Abstract
By binding to SECIS elements located in the 3′-UTR of selenoprotein mRNAs, the protein SBP2 plays a key role in the assembly of the selenocysteine incorporation machinery. SBP2 contains an L7Ae/L30 RNA-binding domain similar to that of protein 15.5K/Snu13p, which binds K-turn motifs with a 3-nt bulge loop closed by a tandem of G.A and A.G pairs. Here, by SELEX experiments, we demonstrate the capacity of SBP2 to bind such K-turn motifs with a protruding U residue. However, we show that conversion of the bulge loop into an internal loop reinforces SBP2 affinity and to a greater extent RNP stability. Opposite variations were found for Snu13p. Accordingly, footprinting assays revealed strong contacts of SBP2 with helices I and II and the 5′-strand of the internal loop, as opposed to the loose interaction of Snu13p. Our data also identifies new determinants for SBP2 binding which are located in helix II. Among the L7Ae/L30 family members, these determinants are unique to SBP2. Finally, in accordance with functional data on SECIS elements, the identity of residues at positions 2 and 3 in the loop influences SBP2 affinity. Altogether, the data provide a very precise definition of the SBP2 RNA specificity.
INTRODUCTION
Based on ribosomal subunit 3D-structure analysis, K-turn motifs were found to be frequent protein-recognition motifs in ribosomal RNAs (1). A total of 8 K-turn motifs were detected in the 23S rRNA from Haloarcula marismortui and the 16S rRNA from Thermus thermophilus (1–4). K-turn motifs are all characterized by a helix I-loop-helix II structure, and the formation of two non-Watson–Crick base pairs (most frequently G.A and A.G) within the internal loop extends helix II (1,5). Due to the stacking onto helix I or helix II of residues in the internal loop, one of the RNA strand forms a sharp angle (1,5). Only one of the residues in the loop is projected out of the K-turn structure and is located in a pocket of the protein in RNA–protein complexes. In addition to their presence in rRNAs, K-turn motifs are also found in the U4 and U4atac spliceosomal snRNAs (5,6) and in the numerous C/D box snoRNAs (7), that guide 2′-O-methylation and cleavages in pre-ribosomal RNA (for review, 8). K-turn motifs were also recently found in both C/D and H/ACA sRNAs, that guide rRNA modifications in archaea (9–11). They are thus very ancient RNA-binding motifs. Both in eukarya and in archaea, small RNAs containing K-turn motifs assemble into RNP particles and the K-turn motifs play a central role in protein assembly (7,9–15). More specifically, the ribosomal L7Ae protein in archaea or its eukaryal homolog, the Snu13p (yeast)/15.5K (human) protein, first recognizes the K-turn structure and the complex formed then serves as a platform for assembly of the other proteins (9,10,12–19).
The Snu13p/15.5K and L7Ae proteins belong to the L7Ae/L30 protein family, which is characterized by the presence of an L7Ae/L30 RNA-binding domain (6,20). The founding member of this protein family, the yeast L30 ribosomal protein recognizes a peculiar K-turn motif in its own pre-mRNA (21–23). One difference between the yeast L30 RNA–protein complex, and the Snu13p/15.5K or L7Ae RNA–protein complexes is the identity of the nucleobase located in the protein pocket. Whereas, a strong preference for an U residue is observed for proteins Snu13p/15.5K and L7Ae (7,24–26), C and A residues are preferentially accommodated in the yeast L30 protein pocket (27). The possibility to bind a G residue was however recently observed (28).
In vertebrates, SECIS-binding protein 2, another member of the L7Ae/L30 protein family, binds SECIS elements in mRNAs (29,30). The SECIS elements contain determinants needed for selenocysteine incorporation into selenoproteins (31,32). Selenocysteine incorporation involves reprogramming of a nonsense UGA codon into a codon recognized by the selenocysteine specific tRNASec. Understanding the mechanism of selenocysteine incorporation into proteins is important as they are key players in the antioxidant defense system (for review, 33). They are also key participants in a variety of other systems including thyroid hormone metabolism, muscle function, transportation and distribution of selenium to remote tissues and can have roles as structural proteins (for reviews, 34–37). In eukarya, the SECIS elements and SBP2 are two essential components of the selenocysteine incorporation machinery. All SECIS elements consist of a hairpin structure composed of two helices I and II, separated by an internal loop. A highly conserved cluster of four non-Watson–Crick base pairs is located in helix II. It contains a tandem of G.A and A.G pairs, which is needed for SBP2 binding (29,30). This cluster of non-Watson–Crick pairs is an essential determinant for selenocysteine incorporation (31,32). A highly conserved AAR sequence present in a loop of all SECIS elements is also important for selenoprotein synthesis in vivo, but not for binding of SBP2 in vitro (30,38). As SBP2 also binds the specific mSelB/EFSec elongation factor, it is proposed to recruit this dedicated elongation factor in a complex formed with the selenocysteyl-tRNASec to the ribosomes (39–41). Additionally, according to a recent investigation on the selenocysteine incorporation machinery (42), the ribosomal protein L30 is able to bind the SECIS motif by displacing protein SBP2. This substitution would facilitate the interaction of the Sec-tRNASec with ribosomes.
A prerequisite to fully understand the SBP2 activity is thus to obtain a more complete picture of the RNA sequence and structural determinants required for SBP2 binding. To this end, we combined the SELEX approach and site-directed mutagenesis experiments. As the RNAs recovered after SELEX experiments could form canonical K-turn motifs with a protruding U residue, we compared the RNA-binding properties of the human SBP2 protein with those of a well-characterized member of the L7Ae/L30 protein family, the S. cerevisiae Snu13p protein. This protein recognizes K-turn motifs in U4 snRNA, the C/D box snoRNAs and U3 snoRNA. Altogether, we show here that in contrast to protein Snu13/15.5K, SBP2 preferentially binds RNA motifs with a large internal loop. In addition, we demonstrate the existence of previously undetected important determinants for RNA recognition by SBP2 that are located in helix II.
MATERIALS AND METHODS
Strains and growth conditions
The Escherichia coli TG1 strain was used as the host strain for plasmid construction. Growth was performed at 37°C in Luria Broth medium, complemented with 100 μg/ml of ampicillin when necessary. The E. coli strain BL21-CodonPlus (Stratagene) was the host strain for production of the recombinant GST/Snu13p, GST/L7Ae and GST/C-SBP2 proteins.
Recombinant DNA
Plasmids pT7SelN (40), pUC18::U3AΔ2,3,4 (26) and pyU4 (43) were used for the production of matrices for in vitro transcription of the SelN, yU3B/C and yU4 RNAs, respectively. The yU3B/C and yU4 matrices were obtained by PCR amplification, under conditions previously described (26). Oligonucleotides yU3B/C-5′, yU3B/C-3′, yU4-5′ and yU4-3′, given in Table 1 of the Supplementary Data, were used as primers. Plasmids pGEX-6P-1::SNU13, pGEX-6P-1::L7AE (44) and pGEX-6-P1::C-SBP2 (this work) were used for production of the recombinant GST/Snu13p, GST/L7Ae and GST/C-SBP2 proteins, respectively. Plasmid pA11 was used for amplification of the PCR fragment coding for region 515–854 of human SBP2 protein (45). DNA fragments amplified by RT-PCR from RNAs obtained after the fourth cycle of the SELEX experiment were cloned into plasmid pCR2.1 (Invitrogen). Mutagenesis of the RNA Se1 coding sequence was performed by the PCR-based site-directed strategy (primers are listed in Table 1 in the Supplementary Data).
In vitro transcription
The EcoRI linearized pT7::SelN plasmid was used as the template for SelN RNA transcription. The yU3B/C, yU4, Se1-Se7 and Se1 variant RNA-coding sequences were transcribed from PCR amplified DNA fragments obtained as described above. Transcriptions were carried out on 1 μg of plasmid DNA linearized with EcoRI or 500 ng of PCR product, in a 15 μl reaction as described in Marmier-Gourrier et al. (26).
RNAs were 5′-end labeled using 10 units of T4 polynucleotide kinase (MBI-Fermentas), 20 pmol of RNA, 5 pmol of [γ-32P] ATP, in a 10-μl reaction mixture containing 10 mM MgCl2; 5 mM DTT; 0.1 mM spermidine; 0.1 mM EDTA; 50 mM Tris-HCl pH 7.6 at 37°C. The 5′-end labeled RNAs were purified on a 10% denaturing polyacrylamide gel.
Recombinant protein preparation
The recombinant GST/Snu13p and GST/L7Ae fusion proteins were produced in E. coli as described in Marmier-Gourrier et al. (26). The same procedure was used for the production of C-SBP2. For purification of untagged proteins, they were bound on glutathione-sepharose 4B as previously described (44) and cleaved on the beads using 80 U of PreScission protease (Pharmacia) per ml of glutathione-sepharose bead suspension, under published conditions (44). The purified proteins were dialyzed against buffer D (150 mM KCl; 1.5 mM MgCl2; 0.2 mM EDTA; 20 mM HEPES, pH 7.9; 10% glycerol) and aliquots were stored at −80°C.
