Abstract
The luxCDABE operon of the bioluminescent bacterium Photorhabdus luminescens has proven to be a superb transcriptional reporter. It encodes a luciferase (LuxA and LuxB) and the enzymes that produce its substrate (LuxC, LuxD and LuxE) so cells that express the cluster emit the 490-nm light spontaneously. The sequence of these genes is AT-rich (>69%) and for this and other reasons, they are not expressed efficiently in high-GC bacteria like Streptomyces coelicolor. We therefore constructed a synthetic luxCDABE operon encoding the P. luminescens Lux proteins optimized for expression in high-GC bacteria. We tested the genes using transcriptional fusions to S. coelicolor promoters having well-established expression profiles during this organism's life cycle. The hrdB gene encodes a housekeeping sigma factor; while ramC is important for the formation of the spore-forming cells called aerial hyphae and whiE is required for the production of a grey, spore-associated pigment that is deposited in the walls of developing spores. Using these fusions we demonstrated that our synthetic lux genes are functional in S. coelicolor and that they accurately report complex developmental gene expression patterns. We suggest that this lux operon and our procedure for generating synthetic high-GC genes will be widely useful for research on high-GC bacteria.
INTRODUCTION
Reporter genes that permit facile detection of promoter activity are of central importance to molecular genetic research. One especially useful transcriptional reporter is the luxCDABE operon which encodes both luciferase (a heterodimer of LuxA and LuxB) and enzymes required for the production of its substrate tetradecanal (LuxC, LuxD and LuxE). The biochemical requirements for the production of light by these gene products are available in the cytoplasm of all aerobically growing organisms, so cells that express the luxCDABE operon emit light spontaneously (1).
Codon usage can limit the applicability of naturally occurring reporter genes to some species. For example, genes of the bacterial genus Streptomyces have a GC content that is typically >70% and there is evidence that translation in these organisms favours G and C residues in the wobble position of most codons (2). Furthermore, in Streptomyces coelicolor there is only one tRNA for the leucine-encoding codon TTA and it is encoded by the gene bldA (3). This bldA is dispensable for viability as it is only required for the expression of a subset of non-essential genes: several of these are required for antibiotic synthesis and the morphogenetic events that culminate with sporulation in this organism. Of greatest relevance to the work we report here, bldA is developmentally regulated such that its product is present at very low abundance during the first 24 h of colony growth and expressed poorly or not at all in liquid culture where sporulation does not occur. For this reason, genes bearing TTA codons are poorly expressed and developmentally regulated. Evidence suggests that this may also be the case in Streptomyces clavuligerus, S. halstedii, S. exfoliatus and S. griseus (4–7). The luxCDABE operon of P. luminescens is relatively rich in codons having either A or T in the wobble position (69%) suggesting that these genes would not be efficiently expressed in Streptomyces or many other high-GC organisms. Furthermore, there are 63 TTA codons in this operon: even in the event that these genes were translated efficiently they would be subject to developmental regulation via bldA that would largely eliminate their experimental utility.
A number of reporter systems have been developed for use in the streptomycetes; however, to date none has been adopted for routine analysis of gene expression. The most successful of these is probably the egfp gene, which encodes the enhanced green fluorescent protein (8). While the green fluorescent protein has revolutionized cell biology in this organism (9–13), it is not a particularly good transcriptional reporter. Its drawbacks are that it tends to photo-bleach rapidly, often compromising the weak signals from developmentally regulated promoters and furthermore S. coelicolor exhibits significant autofluorescence when illuminated with blue light (8). Other reporter genes that have been developed for use in Streptomyces include amy, xylE and melC, each with its own advantages and disadvantages (14–16). More recently, the luxAB genes from the bacterium Vibrio harveyii have been employed in Streptomyces (17). One drawback to these genes is that in order to visualize bioluminescence it is necessary to add the n-decanal substrate to cells and it is not clear that this substrate can pass through the cell wall of all S. coelicolor cell types with equivalent efficiency. Furthermore, if S. coelicolor is to be adapted for high throughput research, a likely and important direction for the field in our opinion, a simple reporter that does not require an added substrate would be advantageous. At present, the vast majority of molecular analysis of gene expression involves direct assessment of transcript levels using S1 nuclease analysis, primer extension, northern blotting or RT-PCR. Each of these has important virtues but all are excessively labour-intensive for high throughput experiments.
The streptomycetes are important subjects of research for several reasons. They produce most of the antibiotics in current use, an industry valued at >$25 billion per annum, as well as chemotherapeutic drugs, immune suppressants and other clinically useful compounds (18). They include at least one important agricultural pathogen, Streptomyces scabies (19) and are related to human pathogens such as the Corynebacteria and Mycobacteria. They also undergo a fascinating and experimentally tractable mechanism of cell differentiation and spore formation that is widely studied as a model for bacterial morphogenesis (20–22).
The Streptomyces life cycle commences with spore germination and the growth of filamentous cells called ‘substrate hyphae’. These grow by tip elongation and branching to produce a colony referred to as a ‘substrate mycelium’. After about two days, a second cell type, the ‘aerial hyphae’, appear on the colony surface and grow without branching up into the air. The aerial hyphae are specialized cells where spore formation takes place through a mechanism that involves, among other things, regulated cell division (9,23,24). Genes that are important for development include the bld genes, which are required for the erection of the aerial mycelium (20), the whi genes, which are required for spore maturation (21) and the ram genes, which are required for production of the molecule SapB that drives aerial growth of the spore-forming cells (22).
