blotting. Indeed, Tat prevented HEXIM1 from interacting with P-TEFb in a dose-dependent manner (Figure 3C, right panel). Taken together, these in vivo data corroborate well with the above in vitro analyses by showing that Tat disrupted the binding between HEXIM1 and P-TEFb via its N-terminal P–TEFb–binding domain.
Tat disrupts the endogenous 7SK snRNP in vivo
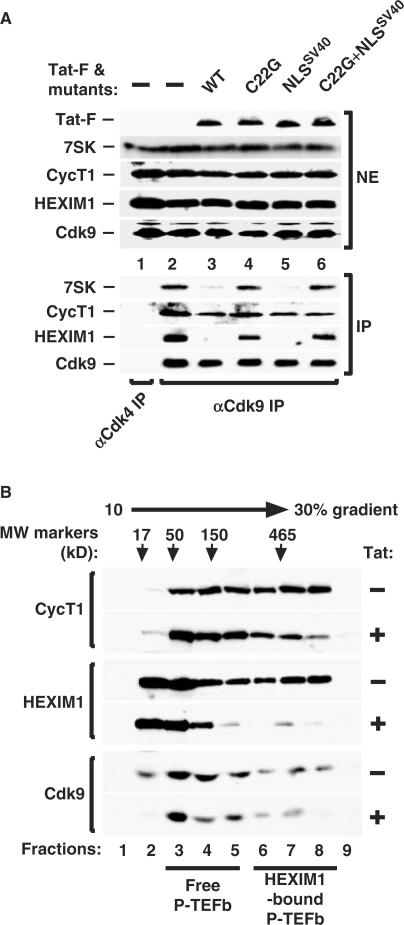
To further our in vivo findings, we next investigated whether Tat could also disrupt the pre-existing 7SK snRNP in HeLa cells. Indeed, the expression of the wild-type Tat and mutant Tat-NLSSV40 proteins, both of which bound P-TEFb, efficiently disrupted the endogenous 7SK snRNP, as evidenced by the marked reduction in the amounts of 7SK and HEXIM1 associated with the immunoprecipitated Cdk9 (Figure 4A, lanes 3 and 5). In contrast, the C22G and C22G–NLSSV40 mutants, which did not bind P-TEFb, largely failed to affect the integrity of 7SK snRNP (Figure 4A, lanes 4 and 6).
Figure 4.
Tat disrupts the endogenous 7SK snRNP to release P-TEFb. (A) NEs prepared from HeLa cells either untransfected (−) or transfected with the indicated plasmids (20 µg per 15–cm dish) encoding wild-type or mutant Tat-F proteins were subjected to anti-Cdk9 or, as a negative control, anti-Cdk4 immunoprecipitation. The levels of endogenous 7SK, CycT1 and HEXIM1 bound to Cdk9 as revealed by northern or western blotting are indicated (bottom panels). The upper panels show the levels of the endogenous 7SK, CycT1, HEXIM1 and Cdk9 as well as the various transfected Tat-F proteins in HeLa NEs. (B) NEs prepared from HeLa cells transfected with an empty vector (−) or the Tat-F-encoding plasmid (20 µg/15-cm dish) were subjected to glycerol gradient sedimentation analysis. The panels show the western detection of CycT1, HEXIM1 and Cdk9 in gradient fractions (left). Molecular size standards were analyzed in a parallel gradient and their positions indicated by arrows.
The notion of a Tat-mediated disruption of pre-existing 7SK snRNPs was further addressed by the glycerol gradient sedimentation analysis of the distributions of P-TEFb complexes in HeLa cells that expressed Tat or not. In NE that did not contain Tat, about half of P-TEFb and 20–30% of total cellular HEXIM1 were sequestered in the 7SK snRNP (Figure 4B, lanes 6–8). The other half of P-TEFb, which bound to Brd4 in a salt-sensitive manner (11), were converted into the smaller, free P-TEFb form under our sedimentation conditions (lanes 3–5). In contrast, this HEXIM1 and P-TEFb distribution pattern was changed considerably in NE that contained Tat. Levels of HEXIM1 in the 7SK snRNP were diminished greatly and this effect was accompanied by a simultaneous reduction in the amounts of both CycT1 and Cdk9 in the same fractions (fractions 6–8) and by the accumulation of free HEXIM1 proteins (fractions 2 and 3). Thus, these observations are consistent with the co-immunoprecipitation data shown above and suggest strongly that Tat not only prevents the formation of new but also disrupts pre-existing 7SK snRNP in cells.