SELEX experiment
The starting DNA matrix containing a 18-nt-long degenerated sequence was produced by PCR amplification, using two partially complementary oligonucleotides (Table 1 in Supplementary Data): SELEX N18 with a 18-nt-long degenerated sequence and SELEX-5′, that generated a T7 RNA polymerase promoter. PCR amplification was as previously described (26), except that MgCl2 was added at a 4 mM concentration in the incubation buffer. About 500 ng of amplified DNA was used for in vitro transcription with T7 RNA polymerase (26). Transcripts were purified by electrophoresis on a 6% denaturing polyacrylamide gel as in Mougin et al. (46). About 0.2 nmol of transcripts were used for the first round of selection. To eliminate RNA molecules having an affinity for the glutathione-sepharose beads, the RNA mixture was first incubated with 30 μl of beads in the absence of the GST/C-SBP2. For RNP complexes, 0.1 nmol of treated RNAs was incubated with 0.01 nmol of purified GST/C-SBP2 for 30 min at 4°C, in 20 μl of buffer D, in the presence of 2 μg of a yeast tRNA mixture (Roche). The mixture was then incubated with 15 μl of glutathione-sepharose beads (Amersham) equilibrated in buffer D. After extensive washing with buffer D, the selected RNAs were released by a 30-min incubation at 37°C, with 20 μg of proteinase K in buffer D. They were extracted with a phenol–chloroform mixture, ethanol precipitated, dissolved in sterile water, hybridized with 50 pmol of SELEX-3′ primer, ethanol precipitated, and finally reverse-transcribed with 25 U of AMV Reverse transcriptase (Q.Biogene) for 30 min at 42°C. Next, 30 cycles of PCR amplification were performed in the presence of primers SELEX-5′ and SELEX-3′ (50 pmol each). The amplified DNA fragments were gel purified and used as the matrix for in vitro transcription. At each cycle of the SELEX experiment, a filter-binding assay was performed after incubation of the uniformly labeled transcripts produced from the DNA pool with the GST/C-SBP2 protein. At the fourth cycle of the amplification-selection experiment, DNA fragments were cloned into plasmid pCR2.1 (Invitrogen). Plasmids were prepared from 30 randomly selected clones and sequenced by the dideoxysequencing method.
Electrophoretic mobility shift assay
About 5 fmol of in vitro transcribed 5′-end labeled RNAs, mixed with 2 μg of yeast tRNAs (Roche), were denatured during 10 min at 65°C in 15 μl of buffer D containing 1.5 mM of MgCl2, followed by a slow cooling to room temperature for renaturation. To test for the effect of Mg++ on complex formation, the Mg++ concentration was adjusted to 1.5, 5, 10, 15 or 20 mM by addition of MgCl2, without modification of the final volume of incubation and a control experiment was performed in the absence of Mg++. The Snu13p or C-SBP2 recombinant proteins were added at various concentrations (from 0 to 4 μM) and the mixture was incubated for 30 min at 4°C. RNA–protein complexes were fractionated by electrophoresis on 6% non-denaturing polyacrylamide gel as in Marmier-Gourrier et al. (26). The amount of radioactivity in the bands, corresponding to the free and complexed RNA, was estimated using a PhosphorImager and the ImageQuant Software. Using these values, apparent Kds were determined with the SigmaPlot Software (SPSS Science Software). For competition assays with an excess of cold RNA or protein, protein–RNA complexes were preformed as mentioned above, and various amounts of cold competitor RNAs or competitor proteins were added, followed by a 30-min incubation at 4°C. The remaining complexes were subjected to gel electrophoresis.
RNA secondary structure analysis
In vitro transcribed 5′-end labeled RNAs (25 fmol) were pre-incubated in buffer D for 5 min at 65°C, in the presence of 2 μg of tRNA followed by a slow cooling for renaturation. The renatured RNAs were then incubated for 30 min at 4°C in the absence or presence of C-SBP2 (100, 50 and 30 pmol, respectively), Snu13p (10, 100 and 30 pmol, respectively) or L7Ae (10 pmol), in 10 μl of buffer D. Digestion was for 6 min at 20°C, in the presence of 0.8 U of T1 RNase (Roche), 2.4 U of T2 RNase (Gibco) or 0.001 U of V1 RNase (Kemotex). V1 RNase reactions were stopped by addition of 100 mM EDTA, followed by phenol extraction. T1 and T2 RNase digestions were stopped by addition of 20 μg of tRNA, followed by phenol extraction and ethanol precipitation. For production of a ladder, an alkaline hydrolysis of the naked RNA was performed for 5 min at 96°C, using 10 fmol of RNA dissolved in 1 μl of 100 mM sodium bicarbonate. The cleavage products were fractionated by electrophoresis on a 10% polyacrylamide–8 M urea gel.
The free energies of the 2D structures of the selected RNAs were calculated at 37°C and in 1 M NaCl with the M-fold software (46).
RESULTS
Protein C-SBP2 does not interact with K-turn motifs recognized by Snu13p
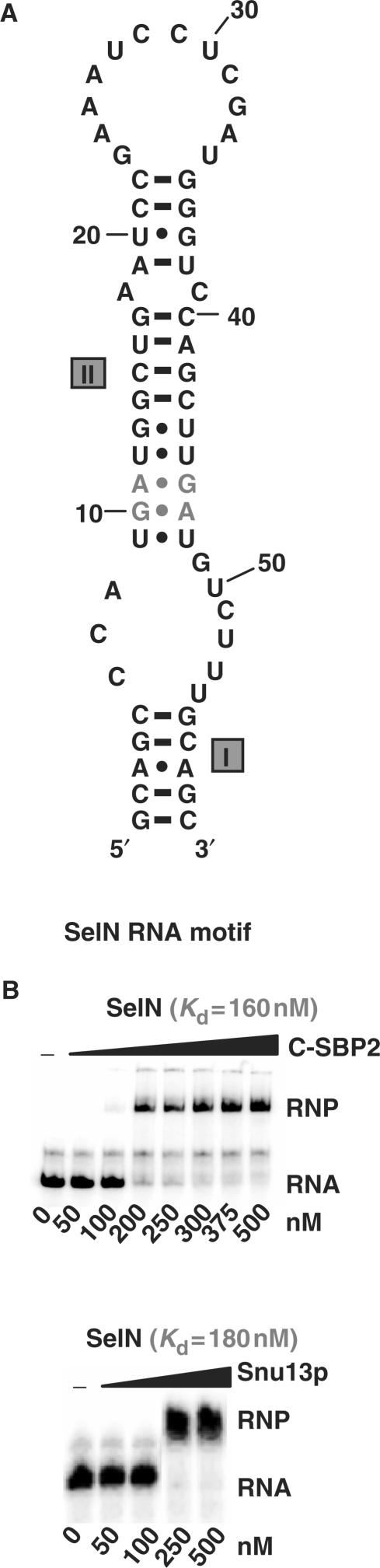
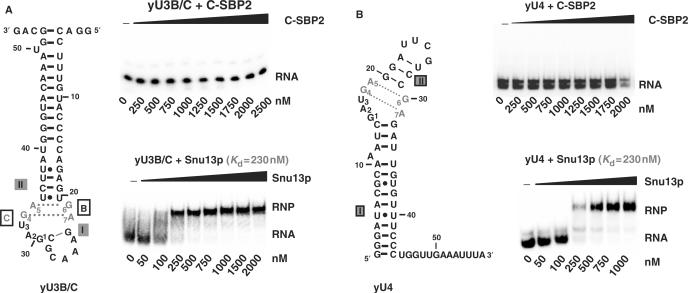
As ribosomal protein L30 was shown to displace SBP2 from SECIS motifs, our first goal was to test whether SBP2 can bind RNA targets of members of the L7Ae/L30 protein family. The large human SBP2 protein (854 aa) has a low solubility. As we wanted to study the RNA-binding property of its L7Ae/L30 domain, we used a truncated version containing this domain. This human SBP2 fragment encompassing residues 515–854 was produced in a soluble form in E. coli. It will be hereafter designated as C-SBP2. To test its capacity to bind SECIS RNAs, we used the well-characterized SECIS RNA motif from the human selenoprotein N mRNA (SelN RNA) (Figure 1A) (30). RNP complexes were formed by incubation of uniformly labeled SelN RNA (5 fmol) with C-SBP2, at concentrations ranging from 50 to 500 nM, in the presence of 2 μg of tRNAs (see the Materials and Methods section for the incubation conditions). As evidenced by gel electrophoresis performed under non-denaturing conditions (Figure 1B), C-SBP2 formed an RNP complex with the SelN RNA and the apparent Kd was of 160 nM. Next, we tested the capacity of this protein to bind K-turn motifs targeted by Snu13p. Two well-characterized RNAs were used: RNA yU3B/C containing the B/C motif of yeast U3 snoRNA (26), and RNA yU4 containing the K-turn motif of yeast U4 snRNA (see the Materials and Methods section for their production). Complexes were formed under the same conditions as for SelN RNA. As a control, the same experiment was performed with Snu13p. Gel electrophoresis revealed the absence of binding of C-SBP2 to both Snu13p RNA targets, even at a high protein concentration (Figure 2). As in contrast, Snu13p was found to bind the SelN RNA with an apparent Kd similar to that of C-SBP2 (Figure 1B), we concluded that to bind C-SBP2, the RNA should have sequence or structure peculiarities, which are not required for association with Snu13p.
Figure 1.
C-SBP2 and Snu13p interact with SelN RNA. (A) The secondary structure of the SelN RNA motif is according to Fagegaltier et al. (52). The G.A sheared base pairs are shown in gray and helices I and II are indicated. (B) The affinity of C-SBP2 and Snu13p for SelN RNA was tested by gel-shift assay using 5 fmol of labeled SelN RNA and protein concentrations ranging from 0 to 500 nM, as indicated below the lanes. Incubation conditions are described in the Materials and Methods section. RNP formation was revealed by electrophoresis on 6% non-denaturing polyacrylamide gels. The apparent Kd values (indicated above the autoradiograms) were calculated with the SigmaPlot Software (SPSS Science Software), by measuring the radioactivity signals corresponding to the free and bound RNAs.
