Abstract
Apoptotic cell death is accompanied by degradation of chromosomal DNA. Here, we established in Drosophila a null mutation in the gene for inhibitor of caspase-activated DNase (ICAD) by P-element insertion. We also identified a loss-of-function mutant in Drosophila for DNase II-like acid DNase. The flies deficient in the ICAD gene did not express CAD, and did not undergo apoptotic DNA fragmentation during embryogenesis and oogenesis. In contrast, the deficiency of DNase II enhanced the apoptotic DNA fragmentation in the embryos and ovary, but paradoxically, the mutant flies accumulated a large amount of DNA, particularly in the ovary. This accumulation of DNA in the DNase II mutants caused the constitutive expression of the antibacterial genes for diptericin and attacin, which are usually activated during bacterial infection. The expression of these genes was further enhanced in flies lacking both dICAD and DNase II. These results indicated that CAD and DNase II work independently to degrade chromosomal DNA during apoptosis, and if the DNA is left undigested, it can activate the innate immunity in Drosophila.
Keywords: apoptosis, DNA fragmentation, mutagenesis, antibacterial peptides, innate immunity
Apoptosis, or programmed cell death, is an evolutionarily conserved process that removes harmful or useless cells during development (Jacobson et al. 1997; Vaux and Korsmeyer 1999). The apoptotic process is accompanied by morphological changes, such as shrinkage and fragmentation of the cells and nuclei, and loss of microvilli from the plasma membranes (Wyllie et al. 1980). Genetic and biochemical studies revealed that caspases, a family of cysteine proteases, are activated during apoptosis (Thornberry and Lazebnik 1998; Earnshaw et al. 1999; Los et al. 1999). More than 100 cellular proteins have been identified in mammalian systems as caspase substrates, the cleavage of which brings about the morphological changes of the cells and nuclei, leading to the cell death (Stroh and Schulze-Osthoff 1998).
In addition to the morphological changes of cell and nuclei, the apoptotic process is often accompanied by the degradation of chromosomal DNA (Wyllie 1980; Nagata 2000). We and others have identified a DNase (caspase-activated DNase, CAD; also called DNA fragmentation factor, DFF40), that is activated in dying cells (Liu et al. 1997; Enari et al. 1998). CAD is complexed with its inhibitor (ICAD, inhibitor of CAD) in proliferating cells. When apoptosis is triggered, caspases—in particular caspase 3—cleave ICAD, and the CAD released from ICAD degrades the chromosomal DNA into nucleosomal units (Sakahira et al. 1998). Accordingly, cells that are deficient in the CAD system do not undergo apoptotic DNA fragmentation in vitro. However, DNA degradation is still observed in vivo in CAD-deficient mice. Because this DNA fragmentation is detected in apoptotic cells inside macrophages, we proposed that the DNA of apoptotic cells can be cleaved by lysosomal acid DNase (DNase II) after the cells are engulfed by macrophages (McIlroy et al. 2000).
Programmed cell death plays an important role in the development of Drosophila melanogaster (Abrams 1999). During embryogenesis, metamorphosis, and oogenesis, many cells die showing the characteristics of apoptosis. Therefore, Drosophila has been widely used to dissect the molecular mechanisms of apoptosis and to understand its physiological role (Bergmann et al. 1998). The programmed cell death in Drosophila is triggered by the expression of a set of genes: reaper, hid, and grim, which bring about the activation of a family of caspases. Loss-of function mutations of these genes impair the apoptotic process, and often prevent embryogenesis and/or lead to tumorigenesis (White et al. 1994; Song et al. 1997). The apoptotic cell death in Drosophila is also accompanied by DNA fragmentation (Nagano et al. 1998). Previously, we identified Drosophila homologs (dCAD and dICAD) for CAD and ICAD, and showed that the apoptotic DNA fragmentation in a BG-2 Drosophila neural cell line is mediated by the dCAD/dICAD system (Mukae et al. 2000; Yokoyama et al. 2000).
In this report, a Drosophila line that is deficient in dICAD was established by P-element-mediated mutagenesis. In the dICAD-null flies, neither the dICAD nor the dCAD protein was expressed. The apoptotic DNA fragmentation into nucleosomal units that normally occurs during embryogenesis and oogenesis was completely suppressed in the mutant flies. In addition, an ethyl-methyl-sulfonate (EMS)-induced mutant line in the Drosophila stock center was found to carry a loss-of-function mutant in lysosomal acid DNase (dDNase II). These dDNase II-deficient flies showed enhanced apoptotic DNA fragmentation, yet accumulated a large amount of DNA, particularly in ovaries, and constitutively expressed the genes for antibacterial peptides. This activation of the antibacterial peptide genes was enhanced in Drosophila mutants that lacked both dICAD and dDNase II. These results indicate that CAD and DNase II work independently to degrade chromosomal DNA during apoptosis, and this process plays an important role in maintaining the homeostasis of these animals.
Results
Establishment of a dICAD-null fly by P-element mutagenesis
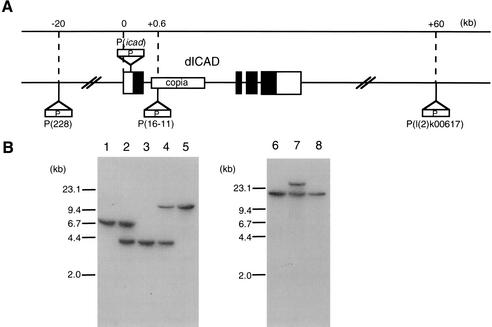
To study the physiological roles of the CAD-ICAD system in Drosophila, a Drosophila line carrying a mutation in the dICAD gene was generated by a local hop of a nearby P-element. The dICAD gene is located on the 48E2 locus of the Drosophila chromosomes. A search of the FlyBase indicated that Drosophila line l(2)k00617 carries a P-element 60 kb downstream of the dICAD gene. This P-element was mobilized in a stepwise manner into the dICAD gene locus (Fig. 1A). The movement of the P-element at each step was followed by a long PCR procedure, and confirmed by Southern hybridization (Fig. 1B). After repeating this local hop procedure three times, a fly line, P(icad), which carries a P-element 17 bp upstream of the ATG initiation codon, was established.
