Abstract
Escherichia coli 6S RNA represents a non-coding RNA (ncRNA), which, based on the conserved secondary structure and previous functional studies, had been suggested to interfere with transcription. Selective inhibition of sigma-70 holoenzymes, preferentially at extended −10 promoters, but not stationary-phase-specific transcription was described, suggesting a direct role of 6S RNA in the transition from exponential to stationary phase. To elucidate the underlying mechanism, we have analysed 6S RNA interactions with different forms of RNA polymerase by gel retardation and crosslinking. Preferred binding of 6S RNA to Eσ70 was confirmed, however weaker binding to Eσ38 was also observed. The crosslinking analysis revealed direct contact between a central 6S RNA sequence element and the β/β′ and σ subunits. Promoter complex formation and in vitro transcription analysis with exponential- and stationary-phase-specific promoters and the corresponding holoenzymes demonstrated that 6S RNA interferes with transcription initiation but does not generally distinguish between exponential- and stationary-phase-specific promoters. Moreover, we show for the first time that 6S RNA acts as a template for the transcription of defined RNA molecules in the absence of DNA. In conclusion, this study reveals new aspects of 6S RNA function.
INTRODUCTION
6S RNA, first discovered in Escherichia coli in the late 1960s, has in the meantime achieved considerable attention, supported particularly by the obvious widespread distribution of this molecule among diverse bacteria. More than 100 potential 6S RNAs have been identified by bioinformatics procedures, many of which have been verified experimentally as stably expressed RNAs (1–3). One unifying element of 6S RNAs is the capacity to fold into a characteristic secondary structure. This secondary structure consists of a central region, characterized by a largely single-stranded internal loop, which is flanked by two long irregular double-stranded stem regions, which are interrupted by small bulge loops. This structure, initially predicted for 6S RNA from E. coli by theoretical folding programs, and recently demonstrated by biochemical structural analysis to be largely correct, has been of great advantage to screen for potential 6S RNA molecules from sequence databases (1,4). The secondary structure, which bears great similarity with a partially single-stranded DNA bubble, characteristic for transcribing RNA polymerase–DNA complexes, has immediately led to a hypothesis for the potential function of 6S RNA (5,6). Supported by the observation that 6S RNA, which exists in the cell as nucleoprotein complex (7), forms a stable complex with RNA polymerase, it was concluded that 6S RNA acts as an open promoter DNA mimicry, interfering with the formation of transcription initiation complexes. Together with the observation that 6S RNA levels increase ∼10-fold during stationary phase (5) it was plausible to suggest a function of 6S RNA in the specificity switch of RNA polymerase from exponential to stationary phase. This view has been strengthened by the finding that 6S RNA interacts preferentially with RNA polymerase holoenzymes formed with the exponential-phase-specific sigma factor σ70 (Eσ70). No such interactions could be demonstrated so far to occur with the corresponding holoenzyme containing the stationary-phase-specific sigma factor σ38, which is responsible for the transcription during stationary growth. Moreover, hitherto existing transcription analysis had shown that σ70-specific promoters, exhibiting an extended −10 motif are especially susceptible for 6S RNA inhibition, while for certain σ38-dependent promoters an activation had been measured (8). 6S RNA has since then been considered to participate in shifting global gene expression from exponential to stationary phase. Although this is an attractive hypothesis, the molecular details for this selective regulation have not yet been worked out.
In this study, we have conducted experiments for a better understanding of the molecular mechanisms underlying 6S RNA specificity and function. In particular we wished to learn how 6S RNA binds to, and discriminates between different RNA polymerase holoenzymes. To this aim, binding studies of 6S RNA to the different Eσ70 and Eσ38 RNA polymerase holoenzymes, RNA polymerase core or the isolated sigma subunits were performed by gel retardation and crosslinking studies. Structural details of the complexes were determined by identifying 6S RNA nucleotides in direct contact with RNA polymerase. Moreover, 6S RNA function was analysed in vitro by transcription interference assays, employing exponential- and stationary-phase-specific promoters on linear and superhelical templates with isolated Eσ70 and Eσ38 holoenzymes. In extension to previous reports, our results show that 6S RNA binds to all forms of RNA polymerase. It has, however, a clear preference for the Eσ70 holoenzyme. We show that the downstream strand of the central loop and parts of the flanking stem regions are involved in RNA polymerase binding, presumably to the β/β′ and σ subunits. The in vitro transcription studies reveal that 6S RNA is capable of inhibiting the formation of initiation complexes with both, exponential- and stationary-phase-specific promoters. Hence, the results clearly indicate that 6S RNA does not generally distinguish between exponential- and stationary-phase-specific transcription complexes. Apparently, additional promoter characteristics or different mechanisms for this specificity switch must be involved.
During in vitro transcription, we made the interesting observation that in the absence of any DNA template 6S RNA causes the de novo transcription of defined RNA molecules. Apparently, 6S RNA itself is able to act as a template, which clearly supports the promoter DNA mimicry model. Whether or not these transcripts are of functional importance remains to be shown. Taken together, our study suggests that the function of 6S RNA in the cell may be of much higher complexity as originally envisaged.
MATERIALS AND METHODS
Plasmid construction
A plasmid vector pUC18-T7-6S for the rapid isolation of 6S RNA was constructed in the following way. The complete ssrS gene was obtained as a PCR fragment from MG1655 DNA with the upstream oligonucleotides 5′-ATT TCT CTG AGA TGT TCG CAA GCG-3′, complementary to the 6S RNA 5′ end and the downstream oligonucleotide, 5′-CCT GGA ATC TCC GAG ATG CCG C-3′, complementary to the 6S RNA 3′ end with three additional nucleotides creating a StuI site for blunt-end restriction hydrolysis. The PCR fragment was cloned into the StuI site of vector pUC18-T7. This vector contained a 388 bp BglII/HindIII fragment with the phage T7 ϕ 10 promoter cloned into pUC18 (9).
Plasmids pRT3HΔP2, a pBR322 derivative with the rrnB P1 promoter, directing transcription of a truncated part of the 16S and the 5S rRNA followed by the tandem rrnB terminators T1, T2 was used as a σ70-dependent template. Plasmid pbolAT1T2, which carries the E. coli bolA promoter upstream of the tandem rrnB terminators, was used as a template for σ38-dependent in vitro transcription.
Preparation of purified 6S RNA
Large amounts of 6S RNA were obtained by multiple round run-off transcription with StuI linearized plasmid pUC18-T7-6S and T7 RNA polymerase. Here, 10–50 μCi α-[32P]UTP was employed to obtain statistically labelled transcripts. 3′-end-labelled 6S RNA was obtained by ligase catalysed addition of [32P]pCp according to Göringer et al. (10). 6S RNA was routinely purified on denaturing polyacrylamide gels as described (10).
Gel retardation assay
Binding of 9 nM radiolabelled 6S RNA to different RNA polymerase preparations was analysed on native 5% polyacrylamide gels. Reaction mixtures contained 3 nM of the respective purified RNA polymerase core, holoenzymes or σ subunits in a total volume of 10 μl 80 mM K-glutamate, 50 mM Tris-acetate, pH 8.0, 10 mM Mg-acetate, 0.5 mM DTT, 0.1 mM EDTA and 100 μg/ml acetylated BSA. Binding reactions were incubated for 20 min at 30°C and heparin was added at the indicated concentrations. Samples were separated on non-denaturing 5% polyacrylamide gels and complexes, and free 6S RNAs were visualized by autoradiography.