Tat suppresses HEXIM1's inhibition of P-TEFb-dependent transcription
Thus far, our data obtained in vitro and in vivo illustrate the ability of Tat to disrupt the interaction between P-TEFb and HEXIM1. Based on this Tat-mediated release of an inhibitor from P-TEFb, one could envision that Tat might be able to antagonize the inhibitory action of HEXIM1 on P-TEFb-dependent transcription. To test this hypothesis, we performed transcriptional assays in HeLa cells, in which the ability of P-TEFb to stimulate transcription was monitored by using an RNA-tethering system consisting of the plasmid reporter pSLIIBCAT and the hybrid Rev-CycT1 protein. The recruitment of CycT1 and its associated Cdk9 protein to pSLIIBCAT via the Rev–RRE interaction activates transcription that depends on the kinase activity of P-TEFb (16).
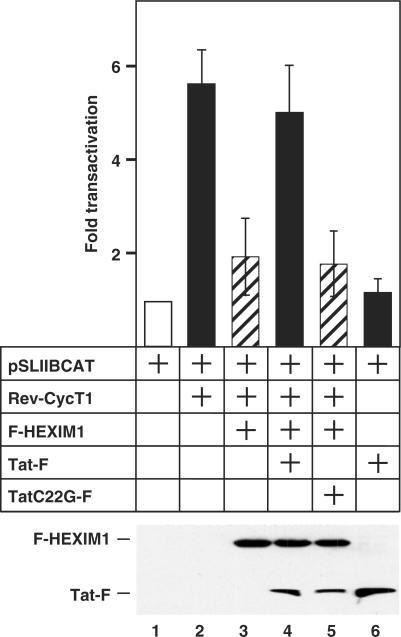
When Rev-CycT1 was co-expressed with pSLIIBCAT in HeLa cells, levels of CAT activity increased ∼6-fold over the basal level (Figure 5, compare bars 1 and 2). As expected, the co-expression of F-HEXIM1 decreased the Rev-CycT1-dependent transcription to ∼2-fold (Figure 5, bar 3). Importantly, the expression of wild-type Tat-F largely reversed the inhibitory effect of F-HEXIM1, whereas the mutant Tat C22G protein failed to do so (bars 4 and 5). It is important to point out that this Tat-mediated increase in transcription was indeed due to the ability of Tat to neutralize HEXIM1's negative effect, as the expression of Tat alone in the absence of Rev-CycT1 and HEXIM1 only minimally increased transcription over the basal level (compare bars 1, 4 and 6). Thus, the release of HEXIM1 from P-TEFb by Tat has a clear functional consequence and allows Tat to antagonize the HEXIM1-mediated inhibition of P–TEFb's transcriptional activity.
Figure 5.
Tat reverses the HEXIM1-mediated inhibition of P-TEFb transcriptional activity. HeLa cells were co-transfected with the reporter plasmid pSLIIBCAT (0.2 μg) and the plasmids coding for the various effectors (Rev-CycT1: 0.6 μg; F-HEXIM1: 0.6 μg; Tat-F: 0.8 μg; TatC22G-F: 0.8 μg) as indicated. The chloramphenicol acetyltransferase (CAT) enzyme activities in cell lysates were measured and the error bars represent the mean +/− SD. Lower panel shows the expression levels of the co-transfected F-HEXIM1 and Tat-F as revealed by western blotting.