Figure 1.
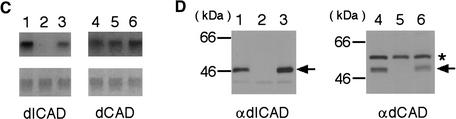
Establishment of a dICAD mutant by the insertion of a P-element. (A) The dICAD chromosomal gene locus and the position of P-element in each Drosophila strain are schematically shown. The dICAD gene consists of four exons and is depicted as boxes in which the open and filled areas represent the noncoding and coding regions, respectively. The Drosophila l(2)k00617 line contains a copia element in intron 1 of the dICAD gene. Positions of the P-element are shown at the top in kb, starting from the 5′ end of the dICAD gene. (B) Southern hybridization analysis of the Drosophila strains carrying the P-element. Genomic DNAs (10 μg) from P(l(2)k00617) (lane 1), P(l(2)k00617)-P(228) (lane 2), P(228) (lane 3), P(228)-P(16-11) (lane 4), and P(16–11) (lane 5) were digested with EcoRI (lanes 1–5), and separated by electrophoresis on a 0.8% agarose gel. Hybridization was carried out with a 32P-labeled 1.0-kb DNA fragment carrying the P-element sequence as a probe. DNA was also prepared from P(16-11) (lane 6); P(icad), which carries two P-elements at 16-11 and at the 5′ noncoding region of the dICAD gene (lane 7); and the revertant of P(icad) (lane 8) in which the P-element at the 5′ noncoding region was removed by excision. DNAs were digested with XbaI and subjected to Southern hybridization as above. (C) Northern hybridization analysis. Poly(A) RNA (2 μg) prepared from adult flies of the wild-type (lane 1), dicad mutant (lane 2), and its excision revertant was analyzed by Northern hybridization with dICAD (left panel) or dCAD (right panel) cDNA as the probe. In the lower panels, the membranes used for hybridization were stained with methylene blue. (D) Western blot analysis. The cell lysates (30 μg of protein) prepared from adult wild-type (lane 1), dicad mutant (lane 2), and its excision revertant (lane 3) flies were subjected to Western blot analysis using a rabbit anti-dICAD (left panel) or anti-dCAD (right panel) antibody. The relative molecular masses of the standard proteins are shown in kD at left. The positions of dICAD and dCAD are indicated by arrows. The bands indicated by asterisks appeared to be nonspecific.
Northern hybridization analysis of the poly(A) RNA and Western blot analysis of the cell lysates from the adult flies indicated that the P(icad) flies expressed neither dICAD mRNA nor dICAD protein (Fig. 1C,D), indicating that P(icad) was a null allele for the dICAD gene. The excision of the P-element from the 5′-noncoding region of the dICAD gene (Fig. 1B) permitted the expression of dICAD mRNA and the dICAD protein, confirming the specific mutation of the dICAD gene in the P(icad) flies. The functional CAD protein cannot be properly folded without ICAD (Sakahira et al. 2000; Yokoyama et al. 2000). Accordingly, the 52-kD dCAD protein was not detected in the dICAD mutant flies, although they expressed the CAD mRNA as abundantly as did the wild-type flies (Fig. 1C,D). From these results, we concluded that the P(icad) allele works as a null allele not only for dICAD but also for dCAD. As reported for ICAD (DFF45)-null mice (Zhang et al. 1998) and transgenic mice expressing a caspase-resistant form of ICAD (McIlroy et al. 2000), the dICAD-mutant flies were fertile, and there was no apparent adverse phenotype.
Identification of a Drosophila mutant of the DNase II gene
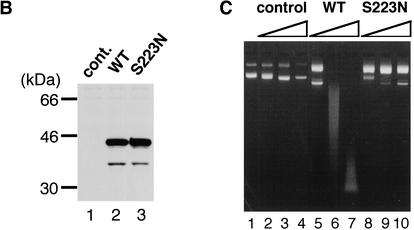
An involvement of DNase II in the apoptotic process has been suggested for Caenorhabditis elegans (Wu et al. 2000) and mouse (McIlroy et al. 2000). Recently, Vernooy et al. (2000) reported that a gene called CG7780 (FlyBase accession no. AY075328) in the Drosophila database codes for a protein related to mammalian DNase II. In fact, the amino acid sequence encoded by CG7780 had 25% identity and 43% similarity with mouse or human DNase II (Fig. 2A). In particular, three histidine residues that may work as the active site for the enzymatic function of DNase II were well conserved. To confirm that the protein encoded by the CG7780 gene had DNase II-like activity, the full-length cDNA for CG7780 was isolated from adult flies by reverse-transcription polymerase chain reaction (RT-PCR). The cDNA was tagged with Flag at the C terminus, expressed in COS cells (Fig. 2B), and purified using an anti-Flag antibody. As shown in Figure 2C, the purified protein showed DNase activity under acidic conditions. Little DNase activity was observed under neutral conditions, and metal ions such as Ca2+ or Mg2+ were not required for the DNase activity (data not shown). Because these properties were similar to those of mammalian DNase II (Bernardi 1971), we concluded that the CG7780 gene codes for dDNase II.
Figure 2.