UV crosslinking
Here, 100 ng of radiolabelled 6S RNA (1 × 106 cpm) was incubated with 3 μg RNA polymerase holoenzymes (Eσ70, Eσ38, core enzyme or isolated sigma factors) in 80 mM K-glutamate, 50 mM Tris-acetate, pH 8.0, 10 mM Mg-acetate, 0.5 mM DTT, 0.1 mM EDTA for 30 minutes at 30°C. Heparin was added to a final concentration of 200 ng/μl. Samples were put on ice and irradiated with a Stratalinker (Stratagene, Inc.) between 0.9 and 2.7 J at 254 nm. Samples were treated with 1 μg RNaseA for 45 min at 37°C, dissolved in SDS-containing sample buffer and separated on a 15% SDS gel. Crosslink bands and marker proteins were visualized by autoradiography and Coomassie staining.
Identification of 6S RNA positions crosslinked to RNA polymerase
For the analysis of 6S RNA positions involved in crosslinking, 100 ng of non-labelled 6S RNA was used for complex formation and UV irradiation. Samples were incubated for 1 h with 500 ng proteinase K at 37°C, phenol extracted and ethanol precipitated. The RNA samples were further analysed by primer extension with AMV reverse transcriptase and a primer oligonucleotide complementary to 6S RNA positions 165–184, as described (11).
Analysis of transcription initiation complexes
The effect of 6S RNA on the formation of transcription initiation complexes was analysed by adding indicated amounts of 6S RNA to 3 nM reconstituted RNA polymerase holoenzymes, which had been incubated with 1 nM of the radiolabelled 260 bp rrnB P1 promoter DNA fragment or a 274 bp bolA promoter fragment, respectively. Complex formation was performed for 10 min at 30°C in 80 mM K-glutamate, 50 mM Tris-acetate, pH 8.0, 10 mM Mg-acetate, 0.5 mM DTT, 0.1 mM EDTA and 100 μg/ml acetylated BSA. Reaction mixtures contained 500 μM ATP and CTP as initiating nucleotides in the case of the rrnB P1 promoter to stabilize open promoter complexes (12). Heparin was added at the indicated concentrations to suppress unspecific binding. Samples were mixed with 5% glycerol, separated on non-denaturing 5% polyacrylamide gels and visualized by autoradiography.
In vitro transcription assay
Escherichia coli RNA polymerase core enzyme and the specificity factors σ70 and σ38 were purified by published procedures (13–17). The different holoenzymes were reconstituted from core RNA polymerase and excess σ factors prior to transcription. A linear 256 bp DNA fragment, directing a 64 nt run-off transcript from rrnB P1 was obtained by HincII/Ecl136II restriction of pUC18-1, which was obtained before by ligation of a 235 bp SspI/DdeI rrnB fragment into the SmaI site of pUC18. The 284 bp fragment with the bolA promoter was obtained by HincII digestion of pUC18-bolAT1T2, giving rise to a 124-nt run-off transcript. Plasmids pbolAT1T2 and pRT3H were used as superhelical templates, giving rise to transcripts of 120 or 320 nt length, respectively.
Transcription reactions with linear templates were performed in 10 μl with 1 nM template, 3 nM reconstituted RNA polymerase holoenzymes in a buffer containing 80 mM K-glutamate, 50 mM Tris-acetate, pH 8.0, 10 mM Mg-acetate, 0.5 mM DTT, 0.1 mM EDTA, 100 μg/ml acetylated BSA, 65 mM each, ATP, GTP, CTP and 133 nM α-[32P] UTP. Transcription reactions were performed at 30°C for 10 min in the presence or absence of 6S RNA as indicated. Reactions were terminated by the addition of 1.5 μl chase solution (1 mM Tris-HCl, pH 7.0, 2 mM each ATP, GTP, CTP and UTP, 2 mg/ml heparin). Samples were mixed with formamide sample buffer and separated on denaturing 10% polyacrylamide gels. Transcription reactions with superhelical templates were performed in an analogous way, except that 5 nM template and 3 nM reconstituted RNA polymerase and a final salt concentration of 160 mM K-glutamate was employed for the Eσ38-dependent transcription.
RESULTS
Interactions of 6S RNA with the components of the transcription machinery
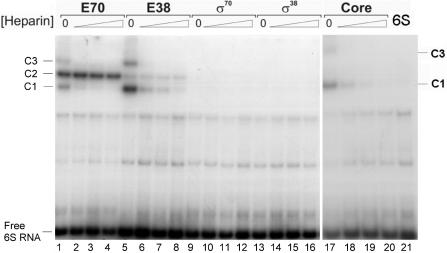
6S RNA was transcribed from StuI linearized plasmid vector pUC18-T7-6S, which carries the complete ssrS gene under the control of the T7 phage RNA polymerase ϕ 10 promoter. 6S RNA was isolated as run-off transcript and 3′ end labelled with [32P] pCp followed by gel purification. Purified 6S RNA (9 nM) was used for complex formation with different RNA polymerase preparations. RNA polymerase holoenzymes were reconstituted from purified core enzyme and isolated sigma factors prior to the binding reaction. Complex formations were performed for 20 min at 30°C with the reconstituted RNA polymerase holoenzymes Eσ70, Eσ38 (3 nM active enzyme each), free σ70 and σ38 subunits (60 nM each) as well as with the core RNA polymerase (3 nM) (see Materials and methods section). Non-specific complex formation was challenged by the addition of increasing amounts of heparin (final concentrations 50, 100 and 200 ng/μl). As can be seen in Figure 1, 6S RNA is able to undergo complex formation with all forms of RNA polymerase (core and holoenzymes). Exceptions are both free sigma factors, which do not interact, even in the absence of competitor.
Figure 1.
Complex formation of 6S RNA with different forms of RNA polymerase. A gel retardation analysis of 6S RNA complexed to different RNA polymerase preparations is shown. Different forms of RNA polymerase were complexed with 9 nM [32P] 3′- end- labelled 6S RNA. Lanes 1–4: 3 nM reconstituted holoenzyme Eσ70; lanes 5–8: 3 nM reconstituted holoenzyme Eσ38; lanes 9–12: 60 nM free σ70; lanes 13–16: 60 nM free σ38; lanes 17–20: 6 nM core RNA polymerase. Complex formation for each enzyme preparation was challenged with increasing heparin concentrations, from left to right: 0, 50, 100 or 200 ng/μl. Lane 21 on the extreme right shows 6S RNA in the absence of protein. The positions of free 6S RNA and of the three complexes with different mobilities are indicated at the margin and labelled 6S RNA, C1, C2 and C3, respectively.
