Abstract
Genome segmentation facilitates reassortment and rapid evolution of influenza A virus. However, segmentation complicates particle assembly as virions must contain all eight vRNA species to be infectious. Specific packaging signals exist that extend into the coding regions of most if not all segments, but these RNA motifs are poorly defined. We measured codon variability in a large dataset of sequences to identify areas of low nucleotide sequence variation independent of amino acid conservation in each segment. Most clusters of codons showing very little synonymous variation were located at segment termini, consistent with previous experimental data mapping packaging signals. Certain internal regions of conservation, most notably in the PA gene, may however signify previously unidentified functions in the virus genome. To experimentally test the bioinformatics analysis, we introduced synonymous mutations into conserved codons within known packaging signals and measured incorporation of the mutant segment into virus particles. Surprisingly, in most cases, single nucleotide changes dramatically reduced segment packaging. Thus our analysis identifies cis-acting sequences in the influenza virus genome at the nucleotide level. Furthermore, we propose that strain-specific differences exist in certain packaging signals, most notably the haemagglutinin gene; this finding has major implications for the evolution of pandemic viruses.
INTRODUCTION
Influenza A virus poses a major threat to public health with the potential to cause global pandemics with mortality figures in the millions, despite effective vaccines and antiviral drugs (1). This danger primarily results from the generation of new viral variants through reassortment of the segmented RNA genome. The eight single-strand RNA segments that comprise the genome of influenza A viruses are encapsidated into separate ribonucleoprotein (RNP) structures by a nucleoprotein (NP) and RNA polymerase (2,3). After dual infection, progeny viruses bearing mixtures of segments from the parental viruses are readily found, suggesting that mixing of individual RNPs occurs prior to their packaging. Although this phenomenon of genome reassortment is hugely significant in the evolution of influenza A virus (4), the mechanisms underlying RNP incorporation into virions are incompletely understood. Understanding this more fully is crucial to assessing the pandemic threat of emerging strains of influenza virus.
It has been disputed whether a mechanism exists to ensure that each virion contains a full complement of the eight segments. The terminal promoter sequences for the viral RNA polymerase are the minimal RNA sequences required for packaging of vRNA molecules (5) and are conserved in all segments. This, together with genetic evidence for virus particles containing nine or more segments, supports a model in which packaging is at least partially random (6,7). However, electron microscopy of virus particles showed that most contain eight structures plausibly representing RNPs organized as an array of seven around a central member (8). The odds of obtaining an infectious genome if eight segments were incorporated at random are punitive (0.0024 at best), suggesting a specific mechanism to package a full complement of vRNAs would be evolutionarily favoured. Supporting this hypothesis, internally deleted defective-interfering (DI) vRNAs specifically compete with their larger parents for incorporation into virus particles, suggesting the existence of segment-specific packaging signals (9–12). Furthermore, studies employing reverse genetic techniques to construct mutant vRNA segments with defined deletions have confirmed that sequences extending into the unique coding regions are necessary for the efficient packaging of segments 1–6 and 8 (13–20).
Thus most if not all influenza A virus vRNA segments contain sequences that act as specific packaging signals. However, how these signals function remains unclear, partly because in the majority of cases, the RNA signals have only been coarsely mapped. Clearly, further experimental analysis will refine this. Alternatively, it is reasonable to hypothesize that functionally important cis-acting sequences will be evolutionarily conserved. However, since many packaging signals extend into the protein-coding regions of the RNAs (11–20), these RNA sequences will be subjected to evolutionary pressures (resulting in sequence constraint) both through their translation product(s) and any embedded cis-acting functions. However, redundancy in the genetic code makes it possible to partially untangle these forces. We reasoned that given sufficient evolutionary time, conservation of a protein sequence will not necessarily be matched at the nucleotide level for those amino acid with multiple codons. Furthermore, we hypothesized that functional conservation of RNA would occur at codons grouped closely together and thus could be distinguished from an overall codon bias resulting from skewed base composition of the virus genome or from a dataset of limited evolutionary time.