Identification of a Drosophila mutant in the DNase II gene. (A) The amino acid sequence of Drosophila DNase II and its mutant. The amino acid sequences of Drosophila DNase II (D), murine DNase II (M), and human DNase II (H) are aligned to give maximum homology by introducing several gaps (−). The amino acid residues that are conserved in all three proteins are shown in bold, and sets of three residues regarded as favored substitutions are indicated by underlines. Three histidine residues conserved in Drosophila, human, and mouse DNase II are indicated by asterisks. The serine residue (S) at amino acid position 223 of Drosophila DNase II was replaced with asparagine (N) in the DNase-1lomutant. (B) Expression of dDNase II in COS cells. COS cells were transfected with the expression vector (lane 1), the expression vector carrying the Flag-tagged wild-type Drosophila DNase II cDNA (lane 2), or the Flag-tagged S223N mutant DNase II cDNA (lane 3). The cell lysates (40 μg of protein) from the transfected cells were separated by electrophoresis on a 10%–20% gradient polyacrylamide gel, and analyzed by Western blot using an anti-Flag antibody. Molecular masses of marker proteins are shown in kD at left. (C) Lack of DNase activity in the mutant DNase II. COS cells were transfected with the empty vector (lanes 2–4), or the expression vector for the wild-type DNase II (lanes 5–7), or its S223N mutant (lanes 8–10). The cell lysates (2 mg of protein) were applied to M2 agarose beads (30 μL bed-volume), and the Flag-tagged DNase II was eluted with 90 μL of PBS containing 100 μg/mL Flag peptide. Using aliquots of 1.0 μL (lanes 2,5,8), 3.0 μL (lanes 3,6,9), and 9.0 μL (lanes 4,7,10) of the eluate, the acid DNase activity was determined with 1.0 μg of plasmid DNA as a substrate. In lane 1, the plasmid DNA was incubated in the assay buffer without the eluate.
The CG7780, dDNase II gene is located at the 90D6 locus of the Drosophila chromosomes (FlyBase, http://flybase.bio.indiana.edu). A Drosophila mutant line (DNase-1lo) obtained by mutagenesis with EMS was reported to carry a low level of acid DNase activity, and this allele was mapped to the 90C2–91A2 locus (Grell 1976). Because this region includes the locus of the dDNase II gene, we examined the possibility that the DNase-1lo mutation is a mutation of dDNase II. The dDNase II cDNA was isolated by RT-PCR from adult DNase-1lo flies. A comparison of its sequence with the sequence of wild-type dDNase II showed that the cDNA from DNase-1lo had one point mutation. The AGC codon at position 223 was changed to AAC, causing a replacement of serine with asparagine (Fig. 2A). When this cDNA was expressed in COS cells, the recombinant protein showed very little DNase activity (Fig. 2C), indicating that the DNase-1lo allele is a loss-of-function allele for dDNase II. The flies carrying the DNase-1loallele were fertile, but their egg production rate was significantly reduced (data not shown).
The apoptotic DNA fragmentation in Drosophila development, and accumulation of DNA
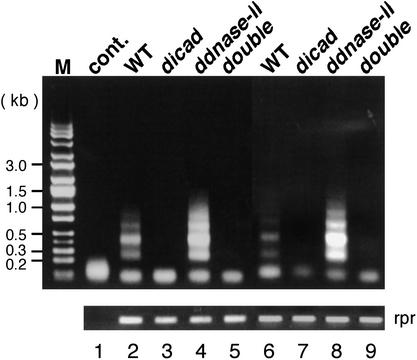
During Drosophila embryogenesis, cells die via the apoptotic process that is activated by reaper, grim, and hid (White et al. 1994; Abrams 1999). Drosophila oogenesis proceeds through asymmetrical division of a germ-line stem cell, giving rise to a cluster of 16 cells. Of the 16 cells, one develops into the oocyte, while the other 15 work as nurse cells. At the late stage of oogenesis, the cytoplasmic contents of the nurse cells are rapidly transferred into the oocyte and their nuclei are degraded in a caspase-dependent manner (Cavaliere et al. 1998; McCall and Steller 1998). We examined the role of CAD and DNase II in the DNA fragmentation in these processes by the linker-mediated PCR method, which amplifies the 5′-phosphorylated DNA fragments (Staley et al. 1997), using the flies deficient in ICAD or DNase II. As shown in Figure 3, the DNA from the wild-type embryos and ovaries showed a clear DNA ladder. On the other hand, no DNA ladder was detected with the DNA from the dICAD-null flies, indicating an indispensable role for CAD in the apoptotic DNA fragmentation that occurs during embryogenesis and oogenesis. In contrast to the dICAD mutation, the mutation of the dDNase II gene did not impair the formation of the apoptotic DNA ladder (Fig. 3). Rather, the DNA ladder in the DNase-1lo flies was more pronounced than that observed in the wild-type flies, indicating an enhanced level of this type of degradation. The DNA ladder was not detected in double-mutant flies lacking both ICAD and DNase II. These results indicated that CAD and DNase II work independently to degrade the chromosomal DNA during embryogenesis and oogenesis.
Figure 3.
The apoptotic DNA fragmentation in Drosophila embryos and ovaries. Genomic DNA was prepared from the 7–11 h embryos (lanes 2–5) or ovaries of day eight adult female flies (lanes 6–9) of the wild-type (CS; lanes 2,6), dicad-mutant (lanes 3,7), ddnase II-mutant of DNase-1lo(lanes 4,8), and the double mutant for dicad and ddnase II (double; lanes 5,9). An aliquot of DNA (40 ng) was subjected to LM-PCR, and the products were separated by electrophoresis on a 1.0% agarose gel. In lane 1, the LM-PCR was carried out without genomic DNA. The lower panel shows the control PCR for the reaper gene, confirming the equivalent amounts of DNA used for LM-PCR. The molecular mass size marker DNAs were run in lane M, and their sizes are indicated in kb at left.