Figure 2.
C-SBP2 does not interact with yU3B/C and yU4 RNAs. The binding of C-SBP2 and Snu13p on yU3B/C or yU4 RNAs was tested by gel-shift assays. The secondary structures of yU3B/C (Panel A) and yU4 (Panel B) are according to Marmier-Gourrier et al. (26) and Mougin et al. (43), respectively. The G.A sheared pairs are in gray and helices I and II are indicated. Nucleotides involved in the K-turn folding are numbered from 1 to 7. Complexes were formed between 5 fmol of uniformly labeled yU3B/C (Panel A) or yU4 (Panel B) RNAs, and C-SBP2 or Snu13p at concentrations ranging from 0 to 2500 nM, as indicated below the lanes. Incubation conditions were as described in the Materials and Methods section. The autoradiograms obtained after electrophoresis on 6% polyacrylamide gels are shown. For the Snu13p–RNP complexes shown as controls, the apparent Kd values were calculated as described in the Materials and Methods section.
A limited diversity of RNAs selected by C-SBP2 in SELEX experiments
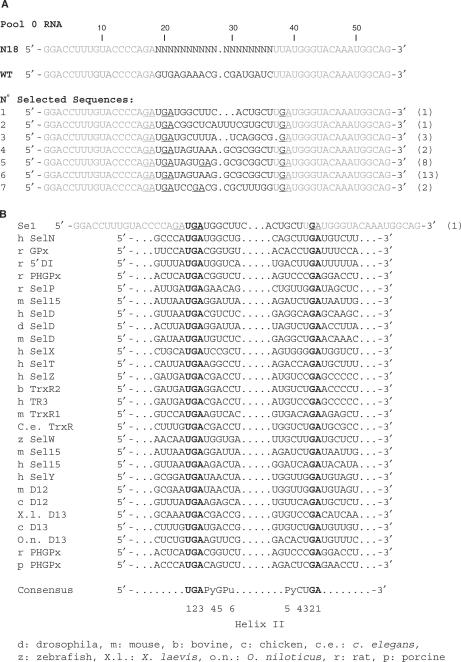
To progress in the understanding of how the SBP2 L7Ae/L30 domain recognizes RNA, we used the yU3B/C RNA, and tried to define by SELEX experiments which kinds of mutations can convert this RNA into a C-SBP2 target. To this end, we degenerated a 18-nt long fragment in the central part of the yU3B/C coding region. The transcripts produced from this degenerated matrix (N18 RNA) were subjected to selection with a GST/C-SBP2 protein fusion that was bound to glutathione-sepharose beads. In spite of the degenerated sequence, all the RNAs were expected to contain the long-terminal stem of RNA yU3B/C (Figure 2A). As the same kind of experiment has previously been performed with Snu13p (47), we also expected to compare the RNA motifs selected by C-SBP2 and by Snu13p. To initiate the selection cycles, we used 5 μg of degenerated RNA mixture, so that each possible RNA sequence was expected to be present 2300 times (47). As a first step, RNAs that might have an affinity for the matrix were eliminated from the RNA pool by incubation with the glutathione-sepharose beads in the absence of protein. Following each selection cycle, the interaction of the pool of selected RNAs with the GST/C-SBP2 fusion was tested by gel-shift assays (conditions for the amplification–transcription–selection cycles are described in the Materials and Methods section). A strong increase of the amount of RNAs showing an affinity for the fusion protein was observed after the fourth cycle of selection. After this cycle, the totality of the selected RNAs was subjected to gel electrophoresis under non-denaturing conditions, and the RNA mixture contained in the slice of gel corresponding to RNPs was extracted, converted into cDNAs, and cloned into plasmid pCR2.1. After transformation of E. coli TG1 cells, thirty colonies were randomly selected among >100 colonies obtained.
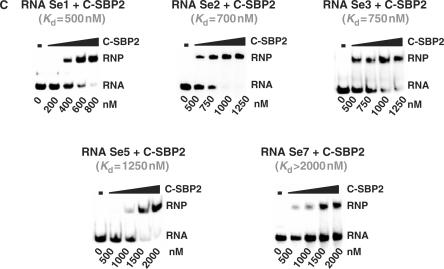
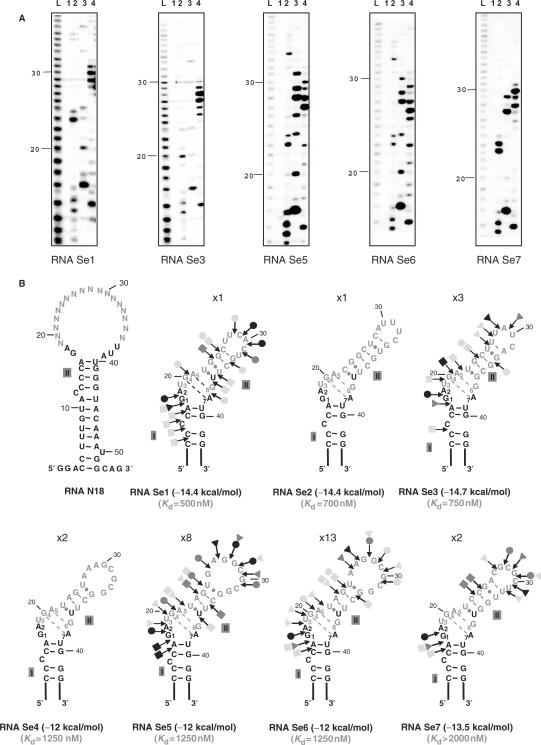
Several of them contained plasmids with identical inserts (Figure 3A). Only seven distinct sequences were found (RNAs denoted Se1 to Se7) (Figure 3A). In addition, three of the sequences that corresponded to the most abundant clones were very similar, suggesting that their small differences were most probably generated by RT-PCR errors in the course of the amplification. Therefore, only four main classes of Se RNAs were selected in the experiment (Figure 3A). This limited diversity of the selected sequences was one major difference, compared to the SELEX experiment performed with Snu13p (31 very different sequences were obtained starting from the same initial RNA mixture). This suggested the existence of strong sequence and/or structural constraints for SBP2 recognition. Three of the less frequent Se RNAs had a different length as compared to the initial RNAs: RNA Se2 (Figure 3A) contained an additional residue in the degenerated sequence, whereas RNA Se1 and Se3 lacked two residues compared to the initial RNAs. In addition, all the Se RNAs had a G instead of a U residue at position 38 in the conserved sequence. The same U38G base substitution was also found in several of the RNAs selected by the GST/Snu13p protein (47). By using gel-shift experiments, we verified that the seven distinct Se RNAs, that were selected, showed an affinity for the untagged C-SBP2 protein (Figure 3C). A wide range of apparent Kd values was observed (from 500 nM to >2000 nM) (Figure 3C).
Figure 3.
Sequences of the RNAs recovered from the SELEX experiment and test of their affinities for C-SBP2. (A) Alignment of the WT yU3B/C RNA sequence with the degenerated N18 RNA and the selected Se1-Se7 RNAs sequences. Nucleotides in Se1-Se7 RNA, are numbered according to the positions of the homolog nucleotides in the WT yU3B/C RNA. The number of sequenced plasmids encoding each selected RNA is indicated in brackets on the right of the sequences. The nucleotides corresponding to the constant sequence are shown in gray, nucleotides in the degenerated sequence and nucleotides mutated during the RT-PCR cycles are shown in black. The GA dinucleotides are underlined. (B) The nucleotide sequences of a series of SECIS motifs from various genes and species (30,52) were aligned with the Se1 RNA sequence taking as references the UGA and GA conserved nucleotides of the K-turn structure (bold characters). A consensus sequence of the SECIS K-turn motifs is deduced from the alignment and indicated below. The positions of the conserved nucleotides in the two strands of helix II are indicated (C) Estimation of the affinity of C-SBP2 for the Se1, Se2, Se3, Se5 and Se7 RNAs by gel-shift assays. RNA–protein complexes formed with 5 fmol of labeled RNA and increasing concentrations of C-SBP2 (as indicated below the lanes) were fractionated by gel electrophoresis as in Figure 1. The apparent Kd values are indicated above the autoradiograms.