Using this approach, we found highly statistically significant clusters of codons with lower than expected synonymous variation within the influenza virus genome. These clusters were mostly but not exclusively located at the terminal regions of segments, where existing experimental data indicates the presence of specific packaging signals. Synonymous mutational analysis of two of these regions confirmed the ability of our method to identify functionally significant cis-acting elements in the virus genome at the single nucleotide level.
MATERIALS AND METHODS
Sequence preparation
The initial set of sequences was obtained from GenBank (as at April 2005), searching for entries with ‘influenza’ in the title field and shorter than 3000 nt (the longest influenza segment is 2341 nt). Influenza B and C were then excluded and the remainder were sorted into segments. Following GenBank convention, all sequences were considered in plus (mRNA) sense although it should be borne in mind that the virus packages the complementary strand. For all predicted protein products bar the two surface glycoproteins, it was straightforward to establish a clear consensus sequence. Each sequence was either easily aligned (then included into dataset), or was not full length, or had insertions or deletions (these were double checked with the Influenza Sequence Database (21), then rejected from dataset). Exon 2 sequences for the spliced influenza virus gene products M2 and NS2/NEP (22,23) were identified using the A/Udorn/72 virus sequences as a template. The presence of subtypes and thus complicated alignments combined with large numbers of incomplete sequences on GenBank rendered HA and NA hard to analyse. To simplify the problem, analysis was therefore restricted to a single antigenic subtype for each. In the case of HA, the H1 subtype was chosen because it provided the best balance between number of available sequences and time span of virus isolation, and because experimental data already existed for the location of the packaging sequence for an H1 HA (14). For similar reasons, we chose the N1 subtype of NA. For the NA, the most frequent alignable ORF length resulted in a dataset of 89 sequences. In addition, a further 41 sequences could be easily aligned except for an insertion of ‘ACA’ at codon position 436. By ignoring that codon in those sequences for the mathematical analysis, the dataset increased to 130. A list of the accession numbers of the sequences used sorted (where the information was extractable) according to the year of isolation and HN subtypes are contained in Supplementary Data S1.
Normalized mean pairwise distance
First, a simple measure of variability of each codon position in each segment was calculated. The distance between two codons we defined as the number of nucleotide differences, so it can be 0, 1, 2 or 3. A frequency distribution of codons used at each position was derived from the dataset above. The distance between each pair of observations was calculated and summed, and this was divided by the number of pairs of observation (n sequences give n(n − 1)/2 possible pairs). This number is the ‘mean pairwise distance’ (MPD). Overall, the MPD score for each position acted as a simple proxy for variation in which low scores correspond to high conservation at the RNA level.
As calculated, the observed MPD values theoretically also reflected three undesired factors: amino acid constraint, codon bias and also the diversity of the dataset. For instance, conserved methionine or tryptophan residues will generate a low MPD score because they are encoded by single codons, and it is impossible with the sequence data alone to identify if this is due to amino acid or RNA constraint, or just pure chance. Furthermore, even for amino acid with redundant coding possibilities, the long established phenomenon of codon bias (24) would be expected to lower the observed MPD values. Previous analyses have concluded that influenza viruses do not show marked codon bias (25,26). However, on the side of caution, this analysis assumed that there might be some segment-specific bias.
To take into account amino acid usage and codon bias, we calculated the expected distribution of codons at each amino acid position given both the distribution of amino acids used at that position and the distribution of codons used for those amino acids in the whole segment. In the case of the two smallest segments, aggregate codon usage tables were calculated for both spliced and unspliced translation products. Observed MPD values were divided by computed MPD scores for this expected distribution: normalized MPD. A low score indicates less RNA variability than expected, after considering amino acid usage and segment codon bias. At positions with invariant tryptophan and methionine codons, both the expected and the observed MPD scores will be zero. Any RNA constraint is totally masked by amino acid constraint. We therefore defined the normalized MPD for these cases to be 1, signifying RNA variability was as expected. This ensured these sites were not flagged as low scoring. They were not included in the moving average described below so that their impact was neutral. When the effective codon number [Nc; a measure of codon bias (27)] of the datasets was calculated, the virus genome had an Nc of 52.4, agreeing with previous analysis (25,26). On aggregate, the set of codons with an MPD score of <0.05 (∼8% of all positions) had an Nc of 44.6. This is well within the range of RNA virus codon usage (25), thus low variability codons are not strongly biased towards a particular subset of triplets.