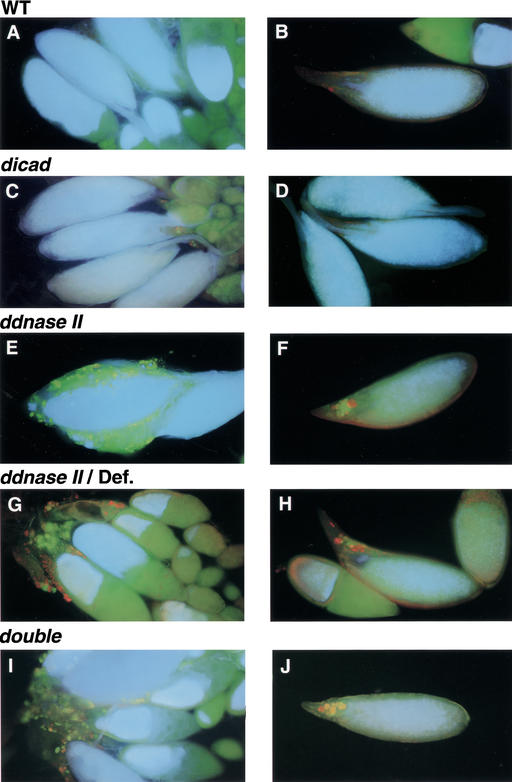
Stone et al. (1983) previously detected Feulgen-positive materials in the egg chambers of the Cy/M(2)S2lo Drosophila strain, which they found to carry a hypomorphic allele of the gene affected by the DNase-1lomutation, that is, DNase II. In fact, the ovaries of DNase-1lo flies showed materials that could be strongly stained with acridine orange in the dorsal appendage of the mature eggs (Fig. 4). By day eight, many acridine orange-positive vesicles were present near the lateral oviduct and these vesicles could be stained with Feulgen, suggesting that the DNA of the nurse cell nuclei were left undigested during oogenesis in the DNase-1lomutant flies. A similar accumulation of acridine-positive materials was observed in the heterozygous flies that were generated by crossing DNase-1lo flies with a deficiency line of Df(3R)P14, indicating that this was a specific phenotype for the loss-of-function mutation of the dDNase II gene. The ovaries of the double mutants deficient in dICAD and dDNase II also accumulated a large amount acridine-positive material. In contrast, there was no abnormal disposition of acridine-positive materials in the ovaries of dICAD mutant flies.
Figure 4.
Accumulation of DNA in ovaries. Ovaries from the adult flies (8 d) of wild-type (A,B), dicad-mutant (C,D), ddnase II-mutant (E,F), the heterozygous mutant line DNase-1lo(ddnase II)/Df(3R)P14 (G,H), and the double mutant for dICAD and dDNase II (I,J) were stained with acridine orange as described in Materials and Methods.
Constitutive activation of antibacterial genes
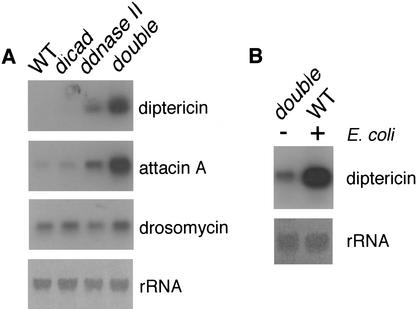
In mammals, nonmethylated CpG DNA activates the innate immunity through the toll-like receptor (TLR) in lysosomes (Hemmi et al. 2000; Leadbetter et al. 2002). Because the dDNase II mutant flies accumulated chromosomal DNA, we examined whether this accumulated DNA could activate the innate immunity in Drosophila. RNA was prepared from adult flies at day eight, and analyzed by Northern hybridization using as probes DNAs for antibacterial or antifungal peptides that are known to be induced during bacterial or fungal infection. As shown in Figure 5A, wild-type flies did not express the genes for the antibacterial peptides, diptericin or attacin, and the mutation in the dICAD gene had little effect on the expression of these genes. In contrast, the mutation in the dDNase II gene caused the constitutive expression of the diptericin and attacin genes. The expression level was enhanced in the double-mutant flies that were deficient in both dICAD and dDNase II, but it was significantly lower than that observed by infection of flies with Escherichia coli (Fig. 5B). In contrast, the gene for the antifungal peptide drosomycin was weakly expressed in the wild-type flies as reported previously (Lemaitre et al. 1997), and its expression was little influenced by the mutation in dDNase II or dICAD. These results indicate that endogenous Drosophila DNA can activate a specific signaling pathway that leads to the induction of genes for antibacterial peptides if it is left undigested during the apoptotic process.
Figure 5.
Expression of antibacterial peptide genes. (A) Constitutive activation of antibacterial peptide genes. Poly(A) RNA (3 μg) from the adult Drosophila flies at day eight of the wild-type (WT), ddicad-mutant (dicad), DNase-1lo (ddnase II), and the double mutant for dICAD and dDNase II (double) was subjected to Northern hybridization using cDNA for diptericin, attacin A, and drosomycin. At bottom, the filter was stained with methylene blue. (B) The endogenous DNA- or bacteria-induced expression of the diptericin gene. The adult flies of the double mutant for dICAD and dDNase II (double) were kept uninfected, and the wild-type flies (WT) were infected for 16 h with E. coli. Total RNA (15 μg each) was analyzed by Northern hybridization with diptericin cDNA as a probe.
Discussion
The fragmentation of chromosomal DNA into nucleosomal units is regarded as a hallmark of apoptotic cell death (Earnshaw 1995; Nagata 2000), and the TUNEL staining that detects DNA fragments carrying 3′-hydroxyl ends is widely used to detect apoptotic cells in vitro and in vivo (Gavrieli et al. 1992). The recently developed linker-mediated PCR (LM-PCR) detects DNA fragments with 5′-phosphate ends, with high sensitivity (Staley et al. 1997). We reported previously that CAD, a DNase that produces DNA fragments with 5′-phosphate and 3′-hydroxyl ends (Widlak et al. 2000), is activated in apoptotic cells and can cleave the chromosomal DNA into nucleosomal units, giving rise to a DNA ladder that is typical for apoptosis (Enari et al. 1998). However, many other nucleases have been proposed as the nuclease that cleaves the chromosomal DNA during apoptosis (Li et al. 2001; Oliveri et al. 2001; Parrish et al. 2001; van Loo et al. 2001), and the contribution of CAD to apoptotic DNA fragmentation has been elusive, particularly in vivo. In this report, we established in Drosophila a CAD-null allele by disrupting the ICAD gene with a P-element insertion, and examined the role of the CAD system in the apoptotic processes associated with embryogenesis and oogenesis in Drosophila. In both of these processes, the apoptotic DNA fragmentation detected by LM-PCR did not occur in the mutant fly lacking the CAD system, suggesting that CAD is solely responsible for the apoptotic DNA fragmentation, at least in the embryogenesis and oogenesis of Drosophila. The contribution of other nucleases to this process, if any, would be small.