To date there have been no reports of the construction of high-GC synthetic genes (25–28). Indeed high-GC sequences can be challenging PCR templates as they are subject to frequent mis-priming and competitive side reactions, so there is reason to believe that the construction of high-GC synthetic genes might be especially difficult. In this work, we have constructed an entirely synthetic 5668 bp luxCDABE operon that lacks TTA codons and in which the majority of codons end in a G or C. We show that the genes are functional in S. coelicolor and that they accurately report the expression profiles of several genes. We suggest that this new synthetic gene cluster will prove applicable to many gene expression experiments in the streptomycetes, as well as other high-GC bacteria. We also suggest that our approach to the assembly of synthetic, high-GC genes could be used to adapt any low-GC genes for expression in high-GC organisms.
MATERIALS AND METHODS
Bacterial strains, plasmids and culture conditions
Bacterial strains and plasmids used in this study are listed in Tables 1 and 2, respectively. Plasmids were propagated in the Escherichia coli strain XL1 Blue. Escherichia coli strains were grown at 37°C in Luria broth media. Antibiotic concentrations were 100 μg/ml of ampicillin, 50 μg/ml of kanamycin, 34 μg/ml of chloramphenicol and 50 μg/ml of apramycin. For introduction of plasmids into S. coelicolor, plasmids were transformed into E. coli strain ET12567 containing the pUZ8002 plasmid allowing for conjugal transfer of the plasmid (29). S. coelicolor strains were grown on solid R2YE or SFM at 30°C. Antibiotic concentrations were 50 μg/ml apramycin and 25 μg/ml nalidixic acid.
Table 1.
Strains used in this work
| Strain | Genotype | Reference or source |
|---|---|---|
| Streptomyces coelicolor A3(2) | ||
| M145 | Prototrophic, SCP1- SCP2- | (47) |
| M600 | Prototrophic, SCP1- SCP2- | (48) |
| bldB | M145 bldB::aphI | (35) |
| ramR | M600 ramR::vph | Marie Elliot |
| whiG | M145 whiG::hyg | (36) |
| Escherichia coli | ||
| XL1 Blue | recA1 endA1 gyrA96 hsdR17 supE44 relA1 lac{F ′ proAB lacIQZΔM15Tn10 (Tetr)] | Stratagene |
| ET12567/pUZ8002 | dam13::Tn9 dcm-6 hsdM hsdR recF143 zjj-201::Tn10 galK2 galT22 ara14 lacY1 xyl-5 leuB6 thi-1 tonA31 rpsL136 hisG4 tsx-78 mtlI glnV44, containing the nontransmissible oriT mobilizing plasmid pUZ8002 | (29) |
Table 2.
Plasmids used in this work
| Plasmid | Description | Reference or source |
|---|---|---|
| Streptomyces coelicolor A3(2) | ||
| pRT801 | Integrative vector for Streptomyces; oriT(RK2) int attP (ΦBT1) aac(3)IV | (30) |
| pMU1 | Promoterless luxCDABE operon, flanked by transcriptional terminators, preceded by in-frame stop codon and ribosome binding site, aac(3)IV | This work |
| phrdBlux | hrdB promoter transcriptional fused to luxCDABE | This work |
| pramClux | ramC promoter transcriptionally fused to luxCDABE | This work |
| pwhiEp1lux | whiEp1 promoter transcriptionally fused to luxCDABE | This work |
| Escherichia coli | ||
| pBluescript II SK+ | pUC18 derivative for cloning in E. coli, bla | Stratagene |
| PBluescript + luxCDABE | Assembled luxCDABE operon | This work |
| pDONR221 | Gateway cloning vector, kan | Invitrogen |
| pDONR221 + luxA22 | luxA allele | This work |
| pDONR221 + luxB23 | luxB allele | This work |
| pDONR221 + luxC4 | luxC allele | This work |
| pDONR221 + luxD2 | luxD allele | This work |
| pDONR221 + luxE5 | luxE allele | This work |
Sequence and oligonucleotide design
Coding sequences for each of the P. luminescens lux genes were designed in silico, substituting codons enriched in GC residues and known to be frequently found in highly expressed S. coelicolor genes. The resulting genes have a GC content of 69%, similar to that of the S. coelicolor chromosome. The length of flanking sequences were shortened such that, following operon assembly (see below) the only non-coding sequence upstream of luxD, luxA, luxB and luxE was the 12 bp required to accommodate a correctly positioned Shine–Dalgarno sequence. The site upstream of the tuf1 gene of S. coelicolor was chosen for the Shine–Dalgarno and added upstream of each gene as we predicted that this would facilitate efficient initiation of translation. The Shine–Dalgarno sequence upstream of luxC was provided by cloning into pIJ8660 (8).
Synthetic DNAs of ∼100 nt were designed based on the top strand of each gene. Each oligonucleotide was analyzed using the ssDNA folding program mfold (see http://www.bioinfo.rpi.edu/applications/mfold/old/dna/). Sequences that exhibited significant intramolecular base pairing in the 20 nucleotides at their 5′ or 3′ end were modified to reduce the strength of, or eliminate, base pairing. This was accomplished by shifting the end by a few base pairs (and making a corresponding shift in the neighbouring oligonucleotide) and/or by making codon changes consistent with translation in Streptomyces that did not change the encoded protein sequence. 40-mer bridging oligonucleotides complementary to the 20 nt at the 5′ and 3′ ends of adjacent top strand oligonucleotides were then designed. The result of this was a set of 53 ∼100-mer oligonucleotides encoding the top strand of the five lux genes and 52 40-mer bridging oligonucleotides (see Supplementary Data). All of these oligonucleotides were run on 10% denaturing polyacrylamide sequencing gels (29:1 acrylamide: bis-acrylamide), visualized by UV-shadow casting, excised, eluted into water and quantified by spectrophotometry. In addition to these assembly sequences, amplification oligonucleotides were designed to amplify each individual lux gene, introducing Gateway recombination sequences at each end and, just inside these recombination sequences, recognition sequences for restriction endonucleases that would be used for subsequent assembly of the genes into an operon (see Supplementary Data).