The majority of codons within the influenza genome have normalized MPD scores of <1 (Supplementary Data S2). This is consistent with patterns expected even for nucleotides with no functional constraints but with insufficient evolutionary time to explore all possible codons. This is the third factor impacting on the MPD listed above: the dataset sequences may be closely related to each other. However, all positions within a segment's dataset are equally subject to this effect, so this factor could be controlled for by considering scores in relation to others in the segment. In particular, we did this here by looking for aggregation of low-scoring codons in nearby positions along the length of a segment. First, the lowest 10% scoring codons were identified for each segment. This method of defining a cut-off was chosen in preference to choosing an absolute MPD value because the overall levels of conservation differed between segments (Table 1, Figure 1). The distances between the low-scoring codons were then plotted as the number of occurrences for each segment against distance (Figure S1). We also calculated the expected outcome for the null hypothesis of conserved codons being randomly positioned along the segment: the observed distribution clearly exceeded this for short distances.
Table 1.
Influenza A virus sequence datasets
| Sega | Geneb | No.c | Cons.d | ORFe | Yearf | Mediang | HA typesh | % ‘Human’i |
|---|---|---|---|---|---|---|---|---|
| 1 | PB2 | 369 | 92 | 759 | 1930 | 2000 | 1–7,9,13 | 54 |
| 2 | PB1 | 351 | 93 | 757 | 1933 | 2000 | 1–7,9,13,16 | 58 |
| 3 | PA | 396 | 89 | 716 | 1930 | 2000 | 1–7,9,13 | 53 |
| 4 | HA (1) | 99 | 67 | 566 | 1918 | 1994 | n/a | 99 |
| 4 | HA (3) | 203 | 82 | 566 | 1968 | 2002 | n/a | 52 |
| 5 | NP | 617 | 85 | 498 | 1918 | 1997 | 1–7,9–11,13,14,16 | 52 |
| 6 | NA (1) | 130 | 75 | 468/9 | 1918 | 1999 | 1–7,9,11 | 51 |
| 7 | M1 | 887 | 89 | 252 | 1902 | 1999 | 1–7,9–13 | 38 |
| M2 | 746 | 73 | (88) | 1902 | 1999 | 1–7,9–13 | 40 | |
| 8 | NS1 | 829 | 51 | 230 | 1902 | 1998 | 1–7,9–13 | 42 |
| NS2 | 962 | 74 | (111) | 1902 | 1999 | 1–7,9–13 | 35 | |
| 8 | NS1(A) | 645 | 62 | 230 | 1902 | 1998 | 1–7,9–11,13 | 58 |
| 8 | NS1(B) | 184 | 80 | 230 | 1949 | 1998 | 1,3–7,9,10,12 | 0 |
aSegment number.
bTranslation product analysed (parentheses specify subtype where applicable).
cNumber of sequences analysed.
d% of amino acid residues within each dataset that are identical in ≥95% of isolates.
eLength of ORF analysed. (Only second exon sequences are considered for M2 and NS2).
fThe earliest year of virus isolation. The latest isolation date was 2004 except for NS1(B), NA (1) (2003) and HA (1) (2002).
gMedian date of virus isolation for each dataset.
hHA subtypes of virus isolates from which sequences were derived; n/a, not applicable.
i% of isolates from potentially human adapted H1N1, H1N2, H2N2 or H3N2 viruses.