The different RNA polymerase forms give rise to complexes with different electrophoretic mobilities. Altogether three complexes (C1, C2 and C3, Figure 1) can be visualized on retardation gels. The fastest and the slowest migrating complexes C1 and C3, respectively are sensitive to heparin and can be assigned to binding of 6S RNA to the core enzyme. Note that the reconstitution of the Eσ70 and Eσ38 holoenzymes is not quantitative and a notable amount of free core RNA polymerase is still present, even if the reconstitution was performed at a large excess of the respective sigma subunit. Hence, complexes C1 and C3 are visible in all samples that contain core RNA polymerase. The stronger bands for C1 and C3 in the samples with the Eσ38 holoenzyme are consistent with the markedly lower reconstitution efficiency of this holoenzyme from the purified core and σ38 subunit, resulting in a higher proportion of free RNA polymerase core. As is evident from Figure 1, neither one of the isolated sigma factors, σ70 and σ38, does bind to 6S RNA, even in the absence of heparin. Moreover we found, that no binding was obtained for phage-specific T7 RNA polymerase under the same conditions (data not shown). Complex formation was not altered whether or not initiating NTPs were present in the binding reactions, a specific requirement for some promoters to form stable RNA polymerase open complexes (data not shown). The only complex that resists heparin challenge and remains stable even after 90 min at 200 ng/μl heparin was C2, which is formed with the RNA polymerase Eσ70 and to a lesser extent with Eσ38. Most likely, this complex therefore represents a significant physiological intermediate. We conclude from this finding that 6S RNA is capable of binding to different forms of RNA polymerase with non-uniform specificity. Weak, probably non-specific binding occurs to the core enzyme. Binding to RNA polymerase holoenzymes is specific but appears to have a clear preference for the Eσ70 over the Eσ38 enzyme.
E. coli 6S RNA makes contact with the β/β′ and σ70 subunits of RNA polymerase holoenzyme
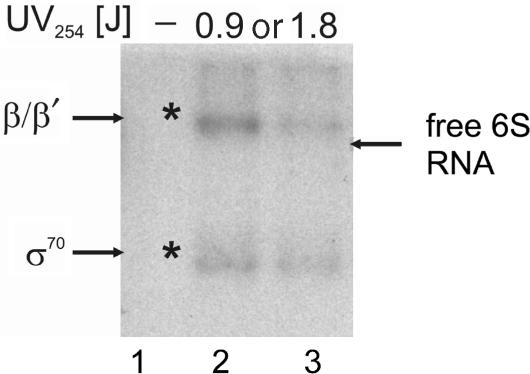
In previous crosslinking studies, contacts between 6S RNA from Haemophilus influencae and RNA polymerase had been identified (5). We wished to test if the same results could be verified for 6S RNA from E. coli and to further identify the RNA contact points. To this aim, a UV crosslinking analysis with radiolabelled E. coli 6S RNA complexed to Eσ70 RNA polymerase was conducted. Crosslink reactions were performed with free 6S RNA and in complex with either Eσ70 or Eσ38 as well as in the presence of the isolated sigma factors σ70 and σ38. After the crosslinking reaction the RNA polymerase complex was digested with RNaseA to remove excess protruding 6S RNA and the individual polymerase subunits were separated on denaturing SDS gel followed by autoradiography.
Clear evidence for crosslinking was only observed in the complex with Eσ70. It remains unclear if the amount of 6S RNA–Eσ38 complex in the reaction mixture was too low to yield notable crosslinking products or whether the molecular architecture of this complex is inadequate for crosslinking. Analysis of the crosslinked RNA polymerase subunits on SDS gels revealed that two bands with characteristic mobility for the β/β′ and σ subunits, containing radioactive nucleotides, can be detected (Figure 2). The formation of these bands clearly depends on UV irradiation and no comparable products are visible in lane 1, where the UV irradiation was omitted. Our analysis did not reveal evidence for crosslinking 6S RNA to any other RNA polymerase subunit. Increasing the UV dosage did not yield higher amounts of crosslinking products. Rather the diminishing band intensity at higher UV dosage indicates UV-dependent 6S RNA aggregation or decomposition. Consistent with the previously identified contacts for H. influencae 6S RNA (5) this experiment reveals 0 Å distance between E. coli 6S RNA and the σ and β/β′ subunits of RNA polymerase.
Figure 2.
UV crosslinking analysis of E. coli 6S RNA complexes formed with Eσ70. Complexes between radiolabelled 6S RNA and Eσ70 holoenzyme were formed and irradiated at 254 nm at increasing UV dosages of 0.9 or 1.8 J. In the lane marked by a dash, irradiation was omitted. Samples were subjected to RNaseA digestion and separated by SDS gel electrophoresis. Radioactive bands were visualized by autoradiography. Asterisks indicate radioactive bands that correspond to marker positions of RNA polymerase subunits. Bands were assigned according to non-radioactive marker proteins separated alongside the digested samples after Coomassie staining. The positions for the β/β′ and σ70 subunits are indicated at the margin. The position of a free 6S RNA sample separated without digestion is indicated by an arrow on the right margin.
Identification of 6S RNA regions in contact with RNA polymerase
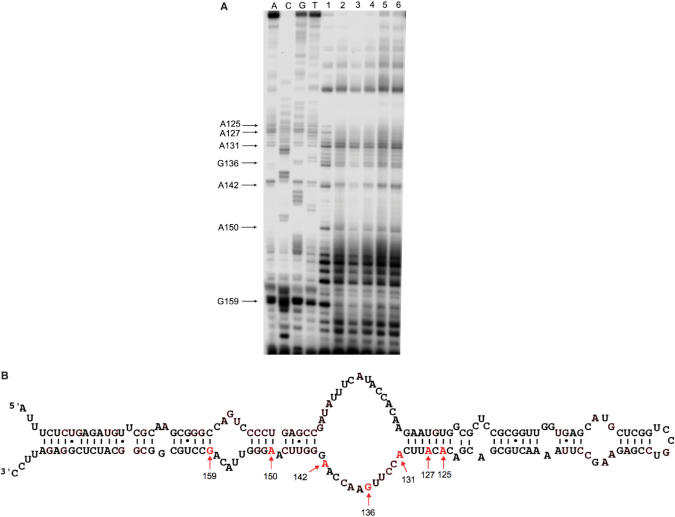
To further identify the 6S RNA nucleotide positions, which are in close contact to the β/β′ and σ subunits the crosslinking reaction was repeated with non-labelled 6S RNA. Following the crosslinking reaction, samples were digested with proteinase K, extracted with phenol and concentrated by ethanol precipitation. Positions of crosslinked amino acids within the 6S RNA structure were identified by primer extension. Two 5′ [32P]-labelled oligonucleotides, complementary to 6S RNA sequence positions 165–184 and 68–89, were employed for the analysis. An UV-irradiated 6S RNA sample in the absence of protein was used as a control to identify possible intermolecular RNA–RNA crosslinks. The example of a primer extension sequencing gel is shown in Figure 3A. Crosslinked positions giving rise to aborted cDNAs in the primer extension reaction could be verified on a consecutive region of the 6S RNA molecule (positions A125, A127, A131, G136, A142, A150 and G159). All identified positions cluster in the 6S RNA secondary structure, comprising the downstream part of the central bulge and the flanking stem sequences (Figure 3B). One has to conclude from this finding that the downstream strand of the central bulge of 6S RNA is in direct contact with the Eσ70 holoenzyme. Since neither core RNA polymerase nor Eσ38 could be crosslinked to 6S RNA it is reasonable to assume that the identified nucleotide positions might be in contact with the σ70 subunit.