Assembly and amplification
Each lux gene was assembled in reactions containing the top strand and corresponding bridging oligonucleotides (both at 25 μM) in Pfu reaction buffer and 5 μM dNTPs. PCR cycle regimens for each gene were as follows: an initial denaturation step was carried out at 94°C followed by 55 cycles of denaturation (94°C for 1 min), annealing (45°C for 40 s) and extension (72°C for 1 min). Following this, 15 μM of the appropriate amplifications primers were used to amplify each lux gene in reactions primed with 5 μl of assembly reaction—conditions for PCR were identical except that 25 cycles of amplification were used instead of 55 and a terminal extension incubation of 4 min, at 72°C was added. All reactions were carried out using a robo-cycler 96 (Stratagene) with zero ramp time between temperatures. Fragments were visualized by agarose gel electrophoresis, excised and purified using QIAEX II gel extraction.
Cloning, correction and operon assembly
Each amplified fragment was cloned into the Gateway entry vector pDONR221 by in vitro recombination as specified by the manufacturer. Transformants containing clones with inserts of the correct length were identified by colony PCR using the amplification primers. Multiple clones were isolated for each lux gene (see Table 3) and subjected to sequencing with the universal primers and, where necessary, primers internal to the cloned insert. Most of the cloned inserts had sequence errors relative to the expected sequence; however some of these had no effect on the sequence of the encoded protein. One allele, luxD2 encoded a perfect LuxD protein and was not subjected to correction mutagenesis. The best alleles of the other four genes: luxC4, luxA22, luxB23 and luxE5 were subjected to quickchange mutagenesis to correct all sequence errors that affected the protein product. Finally, each gene was excised from the pDONR221 vector and assembled into an operon in pBluescript II SK+ (Figure 1C). The sequence of the completed operon was deposited into genbank (accession EF173694). Once assembled, the operon was excised as a 5668 bp NdeI-NotI fragment and inserted into the backbone of pRT801 cut with the same enzymes (30). The phage fd transcriptional terminator tfd and the phage λ transcriptional terminator to were cloned from pIJ8660 (8) to flank the lux operon. The terminator tfd, multiple cloning site and Shine–Dalgarno of pIJ8660 were cloned by adding a HindIII site upstream of luxCDABE at the BamHI site in pRT801+luxCDABE with the oligonucleotides 5′ GATCGGGAAGCTTCCC 3′ and 5′ CTAGCCCTTCGAAGGG 3′ and digesting both pRT801+luxCDABE and pIJ8660 with HindIII and BamHI. The to terminator was removed from pIJ8660 by digesting with NotI and EcoRI and cloned into pRT801+(tfd)luxCDABE by blunting the EcoRI site with Klenow (New England Biolabs) and inserting this fragment downstream of luxCDABE by cutting the pRT801+(tfd)luxCDABE plasmid with NotI and EcoRV, creating the plasmid pMU1. The relevant structural details of pMU1 are shown in Figure 2.
Table 3.
Isolation of individual lux genes after assembly by PCR
| Gene | Clones isolated | Clones with correct size | Best candidate | Mutationsa |
|---|---|---|---|---|
| luxA | 40 | 8 | A22 | 3 bp deletion at 109 |
| 5 bp deletion at 882 | ||||
| luxB | 35 | 14 | B23 | point mutation at 907 |
| point mutation at 912 | ||||
| luxC | 10 | 7 | C4 | 1 bp insertion at 551 |
| 3 bp deletion at 575 | ||||
| luxD | 15 | 6 | D2 | none |
| luxE | 25 | 6 | E5 | point mutation at 726 |
| 1 bp deletions at 970, 988 and 1026 |
athat altered protein sequence
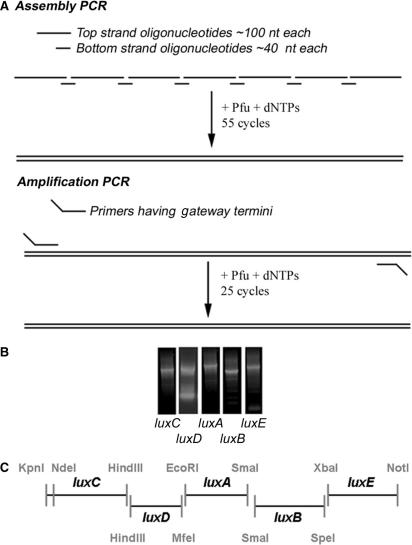
Figure 1.
(A) Schematic diagram of assembly/PCR strategy. In the assembly PCR, the top strand oligonucleotides were mixed with bottom strand oligonucleotides in equimolar amounts. A PCR reaction of 55 cycles annealed the primers to create the full-length product. In the second PCR step, the amplification PCR, the PCR product from the assembly PCR was incubated with Gateway primers which introduced sites for entry into the Gateway cloning vector and restriction sites for subsequent cloning steps. (B) Individual amplified lux genes assembled using the PCR strategy. PCR products resulting from the two rounds of PCR are shown. Expected sizes were luxC 1.5 kb, luxD 1.0 kb, luxA 1.1 kb, luxB 1.1 kb, and luxE 1.1 kb. Fragments of the appropriate size were gel purified and introduced individually into pDONR221 using the Gateway sites for homologous recombination. (C) Strategy for luxCDABE operon assembly. The amplified high-GC lux genes were finally assembled into an operon through restriction sites flanking each individual gene which corresponded to the pBluescript II SK+ multiple cloning site.