The selected RNAs all form canonical K-turn motifs
In order to understand the structural reasons for these different affinities, the possible folding of the seven selected RNAs was investigated. Remarkably, each of them could form a canonical K-turn structure with tandem G.A and A.G base pairs and a 3-nt bulge including a U residue at position 3. Most of the proposed structures were verified by enzymatic probing (Figure 4A). They are represented in Figure 4B, where they are classified according to the values of the established apparent Kds. The free energies of the proposed 2D structures at 37°C in 1 M NaCl were also calculated by using the M-Fold software. Based on these structures, nts 17–21 and 38–39 correspond to residues 1–5 and 6-7 of the K-turn motif. Hence, residues 1 and 2 in the bulge, the A residue of the first G.A pair in stem II and one U residue of the third pair in this stem corresponded to invariant residues in the starting RNA mixture. The G residue of the A.G pair corresponded to the above-mentioned U to G mutation at position 38 in the constant region. This G residue might have been generated by misincorporation in the course of the amplification cycles. Its selection in all the RNAs is in agreement with the high functional importance of the A.G pair in K-turn formation. All the selected RNAs had an identical UGAU sequence from position 19 to 22 in the randomized segment, which demonstrated a strong pressure for the selection of a perfectly canonical K-turn structure with two A.G and G.A pairs, and a U residue at position 3 in the bulge. In all the selected RNAs, except RNA Se3 which contains a G.U pair, the constant U37 residue was always facing a U or a C residue in helix II. Interestingly, a U.U pair was almost always selected at this position of helix II in the SELEX experiment performed with Snu13p (47). Requirement of a non-Watson–Crick pair on top of the G.A and A.G pairs for binding of C-SBP2 may explain the absence of binding of C-SBP2 to yU4 RNA which has a G–C pair at this position in helix II (Figure 2B). Finally, in all the selected Se RNAs, helix II contained at least two Watson–Crick base pairs. They are most frequently (RNAs Se3, 4, 5, 6 and 7) stacked on the three non-Watson–Crick base pairs. In agreement with the absence of binding of C-SBP2 to yU3B/C RNA (Figure 2A), none of the selected RNAs had a bulge in the 3′ strand and a short helix I. In contrast, no restriction on the size of the bulge, or on the length of helices I and II, was found in the SELEX experiment performed with Snu13p (47). Altogether, the data suggested that C-SBP2 binding requires a higher stability of the helices I and II compared to Snu13p binding. Surprisingly, the three selected RNAs, which showed the highest stabilities and also the strongest affinities for C-SBP2, were encoded by DNA sequences that were underrepresented among the cloned DNA sequences. This apparent discrepancy may be explained by the fact that RNAs Se1, 2 and 3 all have different lengths compared to the initial RNAs. They might have been generated in a late step of the selection procedure. The very low affinity found for RNA Se7, which has a stability slightly higher than those of RNAs Se4, Se5 and Se6, might be explained by sequence differences in stem II.
Figure 4.
All the selected RNAs that recognize C-SBP2 can form a K-turn structure. (A) Secondary structure analysis of RNAs Se1, Se3, Se5, Se6 or Se7 by enzymatic probing. The RNAs were 5′-end labeled with 32P, renatured and digested with V1 (0.001 U, lane 2), T1 (0.8 U, lane 3) or T2 (2.4 U, lane 4) RNases, under conditions described in the Materials and Methods section. As a control, undigested RNA was fractionated in parallel (lane 1). Lane L corresponds to the alkaline hydrolysis of the RNA used for localization of the RNase cleavage sites. Electrophoresis was performed on a 10% 8M urea–polyacrylamide gel. Nucleotide positions are indicated on the left. (B) Secondary structure models proposed for the selected RNAs. Models were proposed based on thermodynamic considerations and the results of the enzymatic digestions are shown in A. Regions corresponding to the degenerated sequences are shown by gray characters. For RNAs Se1, 3, 5, 6 and 7, V1, T1 and T2 RNase cleavages are represented by arrows surmounted of squares, dots and triangles, respectively. The color of symbols reflects the intensity of cleavages (gray, dark gray and black for low, medium and strong, respectively). Nucleotide numbering is as in Figure 3A. The apparent Kd values established for each RNA by gel retardation are indicated. The free energies of the proposed secondary structures, expressed in kcal/mol, were calculated by using the M-Fold software.
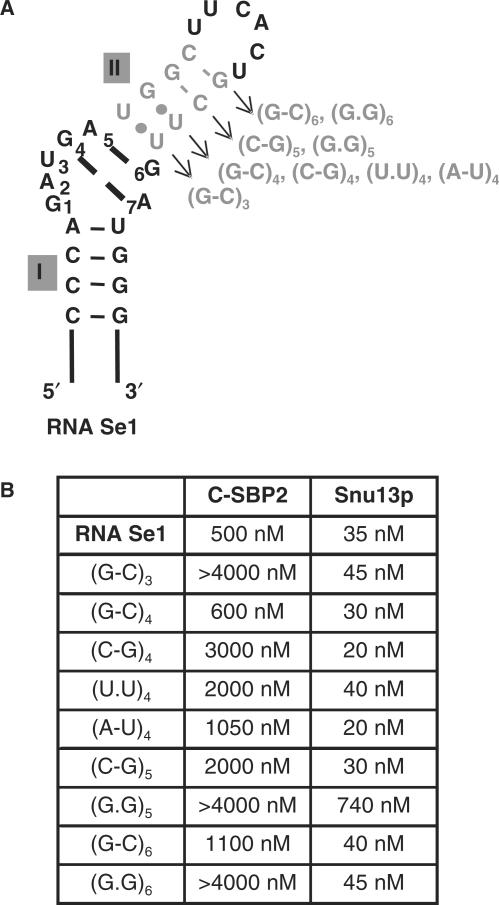
Specific requirements in helix II for efficient binding of protein C-SBP2
Prior to site-directed mutagenesis of Se1 RNA, we tested the influence of Mg++ concentration on C-SBP2 binding to this RNA. Indeed, previous data (42) established the influence of the concentration of this divalent cation on the binding of recombinant SBP2 in vitro. C-SBP2 binding was found to be more sensitive to the presence of Mg++ ions than Snu13p binding. However, the 1.5 mM Mg++ concentration present in the experimental buffer was found to be sufficient to ensure an efficient binding of C-SBP2 on Se1 RNA (Figure 1 in Supplementary Data). Thus subsequent experiments were performed under these conditions. To test the importance of the sequence of helix II for SBP2 binding, we mutated helix II in the winner Se1 RNA. The Se1 RNA variants produced are shown in Figure 5A. Their affinities for C-SBP2 and Snu13p were compared by gel-shift assays. Complexes were formed at different protein concentrations in order to define the apparent Kd values (Figure 5B). Interestingly, Snu13p had a very high affinity for RNA Se1. The estimated Kd (35 nM) was similar to that found for the winner RNAs in the Snu13p SELEX experiment (47). A lower affinity was found for C-SBP2 (Kd of 500 nM). Most of the base substitutions in helix II had no marked effect on Snu13p affinity. Only the strong destabilization of helix II generated by substitution of the fifth Watson–Crick base pair (G-C)5 by a G.G pair had a marked deleterious effect on Snu13p affinity (factor of 20). In contrast, several base substitutions in helix II, (U.U)3 to (G-C)3, (G-C)5 to (G.G)5 and (C-G)6 to (G.G)6 almost abolished C-SBP2 binding. The (G.U)4 to (C-G)4 and, to a lesser extent, the (G.U)4 to (U.U)4 substitutions, also had a marked negative effect. Hence, we concluded that C-SBP2 can interact with canonical K-turn structures, provided that helix II contains a triplet of non-Watson–Crick base pairs including the G.A and A.G sheared pairs and at least two consecutive Watson–Crick base pairs in helix II. In addition, the base pairs on top of the triplet of non-Watson–Crick base pairs should be a Pu.Py pair (G.U, G–C or A–U). This may explain why the Se7 RNA, which has a Py.Pu pair at this position, has a low affinity for protein C-SBP2.
Figure 5.
Mutations in helix II of RNA Se1 are more deleterious for C-SBP2 than for Snu13p binding. (A) Positions of base substitutions in the Se1 RNA are represented in gray on the proposed secondary structure. The nature of the mutations in the variant Se1 RNAs is shown on the right of helix II. (B) The affinities of C-SBP2 and Snu13p for Se1 RNA and its variants were estimated by gel-shift assays using 5′-end labeled RNAs and protein concentrations ranging from 0 to 4000 nM. The apparent Kd values obtained for each of the RNA–protein complexes are indicated.
The presence of a large internal loop instead of the bulge increases C-SBP2 affinity
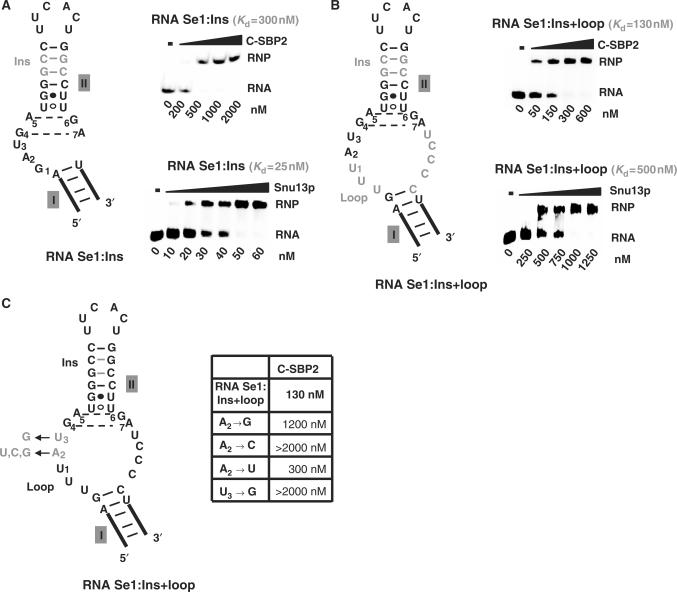
The apparent Kd of the complex formed by C-SBP2 and the winner Se1 RNA was 3-fold lower than that found for the natural SelN RNA (Figures 1 and 5B). Inspection of the 2D structures of these two RNAs suggested two possible explanations for the observed difference of affinity. The presence of a long stem II in SelN RNA, and/or the presence of a large internal loop instead of a bulge in this RNA might increase C-SBP2 affinity. We tested whether the insertion of two Watson–Crick base pairs in helix II of RNA Se1 (RNA Se1:Ins) might increase the affinity of C-SBP2 (Figure 6A). Based on the observed affinities of RNA Se1:Ins for C-SBP2 and Snu13p (apparent Kds of 300 and 25 nM, respectively), the 2 bp insertion only had a limited positive effect on C-SBP2 affinity and no marked effect on Snu13p affinity. When, in addition to the extension of stem II, the bulge of RNA Se1 was converted into a large internal loop (RNA Se1:Ins + loop), the affinity for C-SBP2 was increased by a factor of 4 as compared to RNA Se1. In contrast, the affinity for protein Snu13p was decreased by a factor of 18 (Figure 6B). Hence, the presence of a large internal loop is favorable for C-SBP2 binding, but not for Snu13p interaction.