Despite the lack of DNA fragmentation in CAD-deficient flies, neither the embryos nor the ovaries apparently accumulated DNA. In C. elegans, a three-step process has been proposed for the degradation of DNA in programmed cell death (Wu et al. 2000). At least one of these steps is mediated by Nuc-1, a homolog of DNase II. We have proposed that in mammalian cells, the DNase II in the lysosomes of macrophages can degrade the DNA of apoptotic cells after macrophages engulf the apoptotic cells (McIlroy et al. 2000). However, because DNase II-null mice die prenatally due to severe anemia, the in vivo role of DNase II in the apoptotic process could not be evaluated in mice (Kawane et al. 2001). Here, we identified the DNase-1loallele of Drosophila as a loss-of-function mutation of a DNase in lysosomes (Gaszner and Udvardy 1991) that has significant homology to mammalian DNase II. The DNase-1lo mutants showed enhanced CAD-dependent generation of apoptotic DNA fragments. Paradoxically, these mutants also accumulated acridine orange-positive material, particularly in the ovaries. By hybridizing a labeled polypyrimidine probe to DNA from DNase-1lo flies, Stone et al. (1983) detected a low-melt hybrid with low thermal stability in the ovaries and proposed a two-step model for the degradation of nurse cell nuclei. In the first step, DNA is degraded into AT-rich DNA fragments of <200 bp, and these fragments are further digested to oligo- or mono-nucleotides by acid DNase in the second step. Our results agree with this model and support a scenario in which the first step of DNA degradation is mediated by CAD in the dying cells. CAD is an endonuclease that cleaves naked DNA into oligonucleotides. However, early in the cell-death process, DNA is still wrapped around histones in nucleosomes and CAD can only cleave the spacer regions between the nucleosomes, producing DNA fragments of nucleosomal units (180 bp). When the apoptotic cells are engulfed by phagocytes, the proteins of nucleosomes are digested by lysosomal proteases, and the DNA is exposed to the lysosomal DNase for complete digestion. The fact that the DNA fragmentation is not necessary for phagocytes to recognize apoptotic cells (McIlroy et al. 2000; Hanayama et al. 2002) explains why no accumulation of abnormal DNA was seen in the CAD-deficient flies. This model apparently differs from the model proposed for the apoptotic DNA degradation in C. elegans, in which Nuc-1 works in the dying cells (Wu et al. 2000). Although we cannot rule out this possibility, we prefer the model in which DNase II functions in phagocytes because DNase II is rather specifically expressed in macrophages (Kawane et al. 2001). Mice deficient in either CAD or DNase II have recently been established (Kawane et al. 2001; K. Kawane, H. Fukuyama, and S. Nagata, unpubl.). The coculturing of CAD-null apoptotic cells with DNase II-null macrophages will clarify the relationship between CAD and DNase II in the apoptotic DNA degradation.
The physiological role of the DNA degradation in programmed cell death has been elusive. Here, we found that endogenous DNA that escaped from apoptotic degradation could activate the innate immunity in Drosophila. Infection of Drosophila with E. coli induced a significantly higher level of expression of the antibacterial peptide genes. This may be because bacterial peptidoglycan or lipopolysaccharides (LPS) in addition to DNA activate the antibacterial peptide genes (Gottar et al. 2002), or the cells responding to the endogenous DNA or to bacteria differ. It will be necessary to study when and which cells express the antibacterial peptide genes in DNase II-null flies. There are two signaling pathways for the innate immunity in Drosophila (Hoffmann and Reichhart 2002). In one, infection with gram-positive bacteria or fungi activates the gene for an antifungal peptide, drosomycin, via the Toll/MyD88 pathway. In the other, infection with gram-negative bacteria induces the expression of genes for antibacterial peptides such as diptericin and attacin via a Toll-independent immune deficiency (IMD) pathway. The specific activation of diptericin and attacin but not drosomycin in the DNase II-null flies indicates that the accumulated DNA activates the innate immunity in a Toll-independent manner. This finding seems to contradict the fact that nonmethylated CpG DNA activates the innate immunity through the Toll-like receptor 9 (TLR9)/MyD88 pathway in mammals (Hacker et al. 2000; Hemmi et al. 2000). To clarify this point, it will be necessary to study whether the activation of antibacterial peptide genes in the DNase II-null flies requires MyD88 or IMD signaling, and whether any TLR9-like receptor is involved in recognition of the accumulated DNA in Drosophila. The flies deficient in DNase II as well as the flies deficient in both DNase II and CAD showed no apparent adverse phenotype except for a reduction in egg production. On the other hand, the activation of the innate immunity in mammals is likely to have adverse effects on their homeostasis by producing inflammatory cytokines such as TNF and IL-6 (O'Neill 2002). Previously, we reported that mouse embryos lacking the DNase II gene accumulate a large amount of DNA in the fetal liver and die due to a defect in erythropoiesis (Kawane et al. 2001). Whether this defect is due to inflammation caused by deregulated innate immunity remains to be studied.
Materials and methods
Materials and antibodies
The rabbit anti-dICAD antibody was described previously (Mukae et al. 2000). A rabbit dCAD antibody was produced by immunizing rabbits with recombinant dCAD. In brief, SF9 cells were infected with Baculovirus carrying a cDNA for His-tagged dCAD as described previously (Sakahira et al. 1999). The infected cells were disrupted by three cycles of freezing and thawing in buffer A [10 mM Hepes-KOH at pH 7.2, 5 mM MgCl2, 5 mM EGTA, 1 mM DTT, and 1 mM (p-amidinophenyl) methanesulfonyl fluoride hydrochloride (pAPMSF)], and the lysates were spun for 20 min at 15,000 rpm. The precipitates containing dCAD were washed with buffer B (10 mM Tris-HCl buffer at pH 8.0 and 100 mM NaH2PO4) containing 4 M urea, and dissolved in buffer B containing 7 M guanidine-HCl. The His-tagged dCAD was then loaded onto a Ni-NTA agarose (QIAGEN), eluted from the beads with 10 mM Tris-HCl buffer (pH 4.5) containing 100 mM Na2PO4 and 8 M urea, and used to immunize rabbits. The anti-dCAD antibodies were partially purified from the sera using a Protein A-Sepharose column. The anti-Flag antibody was purchased from Sigma.