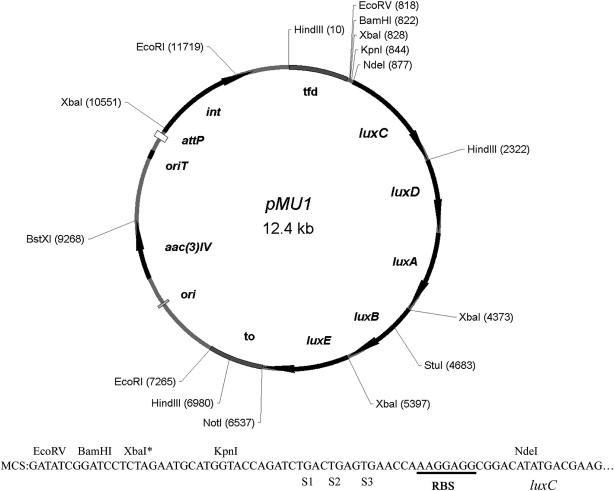
Figure 2.
pMU1, an integrating lux reporter plasmid. tfd, the major transcriptional terminator of phage fd; to, transcriptional terminator of phage λ; ori, the origin of replication from pUC18; aac3(IV) apramycin-resistance cassette selectable in E. coli and Streptomyces; oriT, origin of transfer from the RK2 plasmid; int and attP, the integrase gene and attachment site of the ΦBT1 phage, respectively. The multiple cloning site consists of EcoRV, BamHI and KpnI site (non-unique sites are marked with*). The multiple cloning site is followed by translational stop codons in all three reading frames (S1, S2, S3) and a ribosome binding site (RBS).
Molecular cloning
Promoter fragments for hrdB, ramC and whiEp1 were amplified from S. coelicolor M145 chromosomal DNA using Pfu and primers listed in the Supplementary Data. These fragments were cloned into pMU1 using BamHI and KpnI in the multiple cloning site to create the plasmids, phrdBlux, pramClux and pwhiEp1lux.
Growth curves
Using 96-well white polystyrene plates (Fisher), strains were inoculated to a confluent lawn into wells containing 200 μl of R2YE or SFM with 12 replicates inoculated for each promoter-lux fusion. Plates were grown at 30°C with luminescence read every 8 h using the EnVision Multilabel Reader (PerkinElmer).
RESULTS
Assembly of a high-GC luxCDABE operon
We initiated this work by designing luxC, luxD, luxA, luxB and luxE sequences in silico that were optimized for expression in S. coelicolor and flanked by restriction endonuclease recognition sites for assembly into an operon (Figure 1C). The Shine–Dalgarno site upstream of the S. coelicolor tuf1 gene was included 8 bp upstream of the methionine-encoding start codon of each gene.
PCR-mediated amplification of DNA sequences that are rich in G and C base pairs is known to be more difficult than with balanced sequences for several reasons. First, the reduced complexity of the DNA sequence lowers the selectivity of base-pairing resulting in more frequent mis-priming. Second, as mis-primed DNA sequences are usually the most GC rich, they tend to be highly stable duplexes. Third, primers frequently have the capacity to form highly stable intramolecularly base-paired structures. The action of DNA polymerase on any of these mis-primed structures results in competing side reactions that can dramatically lower the yield of, or prevent altogether, the production of the desired fragment. We therefore took precautions to minimize the formation of mis-annealed duplexes, focusing mostly on the third caveat mentioned above: the capacity of oligonucleotides to form intramolecular rather than intermolecular duplexes (see the Materials and methods section).
We designed oligonucleotides of 90–110 nt corresponding to the top strand of each sequence. We also designed 40-nt bridging oligonucleotides complementary to the 20 nt at either end of adjacent top strand oligonucleotides. The top strand DNAs were designed to have minimal capacity to form intramolecular secondary structures in the 20-nt stretches complementary to the bridge DNAs (see the Materials and methods section): trial sequences were folded in silico and, if possible, such duplexes were eliminated or weakened by altering codons in the wobble position. In this way, we tried to favour the formation of intermolecular duplexes between the top strands and the bridging oligonucleotides over intramolecular snap-back structures. The resulting oligonucleotide sequences are shown in the Supplementary Data.
Genes were then assembled in a two-step process. First, equimolar mixtures of top strand and bridging DNAs for each lux gene were subjected to a 55-cycle assembly reaction with Pfu DNA polymerase (Figure 1A—Assembly PCR). Second, the products of assembly reactions were gel purified, incubated with PCR primers complementary to the 5′ and 3′ ends of the genes, each including the sequences necessary for Gateway-mediated recombinational cloning and for subsequent operon assembly, and subjected to a 25-cycle amplification reaction (Figure 1A—Amplification PCR). The products of amplification reactions for luxC, luxD, luxA, luxB and luxE are shown in Figure 1B. Fragments of the appropriate size were then gel purified and introduced into the Gateway entry vector pDONR221 using bacteriophage λ Integrase.
We determined the sequence of a number of alleles for each cloned lux gene to establish whether the assembly and amplification reactions had yielded the desired sequences. We identified one perfect allele of luxD as well as alleles of the other genes that contained a small enough number of sequence alterations for correction by site-directed mutagenesis (Table 3). Once we obtained perfect clones for each gene, we assembled a synthetic luxCDABE operon (genbank accession EF173694) in the vector pBluescript II SK+ (Figure 1C). The assembled operon was excised as a 5668 bp NdeI-NotI fragment and ligated to the backbone of the vector pRT801 (30). The transcriptional terminators flanking the multiple cloning site of pIJ8660 (8) were transferred as restriction fragments (see the Materials and methods section) to flank luxCDABE, creating the vector pMU1 (Figure 2). This vector can be conjugated directly from E. coli to S. coelicolor and integrates site-specifically into the S. coelicolor chromosome to ensure single copy gene expression (30).