Figure 6.
A K-turn motif with an extended internal loop increases C-SBP2 affinity. The variant Se1:Ins (A) and Se1:Ins + loop RNAs (B and C) are shown. The additional residues in these variant RNAs compared to Se1 RNA are shown in gray. The affinities of C-SBP2 and Snu13p for Se1:Ins (A) and Se1:Ins + loop (B) were tested by gel-shift assays. Complex formation was performed as described in Figure 1, using 5 fmol of 5′-end labeled RNA and increasing concentrations of C-SBP2 or Snu13p proteins. In Panels A and B, the apparent Kds are indicated above the autoradiograms. (C) The base substitutions generated at positions 2 and 3 in the internal loop of the Se1:Ins + loop RNA are indicated in gray. The table gives the apparent Kd values established by gel-shift assays for complexes formed between C-SBP2 and the variant Se1:Ins + loop RNAs.
Having selected an RNA (Se1:Ins + loop RNA) with an affinity for C-SBP2 similar to that of the authentic SBP2 RNA target (SelN RNA) (Figure 1B), we then tested the effect on C-SBP2 affinity of mutations at positions 2 and 3 in the internal loop of this RNA (Figure 6C). The results obtained revealed a preference for an A and to a lesser extent a U residue at position 2. The strongest negative effect on C-SBP2 affinity was observed for an A to C substitution at position 2 and a U to G substitution at position 3 (Figure 6C). Therefore, the identity of residues at positions 2 and 3 in the internal loop has a strong influence on C-SBP2 affinity.
A large internal loop in the RNA confers a higher stability to C-SBP2–RNA complexes
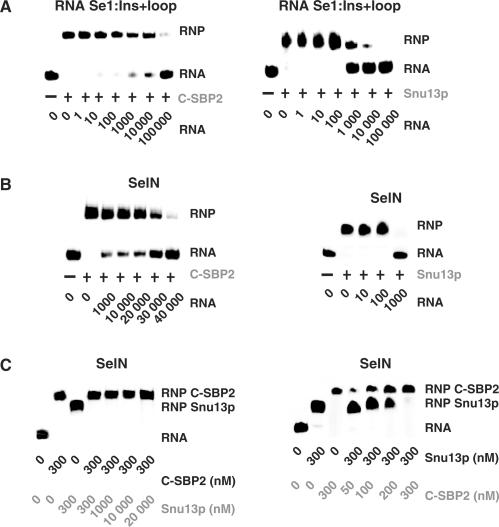
Based on gel-shift experiments, Snu13p and C-SBP2 were found to have similar affinities for RNA SelN (Kds of 180 and 160 nM, respectively) (Figure 1B). However, such apparent Kds, established by gel-shift assays, mostly reflect the capacity of the RNA and protein partners to form a complex which is stable under electrophoresis conditions. Thus, for a better estimation of the stability of the RNP complexes, we used competition experiments. Complexes were formed, as above, with radiolabeled RNA and a protein concentration about twice that of the apparent Kds (300 nM for C-SBP2 and 1000 nM for Snu13p, for assays on Se1:Ins + loop RNA, and two identical protein concentrations, 300 nM, for assays on SelN RNA). Cold RNA was added in excess to destabilize the complex. When complexes were formed with the Se1:Ins + loop RNA (Figure 7), a larger excess of cold Se1:Ins + loop RNA was required to dissociate C-SBP2–RNA complexes compared to Snu13p–RNA complexes and this in spite of the higher Snu13p concentration used to form the initial complex (Figure 7A). Furthermore, a much stronger difference was observed when complexes were formed with the SelN RNA: whereas a 1000-fold molar excess of SelN RNA was sufficient to destabilize the SelN–Snu13p complexes, dissociation of the SelN–C-SBP2 complexes required as much as a 40 000-fold excess of cold SelN RNA (Figure 7B). These observations revealed the high stability of complexes formed with C-SBP2.
Figure 7.
C-SBP2 forms highly stable complexes with RNAs containing an extended internal loop. The stabilities of the complexes formed between C-SBP2 and Snu13p and the Se1:Ins + loop (A) and SelN (B) RNAs were tested by competition experiments. RNA–protein complexes were formed by using 5 fmol of 5′-end labeled Se1:Ins + loop or SelN RNAs and C-SBP2 (300 nM) or Snu13p (1000 or 300 nM). The RNA–protein complexes were challenged with increasing concentrations of cold Se1:Ins + loop or SelN RNAs (10–100 000- and 10–40 000-fold molar excess, respectively, as indicated below the lanes). The remaining complexes were fractionated by gel electrophoresis. (C) Comparison of the relative stabilities of the Snu13p–SelN and C-SBP2–SelN complexes. RNP complexes formed with C-SBP2 at 300 nM were challenged by addition of an excess of Snu13p protein and vice versa. Complexes formed with Snu13p at 300 nM were challenged by addition of an excess of C-SBP2. The remaining complexes were fractionated by gel electrophoresis. The identities and concentrations of the protein competitors used in the assays are indicated below the lanes.
Another approach to verify the high stability of the SelN RNA–C-SBP2 complexes was to destabilize the RNA–protein complex by addition of an excess of a competitor protein (C-SBP2 for complexes formed with Snu13p and vice versa). As seen in Figure 7C, even when added in large excess (65-fold) to the preformed SelN–C-SBP2 complex, Snu13p could not dissociate this complex. In contrast, when C-SBP2 was added at the same concentration as the Snu13p protein used to form the SelN–Snu13p complex, this complex was completely converted into a SelN RNA–C-SBP2 complex. This observation argues in favor of a strong specificity of C-SBP2 for the SECIS RNAs.
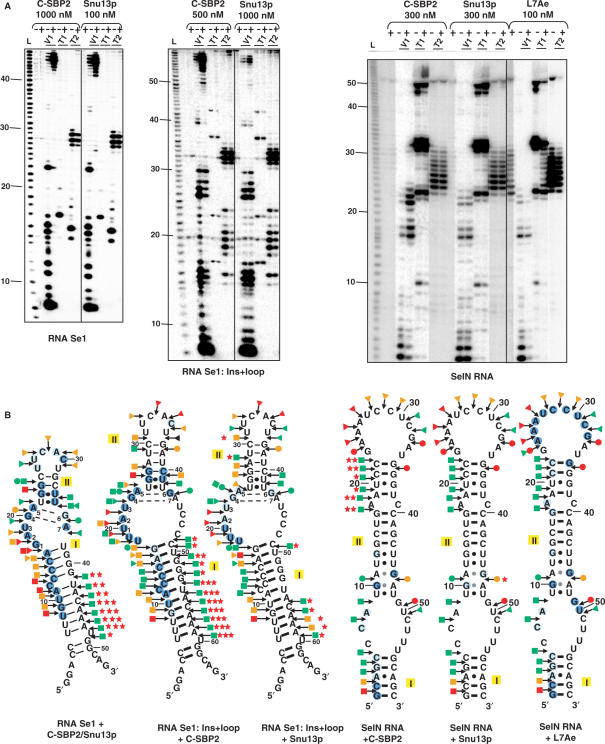
C-SBP2 protects a larger region of the Se1:Ins + loop and SelN RNAs than Snu13p
One possible explanation for the strong stability of complexes formed by protein C-SBP2 and the Se1:Ins + loop and SelN RNAs was the occurrence of more extended RNA–protein contacts in these complexes compared to those formed with Snu13p. To answer this question, we probed the RNA accessibilities in the six RNP complexes formed by the Se1, Se1:Ins + loop and SelN RNAs and each of the C-SBP2 and Snu13p proteins. We used T1 and T2 RNases under conditions such that they cleaved single-stranded regions, and V1 RNase that cleaves specifically double-stranded and stacked RNA regions. Very similar RNA protections were obtained for complexes formed by RNA Se1 and each of the proteins (Figure 8). Both proteins protected the bulge sequence, part of helix II and the 5′ strand of helix I. In contrast, protections of RNAs Se1:Ins + loop and SelN by Snu13p were very limited compared to those found for C-SBP2. Thus, with RNAs containing an extended internal loop, the architecture of C-SBP2 allows tight RNA–protein contacts with both helices and the 5′ strand of the internal loop, which is not the case for Snu13p. Interestingly also, the sensitivity to V1 RNase of the 3′ strand of helix I was strongly increased by binding of C-SBP2 or Snu13p to RNA Se1. The same situation was observed upon binding of C-SBP2 to RNA Se1:Ins + loop (Figure 8). This effect was quite less marked upon Snu13p binding on this RNA. Altogether, this suggested the occurrence of a profound RNA conformational change when Snu13p or C-SBP2 bind RNA Se1 and when C-SBP2 binds RNA Se1:Ins + loop. This strong RNA conformational change is probably not induced upon binding of Snu13p to an RNA with a large internal loop. Binding of C-SBP2 to SelN RNA also induced a hypersensitivity to V1 RNase, but the RNA segment concerned was different (extremity of the 5′ strand of helix II). No significant hypersensitivity to V1 RNase was observed upon Snu13p binding to SelN RNA, which reinforces the idea that only C-SBP2 can establish tight contacts with RNAs containing a large internal loop and as a consequence remodel their conformation. The archaeal protein L7Ae is known to interact with both canonical K-turn and K-loop structures formed in terminal loops (9–11,15,25,48). Thus, by footprinting assays, we tested whether L7Ae can establish a tight interaction with the SelN RNA, as does C-SBP2 (Figure 8). The apparent Kd established by gel-shift assays for the SelN–L7Ae complex revealed a high affinity (Kd of 35 nM, not shown). According to enzymatic footprinting assays (Figure 8), this high affinity may be due to the presence of two L7Ae-binding sites in SelN RNA: one of them corresponds to the quartet of non-Watson–Crick base pairs, the other one to the terminal loop. Due to the presence of two G.A dinucleotides in this loop, a K-loop recognized by protein L7Ae can be formed. Interestingly, the protections found in the 5′ strand of the internal loop, helix I, and the quartet of non-Watson–Crick base pairs, are very similar in the C-SBP2–SelN and L7Ae–SelN complexes. Protein L7Ae protects two additional residues in the 3′ strand of the internal loop as compared to C-SBP2. Hence, concerning the recognition of RNAs with an internal loop, the behavior of protein L7Ae is closer to that of C-SBP2 than that of Snu13p.