Tat-dependent HIV-1 transcription is insensitive to the inhibition by HEXIM1
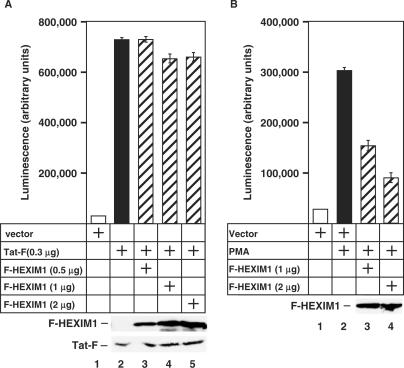
Our demonstration that Tat antagonizes HEXIM1 by disrupting the 7SK snRNP raises an intriguing possibility that Tat transactivation of the HIV-1 LTR would be largely refractory to HEXIM1's inhibitory action. To test this hypothesis, we examined effects of HEXIM1 on Tat transactivation of the integrated HIV-1 LTR that controls the expression of the luciferase reporter gene in the HeLa-based HL3T1 cells. As expected, the expression of Tat-F stimulated the HIV-1 transcription potently (Figure 6A, compare bars 1 and 2). However, the expression of increasing amounts of F-HEXIM1 did not cause a significant decrease in Tat-dependent HIV–1 transcription (bars 3–5), suggesting that HEXIM1 failed to inhibit Tat transactivation.
Figure 6.
HEXIM1 inhibits Tat-independent but not Tat-dependent transcriptional activation from the HIV-1 LTR. (A,B) HL3T1 cells containing an integrated HIV-1 LTR-driven luciferase reporter construct were transfected with an empty vector or the plasmids encoding Tat-F and/or F-HEXIM1. Where indicated, the cells were also stimulated by PMA. Luciferase activities in cell lysates were measured and the error bars represent the mean +/− SD. Lower panels show the expression levels of the co-transfected F-HEXIM1 and Tat-F as revealed by western blotting.
Since transcriptional activation of the HIV-1 LTR could also be achieved in a Tat–independent manner by inducing the transcriptional activity of NF-κB with PMA (20), we next tested whether HEXIM1 could affect the PMA-stimulated HIV-1 transcription. In contrast to Tat-dependent activation, the robust stimulation of the integrated HIV-1 LTR by PMA was decreased progressively by F–HEXIM1 in a dose-dependent manner (Figure 6B, bars 1–4). This finding is consistent with a previous report showing that HEXIM1 expression reduced the transcription from an NF-kB-dependent reporter plasmid (21). Of note, PMA treatment of cells failed to disrupt the endogenous 7SK snRNP (data not shown). Thus, the Tat-dependent and -independent activation of HIV-1 transcription displayed distinct sensitivities to HEXIM1 inhibition. It is likely that the Tat-mediated disruption of the HEXIM1-P-TEFb interaction was responsible for the inability of HEXIM1 to inhibit Tat transactivation.
HIV-1 infection of primary blood lymphocytes results in a reduced amount of the endogenous 7SK snRNP
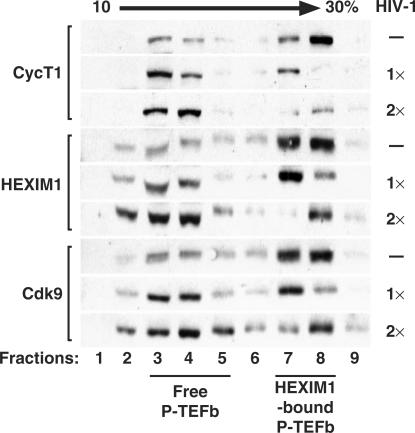
Since Tat prevents the formation of and also releases P-TEFb from the 7SK snRNP, we were prompted to investigate whether the amount of endogenous 7SK snRNP was also affected in HIV-1-infected cells. To address this question, we infected the primary blood lymphocytes (PBLs) with the HIV-1 Lai strain at two different concentrations that yielded 3.22 ng/ml (1×) and 6.32 ng/ml (2×) of HIV p24 in the supernatant after five days of infection. The distributions of P-TEFb complexes between infected and uninfected cells were then compared by the glycerol gradient sedimentation analysis (Figure 7). In total cell lysates of PBLs that were not exposed to HIV-1, ∼80% of total cellular Cdk9, CycT1 and HEXIM1 were present in the large 7SK snRNP (Figure 7, lanes 7 and 8). However, these distribution patterns were changed in cell lysates of infected PBLs. We observed that with increasing virus concentration and thereby more Tat expression, there was a dosage-dependent reduction in the levels of HEXIM1, Cdk9 and CycT1 in the 7SK snRNP (fractions 7 and 8). Meanwhile, the levels of Cdk9 and CycT1 in the smaller size, 7SK/HEXIM1-free P-TEFb (fractions 3–5) as well as the amounts of released HEXIM1 (fractions 2–4) also showed a concomitant increase. Although it is unclear at present whether these changes could be entirely attributed to the action of Tat, the dosage-dependent reduction in 7SK snRNP in HIV-1-infected PBLs are nevertheless consistent with our extensive in vitro and in vivo findings presented above and suggest that Tat could disrupt the 7SK snRNP when expressed in the course of HIV-1 infection.