Bioluminescence directed by the ramC, whiEp1 and hrdB promoters
To test the expression of our synthetic lux operon in S. coelicolor, we introduced DNA fragments upstream of luxCDABE containing the promoters for the gene hrdB, which encodes an S. coelicolor housekeeping sigma factor and those of two genes involved in morphogenesis: ramC and whiE. ramC, which is the first gene in the ramCSAB operon, encodes an enzyme required for production of the morphogenetic peptide SapB (22,31,32). The expression of this operon depends on a number of other developmental genes including bldB and ramR (13,33). The whiE gene cluster expresses a polyketide synthase that produces a grey pigment that is deposited on the spore surface late in development and depends on the activity of earlier developmental genes. whiEp1 is the stronger of two divergent promoters within the whiE gene cluster (34). We then introduced the resulting plasmids phrdBlux, pramClux and pwhiEp1lux into the morphogenetically wild-type strains M145 and M600 as well as developmental mutants bldB (35), ramR (kindly provided by Marie Elliot) and whiG (36).
Strains containing pMU1 and the promoter-lux fusions were then tested for luminescence on solid media during a 3-day time course. Strains were inoculated to a confluent lawn on 200 μl of solid R2YE or SFM media in 96-well tissue culture plates with 12 replicates designated for each promoter fusion and plates incubated at 30°C. Luminescence was measured at 8 h time points using an Envision multilabel plate reader. We investigated normalizing the luminescence data to the mycelial mass but found that this had no effect on the resulting expression profiles—as long as a confluent lawn was growing in the well, simply measuring luminescence without normalizing to mass gave a reading that was reproducible from well to well. Luminescence patterns in the wild-type strains M145 and M600 were very similar and as a result only the data for M145 are shown.
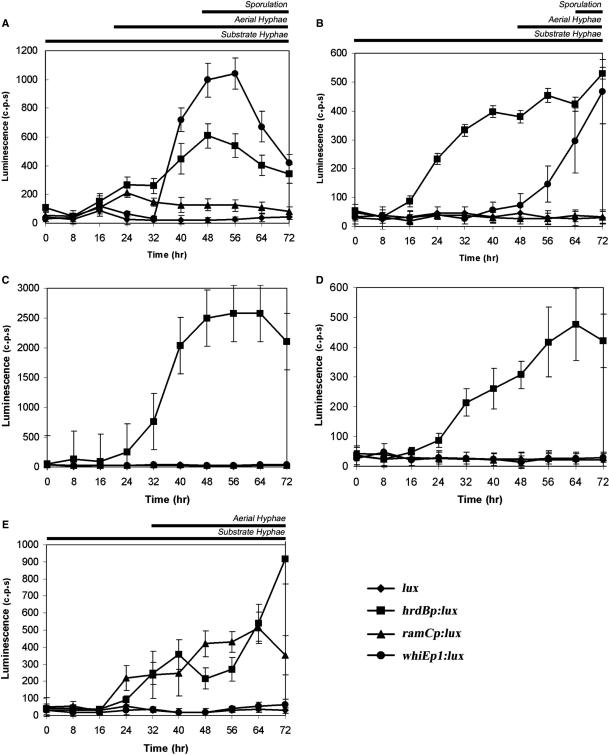
We observed no background luminescence in strains containing the promoterless lux operon. In contrast, the hrdB promoter fusion was clearly active in wild-type and the three developmental mutants (Figure 3A–E). Little to no expression was detected during the first 8 h of growth. However by 16 h, hrdB promoter-dependent luminescence could be detected that was reproducible and well above that of the promoterless control. Luminescence levels directed by this promoter were similar in M145, the ramR and whiG mutants, reaching values of ∼500 c.p.s. Interestingly, hrdB promoter-directed luminescence was reproducibly ∼5× higher in the bldB mutant than in the other strains, routinely reaching 2500 c.p.s. (Figure 3C).
Figure 3.
Activity of the lux operon in S. coelicolor when grown on solid media. phrdBlux, pramClux, pwhiEp1lux and pMU1 were introduced into wild-type M145 and developmental mutants bldB, ramR and whiG. (A) M145 containing the lux promoter fusions grown on R2YE. (B) M145 containing the lux promoter fusions grown on SFM. (C) bldB mutant strain containing the lux promoter fusions grown on R2YE. (D) ramR mutant strain containing the lux promoter fusions grown on R2YE. (E) whiG mutant strain containing the lux promoter fusions grown on R2YE.
In wild-type cells, detectable luminescence from whiEp1 was achieved after 40 h of growth and reached a peak of ∼1000 c.p.s. at 56 h (Figure 3A). In agreement with the established requirements for activation of the whiE cluster, no significant luminescence was detected from the whiEp1 promoter in any of the developmental mutants tested as they are blocked in either aerial hyphae formation (bldB and ramR) or maturation of the aerial hyphae prior to division (whiG) (Figure 3C–E).
The formation of aerial hyphae on rich media depends on the ramCSAB gene cluster as it is the source of a surfactant molecule, SapB that facilitates upward growth of the spore-forming cells. We measured bioluminescence directed by pramClux on the rich medium R2YE and observed significant luminescence at 16 h, which then peaked at 24 h and decreased at subsequent time points (Figure 3A). We detected no ramC promoter-dependent luminescence in either the bldB and ramR mutant strains, consistent with previous results (13,33) (Figure 3C and D). In the whiG null strain, no observable luminescence was detected for the first 16 h of growth from the ramC promoter, after which expression increased throughout the rest of development reaching a peak value of ∼500 c.p.s. at 64 h (Figure 3E). This is consistent with western blot analysis demonstrating that RamC production is up-regulated in many of the whi mutants, including whiG, such that RamC accumulates at later time points instead of peaking at aerial hyphae formation (O’Connor,T., Zhang,D. and Nodwell,J. unpublished data).