Figure 8.
C-SBP2 protects a larger region of the Se1:Ins + loop and SelN RNAs than Snu13p. (A) The in vitro transcribed 5′-end labeled Se1, Se1:Ins + loop and SelN RNAs (25 fmol) were incubated in the absence (−) or presence (+) of C-SBP2, Snu13p or L7Ae. The protein concentrations used in the assays are indicated above each panel, 2 µg of tRNA were added in each assay and the digestion was carried out for 6 min at 20°C, in buffer D, in the presence of 0.8 U RNase T1, 2.4 U RNase T2 or 0.001 U RNase V1, as described in the Materials and Methods section. The cleavage products were fractionated on a 10% polyacrylamide–8M urea gel. L: alkaline hydrolysis. Nucleotide positions are indicated on the left. (B) Schematic representation of the results shown in panel A on the secondary structures proposed for the three studied RNAs. Helices I and II are indicated. V1, T1 and T2 RNase cleavages are represented by arrows surmounted of squares, dots and triangles, respectively. The color of symbols reflects the intensity of cleavages (green, orange and red for low, medium and strong, respectively). Nucleotides with decreased sensitivity to RNase in the presence of the proteins are circled in blue (pale and dark for low and strong protection, respectively). Nucleotides with increased sensitivity to RNase in the presence of the proteins are indicated by a red star. The number of stars reflects the increased sensitivity to cleavage (one, two and three representing low, medium and strong, respectively).
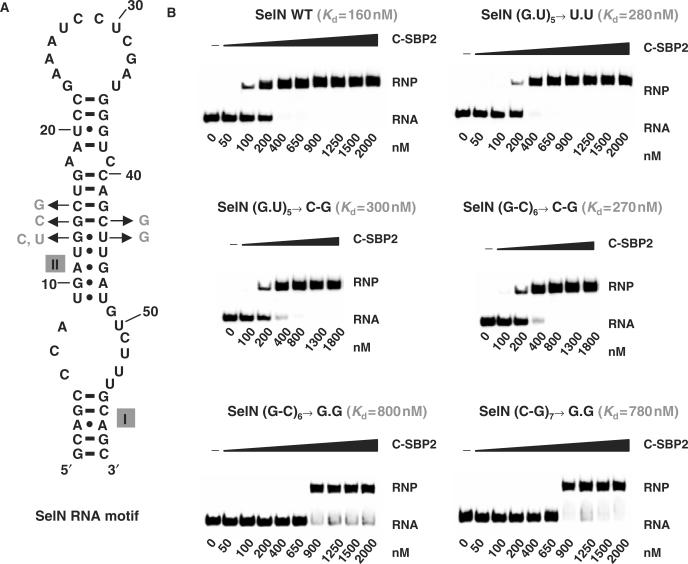
Mutations in helix II of SelN RNA limit C-SBP2 affinity
Since our data suggested a functional importance of helix II for C-SBP2 binding, we tested the effects of mutations in helix II of the authentic SelN SECIS motif on C-SBP2 binding. Substitution of the fifth G.U pair in helix II by a C–G pair as well as substitution of the sixth G–C pair by C–G pair, had less negative effects on C-SBP2 binding (factor of 2) (Figure 9) compared to those found for the corresponding substitution in RNA Se1 (factor of 4) (Figure 5). However, substitutions of the sixth G–C pair and of the seventh C–G pair by G.G pairs had strong negative effects on C-SBP2 binding (Kds of 800 and 780 nM instead of 160 nM for the WT RNA). Therefore, mutations in an authentic SECIS RNA confirmed our observation of the importance of the stability and the sequence of helix II for C-SBP2 binding. Accordingly, Pu–Py pairs are the most frequently observed base pairs at the fifth and sixth positions in helix II of SECIS elements (Figure 3B).
Figure 9.
The sequence and stability of helix II are important for C-SBP2 binding onto SelN RNA. (A) The base-pair substitutions generated at positions 5, 6 and 7 in helix II of the SelN RNA are shown. (B) Complexes were formed with 5 fmol of radiolabeled WT or mutated SelN RNA and increasing concentrations of the C-SBP2 protein (from 50 to 2000 nM). The RNP complexes were fractionated on 6% polyacrylamide 8–M urea gel and apparent dissociation constants were determined by measuring the radioactivity in the bands of gel corresponding to free RNA and the RNP. The determined Kds are indicated above each autoradiogram.
DISCUSSION
The present data based on SELEX and site-directed mutagenesis experiments improve our understanding of the sequence and structural features required for efficient interaction of SBP2 with RNA. These findings bring new insights that will facilitate the understanding of its mechanism of action in the selenocysteine incorporation machinery.
When used for studying RNA–protein interactions, the SELEX approach most generally leads to the establishment of an RNA consensus sequence. Here, despite the wide diversity of the initial RNA mixture (184), only seven different sequences were selected, and several of them were very similar. All of them folded into very similar 2D structures that contained a canonical K-turn motif. This limited diversity of the selected sequences indicated narrow RNA structure requirements for efficient binding of SBP2. We confirmed this hypothesis by several experimental approaches.
Dual Mg++ dependence of SBP2 binding to different RNA substrates
Earlier work (49) established that SBP2 contained in testis extracts displayed high sensitivity to Mg++ concentration for SECIS binding, the IC50 being around 4 mM. This sensitivity was however less pronounced (IC50 > 20 mM) with a shorter, recombinant version of SBP2, and PHGPx SECIS RNA as the RNA partner (42). Interestingly, here we found that binding of C-SBP2 to the Se1 RNA, which forms a canonical K-turn structure, requires a 1.5 mM Mg++ concentration, higher concentrations being innocuous. At first glance, the two series of results may appear contradictory. Nevertheless, these differential behaviors toward Mg++ are likely explained by the use of different RNA partners. Se1 RNA is a genuine K-turn RNA, and it is known that divalent cations favor the closed conformation of canonical K-turn motifs (50). SECIS RNAs possess a large internal loop and thus contain a K-turn like motif (32). A high Mg++ concentration may induce a conformational change into SECIS RNAs, which is not favorable for SBP2 binding. For instance, based on our data, we can imagine that a high Mg++ concentration promotes closing of the internal loop, and we show that a large internal loop is needed for maximum binding efficiency of SBP2. The Se1 RNA is a typical Snu13p partner. As expected, no marked Mg++ requirement is observed for Snu13p binding to this RNA. In contrast, as Se1 RNA does not contain an internal loop, a prior stabilization of the kink structure may be needed to reinforce SBP2 binding. Altogether, the previous and present data strongly suggest that each member of the L7A/L30 family is perfectly suited for binding to its authentic partner at the physiological concentration of divalent cations. When RNA partners are exchanged in in vitro experiments, the Mg++ ion concentration has to be adapted in order to form the heterologous interaction. Accordingly, a high Mg++ ion concentration was found to be required for efficient in vitro binding of protein L30 to a SECIS element, in the presence of SBP2 (42).
Specific sequence requirements in helix II
Site-directed mutagenesis, performed on the winner Se1 RNA obtained by SELEX experiments, demonstrated that binding of C-SBP2 requires the presence of a stable helix II containing at least two Watson–Crick base pairs. In agreement with this observation, all the SECIS elements identified so far contain a series of Watson–Crick base pairs on top of the non-Watson–Crick base-pair quartet (30,51–53; A.K., unpublished data). Accordingly, we showed that their individual disruption in SelN RNA decreases C-SBP2 affinity. Not only helix II stability but also its sequence has an influence on C-SBP2 affinity. The presence of a Pu–Py pair at the fourth position in helix II was found to be of high importance for C-SBP2 binding to Se1 RNA, a Pu–Py pair at this position being also more favorable for C-SBP2 binding to SelN RNA. This is in contrast with the absence of sequence requirement in helix II, except for the G.A and A.G base pairs and the adjacent U.U pair found for proteins Snu13p/15.5K and L7Ae (6,7,16,24,47,54–56). Up to now, little attention was given to the importance of the identity of base pairs in the upper part of helix II of SECIS elements. However, at position 4 of helix II, a Pu residue (most frequently a G residue) is almost always found in the 5′ strand and a Py residue (most frequently a C residue) is observed in the 3′ strand (30,51–53; A.K., unpublished data). Although less strictly conserved, the fifth base pair in helix II is predominantly a Pu–Py pair (Figure 3B). Together with our experimental data, these phylogenetic observations strongly suggest a functional importance of these conserved Pu.Py base pairs at positions 4 and 5 in helix II. In accordance with this hypothesis, the G–C pair at the fourth position in RNA Se1 was protected against V1 RNase digestion in the complex formed with C-SBP2, but not in the complex formed with Snu13p (Figure 8). Accordingly, the very limited V1 RNase cleavage, which is located between residues G13 and G14 in free SelN RNA, disappeared in the presence of C-SBP2, but not with Snu13p. Remarkably, this V1 RNase cleavage was also abolished in the presence of protein L7Ae.