Figure 7.
HIV-1-infected PBLs contain a reduced amount of 7SK snRNP. Total cell lysates prepared from uninfected (−) or HIV-1-infected PBLs at two different viral concentrations (1× or 2×) were subjected to glycerol gradient sedimentation analysis in a 10–30% gradient. The panels show western detection of HEXIM1, Cdk9 and CycT1 in gradient fractions with the positions of the HEXIM1-bound P-TEFb and the 7SK/HEXIM1-free P-TEFb marked at the bottom. For HIV infection, 1× equals to 3.22 ng/ml and 2× to 6.32 ng/ml of HIV-1 p24 detected in the supernatants at day 5 post infection.
DISCUSSION
Tat has long been recognized as a critical HIV-1 regulatory protein that recruits the cellular co-factor P-TEFb to the viral LTR to stimulate transcription elongation. In this report, we present evidence that Tat not only recruits but also increases the active pool of P-TEFb for HIV-1 transcription. Our in vitro, in vivo and functional studies indicate that Tat accomplishes this task by competing with HEXIM1 for binding to CycT1. Compared to HEXIM1, Tat binds to the same region of CycT1 but with higher affinity, allowing it to displace HEXIM1 from P–TEFb. As a result, Tat prevents the formation of new 7SK snRNPs as well as disrupts pre–existing ones. Thus, approximately half of the total P–TEFb that would otherwise be sequestered and inactive in the 7SK snRNP, are now available for Tat transactivation.
Our in vitro and in vivo data confirmed and extended a recent report showing that Tat and the HEXIM1-TBD bound to CycT1 in vitro in a mutually exclusive manner (7). Despite the fact that the HIV-1 TAR RNA is involved in cooperative interactions with both Tat and P-TEFb in vitro (19,22), it did not play any significant role in the Tat displacement of HEXIM1 from P-TEFb in vivo (Figures 3 and 4). Likewise, ectopic expression of HIV-1 TAR RNA did not further enhance this displacement (data not shown). Rather, the N-terminal activation/P-TEFb-binding domain of Tat was critical for this effect. However, our in vitro fluorescence competition experiments containing recombinant proteins and protein fragments suggest that the presence of the HIV-1 TAR RNA enhanced the ability of Tat to displace HEXIM1-TBD from CycT1 (Figure 2D). The apparent discrepancy between our in vivo and in vitro analyses may be explained by the fact that bacterially expressed Tat proteins are often folded improperly and the presence of HIV-1 TAR RNA may act as a molecular chaperone to induce a higher order structure of Tat in vitro. In contrast, Tat proteins expressed in human cells are likely to be modified correctly and display improved structural integrity, which may be responsible for our observed TAR-independent displacement of HEXIM1 from P-TEFb in vivo.
In contrast to our results, a recent report suggested that overexpression of HEXIM1 results in a decreased Tat transactivation (23). However, the seemingly discrepancy between these two studies could stem from the different experimental conditions. For example, Fraldi et al. (23) used a much higher HEXIM1 versus Tat plasmid ratio in their experiments, which may have caused a relatively small amount of Tat to be overwhelmed by a large excess of HEXIM1. As a result, the shift of the P-TEFb equilibrium from the Tat-bound toward the HEXIM1–bound state may have led to the observed inhibition of Tat-transactivation by HEXIM1. Notably, even under such conditions, HEXIM1 displayed only a modest, 2-fold inhibitory effect (23). In addition, the fact that their study employed an episomal HIV-1-LTR promoter but not an integrated one as in our experiments could have also contributed to the different observations.