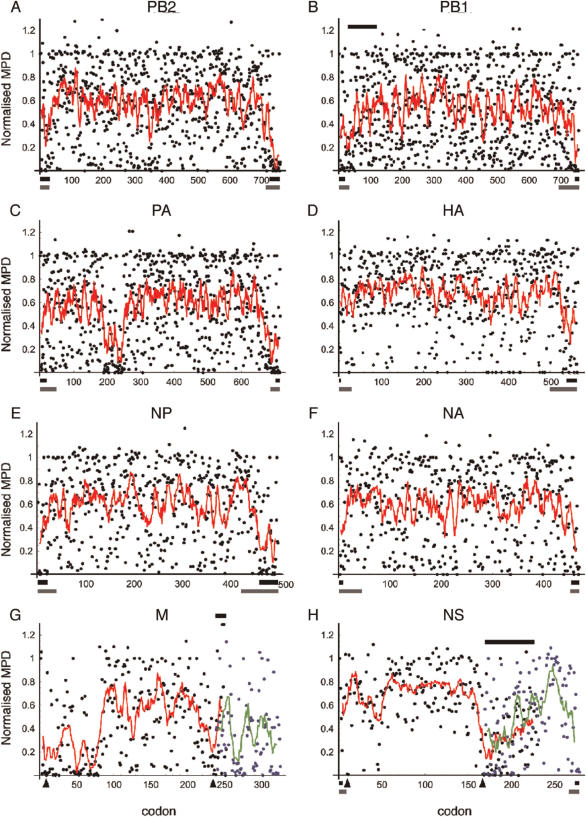
Figure 1.
Normalized codon MPD plots for influenza A virus segments. Dots indicate individual codon scores while lines delineate a moving average taken over a window of 10 residues. For (G) M and (H) NS segments, data for the spliced gene products (blue dots and green lines) are plotted on the same scale with the appropriate degree of overlap. Bars above plots represent areas of dual coding capacity. Bars below plots represent packaging signals known from reverse genetics experiments (black lines) or inferred from sequencing studies of DI RNAs (grey lines). Arrow heads indicate splice donor and acceptor sites.
We calculated the moving average normalized MPD score over a sliding window of 10 codons, which made it easy to identify low-scoring regions by eye. The lowest point of the moving average was used as a further statistic to test whether low-scoring codons are aggregated. For each gene product, the normalized MPD was sampled with replacement as many times as there are codon positions and the minimum of the moving average was computed. This was repeated 100 000 times. For NS2 and M2, no significant result was found, however these ORFs are notably shorter than the others analysed (Table 1). For all other gene products, the actual minimum was lower than nearly all of the random minima (PB2, PB1, PA, NP, M1, NS1 actuals were lower than all but 0, 14, 34, 34, 8, 22 random runs out of 100 000 respectively, corresponding to P < 0.001). For HA and NA, the actual minimum was lower than all but 646 and 3442 of the 100 000 (corresponding to P < 0.01 and P < 0.05, respectively). These weaker results are perhaps due to the fewer sequences aligned without insertions and deletions. Overall however, the moving average dipped significantly lower in the actual ordering of most genes than in random ordering, again signifying aggregation of low scores. Generalized low diversity of the dataset would apply evenly through each segment, so this cannot explain our observations, and thus we conclude from this analysis that we have detected regions with some particular constraint at the RNA level.
RNA packaging assays
The packaging efficiency of artificial vRNA segments engineered to contain reporter genes flanked by the terminal-coding regions of the influenza ORFs was carried out essentially as previously described (16). Briefly, cells were transfected with plasmids that expressed the vRNA segments from RNA polymerase I promoters and superinfected with A/WSN/33 virus. In the case of segment 1, packaging efficiency was assessed by the ability of progeny viruses to transduce second sets of cells with gene fluorescent protein (GFP), as measured by flow cytometry (16). In the case of segment 6, packaging efficiency was similarly measured by assaying chloramphenicol acetyl transferase (CAT) levels by ELISA (CAT ELISA kit, Roche). To control for an apparent reduction in segment packaging simply resulting from less available material for incorporation, accumulation of segment 6 vRNAs in the primary cells was assessed by measuring expression of the CAT reporter gene. Expression of segment 1 vRNAs was determined using a plasmid-based system for reconstituting active viral RNPs in transiently transfected COS-1 cells; vRNA levels were detected by primer-extension reactions using the oligonucleotide ATCACTCTCGGCATGGAC and quantified by densitometry (28). In this system, RNA Pol I-mediated transcription results in low levels of the model vRNA that are substantially amplified by the influenza virus polymerase (28).