Comparison of the Se1 to Se7 RNAs and site-directed mutagenesis of the Se1 RNA also show the importance for a non-Watson–Crick base pair on top of the A.G and G.A pair tandem (Figures 4 and 5). Accordingly, U.U pairs are frequently encountered pairs at this position in SECIS elements (30,51–53; A.K., unpublished data) and a U.U pair was also preferentially selected at this position of helix II, in the SELEX experiment performed with Snu13p. The presence of a U.U base pair at this position, with a C1′–C1′ distance of the ribose ring close to that in G.A pairs, is very likely required to favor the smooth transition from the non-Watson–Crick to the Watson–Crick section of helix II. Noticeably also, in K-turn structures found in ribosomal RNAs, the nucleobase of one of this unpaired couple of nucleotides was proposed to interact with one nucleobase in helix I, and thus to reinforce the inter-helical angle between helix I and helix II (57).
Importance of a large SECIS internal loop
Increasing the size of both helix II and the internal loop of the winner Se1 RNA obtained by SELEX, yielded an RNA with an affinity similar to that of SelN RNA (Figure 6B). Such an RNA could not be obtained in the SELEX experiment, because of limitation in size of the degenerated sequence that can be used (18 nt) in these experiments. Indeed, due to the necessity to cover all the possible sequences during the screening, one cannot use largely extended degenerated sequences (58,59). In agreement with the importance of the size of helix II, all the identified SECIS elements contain a long helix II. Based on our footprinting data, the high affinity of C-SBP2 for RNAs with an internal loop, as well as the stability of the complexes formed, are due to its capacity to contact helices I and II and the 5′ strand of the internal loop in these RNAs (Figure 8). Interestingly, Martin et al. (38) showed that closing of the internal loop of the rat D1 SECIS element almost completely abolished selenocysteine incorporation in vivo. In agreement with the observed requirement of at least one base pair closing the 3-nt bulge loop of K-turn motifs for efficient binding of Snu13p (44), Snu13p establishes very loose contacts with RNAs containing an internal loop. The presence of a closing base pair is not required for L7Ae and this protein is able to bind open K-loop structures (9,44,48). The presence of an arginine at position 95 in the 15.5K/Snu13p protein, that forms a salt bridge with the 5′ phosphate of the residue at position 1 in the bulge, and its replacement by a valine in L7Ae, were proposed to explain this difference between proteins 15.5K/Snu13p and L7Ae (60). Interestingly, like L7Ae, SBP2 contains a valine at the corresponding position in the L7A/L30 domain (29). This may explain our observation of similar binding properties of proteins SBP2 and L7Ae on RNAs containing a large internal loop.
In free RNAs containing a canonical K-turn structure with a bulge loop, helices I and II form a 76° angle. Upon Snu13p/15.5K binding, the RNA undergoes further folding, so that the helix I–helix II angle is reduced to 48° (56,61). This folding likely explains the tight contact of Snu13p with both helices of RNA Se1 that we detected by footprinting assay. As very similar footprinting results were obtained with C-SBP2, it probably also induces a folding of this RNA. However, C-SBP2 but not Snu13p may induce a similar folding in both the Se1:Ins + loop and SelN RNAs.
Sequence requirement in the internal loop
Our site-directed mutagenesis experiments on the Se1:Ins + loop RNA revealed the importance of the identity of residues at positions 2 and 3 in the internal loop for C-SBP2 binding. The most deleterious base substitution at position 2 was the A to C replacement. Interestingly, a 66% decrease of selenocysteine incorporation was observed when the same A to C substitution was generated in the SECIS element of the rat GPx mRNA while A to G and A to U changes only led to a loss of 30 and 22% of the incorporation, respectively (52). In accordance with the decrease of the C-SBP2 affinity upon U to G substitution at position 3 in RNA Se1:Ins + loop, selenocysteine incorporation was decreased by 88% when this base substitution was generated in the SECIS element of the rat GPx mRNA (38). In addition, a U to C substitution at this position abolished the binding of SBP2 to SelN RNA and is responsible for a human genetic disease, the rigid spine muscular dystrophy (62). As the residue at position 3 in canonical K-turns is located in the protein pocket, its mutation also has a deleterious effect on 15.5K/Snu13p and L7Ae binding (6,24–26). Residues E61 and K86 in 15.5K, and D54 and K79 in Archaeoglobus fulgidus L7Ae, are involved in the interaction with the nucleobase at position 3. Their counterparts in SBP2 (E699 and R730) probably play a similar role, since they are crucial for binding to SECIS RNAs (5,29,56). The specificity of L7Ae/L30 protein members towards the residue at position 2 in the K-turn motif is variable. Whereas an A or G residue at position 2 increases 15.5K/Snu13p affinity, substitutions at position 2 have no marked effect on L7Ae affinity (25). Concerning position 2, SBP2 exhibits a behavior closer to that of 15.5K/Snu13p than to L7Ae.
The ribosomal protein L30 was recently shown to compete with SBP2 for binding to SECIS RNA (42). L30 was found to recognize a K-turn structure of its pre-mRNA that contains a protruding A residue in a small internal loop (21–23). SELEX experiments performed with L30 revealed its preference for K-turn motifs with protruding C or A residues (27). Later, it was shown that L30 also has the ability to accommodate K-turn structures with a protruding G (28). However, its interaction with K-turn motifs containing a protruding U residue has not been demonstrated yet. Consequently, the binding of L30 to SECIS elements, that all contain a U residue at position 3, raises the question of how it can achieve this.
CONCLUSION
Assembly of the selenocysteine incorporation machinery is proposed to be initiated by SBP2 association to SECIS elements in the nucleus, and more likely in the nucleolus (63). Protein 15.5K/Snu13p is abundant in the nucleolus and SBP2 shares several common features with Snu13/15.5K. However, our data reveal important differences in RNA specificities that may ensure the specific association of SBP2 to SECIS elements in the nuclear compartment.
Supplementary Material
ACKNOWLEDGEMENTS
V. Senty-Ségault and S. Massenet are acknowledged for helpful discussions. S. Sonkaria is thanked for careful reading of the manuscript. A. Cléry was a fellow from the French ‘Ministère de la Recherche et des Nouvelles Technologies’. The work was supported by the Centre National de la Recherche Scientifique, the French ‘Ministère de la Recherche et des Nouvelles Technologies’, the ACI ‘Biologie Cellulaire, Moléculaire et Structurale’ n°BCMS226, the PRST ‘Bioingénierie’ of the ‘Conseil Régional Lorrain’ and the ToxNuc-E Programme ‘Toxicologie Nucléaire Environnementale’. Funding to pay the Open Access publication charge was provided by CNRS-Sciences de la vie.
Conflict of interest statement. None declared.