Fly stocks, mutagenesis, and septic injury
All fly stocks were maintained at 25°C on standard fly food. Canton S was used as the wild-type strain. The DNase-1lomutant line (Grell 1976) and Df(3R)P14 deficiency line were obtained from the Bloomington Stock Center. The flies carrying transposase and balancer lines were provided by Drs. F. Matsuzaki (Laboratory of Cell Assymetry, Riken Center for Developmental Biology, Chuo-ku, Kobe, Hyogo, Japan) and M. Miura (Laboratory of Cell Recovery Mechanisms, Riken Brain Science Institute, Tsukuba, Ibaraki, Japan).
A fly line carrying a mutation in dICAD gene was generated by the stepwise transposition of a P-element (Tower et al. 1993). In brief, P(l(2)k00617)/CyO flies carrying a P-element at the 48E2 region were crossed to female flies carrying transposase (w/w; +/+; Dr/TMS, Δ2-3, Sb). Mosaic male progeny [w/Y; P(l(2)k00617)/CyO; +/TMS, Δ2-3, Sb] were backcrossed to w/w; Sp/SM1; +/+ virgins. Descendant male flies with more intensely red eyes were crossed to w/w; Sp/SM1 and analyzed by a long PCR method. One line (line 228) was found to carry a new P-element insertion ∼20 kb upstream of the dICAD gene. After removing the P-element at the original position, this line was used to further transpose the P-element, and a line (16-11-36) carrying a P-element in intron 1 of the dICAD gene was established. The local P-element hop was repeated once again to establish line 42-1 [P(icad)], which carried a P-element at the 5′ noncoding region of the dICAD gene. This P-element insertion was found to be a null allele for dICAD.
For screening by long PCR, a set of 30-mer oligonucleotides carrying sequences derived from 10 kb upstream, downstream, and coding regions of the dICAD gene was prepared referring to the FlyBase (http://flybase.bio.indiana.edu) and used as a primer together with a 30-mer oligonucleotide (5′-GACGGGACCAC CACCTTATGTTATTTCATCATG-3′) carrying a sequence in the LTR region of the P-element. PCR was carried out with rTth DNA polymerase (PE Applied Biosystems) using a Gene AmpR XLPCR kit. For plasmid rescue, Drosophila DNA was digested with EcoRI or PstI, ligated with T4 DNA ligase, and introduced into E. coli.
To generate a Drosophila line deficient in both the dICAD and DNase II genes, the lines w/w; P(icad), P(16-11)/CyO; +/MKRS and w/Y; Sco/CyO; DNase-1lo/DNase-1lo were established, and these were crossed to obtain w/w; P(icad), P(16-11)/CyO; DNase-1lo/MKRS flies. These flies were intercrossed to generate the double-mutant flies. The heterozygous flies carrying DNase-1lo/Df(3R)P14 were generated by crossing the female flies DNase-1lo/DNase-1lo with Df(3R)P14 flies Df(3R)P14, sr[1]/T(2;3)ap[Xa],ap[Xa].
The septic injuries were done as described (Rutschmann et al. 2000). In brief, the adult wild-type flies were infected at 25°C by pricking with a tungsten needle that had been dipped in a concentrated culture of E. coli DH10B, and kept at 25°C for 16 h.
Cloning and expression of dDNase II cDNA
Total RNA was isolated by the acid-guanidine thiocyanate method using an Isogen kit from Nippongene. Two oligonucleotides (a sense primer, 5′-ACTTGAAGGTTGTACAATGCG-3′, and an antisense primer, 5′-TTCTATTCCTGTTGGGCGCAC-3′) were designed from Drosophila EST clones (Al114173 and GH10876) and Gene CG7780 in the FlyBase. RNA was reverse-transcribed using Superscript II reverse transcriptase (GIBCO-BRL), and the coding sequence of dDNase II cDNA was amplified by PCR using Taq polymerase. The PCR product of 1.1 kb was inserted into pGEM-T-Easy (Promega), and the nucleotide sequence was determined.
To express dDNase II in monkey COS cells, the coding sequence of dDNase II was tagged with a FLAG-epitope at the C terminus and inserted into the pEF-BOS-EX vector. COS cells in DMEM containing 10% fetal calf serum (GIBCO-BRL) were transfected with the dDNase II expression plasmid by electroporation. After culturing at 37°C for 48 h, the cells were suspended in lysis buffer (20 mM Tris-HCl at pH 7.5, 2 mM EDTA, 1% Triton X-100, 1 mM pAPMSF, and 1 μg/mL aprotinin), and disrupted by three cycles of freezing and thawing. After adding NaCl to a final concentration of 150 mM, the lysates were homogenized by grinding with a pestle, and the nuclei and large cell debris were removed by centrifugation. The FLAG-tagged dDNase II in the lysates was purified using anti-FLAG M2 agarose (Sigma).
The enzymatic activity of dDNase II was measured as described (Kawane et al. 2001). In brief, samples were preincubated on ice for 30 min in 20 μL of 50 mM Citrate-NaOH buffer (pH 4.5) containing 10 mM EDTA and 50 μg/mL RNase A. Plasmid DNA (1.0 μg) was added to the mixture, followed by incubation at 30°C for 2 h. The DNA was analyzed by electrophoresis on a 1.5% agarose gel.
Acridine orange staining and linker-mediated PCR
Acridine orange staining of the ovary was conducted according to Foley and Cooley (1998). In brief, ovaries were dissected in buffer C (10 mM Hepes-HCl buffer at pH 6.9, 130 mM NaCl, 4.7 mM KCl, and 1.9 mM CaCl2), immersed for 3 min in the dark in a 1:1 mixture of 10 μg/mL acridine orange in buffer C and heptane, and placed onto glass slides. After removing the excess dye by aspiration, samples were covered with halocarbon oil and examined with an Olympus IX70 fluorescence microscope using a UV-G excitation filter.