The ram gene cluster is dispensable for aerial mycelium formation when S. coelicolor is grown on some carbon sources such as mannitol and indeed, SapB is not produced under these conditions (37). In agreement with this, when we cultivated wild-type M145 cells containing the lux fusions on SFM (a mannitol-containing media) we detected luminescence directed by both the hrdB and whiEp1 promoters but none from the ramC promoter (Figure 3B). Luminescence from the hrdB promoter reached similar levels of expression as when grown on rich media with values reaching ∼500 c.p.s. Expression from whiEp1 was delayed in comparison to growth on rich media, as was the production of aerial hyphae and grey pigmented spores.
DISCUSSION
We have constructed a luxCDABE gene cluster optimized for expression in high-GC bacteria and established that the cluster is active in S. coelicolor. Fusions of this cluster to promoters for the genes hrdB, ramC and whiE faithfully report their activities via spontaneous bioluminescence. The activity of each promoter behaved precisely as expected: the hrdB promoter fusion was active in wild-type cells and developmental mutants, the ramC promoter required the genes bldB and ramR but not whiG, and whiEp1 was dependent on bldB, ramR and whiG. These fusions also reiterated previously demonstrated medium dependence of the developmental genes: the ramC promoter was active on rich medium but not on SFM while whiEp1 and hrdB were active on both media. We predict therefore that this gene cluster will be widely applicable for routine gene expression studies in S. coelicolor and other high-GC micro-organisms.
The utility of this operon is evident in the fact that there was no background luminescence under any circumstances and strains bearing the promoterless control exhibited similar luminescence readings as strains lacking the genes altogether. This is not the case for reporters such as egfp (S. coelicolor exhibits significant autofluorescence), xylE or amy. We found that this reporter works exceptionally well in 96-well format, in turn permitting a large number of replicates for each measurement. As a result we have been able to calculate standard deviations for all the values we have recorded and have a very high degree of confidence in the data. While S1 nuclease analysis is an excellent tool for mapping 5′ ends of transcripts and can be used for comparative analysis of promoter activities, it is simply too labour-intensive to be used with this density of replicates. In addition to the luminescence we detected from the ramC, whiEp1 and hrdB promoters, we have also successfully observed luminescence from fusions to the promoters of the antibiotic biosynthetic genes actI ORF1, actII ORF2 and actII ORF4. As with the developmental promoters reported here, each behaved in a manner that is biologically relevant. These data suggest that this reporter will greatly facilitate future gene expression studies in Streptomyces. Variation in fatty acid pools needed for biosynthesis of the lux substrate may vary in some growth conditions and we imagine that this could compromise some expression data. Should problematic behaviour arise, a solution would be to resort to added n-decanal substrate such that the luminescence is more consistent. This can also be used to boost the signals of exceptionally weak promoters (data not shown). We have found that most promoter-lux fusions discussed in this and in other work (Xu,Y. and Nodwell, J. unpublished data) are consistent under a variety of conditions and doubt that substrate addition will be necessary under the majority of growth conditions.
We were especially intrigued by the results comparing the activity of the hrdB promoter in wild-type S. coelicolor and in the bldB mutant. The hrdB transcript is often used as a reference in the analysis of promoter activities (11,34,38–41) and implicit in this is the assumption that it is expressed more or less invariantly in all strains. Our results suggest that this may not be true in all cases. We found that lux activity directed by the hrdB promoter is almost identical in wild-type cells and in ramR and whiG mutants. In contrast however, the promoter appeared to be ∼5× more active in the bldB mutant strain. Aerial hyphae formation and sporulation is known to involve the action of at least four sigma factors, bldN (42), whiG (43), sigF (44) and sigH (45); while the sigma factor encoded by hrdD has been implicated in antibiotic production (46). The bldB null mutant suffers particularly extreme effects on both antibiotic production and development; this likely includes the failure to express many of these sigma factor genes. Perhaps the absence of these sigma factors frees up RNA polymerase holoenzyme in a way that somehow enhances the expression of hrdB. We do not mean to imply that this invalidates previous studies in which hrdB was used as a standard: as far as we can tell from this work (and a parallel project on genes involved in antibiotic production), the effect of the bldB mutation on hrdB promoter activity appears to be unique.