Figure 3.
Regions of 6S RNA in contact with RNA polymerase. (A) The example of an autoradiogram of a primer extension analysis of isolated 6S RNA samples after crosslinking is shown. Samples have been separated on 10% denaturing polyacrylamide gels. A, C, G and T indicate primer extension sequencing reactions, which have been performed with a 6S RNA template. In lanes labelled 1–6, different 6S RNA-polymerase complexes were analysed after crosslinking; 1: 6S RNA complexed with Eσ70, 2: 6S RNA complexed with Eσ38, 3: 6S RNA complexed with core enzyme, 4: 6S RNA complexed with the isolated σ70 subunit, 5: 6S RNA complexed with the isolated σ38 subunit, 6: 6S RNA after UV irradiation in the absence of protein. Characteristic positions that deviate in the primer extension pattern between free 6S RNA and RNA polymerase complexes are indicated at the left margin. (B) Location of crosslink sites within the 6S RNA secondary structure. Positions of the 6S RNA nucleotides, which have been identified to be in contact with the Eσ70 holoenzyme, are indicated by arrows and sequence numbers.
6S RNA affects the formation of transcription initiation complexes at σ70- and σ38-dependent promoters
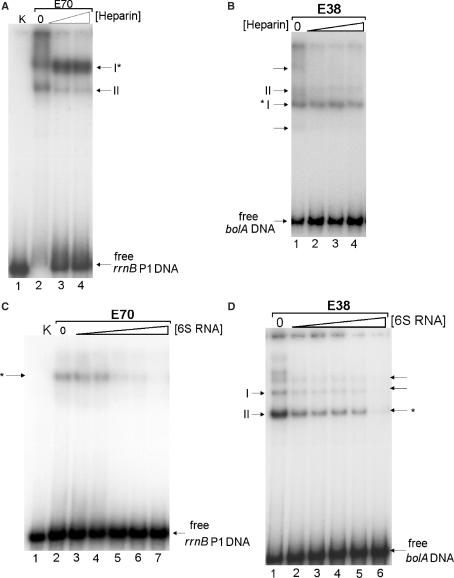
To test whether 6S RNA affects exponential- or stationary-phase-specific transcription in a differential manner we investigated the influence of the regulatory RNA on the initiation complex formation of Eσ70 or Eσ38 holoenzymes with σ70- and σ38-dependent promoters. Binding of RNA polymerase to promoter containing DNA fragments was studied by gel retardation. Two representative promoters, specific for either Eσ70 (rrnB P1) or Eσ38 (bolA) were tested with both holoenzymes. The RNA polymerase specificities had been established before and stable binding was tested by the addition of heparin. Both holoenzymes form specific initiation complexes with the respective promoter fragments, which are resistant to high heparin concentrations (Figure 4A and B). Some heparin-sensitive complexes were also formed by free core enzyme, present in the reaction mixtures. Only weak or no holoenzyme complexes are observed when the heterologous systems were incubated (data not shown).
Figure 4.
Effect of 6S RNA on transcription initiation complex formation. (A) Formation of initiation complexes at the rrnB P1 promoter was analysed by gel retardation. Initiation complexes were formed with a radioactive DNA fragment, containing the rrnB P1 promoter (1.5 nM) and 3 nM active RNA polymerase Eσ70 in the presence of 65 μM ATP and CTP as starting nucleotides. Samples were treated with various heparin concentrations (0, 100 and 200 ng/μl) and separated on a native 5% polyacrylamide gel. The positions of two RNA polymerase complexes (holoenzyme: I and core: II) and the free DNA are indicated. Different concentrations of heparin (lane 3: 100 ng/μl, lane 4: 200 ng/μl) were included. The essentially heparin resistant rrnB P1 ternary initiation complex is indicated by an asterisk. The gel band II corresponds to a core enzyme–DNA complex. In K, only free P1 DNA was separated. (B) Formation of initiation complexes at the bolA promoter is shown for the Eσ38 holoenzyme. The ternary initiation complex at the bolA promoter (I) is indicated by an asterisk. Bands labelled II correspond to the RNA polymerase core complex. Minor complexes are indicated by arrows. Various heparin concentrations were applied: lane 1: 0, lane 2: 50 ng/μl, lane 3: 100 ng/μl, lane 4: 200 ng/μl.(C) Effects of 6S RNA on initiation complex formation at rrnB P1. Complexes were formed as in (A), but in the presence of increasing amounts of 6S RNA (lane 2: 0, lane 3: 5 nM, lane 4: 10 nM, lane 5: 50 nM, lane 6: 100 nM, lane 7: 1 μM). An asterisk denotes the positions of specific rrnB P1 initiation complexes. In K, only free P1 DNA was separated. (D) Complexes were formed as in (B) but in the presence of increasing amounts of 6S RNA (lane 1: 0, lane 2: 5 nM, lane 3: 10 nM, lane 4: 50 nM, lane 5: 100 nM, lane 6: 1 μM). Bands labelled II, and indicated by an asterisk, denote the position of specific bolA initiation complex. Bands labelled I correspond to the RNA polymerase core complex. Minor complexes are indicated by arrows.
Initiation complex formation in the presence of increasing amounts of 6S RNA is markedly reduced with both promoters (Figure 4C and D), indicating that 6S RNA potentially inhibits initiation complex formation with σ70- and σ38-dependent promoters, independent of the respective holoenzyme, Eσ70 or Eσ38. The magnitude of the inhibition is also comparable, though slightly higher for the Eσ38 complexes, reaching ∼50% for both promoters between 10 and 100 nM 6S RNA, respectively. At 1 μM 6S RNA almost complete decomposition of initiation complexes is visible in both systems. For a quantitative evaluation see Supplementary Data (Figure S2).
6S RNA inhibits in vitro transcription of σ70- and σ38-dependent promoters
6S RNA-dependent inhibition of initiation complex formation already strongly indicates the functional involvement of 6S RNA in transcription. To extend the analysis we determined the influence of the regulatory RNA on the complete transcription process. To this aim, we performed multiple round in vitro transcription reactions with σ70- and σ38-dependent promoters present on linear and superhelical templates. Reactions were initiated with either Eσ70 or Eσ38 holoenzymes. Figure 5 exemplifies results obtained with linear DNA fragments containing either the σ70-dependent rrnB P1 or the σ38-dependent bolA promoter. Both holoenzymes, Eσ70 and Eσ38, were employed to evaluate the cross reactivity of the exponential- and stationary-phase-specific transcription systems.
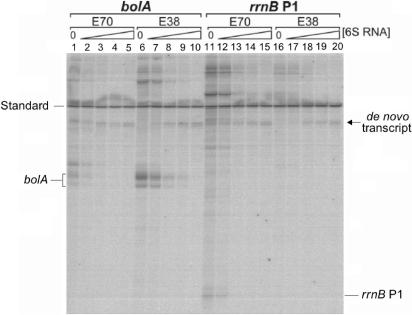
Figure 5.