For linker-mediated PCR (LM-PCR), the genomic DNA prepared by the standard method was further purified by treatment with RNase A followed by precipitation with 7.5% polyethylene glycol. The DNA (40 ng) was subjected to an LM-PCR Ladder assay (Staley et al. 1997) using a kit from Clontech. The PCR was carried out for 26 cycles and the products were analyzed by electrophoresis on a 1% agarose gel.
Southern hybridization, Northern hybridization, and Western blotting
Southern and Northern hybridization were carried out under high stringency (Sambrook and Russell 2001) with 32P-labeled fragment of dICAD or dCAD cDNA (Mukae et al. 2000; Yokoyama et al. 2000). The cDNAs for diptericin (Wicker et al. 1990), attacin A (Lee et al. 1983), and drosomycin (Fehlbaum et al. 1994) were isolated by RT-PCR from adult Drosophila, cloned in the pGEM-T-Easy vector, and used as probes for Northern hybridization.
For Western Blotting, adult flies were homogenized in the lysis buffer using a pestle. The homogenates were spun for 30 min at 30,000g, and the supernatants were used as the S-30 fraction. Proteins were separated by electrophoresis on a 10%–20% gradient polyacrylamide gel (Dai-ichi Pure Chemicals) and transferred to a PVDF membrane (Hybond P, Amersham Biosciences). The membranes were probed with 2000-fold diluted anti-dICAD or 1000-fold diluted anti-dCAD antibody followed by peroxidase-conjugated anti-rabbit IgG (DAKO). The bands were revealed by chemiluminescence using a Renaissance kit from PerkinElmer Life Sciences.
Acknowledgments
We thank Drs. F. Matsuzaki (The Center for Developmental Biology, Riken) and M. Miura (Brain Science Institute, Riken), Bloomington Center, and Szeged Center for providing Drosophila strains; S. Hirose (Department of Developmental Genetics, National Institute of Genetics, Mishima, Shizuoka-ken, Japan) for the protocol for P-element mediated mutagenesis; T. Uemura (Department of Molecular Genetics, Institute for Virus Research, Kyoto University, Kyoto, Japan) for valuable advice; and S. Aoyama and M. Harayama for secretarial assistance. This work was supported in part by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture in Japan. N.M., H.Y., and T.Y. were supported by Research Fellowships from the Japan Society for the Promotion of Science.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL nagata@genetic.med.osaka-u.ac.jp; FAX 81-6-6879-3319.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1022802.
References
- Abrams JM. An emerging blueprint for apoptosis in Drosophila. Trends Cell Biol. 1999;9:435–440. doi: 10.1016/s0962-8924(99)01646-3. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Agapite J, Steller H. Mechanisms and control of programmed cell death in invertebrates. Oncogene. 1998;17:3215–3223. doi: 10.1038/sj.onc.1202586. [DOI] [PubMed] [Google Scholar]
- Bernardi G. Spleen acid deoxyribonuclease. In: Boyer PD, editor. The enzymes. New York and London: Academic Press; 1971. pp. 271–287. [Google Scholar]
- Cavaliere V, Taddei C, Gargiulo G. Apoptosis of nurse cells at the late stages of oogenesis of Drosophila melanogaster. Dev Genes Evol. 1998;208:106–112. doi: 10.1007/s004270050160. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC. Nuclear changes in apoptosis. Curr Biol. 1995;7:337–343. doi: 10.1016/0955-0674(95)80088-3. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Martins LM, Kaufmann SH. Mammalian caspases: Structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- Fehlbaum P, Bulet P, Michaut L, Lagueux M, Broekaert WF, Hetru C, Hoffmann JA. Insect immunity. Septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J Biol Chem. 1994;269:33159–33163. [PubMed] [Google Scholar]
- Foley K, Cooley L. Apoptosis in late stage Drosophila nurse cells does not require genes within the H99 deficiency. Development. 1998;125:1075–1082. doi: 10.1242/dev.125.6.1075. [DOI] [PubMed] [Google Scholar]
- Gaszner M, Udvardy A. Purification of a lysosomal DNase from Drosophila melanogaster. Biochem Biophys Res Commun. 1991;181:44–50. doi: 10.1016/s0006-291x(05)81379-8. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- Grell EH. Genetic analysis of aspartate aminotransferase isozymes from hybrids between Drosophila melanogaster and Drosophila simulans and mutagen-induced isozyme variants. Genetics. 1976;83:753–764. doi: 10.1093/genetics/83.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker H, Vabulas RM, Takeuchi O, Hoshino K, Akira S, Wagner H. Immune cell activation by bacterial CpG-DNA through myeloid differentiation marker 88 and tumor necrosis factor receptor-associated factor (TRAF)6. J Exp Med. 2000;192:595–600. doi: 10.1084/jem.192.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R, Tanaka M, Miwa K, Shinohara A, Iwamatsu A, Nagata S. Identification of a factor that links apoptotic cells to phagocytes. Nature. 2002;417:182–187. doi: 10.1038/417182a. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA, Reichhart JM. Drosophila innate immunity: An evolutionary perspective. Nat Immunol. 2002;3:121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- Kawane K, Fukuyama H, Kondoh G, Takeda J, Ohsawa Y, Uchiyama Y, Nagata S. Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science. 2001;292:1546–1549. doi: 10.1126/science.292.5521.1546. [DOI] [PubMed] [Google Scholar]
- Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- Lee JY, Edlund T, Ny T, Faye I, Boman HG. Insect immunity. Isolation of cDNA clones corresponding to attacins and immune protein P4 from Hyalophora cecropia. EMBO J. 1983;2:577–581. doi: 10.1002/j.1460-2075.1983.tb01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- Liu X, Zou H, Slaughter C, Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- Los M, Wesselborg S, Schulze-Osthoff K. The role of caspases in development, immunity, and apoptotic signal transduction: Lessons from knockout mice. Immunity. 1999;10:629–639. doi: 10.1016/s1074-7613(00)80062-x. [DOI] [PubMed] [Google Scholar]