A simple reporter system for the Streptomyces bacteria will be a very useful research tool. These data demonstrate that our synthetic luxCDABE operon is active in substrate and aerial hyphae and that it is not significantly toxic to either cell type. It provides a means of accurately monitoring expression of several genes as a function of colony growth and furthermore, can provide a simple means for detecting epistasis relationships between genes. We predict that this construct will be highly useful for studies in S. coelicolor and other high-GC bacteria.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to Dr John Capone who supported this project as Associate Dean of Medicine at McMaster University. We also thank the staff of the MOBIX centre for their patient assistance with the construction of oligonucleotides and the sequencing of the synthetic lux genes and Marie Elliot for providing the ramR null strain and its parental strain M600. This work was supported with grants MOP-57684 and MOP-68817 and a New Investigator Award for JN from the Canadian Institutes for Health Research. Funding to pay the Open Access publication charge was provided by CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Meighen EA. Bacterial bioluminescence: Organization, regulation, and application of the lux genes. FASEB J. 1993;7:1016–1022. doi: 10.1096/fasebj.7.11.8370470. [DOI] [PubMed] [Google Scholar]
- 2.Sanli G, Blaber SI, Blaber M. Reduction of wobble-position GC bases in Corynebacteria genes and enhancement of PCR and heterologous expression. J. Mol. Microbiol. Biotechnol. 2001;3:123–126. [PubMed] [Google Scholar]
- 3.Leskiw BK, Lawlor EJ, Fernandez-Abalos JM, Chater KF. TTA codons in some genes prevent their expression in a class of developmental, antibiotic-negative, Streptomyces mutants. Proc. Natl. Acad. Sci. USA. 1991;88:2461–2465. doi: 10.1073/pnas.88.6.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwak J, McCue LA, Kendrick KE. Identification of bldA mutants of Streptomyces griseus. Gene. 1996;171:75–78. doi: 10.1016/0378-1119(96)00066-2. [DOI] [PubMed] [Google Scholar]
- 5.Garda AL, Fernandez-Abalos JM, Sanchez P, Ruiz-Arribas A, Santamaria RI. Two genes encoding an endoglucanase and a cellulose-binding protein are clustered and co-regulated by a TTA codon in Streptomyces halstedii JM8. Biochem. J. 1997;324(Pt 2):403–411. doi: 10.1042/bj3240403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Servin-Gonzalez L, Castro C, Perez C, Rubio M, Valdez F. bldA-dependent expression of the Streptomyces exfoliatus M11 lipase gene (lipA) is mediated by the product of a contiguous gene, lipR, encoding a putative transcriptional activator. J. Bacteriol. 1997;179:7816–7826. doi: 10.1128/jb.179.24.7816-7826.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trepanier NK, Jensen SE, Alexander DC, Leskiw BK. The positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus is mistranslated in a bldA mutant. Microbiology. 2002;148:643–656. doi: 10.1099/00221287-148-3-643. [DOI] [PubMed] [Google Scholar]
- 8.Sun J, Kelemen GH, Fernandez-Abalos JM, Bibb MJ. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2) Microbiology. 1999;145(Pt 9):2221–2227. doi: 10.1099/00221287-145-9-2221. [DOI] [PubMed] [Google Scholar]
- 9.Grantcharova N, Lustig U, Flardh K. Dynamics of FtsZ assembly during sporulation in Streptomyces coelicolor A3(2) J. Bacteriol. 2005;187:3227–3237. doi: 10.1128/JB.187.9.3227-3237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flardh K. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2) Mol. Microbiol. 2003;49:1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- 11.Jakimowicz D, Mouz S, Zakrzewska-Czerwinska J, Chater KF. Developmental control of a parAB promoter leads to formation of sporulation-associated ParB complexes in Streptomyces coelicolor. J. Bacteriol. 2006;188:1710–1720. doi: 10.1128/JB.188.5.1710-1720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruban-Osmialowska B, Jakimowicz D, Smulczyk-Krawczyszyn A, Chater KF, Zakrzewska-Czerwinska J. Replisome localization in vegetative and aerial hyphae of Streptomyces coelicolor. J. Bacteriol. 2006;188:7311–7316. doi: 10.1128/JB.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Connor TJ, Kanellis P, Nodwell JR. The ramC gene is required for morphogenesis in Streptomyces coelicolor and expressed in a cell type-specific manner under the direct control of RamR. Mol. Microbiol. 2002;45:45–57. doi: 10.1046/j.1365-2958.2002.03004.x. [DOI] [PubMed] [Google Scholar]
- 14.Ingram C, Brawner M, Youngman P, Westpheling J. xylE functions as an efficient reporter gene in Streptomyces spp.: Use for the study of galP1, a catabolite-controlled promoter. J. Bacteriol. 1989;171:6617–6624. doi: 10.1128/jb.171.12.6617-6624.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flores FJ, Rincon J, Martin JF. Characterization of the iron-regulated desA promoter of Streptomyces pilosus as a system for controlled gene expression in actinomycetes. Microb. Cell. Fact. 2003;2:5. doi: 10.1186/1475-2859-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paget MS, Hintermann G, Smith CP. Construction and application of streptomycete promoter probe vectors which employ the Streptomyces glaucescens tyrosinase-encoding gene as reporter. Gene. 1994;146:105–110. doi: 10.1016/0378-1119(94)90842-7. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Garcia A, Combes P, Perez-Redondo R, Smith MC, Smith MC. Natural and synthetic tetracycline-inducible promoters for use in the antibiotic-producing bacteria Streptomyces. Nucleic Acids Res. 2005;33:e87. doi: 10.1093/nar/gni086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berdy J. Bioactive microbial metabolites. J. Antibiot. (Tokyo) 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 19.Loria R, Kers J, Joshi M. Evolution of plant pathogenicity in Streptomyces. Annu. Rev. Phytopathol. 2006;44:469–487. doi: 10.1146/annurev.phyto.44.032905.091147. [DOI] [PubMed] [Google Scholar]
- 20.Kelemen GH, Buttner MJ. Initiation of aerial mycelium formation in Streptomyces. Curr. Opin. Microbiol. 1998;1:656–662. doi: 10.1016/s1369-5274(98)80111-2. [DOI] [PubMed] [Google Scholar]
- 21.Chater KF. Regulation of sporulation in Streptomyces coelicolor A3(2): A checkpoint multiplex? Curr. Opin. Microbiol. 2001;4:667–673. doi: 10.1016/s1369-5274(01)00267-3. [DOI] [PubMed] [Google Scholar]