Effect of 6S RNA on the in vitro transcription from σ70 and σ38-specific promoters on linear DNA fragments. Products from in vitro transcription reactions were separated on denaturing polyacrylamide gels and visualized by autoradiography. Reactions with the bolA or rrnB P1 promoters are shown on the left or right side, respectively. The different holoenzymes employed (E70, E38) are indicated above the lanes. For each system the amount of 6S RNA present in the reaction was varied (lanes 1, 6, 11, 16: 0 nM, lane 2, 7, 12, 17: 10 nM, lane 3, 8, 13, 18: 50 nM, lane 4, 9, 14, 19: 100 nM, lane 5, 10, 15, 20: 250 nM). A 260 bp radiolabelled DNA fragment, indicated at the margin, was included as internal standard for quantification. The positions of the run-off transcripts for the bolA (∼124 nt) and rrnB P1 (64 nt) promoters are marked. An arrow denotes a de novo product that consistently arises when 6S RNA is incubated with E. coli RNA polymerase, even in the absence of any template DNA.
The analysis shows that transcription from both promoters is strongly affected by the presence of 6S RNA. A quantitative evaluation of the intensities for the two run-off products, characteristic for the stationary-phase-specific bolA promoter, revealed >50% reduction of their initial intensity at <30 nM 6S RNA (see Supplementary Data, Figure S3). A similar reduction is obtained for the homologous system with Eσ70 and the rrnB P1 promoter. It should be noted that reactions performed in the presence of tRNA as non-specific control revealed only weak, probably non-specific inhibition. Interestingly, a bolA-specific transcript can be detected in the heterologeous transcription system, employing the exponential-phase-specific holoenzyme Eσ70. It is much weaker, however, as in case of the stationary-phase-specific enzyme Eσ38. This heterologous transcript is subject to the same degree of inhibition by 6S RNA as the transcript derived by Eσ38. As expected, no comparable heterologous transcript can be observed for the linearized P1 promoter and the Eσ38 holoenzyme. It should be noted in this respect, that under those conditions the P1 transcript is already rather weak with the homologous Eσ70 enzyme.
In summary, the in vitro transcription analysis demonstrates that 6S RNA strongly inhibits formation of transcripts from exponential- and stationary-phase-specific promoters, independent of whether RNA polymerase Eσ70 or Eσ38 holoenzymes were employed. Hence, the analysis completely confirms the results obtained from the promoter binding studies.
Effect of the order of 6S RNA addition to the in vitro transcription reaction
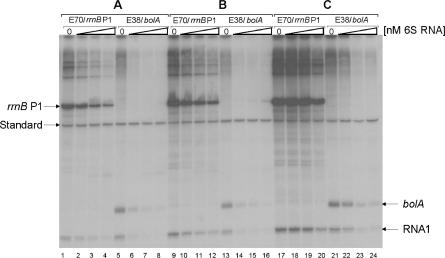
Transcription initiation is a multi-step procedure with several functionally important intermediates, each of which might be affected by specific regulators. To clarify any potential mechanism of 6S RNA-dependent transcription inhibition, it is essential to dissect individual steps of the transcription cycle and to determine the effect of the regulator separately. We therefore conducted experiments in which the potential action of 6S RNA was controlled by the order of addition to the in vitro transcription reaction. To better match the in vivo conditions we used supercoiled DNA templates, harbouring either one of the rrnB P1 or bolA promoters in addition to the Eσ70-dependent RNAI promoter. Transcription starting at the rrnB P1 and bolA promoters was terminated by the presence of the two strong rho-independent terminators from the rrnB operon, cloned downstream of the transcription start sites. We tested the addition of 6S RNA to the reaction at three different steps of the transcription initiation pathway (Figure 6). In A, 6S RNA was included in the reconstitution reaction with RNA polymerase core and the specific sigma subunits to test whether holoenzyme formation might be affected. In B, 6S RNA was added to the reconstituted holoenzyme and the transcription reaction was started by the addition of template DNA in order to allow 6S RNA binding to RNA polymerase prior to the formation of closed and open promoter complexes. Finally, in C, 6S RNA was included with the complete transcription mixture except for the enzyme. The reaction was then initiated by the addition of reconstituted RNA polymerase. In this case, the time frame for 6S RNA to interact with RNA polymerase is limited, and competition has to occur with productive transcription. The result of such an ‘order of addition’ experiment is presented in Figure 6.
Figure 6.
Effect of 6S RNA addition on the in vitro transcription with supercoiled templates. Three different reactions, corresponding to different steps of the transcription cycle, were directed by the controlled addition of 6S RNA. In A (lanes 1–8), 6S RNA was added to the RNA polymerase reconstitution reaction. In B (lanes 9–16), 6S RNA was applied to the transcription reaction, which was initiated by the addition of template DNA as the last component. In C (lanes 17–24), 6S RNA was present in the reaction mixture before transcription was started by the addition of the enzyme. The amount of 6S RNA was varied in each series (lanes: 1, 5, 9, 13, 17, 21: 0 nM, lanes: 2, 6, 10, 14, 18, 22: 50 nM, lanes: 3, 7, 11, 15, 19, 23: 100 nM, lanes: 4, 8, 12, 16, 20, 24: 250 nM, respectively). The different holoenzymes (Eσ70, Eσ38) and supercoiled template DNAs (rrnB P1, bolA) are indicated above the lanes. Terminated transcripts started from either rrnB P1 or bolA and the σ70-dependent plasmid-encoded RNA1 promoter are marked at the margin. A 260 bp radiolabelled reference DNA fragment (standard) was included in each lane for quantification.
Clearly, in all three sets of experiments the amount of homologous transcripts decreases with increasing concentrations of 6S RNA for all promoters tested. The reduction in activity is not uniform, however. The strongest inhibition is observed when 6S RNA is already present during the reconstitution reaction (A) while inhibition is smallest for reactions where 6S RNA has to compete with RNA polymerase and the promoter DNA (C). This is also reflected by the absolute yields of transcription products, which are highest for C and lowest for A. This observation is valid for holoenzymes, Eσ70 or Eσ38 and both promoters rrnB P1 and bolA. For a quantitative evaluation of the above experiments see Supplementary Data Figure S4. The RNAI promoter, present on both plasmid templates, can be taken as a σ70-specific reference promoter. Transcripts directed by this promoter show the same gradual inhibition, though to a slightly smaller degree, as for rrnB P1, reinforcing the notion of a general inhibitory activity of 6S RNA. Interestingly, inhibition of the heterologous transcript from the RNAI promoter by Eσ38 is stronger compared to the corresponding homologous Eσ70 holoenzyme. We conclude from this experiment that at least under in vitro conditions inhibition of 6S RNA is neither restricted to a single form of RNA polymerase nor to transcription from exponential-phase-specific promoters. Moreover, inhibition is probably not only acting at one defined point of the transcription initiation cycle. It should be noted that not all the steps leading to productive transcription are readily reversible. On the other hand, there is no step beyond which we do not see inhibition, pointing to a function of 6S RNA as an inhibitor, which is strongest when the time available for inhibition is large. The fact that inhibition is strongest when 6S RNA is present during the RNA polymerase reconstitution reaction is consistent with the existence of a binary complex between 6S RNA and core enzyme as a functional intermediate, although, according to the binding studies (Figure 1) interaction of 6S RNA with the Eσ70 holoenzyme seems to be preferred.