Importantly, the analyses of the endogenous 7SK snRNP in HIV-1-infected primary cells have furthered our extensive in vitro and in vivo competition studies and revealed the physiological significance of our findings. We demonstrate that an efficient and dosage–dependent disruption of the inactive 7SK snRNP occurred in the HIV-1-infected PBLs. Although indirect effects of HIV-1 infection cannot be completely ruled out at this stage, our findings are nevertheless consistent with the notion that the local Tat recruitment of P-TEFb from 7SK snRNP can take place, especially since abundant P-TEFb complexes are found on the HIV-1 LTR in the presence of Tat (1).
Because very few free P-TEFb exist in the cell and most of them are sequestered in two main complexes (10,11), it is not too surprising that Tat has to disrupt the 7SK snRNP for efficient HIV transcription. Besides this inactive P-TEFb complex, it has been suggested that the disruption of the active Brd4–P-TEFb complex by Tat also favors HIV-1 transactivation (11). Since the bindings of Brd4 and Tat to P-TEFb are mutually exclusive, the Brd4–P-TEFb interaction that normally recruits P–TEFb to chromatin templates for general elongation has to be disrupted by Tat for activated HIV transcription (11). These findings, together with the Tat–mediated disruption of the 7SK snRNP presented here, reveal a clear preference of Tat to bind and recruit the free form of P–TEFb to the HIV-1 LTR.
Our demonstration that Tat releases P-TEFb from 7SK snRNP reveals important insights into how this transcriptional kinase contributes to Tat function. Despite the fact that it originates from the inactive pool, this P-TEFb is nevertheless expected to be fully active. This is because P–TEFb within the 7SK snRNP is phosphorylated on threonine 186 at the tip of the Cdk9T-loop (18,24). As a hallmark of any activated Cdks, this event is expected to render the catalytic pocket in Cdk9 in an open conformation. Thus, the Tat-bound P-TEFb is pre-activated and ready to phosphorylate its substrates once liberated from 7SK snRNP.
Based on the data presented in this study, we propose a simple ‘coupling’ model for Tat transactivation of HIV transcription. This model suggests that the inactive 7SK snRNP and active Tat-bound P-TEFb reside in close proximity within the cell nucleus and are exchanging rapidly during viral replication. Indeed, HEXIM1 and P-TEFb have been found in or near nuclear speckles (8,25,26). Notably, Tat redirects CycT1 out of this sub-nuclear compartment (26), which could reflect the release of P-TEFb from 7SK snRNP. Thus, a role for Tat in the dynamic and rapid deployment of P-TEFb to sites of active viral replication is envisioned. In this scenario, Tat not only recruits P-TEFb to the initiating RNAPII via TAR, but also ensures an ample supply of the kinase for the rapid replication and spread of HIV in the infected host. Future studies are necessary to investigate the importance of this newly discovered Tat function to HIV infection, replication and the escape from viral latency.
ACKNOWLEDGEMENTS
We thank Vikash Jethwani and Dr Romero for providing us with the HL-3T1 cell line, Karin Vogel-Bachmayr and Vivien Lee for expert technical assistance, and Roger Goody and members of Peterlin and Zhou laboratories for stimulating discussions and continuous support. This work was supported by grants from the National Institutes of Health to B.M.P. (AI058708) and Q.Z. (AI41757). M.B. was supported by a grant (106584–36–RFNT) from the American Foundation for AIDS Research (amfAR). J.H.N.Y is supported by a Ruth L. Kirschstein NRSA post-doctoral fellowship (AI058400) from the National Institutes of Health. N.C. was supported by a grant from the Verband der Chemischen Industrie (VCI). Funding to pay the Open Access publication charge was provided by the grants from the National Intitutes of Health to B.M.P. and Q.Z.
Conflicts of interest statement. None declared.