- 22.Willey JM, Willems A, Kodani S, Nodwell JR. Morphogenetic surfactants and their role in the formation of aerial hyphae in Streptomyces coelicolor. Mol. Microbiol. 2006;59:731–742. doi: 10.1111/j.1365-2958.2005.05018.x. [DOI] [PubMed] [Google Scholar]
- 23.Schwedock J, McCormick JR, Angert ER, Nodwell JR, Losick R. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol. Microbiol. 1997;25:847–858. doi: 10.1111/j.1365-2958.1997.mmi507.x. [DOI] [PubMed] [Google Scholar]
- 24.Flardh K, Leibovitz E, Buttner MJ, Chater KF. Generation of a non-sporulating strain of Streptomyces coelicolor A3(2) by the manipulation of a developmentally controlled ftsZ promoter. Mol. Microbiol. 2000;38:737–749. doi: 10.1046/j.1365-2958.2000.02177.x. [DOI] [PubMed] [Google Scholar]
- 25.Stahler P, Beier M, Gao X, Hoheisel JD. Another side of genomics: Synthetic biology as a means for the exploitation of whole-genome sequence information. J. Biotechnol. 2006;124:206–212. doi: 10.1016/j.jbiotec.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Wu G, Zheng Y, Qureshi I, Zin HT, Beck T, Bulka B, Freeland SJ. SGDB: A database of synthetic genes re-designed for optimizing protein over-expression. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl648. 10.1093/nar/gkl648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Au LC, Yang FY, Yang WJ, Lo SH, Kao CF. Gene synthesis by a LCR-based approach: High-level production of leptin-L54 using synthetic gene in Escherichia coli. Biochem. Biophys. Res. Commun. 1998;248:200–203. doi: 10.1006/bbrc.1998.8929. [DOI] [PubMed] [Google Scholar]
- 28.Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 29.Flett F, Mersinias V, Smith CP. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 1997;155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 30.Gregory MA, Till R, Smith MC. Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J. Bacteriol. 2003;185:5320–5323. doi: 10.1128/JB.185.17.5320-5323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudson ME, Nodwell JR. Dimerization of the RamC morphogenetic protein of Streptomyces coelicolor. J. Bacteriol. 2004;186:1330–1336. doi: 10.1128/JB.186.5.1330-1336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kodani S, Hudson ME, Durrant MC, Buttner MJ, Nodwell JR, Willey JM. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA. 2004;101:11448–11453. doi: 10.1073/pnas.0404220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willey J, Schwedock J, Losick R. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 1993;7:895–903. doi: 10.1101/gad.7.5.895. [DOI] [PubMed] [Google Scholar]
- 34.Kelemen GH, Brian P, Flardh K, Chamberlin L, Chater KF, Buttner MJ. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3 (2) J. Bacteriol. 1998;180:2515–2521. doi: 10.1128/jb.180.9.2515-2521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eccleston M, Ali RA, Seyler R, Westpheling J, Nodwell J. Structural and genetic analysis of the BldB protein of Streptomyces coelicolor. J. Bacteriol. 2002;184:4270–4276. doi: 10.1128/JB.184.15.4270-4276.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flardh K, Findlay KC, Chater KF. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2) Microbiology. 1999;145(Pt 9):2229–2243. doi: 10.1099/00221287-145-9-2229. [DOI] [PubMed] [Google Scholar]
- 37.Willey J, Santamaria R, Guijarro J, Geistlich M, Losick R. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell. 1991;65:641–650. doi: 10.1016/0092-8674(91)90096-h. [DOI] [PubMed] [Google Scholar]
- 38.Yamazaki H, Ohnishi Y, Horinouchi S. An A-factor-dependent extracytoplasmic function sigma factor (sigma(AdsA)) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 2000;182:4596–4605. doi: 10.1128/jb.182.16.4596-4605.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryding NJ, Kelemen GH, Whatling CA, Flardh K, Buttner MJ, Chater KF. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2) Mol. Microbiol. 1998;29:343–357. doi: 10.1046/j.1365-2958.1998.00939.x. [DOI] [PubMed] [Google Scholar]
- 40.Lee EJ, Karoonuthaisiri N, Kim HS, Park JH, Cha CJ, Kao CM, Roe JH. A master regulator sigmaB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol. Microbiol. 2005;57:1252–1264. doi: 10.1111/j.1365-2958.2005.04761.x. [DOI] [PubMed] [Google Scholar]
- 41.Kelemen GH, Brown GL, Kormanec J, Potuckova L, Chater KF, Buttner MJ. The positions of the sigma-factor genes, whiG and sigF, in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2) Mol. Microbiol. 1996;21:593–603. doi: 10.1111/j.1365-2958.1996.tb02567.x. [DOI] [PubMed] [Google Scholar]
- 42.Bibb MJ, Molle V, Buttner MJ. Sigma(BldN), an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2) J. Bacteriol. 2000;182:4606–4616. doi: 10.1128/jb.182.16.4606-4616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendez C, Chater KF. Cloning of whiG, a gene critical for sporulation of Streptomyces coelicolor A3(2) J. Bacteriol. 1987;169:5715–5720. doi: 10.1128/jb.169.12.5715-5720.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potuckova L, Kelemen GH, Findlay KC, Lonetto MA, Buttner MJ, Kormanec J. A new RNA polymerase sigma factor, sigma F, is required for the late stages of morphological differentiation in Streptomyces spp. Mol. Microbiol. 1995;17:37–48. doi: 10.1111/j.1365-2958.1995.mmi_17010037.x. [DOI] [PubMed] [Google Scholar]
- 45.Kormanec J, Sevcikova B, Halgasova N, Knirschova R, Rezuchova B. Identification and transcriptional characterization of the gene encoding the stress-response sigma factor sigma(H) in Streptomyces coelicolor A3(2) FEMS Microbiol. Lett. 2000;189:31–38. doi: 10.1111/j.1574-6968.2000.tb09202.x. [DOI] [PubMed] [Google Scholar]
- 46.Fujii T, Gramajo HC, Takano E, Bibb MJ. redD and actII-ORF4, pathway-specific regulatory genes for antibiotic production in Streptomyces coelicolor A3(2), are transcribed in vitro by an RNA polymerase holoenzyme containing sigma HrdD. J. Bacteriol. 1996;178:3402–3405. doi: 10.1128/jb.178.11.3402-3405.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kieser T, Bibb M, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich: The John Innes Foundation; 2000. [Google Scholar]
- 48.Chakraburtty R, Bibb M. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 1997;179:5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.