6S RNA acts as a template for defined de novo transcripts in the absence of DNA
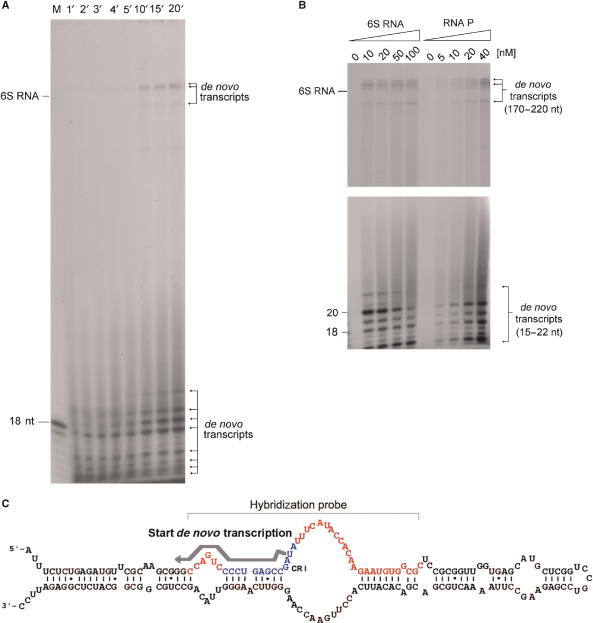
During the analysis of the 6S RNA effect on transcription we noticed the occurrence of new products in reaction mixtures with RNA polymerase and NTPs, independent from the template DNA, but which depend on the presence of 6S RNA. The formation of some of these products can for instance be seen in Figure 5, where they have been marked by an arrow. When 6S RNA was incubated with RNA polymerase and NTPs in the complete absence of DNA templates a number of defined transcripts is reproducibly formed (Figure 7A). These de novo transcripts fall into two clusters of different length. One group of relative high abundance consists of transcripts with an approximate length between 14 and 24 nt, while some longer transcripts between 170 and 220 nt are formed at lower yield. Control experiments after extensive DNase treatment revealed that these de novo transcripts are not the result of small traces of contaminating DNA. The characteristic de novo transcripts were only observed after the addition of pure 6S RNA. All forms of RNA polymerase, Eσ70, Eσ38 or core RNA polymerase were active in de novo transcript formation, although the yield of the products varied slightly (as an example see Figure 5, arrow). Other small RNA molecules, different from 6S RNA, like tRNA, 5S rRNA or PSTVd viroid RNA did not result in comparable products (data not shown). Moreover, the long de novo transcripts do not result from extension of 6S RNA or complex formation between short de novo transcripts and 6S RNA, which survived the denaturating gel separation. When radioactive 6S RNA together with non-radioactive NTPs were used for in vitro transcription, no elongated products were observed. For details of the analysis, see Supplementary Data, Figure S1D. Higher 6S RNA concentrations provoke the formation of de novo transcripts in favour of shorter length, while higher RNA polymerase concentrations cause a general increase in the yield of all de novo transcripts (Figure 7B). Characterization of the smaller products by RNase T1 digestion and hybridization to 6S RNA cDNA fragments is consistent with the assumption that they are complementary to the 6S RNA sequence region between nucleotide positions 20–40 (Figure 7C; Supplementary Data, Figure S1A). RNase T1 analysis revealed that transcription of all small de novo transcripts starts at 6S RNA position U44 (see Supplementary Data, Figure S1B and C). Interestingly, this sequence region has been defined as absolutely conserved element (CR I) in comparative 6S RNA sequence analysis studies (4) and forms the upstream part of the central bulge region, which bears resemblance with an open promoter structure. It is certainly an interesting question, whether or not these de novo transcripts participate in transcription regulation or are otherwise directly or indirectly involved in the function of 6S RNA. The precise analysis of the long de novo transcripts and elucidation of their importance for 6S RNA-dependent regulation has to await more detailed sequence characterization and functional studies, however. Work along those lines is currently underway in our laboratory.
Figure 7.
Formation of 6S RNA-dependent de novo transcripts. (A) The time-dependent formation of 6S RNA-directed formation of de novo transcripts is shown. Reaction mixtures contained 20 nM Eσ70 RNA polymerase holoenzyme and 100 nM 6S RNA, free of any noticeable DNA. Reaction was performed in standard in vitro transcription buffer (see Materials and methods section) for the times in minutes indicated on top of the lanes. Products were separated on 15% denaturing polyacrylamide gels. The positions of a 18-mer DNA oligonucleotide and a 6S RNA marker are indicated. (B) Effect of 6S RNA and RNA polymerase concentrations on the formation of de novo transcripts. The upper and lower part of the figure represent different exposures of the upper and lower part of the same gel in order to better visualize the longer products. Reactions have been performed as in (A) except that 6S RNA or RNA polymerase was varied as indicated on top of the lanes (6S RNA: 0–100 nM; RNA polymerase: 0–40 nM). Reactions with increasing 6S RNA contained constant 20 nM RNA polymerase and reactions with increasing RNA polymerase constant amounts of 6S RNA (50 nM). The lengths of the different de novo transcripts are indicated at the margin. (C) Scheme depicting the de novo transcription start site within the secondary structure of 6S RNA, according to the analysis presented in Supplementary Data (Figure S1A–D). The start site (U44), direction of synthesis and approximate length of the short de novo transcripts are indicated by a grey arrow. The conserved CRI sequence is shown in blue, and the hybridization sequence region is shown in red and indicated by a bracket.
DISCUSSION
The functional importance of the non-coding 6S RNA is supported by numerous recent observations, including its high abundance and widespread distribution among diverse bacteria. First obvious indices for its function as a potential transcriptional regulator became clear when it turned out that 6S RNA, which was known for many years to exist as 11S ribonucleoprotein particle (7), forms complexes with RNA polymerase (5). The highly conserved 6S RNA secondary structure, resembling an open promoter, further supported a functional involvement of this molecule in transcription (1,18). Based on the conserved characteristic secondary structure a promoter mimicry model had been postulated. The hypothesis that 6S RNA functions as an open promoter was further substantiated by a mutagenesis study, analysing 6S RNA variants with altered sequences in the central loop region (18). Moreover, the fact that the number of 6S RNA molecules increases from 1.000 to ∼10.000, when E. coli cells enter stationary phase clearly points to a function in the transition between exponential and stationary growth. This notion is consistent with the previous report where after 6S RNA exclusively interacts with Eσ70 RNA polymerase holoenzyme, but not with RNA polymerase core or Eσ38. From the functional analyses presented up to now it was concluded, that 6S RNA interferes specifically with transcription of σ70-dependent promoters, particularly, extended −10 promoters were proposed to be under 6S RNA regulation. No inhibition was reported to occur with σ38-dependent promoters, in fact, some σ38-dependent transcripts were found to be activated by 6S RNA (8).