REFERENCES
- 1.Barboric M, Peterlin BM. A new paradigm in eukaryotic biology: HIV Tat and the control of transcriptional elongation. PLoS Biol. 2005;3:e76. doi: 10.1371/journal.pbio.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 4.Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, Bensaude O. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription–dependent manner. Mol. Cell Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yik JH, Chen R, Pezda AC, Samford CS, Zhou Q. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol. Cell Biol. 2004;24:5094–5105. doi: 10.1128/MCB.24.12.5094-5105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulte A, Czudnochowski N, Barboric M, Schonichen A, Blazek D, Peterlin BM, Geyer M. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J. Biol. Chem. 2005;280:24968–24977. doi: 10.1074/jbc.M501431200. [DOI] [PubMed] [Google Scholar]
- 8.Barboric M, Kohoutek J, Price JP, Blazek D, Price DH, Peterlin BM. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J. 2005;24:4291–4303. doi: 10.1038/sj.emboj.7600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egloff S, Van Herreweghe E, Kiss T. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol. Cell Biol. 2006;26:630–642. doi: 10.1128/MCB.26.2.630-642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 12.Sano M, Abdellatif M, Oh H, Xie M, Bagella L, Giordano A, Michael LH, DeMayo FJ, Schneider MD. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 13.Wittmann BM, Wang N, Montano MM. Identification of a novel inhibitor of breast cell growth that is down-regulated by estrogens and decreased in breast tumors. Cancer Res. 2003;63:5151–5158. [PubMed] [Google Scholar]
- 14.Turano M, Napolitano G, Dulac C, Majello B, Bensaude O, Lania L. Increased HEXIM1 expression during erythroleukemia and neuroblastoma cell differentiation. J. Cell Physiol. 2006;206:603–610. doi: 10.1002/jcp.20502. [DOI] [PubMed] [Google Scholar]
- 15.He N, Pezda AC, Zhou Q. Modulation of a P-TEFb functional equilibrium for the global control of cell growth and differentiation. Mol. Cell Biol. 2006;26:7068–7076. doi: 10.1128/MCB.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujinaga K, Cujec TP, Peng J, Garriga J, Price DH, Grana X, Peterlin BM. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 1998;72:7154–7159. doi: 10.1128/jvi.72.9.7154-7159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yik JH, Chen R, Pezda AC, Zhou Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive P-TEFb complexes for control of transcription. J. Biol. Chem. 2005;280:16368–16376. doi: 10.1074/jbc.M500912200. [DOI] [PubMed] [Google Scholar]
- 18.Chen R, Yang Z, Zhou Q. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 2004;279:4153–4160. doi: 10.1074/jbc.M310044200. [DOI] [PubMed] [Google Scholar]
- 19.Garber ME, Wei P, Kewal Ramani VN, Mayall TP, Hermann CH, Rice AP, Littman DR, Jones KA. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong-Starkesen SE, Luciw PA, Peterlin BM. Signaling through T lymphocyte surface proteins, TCR/CD3 and CD28, activates the HIV-1 long terminal repeat. J. Immunol. 1989;142:702–707. [PubMed] [Google Scholar]
- 21.Ouchida R, Kusuhara M, Shimizu N, Hisada T, Makino Y, Morimoto C, Handa H, Ohsuzu F, Tanaka H. Suppression of NF-κB-dependent gene expression by a hexamethylene bisacetamide-inducible protein HEXIM1 in human vascular smooth muscle cells. Genes Cells. 2003;8:95–107. doi: 10.1046/j.1365-2443.2003.00618.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Tamilarasu N, Hwang S, Garber ME, Huq I, Jones KA, Rana TM. HIV-1 TAR RNA enhances the interaction between Tat and cyclin T1. J. Biol. Chem. 2000;275:34314–34319. doi: 10.1074/jbc.M006804200. [DOI] [PubMed] [Google Scholar]
- 23.Fraldi A, Varrone F, Napolitano G, Michels AA, Majello B, Bensaude O, Lania L. Inhibition of Tat activity by the HEXIM1 protein. Retrovirology. 2005;2:42–52. doi: 10.1186/1742-4690-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 2005;280:28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 25.Herrmann CH, Mancini MA. The Cdk9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J. Cell Sci. 2001;114:1491–1503. doi: 10.1242/jcs.114.8.1491. [DOI] [PubMed] [Google Scholar]
- 26.Marcello A, Cinelli RA, Ferrari A, Signorelli A, Tyagi M, Pellegrini V, Beltram F, Giacca M. Visualization of in vivo direct interaction between HIV-1 TAT and human cyclin T1 in specific subcellular compartments by fluorescence resonance energy transfer. J. Biol. Chem. 2001;276:39220–39225. doi: 10.1074/jbc.M104830200. [DOI] [PubMed] [Google Scholar]