REFERENCES
- 1.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 3.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 4.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 5.Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 6.Nottrott S, Hartmuth K, Fabrizio P, Urlaub H, Vidovic I, Ficner R, Luhrmann R. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 1999;18:6119–6133. doi: 10.1093/emboj/18.21.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, et al. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 8.Terns MP, Terns RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- 9.Charpentier B, Muller S, Branlant C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 2005;33:3133–3144. doi: 10.1093/nar/gki630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omer AD, Ziesche S, Ebhardt H, Dennis PP. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5289–5294. doi: 10.1073/pnas.082101999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31:869–877. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granneman S, Pruijn GJ, Horstman W, van Venrooij WJ, Luhrmann R, Watkins NJ. The hU3-55K protein requires 15.5K binding to the box B/C motif as well as flanking RNA elements for its association with the U3 small nucleolar RNA in Vitro. J. Biol. Chem. 2002;277:48490–48500. doi: 10.1074/jbc.M206631200. [DOI] [PubMed] [Google Scholar]
- 13.Nottrott S, Urlaub H, Luhrmann R. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J. 2002;21:5527–5538. doi: 10.1093/emboj/cdf544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watkins NJ, Dickmanns A, Luhrmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 2002;22:8342–8352. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker DL, Youssef OA, Chastkofsky MI, Dy DA, Terns RM, Terns MP. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev. 2005;19:1238–1248. doi: 10.1101/gad.1309605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran EJ, Zhang X, Maxwell ES. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J. 2003;22:3930–3940. doi: 10.1093/emboj/cdg368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rashid R, Aittaleb M, Chen Q, Spiegel K, Demeler B, Li H. Functional requirement for symmetric assembly of archaeal box C/D small ribonucleoprotein particles. J. Mol. Biol. 2003;333:295–306. doi: 10.1016/j.jmb.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Bortolin ML, Bachellerie JP, Clouet-d'Orval B. In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNA(Trp) Nucleic Acids Res. 2003;31:6524–6535. doi: 10.1093/nar/gkg860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultz A, Nottrott S, Hartmuth K, Luhrmann R. RNA structural requirements for the association of the spliceosomal hPrp31 protein with the U4 and U4atac small nuclear ribonucleoproteins. J. Biol. Chem. 2006;281:28278–28286. doi: 10.1074/jbc.M603350200. [DOI] [PubMed] [Google Scholar]
- 20.Koonin EV, Bork P, Sander C. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 1994;22:2166–2167. doi: 10.1093/nar/22.11.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vilardell J, Warner JR. Regulation of splicing at an intermediate step in the formation of the spliceosome. Genes Dev. 1994;8:211–220. doi: 10.1101/gad.8.2.211. [DOI] [PubMed] [Google Scholar]
- 22.Chao JA, Williamson JR. Joint X-ray and NMR refinement of the yeast L30e-mRNA complex. Structure. 2004;12:1165–1176. doi: 10.1016/j.str.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 23.Mao H, White SA, Williamson JR. A novel loop-loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nat. Struct. Biol. 1999;6:1139–1147. doi: 10.1038/70081. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn JF, Tran EJ, Maxwell ES. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 2002;30:931–941. doi: 10.1093/nar/30.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charron C, Manival X, Clery A, Senty-Segault V, Charpentier B, Marmier-Gourrier N, Branlant C, Aubry A. The archaeal sRNA binding protein L7Ae has a 3D structure very similar to that of its eukaryal counterpart while having a broader RNA-binding specificity. J. Mol. Biol. 2004;342:757–773. doi: 10.1016/j.jmb.2004.07.046. [DOI] [PubMed] [Google Scholar]
- 26.Marmier-Gourrier N, Clery A, Senty-Segault V, Charpentier B, Schlotter F, Leclerc F, Fournier R, Branlant C. A structural, phylogenetic, and functional study of 15.5-kD/Snu13 protein binding on U3 small nucleolar RNA. RNA. 2003;9:821–838. doi: 10.1261/rna.2130503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, White SA. RNA apatamers for yeast ribosomal protein L32 have a conserved purine-rich internal loop. RNA. 1997;3:245–254. [PMC free article] [PubMed] [Google Scholar]
- 28.White SA, Hoeger M, Schweppe JJ, Shillingford A, Shipilov V, Zarutskie J. Internal loop mutations in the ribosomal protein L30 binding site of the yeast L30 RNA transcript. RNA. 2004;10:369–377. doi: 10.1261/rna.2159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allmang C, Carbon P, Krol A. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA. 2002;8:1308–1318. doi: 10.1017/s1355838202020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fletcher JE, Copeland PR, Driscoll DM, Krol A. The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA. 2001;7:1442–1453. [PMC free article] [PubMed] [Google Scholar]
- 31.Walczak R, Carbon P, Krol A. An essential non-Watson–Crick base pair motif in 3′UTR to mediate selenoprotein translation. RNA. 1998;4:74–84. [PMC free article] [PubMed] [Google Scholar]
- 32.Allmang C, Krol A. Selenoprotein synthesis: UGA does not end the story. Biochimie. 2006;88:1561–1571. doi: 10.1016/j.biochi.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 33.Krol A. Evolutionarily different RNA motifs and RNA-protein complexes to achieve selenoprotein synthesis. Biochimie. 2002;84:765–774. doi: 10.1016/s0300-9084(02)01405-0. [DOI] [PubMed] [Google Scholar]
- 34.Gladyshev VN. Selenium in biology and human health: controversies and perspectives. In: Hatfield DL, editor. Selenium: Its Molecular Biology and Role in Human Health. Boston: Kluwer; 2001. pp. 313–317. [Google Scholar]
- 35.Flohe L, Andreesen JR, Brigelius-Flohe R, Maiorino M, Ursini F. Selenium, the element of the moon, in life on earth. IUBMB Life. 2000;49:411–420. doi: 10.1080/152165400410263. [DOI] [PubMed] [Google Scholar]
- 36.Rayman MP. The importance of selenium to human health. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 37.Rederstorff M, Krol A, Lescure A. Understanding the importance of selenium and selenoproteins in muscle function. Cell Mol. Life Sci. 2006;63:52–59. doi: 10.1007/s00018-005-5313-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin GW, III, Harney JW, Berry MJ. Functionality of mutations at conserved nucleotides in eukaryotic SECIS elements is determined by the identity of a single nonconserved nucleotide. RNA. 1998;4:65–73. [PMC free article] [PubMed] [Google Scholar]
- 39.Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, Driscoll DM, Hatfield DL, Berry MJ. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zavacki AM, Mansell JB, Chung M, Klimovitsky B, Harney JW, Berry MJ. Coupled tRNA(Sec)-dependent assembly of the selenocysteine decoding apparatus. Mol. Cell. 2003;11:773–781. doi: 10.1016/s1097-2765(03)00064-9. [DOI] [PubMed] [Google Scholar]
- 42.Chavatte L, Brown BA, Driscoll DM. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 2005;12:408–416. doi: 10.1038/nsmb922. [DOI] [PubMed] [Google Scholar]
- 43.Mougin A, Gottschalk A, Fabrizio P, Luhrmann R, Branlant C. Direct probing of RNA structure and RNA-protein interactions in purified HeLa cells and yeast spliceosomal U4/U6.U5 tri-snRNP particles. J. Mol. Biol. 2002;317:631–649. doi: 10.1006/jmbi.2002.5451. [DOI] [PubMed] [Google Scholar]
- 44.Charron C, Manival X, Charpentier B, Branlant C, Aubry A. Purification, crystallization and preliminary X-ray diffraction data of L7Ae sRNP core protein from Pyrococcus abyssii. Acta Crystallogr. D Biol. Crystallogr. 2004;60:122–124. doi: 10.1107/s090744490302239x. [DOI] [PubMed] [Google Scholar]
- 45.Lescure A, Allmang C, Yamada K, Carbon P, Krol A. cDNA cloning, expression pattern and RNA binding analysis of human selenocysteine insertion sequence (SECIS) binding protein 2. Gene. 2002;291:279–285. doi: 10.1016/s0378-1119(02)00629-7. [DOI] [PubMed] [Google Scholar]
- 46.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 47.Clery A, Senty-Segault V, Leclerc F, Raue HA, Branlant C. Analysis of sequence and structural features that identify the B/C motif of U3 small nucleolar RNA as the recognition site for the Snu13p-Rrp9p protein pair. Mol. Cell. Biol. 2007;27:1191–1206. doi: 10.1128/MCB.01287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nolivos S, Carpousis AJ, Clouet-d'Orval B. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res. 2005;33:6507–6514. doi: 10.1093/nar/gki962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Copeland PR, Driscoll DM. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J. Biol. Chem. 1999;274:25447–25454. doi: 10.1074/jbc.274.36.25447. [DOI] [PubMed] [Google Scholar]
- 50.Matsumura S, Ikawa Y, Inoue T. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 2003;31:5544–5551. doi: 10.1093/nar/gkg760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 52.Fagegaltier D, Lescure A, Walczak R, Carbon P, Krol A. Structural analysis of new local features in SECIS RNA hairpins. Nucleic Acids Res. 2000;28:2679–2689. doi: 10.1093/nar/28.14.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lescure A, Gautheret D, Carbon P, Krol A. Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J. Biol. Chem. 1999;274:38147–38154. doi: 10.1074/jbc.274.53.38147. [DOI] [PubMed] [Google Scholar]
- 54.Szewczak LB, DeGregorio SJ, Strobel SA, Steitz JA. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 2002;9:1095–1107. doi: 10.1016/s1074-5521(02)00239-9. [DOI] [PubMed] [Google Scholar]
- 55.Szewczak LB, Gabrielsen JS, Degregorio SJ, Strobel SA, Steitz JA. Molecular basis for RNA kink-turn recognition by the h15.5K small RNP protein. RNA. 2005;11:1407–1419. doi: 10.1261/rna.2830905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–818. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 57.Razga F, Spackova N, Reblova K, Koca J, Leontis NB, Sponer J. Ribosomal RNA kink-turn motif – a flexible molecular hinge. J. Biomol. Struct. Dyn. 2004;22:183–194. doi: 10.1080/07391102.2004.10506994. [DOI] [PubMed] [Google Scholar]
- 58.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 59.Klug SJ, Famulok M. All you wanted to know about SELEX. Mol. Biol. Rep. 1994;20:97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- 60.Hamma T, Ferre-D'Amare AR. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 A resolution. Structure. 2004;12:893–903. doi: 10.1016/j.str.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 61.Wozniak AK, Nottrott S, Kuhn-Holsken E, Schroder GF, Grubmuller H, Luhrmann R, Seidel CA, Oesterhelt F. Detecting protein-induced folding of the U4 snRNA kink-turn by single-molecule multiparameter FRET measurements. RNA. 2005;11:1545–1554. doi: 10.1261/rna.2950605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allamand V, Richard P, Lescure A, Ledeuil C, Desjardin D, Petit N, Gartioux C, Ferreiro A, Krol A, et al. A single homozygous point mutation in a 3′untranslated region motif of selenoprotein N mRNA causes SEPN1-related myopathy. EMBO Rep. 2006;7:450–454. doi: 10.1038/sj.embor.7400648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Jesus LA, Hoffmann PR, Michaud T, Forry EP, Small-Howard A, Stillwell RJ, Morozova N, Harney JW, Berry MJ. Nuclear assembly of UGA decoding complexes on selenoprotein mRNAs: a mechanism for eluding nonsense-mediated decay? Mol. Cell. Biol. 2006;26:1795–1805. doi: 10.1128/MCB.26.5.1795-1805.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.