- McCall K, Steller H. Requirement for DCP-1 caspase during Drosophila oogenesis. Science. 1998;279:230–234. doi: 10.1126/science.279.5348.230. [DOI] [PubMed] [Google Scholar]
- McIlroy D, Tanaka M, Sakahira H, Fukuyama H, Suzuki M, Yamamura K-I, Ohsawa Y, Uchiyama Y, Nagata S. An auxiliary mode of apoptotic DNA fragmentation provided by phagocytes. Genes & Dev. 2000;14:549–558. [PMC free article] [PubMed] [Google Scholar]
- Mukae N, Yokoyama H, Yokokura T, Sakoyama Y, Sakahira H, Nagata S. Identification and developmental expression of inhibitor of caspase-activated DNase (ICAD) in Drosophila melanogaster. J Biol Chem. 2000;275:21402–21408. doi: 10.1074/jbc.M909611199. [DOI] [PubMed] [Google Scholar]
- Nagano M, Suzuki H, Ui-Tei K, Sato S, Miyake T, Miyata Y. H-7-induced apoptosis in the cells of a Drosophila neuronal cell line through affecting unidentified H-7-sensitive substance(s) Neurosci Res. 1998;31:113–121. doi: 10.1016/s0168-0102(98)00030-3. [DOI] [PubMed] [Google Scholar]
- Nagata S. Apoptotic DNA fragmentation. Exp Cell Res. 2000;256:12–18. doi: 10.1006/excr.2000.4834. [DOI] [PubMed] [Google Scholar]
- Oliveri M, Daga A, Cantoni C, Lunardi C, Millo R, Puccetti A. DNase I mediates internucleosomal DNA degradation in human cells undergoing drug-induced apoptosis. Eur J Immunol. 2001;31:743–751. doi: 10.1002/1521-4141(200103)31:3<743::aid-immu743>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- O'Neill LA. Toll-like receptor signal transduction and the tailoring of innate immunity: A role for Mal? Trends Immunol. 2002;23:296–300. doi: 10.1016/s1471-4906(02)02222-6. [DOI] [PubMed] [Google Scholar]
- Parrish J, Li L, Klotz K, Ledwich D, Wang X, Xue D. Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature. 2001;412:90–94. doi: 10.1038/35083608. [DOI] [PubMed] [Google Scholar]
- Rutschmann S, Jung AC, Zhou R, Silverman N, Hoffmann JA, Ferrandon D. Role of Drosophila IKKγ in a toll-independent antibacterial immune response. Nat Immunol. 2000;1:342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- Sakahira H, Enari M, Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature. 1998;391:96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- Sakahira H, Enari M, Nagata S. Functional differences of two forms of the inhibitor of caspase-activated DNase, ICAD-L, and ICAD-S. J Biol Chem. 1999;274:15740–15744. doi: 10.1074/jbc.274.22.15740. [DOI] [PubMed] [Google Scholar]
- Sakahira H, Iwamatsu A, Nagata S. Specific chaperone-like activity of inhibitor of caspase-activated DNase for caspase-activated DNase. J Biol Chem. 2000;275:8091–8096. doi: 10.1074/jbc.275.11.8091. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- Song Z, McCall K, Steller H. DCP-1, a Drosophila cell death protease essential for development. Science. 1997;275:536–540. doi: 10.1126/science.275.5299.536. [DOI] [PubMed] [Google Scholar]
- Staley K, Blaschke A, Chun J. Apoptotic DNA fragmentation is detected by a semi-quantitative ligation-mediated PCR of blunt DNA ends. Cell Death Differ. 1997;4:66–75. doi: 10.1038/sj.cdd.4400207. [DOI] [PubMed] [Google Scholar]
- Stone JC, Dower NA, Hauseman J, Cseko YM, Sederoff R. The characterization of a mutant affecting DNA metabolism in the development of D. melanogaster. Can J Genet Cytol. 1983;25:129–138. doi: 10.1139/g83-025. [DOI] [PubMed] [Google Scholar]
- Stroh C, Schulze-Osthoff K. Death by a thousand cuts: An ever increasing list of caspase substrates. Cell Death Differ. 1998;5:997–1000. doi: 10.1038/sj.cdd.4400451. [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Lazebnik Y. Caspases: Enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- Tower J, Karpen GH, Craig N, Spradling AC. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loo G, Schotte P, van Gurp M, Demol H, Hoorelbeke B, Gevaert K, Rodriguez I, Ruiz-Carrillo A, Vandekerckhove J, Declercq W, et al. Endonuclease G: A mitochondrial protein released in apoptosis and involved in caspase-independent DNA degradation. Cell Death Differ. 2001;8:1136–1142. doi: 10.1038/sj.cdd.4400944. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- Vernooy SY, Copeland J, Ghaboosi N, Griffin EE, Yoo SJ, Hay BA. Cell death regulation in Drosophila: Conservation of mechanism and unique insights. J Cell Biol. 2000;150:F69–F76. doi: 10.1083/jcb.150.2.f69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Grether M, Abrams J, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- Wicker C, Reichhart JM, Hoffmann D, Hultmark D, Samakovlis C, Hoffmann JA. Insect immunity. Characterization of a Drosophila cDNA encoding a novel member of the diptericin family of immune peptides. J Biol Chem. 1990;265:22493–22498. [PubMed] [Google Scholar]
- Widlak P, Li P, Wang X, Garrard WT. Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates. J Biol Chem. 2000;275:8226–8232. doi: 10.1074/jbc.275.11.8226. [DOI] [PubMed] [Google Scholar]
- Wu YC, Stanfield GM, Horvitz HR. NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes & Dev. 2000;14:536–548. [PMC free article] [PubMed] [Google Scholar]
- Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- Wyllie AH, Kerr JFR, Currie AR. Cell death: The significance of apoptosis. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- Yokoyama H, Mukae N, Sakahira H, Okawa K, Iwamatsu A, Nagata S. A novel activation mechanism of caspase-activated DNase from Drosophila melanogaster. J Biol Chem. 2000;275:12978–12986. doi: 10.1074/jbc.275.17.12978. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu X, Scherer DC, van Kaer L, Wang X, Xu M. Resistance to DNA fragmentation and chromatin condensation in mice lacking the DNA fragmentation factor 45. Proc Natl Acad Sci. 1998;95:12480–12485. doi: 10.1073/pnas.95.21.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]