6S RNA interaction with RNA polymerase
Comparison of the results presented in this study with former investigations reveals that 6S RNA binding to RNA polymerase and promoter-specific transcriptional regulation appears to be more complex as recently suggested. Hence, the findings we report here are consistent with some but not all the conclusions derived from recent studies. Generally we could not verify strict Eσ70 holoenzyme- or σ70-dependent promoter specificity. Our gel shift experiments reveal, for instance, that binding of 6S RNA is likely to occur to different forms of RNA polymerase, albeit with reduced stability. We show that complexes may also be formed with Eσ38 and the core enzyme, not however with the free sigma subunits. This partly extends recent results obtained by immunoprecipitation where the interaction of Eσ38, Eσ32, core polymerase or free σ70 with 6S RNA was classified as non-specific (18). Heparin challenge with our direct-binding assay suggests that at least for Eσ38, and to some extent also for the core enzyme, heparin-resistant binding occurs. Whether or not these complexes actually represent functional intermediates is not entirely clear. With respect to the transcriptional analysis, however, which clearly demonstrates that 6S RNA also affects Eσ38 holoenzymes, it is at least plausible that some kind of interaction between 6S RNA and Eσ38 holoenzymes has to occur. Moreover, the observation that 6S RNA catalyses the synthesis of de novo transcripts in the absence of DNA templates not only with Eσ70 but also with Eσ38 and core RNA polymerase supports the observed interaction between 6S RNA and all three different enzymes.
Promoter specificity of 6S RNA-dependent transcription regulation
Results observed in this study concerning the promoter specificity of 6S RNA-dependent inhibition are not in complete accord with previous reports. For example, in our in vitro transcription experiments with superhelical templates the σ70-dependent RNAI promoter was shown to be clearly inhibited by 6S RNA (even more by the Eσ38 holoenzyme, see Figure 6). In prior experiments with lacZ fusions the RNAI promoter had been reported as non-responsive to elevated concentrations of 6S RNA, however. Strong inhibition was only observed for fusion constructs with galP1, galP2 and rsdP2 promoters in the same study (8). In addition, the in vitro transcription analyses in this study reveal that the σ70-dependent rrnB P1 promoter is subject to a strong and specific inhibition by 6S RNA. This apparently contrasts with previous reports, where equivalent expression levels in vivo had been measured in ssrS1 and wild-type strain backgrounds for the same promoter as lacZ fusion (8). It should be noted, however, that the activity of the fusion construct was possibly too low to allow detecting reliable differences. When σ38-dependent promoters were analysed in vivo in the same study, activation for bolA and several other σ38-dependent promoters was found, while appY and rsdP1 promoters showed comparable levels of expression in ssrS1 and wild-type cells. In contrast, results from in vitro transcription presented here demonstrate significant inhibition of the bolA promoter in the presence of 6S RNA. This inhibition was observed for both the Eσ38 as well as the Eσ70 holoenzymes. Different findings were also reported recently (19) which showed that the bolA promoter activity in vivo was not reduced but increased in the absence of 6S RNA and that the preferential expression of stationary-phase genes by Eσ38 is unlikely the consequence of selective inhibition of Eσ70 by 6S RNA. The exact reason for the apparent discrepancies is not obvious. It may, in part, be due to the difference of the in vitro and in vivo analysis systems and possibly indicates that in vivo additional trans-acting factors are involved in regulation. In any case it is clear, however, that the promoter and RNA polymerase specificities of 6S RNA are not completely understood and additional comparative studies with more promoter constructs are required.
Mechanism of 6S RNA-dependent regulation
With regard to the 6S RNA-dependent mechanism of transcription regulation the available evidence is largely consistent with the postulated promoter mimicry model, which is based on the highly conserved central loop structure. This model is particularly supported by analysing the activity of 6S RNA mutants (18). The crosslinking analysis presented here to identify the positions of 6S RNA, which are in direct contact with RNA polymerase add further evidence to the importance of the central loop structure and the flanking stem regions. Moreover, the fact, that only the downstream strand of the central bulge structure can be crosslinked indicates that the two single-stranded sequences of the central loop occupy different trajectories on the surface of RNA polymerase, as it is known for the coding and non-coding strand of RNA polymerase–promoter complexes (20).
The role of 6S RNA in facilitating the switch in transcription from exponential- to stationary-phase-specific promoters (RNA polymerase competition) can also be explained by the preferential affinity of 6S RNA to Eσ70 holoenzymes. Almost all σ70 molecules will be sequestered at the stationary phase by 6S RNA–Eσ70 complexes, which inactivate most of the Eσ70-dependent transcription. The remaining fraction of free core RNA polymerase is now available for Eσ38 complex formation and transcription of stationary-phase-specific genes.
6S RNA is not the only small non-coding RNA (ncRNA) known to affect transcription. In the mouse, B2 RNA, a small ncRNA transcribed by RNA polymerase III, has been shown to inhibit RNA polymerase II-dependent mRNA transcription in response to heat shock (21). Analysis of the mechanism of inhibition of B2 RNA has revealed that it binds directly to Pol II and assembles into stable preinitiation complexes on promoter DNA, rendering them inactive for RNA synthesis until B2 RNA is removed again (22). Binding of B2 RNA occurs to a previously identified RNA-docking site on Pol II, which is distinct from the DNA-binding channel and RNA exit groove. The B2 RNA complexes are inactive, even for the synthesis of short abortive transcripts. Attempts to identify the steps of transcription that B2 RNA inhibits leave the question open, whether B2 RNA inhibits initiation by preventing promoter melting, thereby blocking formation of open complexes. Alternatively, the access of incoming substrate nucleotides could be blocked or the formation, release or extension of initial transcripts might be impaired. From the ‘order of addition’ experiments performed here with 6S RNA (Figure 6) one has to conclude that, similar as with B2 RNA, a step early in transcription initiation is affected. It will be a very interesting question, to find out whether the mechanism of eukaryotic B2 RNA to regulate transcription is unique or if it bears parallels with the transcriptional regulation by bacterial 6S RNA.
Are the 6S RNA-directed de novo transcripts involved in transcriptional regulation?
Although highly structured double-stranded RNA molecules have in some rare cases been demonstrated to exert promoter activity by unknown mechanisms transcription from RNA templates by (DNA-dependent) RNA polymerase is a rather unusual reaction (23–25). It would be consistent, however with the proposed promoter mimicry model for 6S RNA function. Since the de novo transcripts are very specific with respect to size and sequence we assume that they may have a specific function. The complete characterization of the sequence and the mechanism of the de novo RNA synthesis are currently under study in the laboratory. The new transcription products could be key molecules for the understanding of the regulatory mechanism associated with 6S RNA. It is also possible that the de novo transcripts themselves are involved as regulators by unknown mechanisms.
Putative alternative functions for 6S RNA
An alternative for a further putative 6S RNA function is related to the stability of the 11S nucleoprotein complex formed with RNA polymerase holoenzyme. This complex, which accumulates at late stationary phase, may sequester RNA polymerase in a functional form and impede with its degradation or turnover. At the return of favourable growth conditions sufficient RNA polymerase can be rapidly released to accomplish the need for efficient transcription. A similar mechanism is known to act during translation. In this case the RMF protein, which transiently inactivates ribosomes by forming 100S dimers under unfavourable conditions for protein synthesis is a representative example (26,27). In line with this hypothesis it has been shown that 6S RNA enhances long-term cell survival and helps cells to persist during nutritional deprivation (8). In addition it was shown that 6S RNA plays a role in cell survival at elevated pH during stationary phase, most likely by regulating transcription of pspF, a transcriptional activator with specific responsiveness to σ54 RNA polymerase in the phage shock protein response (28). This finding supports the function of 6S RNA in stabilization of cellular components under stress and longtime stationary phase and possibly points to role in general stress adaptation.
The enigmatic role of the cotranscribed YgfA protein, a putative 5,10-methenyltetrahydrofolate synthetase, which is found upstream of many enterobacteriae and γ-proteobacterial ssrS transcription units presents an additional open question that will hopefully be answered in near future.
Note Added During Revision
We would like to mention that during the processing of this manuscript a publication by Wassarman and Saecker (Wassarman,K.M. and Saecker,R.M. (2006) appeared in Science (Science 313:1601DOI: 10.1126/science.1134830), in which the phenomenon of 6S RNA-mediated de novo transcription and the analysis of the short transcripts were independently reported. The findings presented in this publication are fully consistent with the analysis reported in this article. The above authors conclude that 6S RNA-directed de novo transcription may be responsible for liberating RNA polymerase from stably bound 6S RNA in response to improved growth conditions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The friendly gift of expression systems for σ70 and σ38 by A. Ishihama is greatly acknowledged. Additional thanks goes to C. Bradaczek and B. Reckendrees for the plasmid constructions used for the analysis of rrnB P1 and bolA promoters. Moreover, we like to thank all members of the group for helpful discussion and G. Rusty for patience and endurance during lab meetings.
Conflict of interest statement. None declared.
REFERENCES
- 1.Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA. 2005;11:774–784. doi: 10.1261/rna.7286705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuma S, Asari S, Bunai K, Yoshino K, Ando Y, Kakeshita H, Fujita M, Nakamura K, Yamane K. Identification and characterization of novel small RNAs in the aspS-yrvM intergenic region of the Bacillus subtilis genome. Microbiology. 2002;148:2591–2598. doi: 10.1099/00221287-148-8-2591. [DOI] [PubMed] [Google Scholar]
- 3.Ando Y, Asari S, Suzuma S, Yamane K, Nakamura K. Expression of a small RNA, BS203 RNA, from the yocI-yocJ intergenic region of Bacillus subtilis genome. FEMS Microbiol. Lett. 2002;207:29–33. doi: 10.1111/j.1574-6968.2002.tb11023.x. [DOI] [PubMed] [Google Scholar]
- 4.Brown JW, Ellis JC. Comparative Analysis of RNA Secondary Structures: 6S RNA. Weinheim: Wiley-VCH GmbH & Co; 2005. pp. 491–512. [Google Scholar]
- 5.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 6.Vogel DW, Hartmann RK, Struck JC, Ulbrich N, Erdmann VA. The sequence of the 6S RNA gene of Pseudomonas aeruginosa. Nucleic Acids Res. 1987;15:4583–4591. doi: 10.1093/nar/15.11.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SY, Bailey SC, Apirion D. Small stable RNAs from Escherichia coli: evidence for the existence of new molecules and for a new ribonucleoprotein particle containing 6S RNA. J. Bacteriol. 1978;133:1015–1023. doi: 10.1128/jb.133.2.1015-1023.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trotochaud AE, Wassarman KM. 6S RNA function enhances long-term cell survival. J. Bacteriol. 2004;186:4978–4985. doi: 10.1128/JB.186.15.4978-4985.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanisch-Peron C, Vieira J, Messing J. Improved M13 mp18 pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 10.Göringer HU, Bertram S, Wagner R. The effect of tRNA binding on the structure of 5S RNA in E. coli. A chemical modification study. J. Biol. Chem. 1984;259:491–496. [PubMed] [Google Scholar]
- 11.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 12.Gourse RL. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 1988;16:9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burgess RR, Jendrisak JJ. A procedure for the rapid, large scale purification of E. coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA cellulose chromatography. Biochemistry. 1975;14:4636–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 14.Gonzales N, Wiggs J, Chamberlin MJ. A simple procedure for resolution of E. coli RNA polymerase holoenzyme from core polymerase. Arch. Biochem. Biophys. 1977;182:404–408. doi: 10.1016/0003-9861(77)90521-5. [DOI] [PubMed] [Google Scholar]
- 15.Igarashi K, Fujita N, Ishihama A. Identification of a subunit assembly domain in the alpha subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 1991;218:1–6. [PubMed] [Google Scholar]
- 16.Borukhov S, Goldfarb A. Recombinant Escherichia coli RNA polymerase: purification of individually overexpressed subunits and in vitro assembly. Protein Expr. Purif. 1993;4:503–511. doi: 10.1006/prep.1993.1066. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka K, Takayanagi Y, Fujita N, Ishihama A, Takahashi H. Heterogeneity of the principal σ factor in Escherichia coli: the rpoS gene product, σ38, is a second principal σ factor of RNA polymerase in stationary-phase Escherichia coli. Proc. Natl. Acad. Sci. USA. 1993;90:3511–3515. doi: 10.1073/pnas.90.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trotochaud AE, Wassarman KM. A highly conserved 6S RNA structure is required for regulation of transcription. Nature Structural Biol. 2005;12:313–319. doi: 10.1038/nsmb917. [DOI] [PubMed] [Google Scholar]
- 19.Kim EY, Shin MS, Rhee JH, Choy HE. Factors influencing preferential utilization of RNA polymerase containing sigma-38 in stationary-phase gene expression in Escherichia coli. J. Microbiol. 2004;42:103–110. [PubMed] [Google Scholar]
- 20.Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 21.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 22.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 23.Pelchat M, Perreault JP. Binding site of Escherichia coli RNA polymerase to an RNA promoter. Biochem. Biophys. Res. Commun. 2004;319:636–642. doi: 10.1016/j.bbrc.2004.05.044. [DOI] [PubMed] [Google Scholar]
- 24.Pelchat M, Grenier C, Perreault JP. Characterization of a viroid-derived RNA promoter for the DNA-dependent RNA polymerase from Escherichia coli. Biochemistry. 2002;41:6561–6571. doi: 10.1021/bi025595k. [DOI] [PubMed] [Google Scholar]
- 25.Wettich A, Biebricher CK. RNA species that replicate with DNA-dependent RNA polymerase from Escherichia coli. Biochemistry. 2001;40:3308–3315. doi: 10.1021/bi002756g. [DOI] [PubMed] [Google Scholar]
- 26.Wada A. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells. 1998;3:203–208. doi: 10.1046/j.1365-2443.1998.00187.x. [DOI] [PubMed] [Google Scholar]
- 27.Yamagishi M, Matsushima H, Wada A, Sakagami M, Fujita N, Ishihama A. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor – growth phase-dependent and growth rate-dependent control. EMBO J. 1993;12:625–630. doi: 10.1002/j.1460-2075.1993.tb05695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trotochaud AE, Wassarman KM. 6S RNA regulation of pspF transcription leads to altered cell survival at high pH. J. Bacteriol. 2006;188:3936–3943. doi: 10.1128/JB.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.