RESULTS AND DISCUSSION
Sequence conservation within influenza virus ORFs
Development of an algorithm to detect sequence conservation acting at the level of RNA whilst controlling for constraints due to coding capacity is described in the Materials and methods section. The approach required a large and evolutionarily diverse dataset of aligned influenza A virus sequences, acquired from GenBank. The numbers of sequences present in the datasets of each non-glycoprotein ORF ranged between 351 (PB1) and 962 (NS2) (Table 1). The year of isolation of the viruses spanned 102 years, but with a bias towards recent times. The median dates for the datasets of individual ORFs ranged from 1994 (HA) to 2000 (polymerase genes) (Table 1). Overall sequence conservation in the datasets was high, with between 51 (NS1) and 93% (PB1) of amino acid residues identical in >95% of sequences (Table 1). With respect to the diversity of the sequences, examination of the HA subtypes of the viruses from which sequences were derived showed that subtypes 1–7, 9–14 and 16 were represented (Table 1). Influenza A viruses are known to infect a wide variety of avian and mammalian species (4) and visual inspection of the datasets confirmed that virus sequences isolated from a diverse mixture of host species were present. However, attempts to extract a definitive list of hosts from the datasets were defeated by a combination of the size of the database, the nature of the influenza naming convention and the occasionally erratic nature with which it has been applied. Therefore, as a more amenable approach to assessing the diversity of species the viruses were isolated from, we examined the haemagglutinin (H) and neuraminidase (N) subtypes of the parental viruses as only a limited number of these subtypes are known to routinely infect man (4). Between 88 (NS1) and 98% (PA) of the non-glycoprotein influenza sequences in our database had subtype information embedded in a readily interpretable format (Supplementary data S1) and for all internal genes, H subtypes 1–7, 9 and 13 were represented (Table 1). With the exception of the polymerase segments, all N subtypes were represented (data not shown). As expected, the numbers of each H and N subtype were not equal, so to provide a measure of this, the datasets were analysed for the proportion of sequences from viruses with HN types that could potentially (but not necessarily) reflect efficient circulation in the human population. The proportion of internal gene sequences from viruses with H1N1, H1N2, H2N2 or H3N2 antigenic types varied from segment to segment in the 35–58% range (Table 1) but on average made up less than half of the available data. Therefore at a minimum, half of the sequences were obtained from viruses isolated from non-human hosts. Overall, the available sequences comprise a virus database whose size, age range and diversity of host species is unmatched.
Our analysis generated a MPD score for each codon of an ORF in which low values reflect higher conservation of a specific triplet than expected from amino acid conservation or codon bias. Plotting these scores for each segment against codon number gave a set of graphs resembling scatterplots (Figure 1). Visual inspection suggested that for most segments, there was non-random clustering of low-scoring codons. This was further evident when a moving average of the MPD scores (summed over 10 codons, corrected for end effects) was plotted (Figure 1). The clusters of conserved codons were mostly located towards the start and/or ends of ORFs, although some internal conserved regions were noted (e.g. PA; Figure 1C, M1; Figure 1G). In general, the 3′-ends of the ORFs appeared to be slightly more conserved. To assess the likelihood of these clusters occurring by chance, we compared the observed distances between low-scoring codons to that expected by chance. Plots of the gap size between low-scoring codons against the number of occurrences within each segment showed a tendency for conserved codons to aggregate at close distances (Figure S1A–H). For all but the glycoprotein segments, this was significant at <0.01 level (see Supplementary Data S1 for more details). Analysis of the whole coding genome as a pooled dataset further highlighted the short-range aggregation of low-scoring codons (Figure S1I). In this pooled dataset, the distances between conserved codons exceeded the 99% confidence limits for that predicted by chance for distances of up to ∼20 codons. Further, the point at which the observed distribution crosses the random expected distribution gives an estimate for the length of conserved elements. By visual estimate from Figure S1I, this appears to be ∼30 codons or 90 nt, though this is clearly very approximate. We also considered the statistical significance of the degree of aggregated conservation as well as the clustering itself by analysing the minima in the moving point average. For non-glycoprotein segments, the observed MPD minimum was lower than that obtained by 100 000 iterations of sampling by replacement at the P < 0.001 level, and P < 0.05 level for the glycoproteins (see Materials and methods section). Thus each segment in the influenza A virus genome contains one or more regions of sequence conservation over and above that expected for their protein-coding function.
Inspection of the datasets for individual codons revealed interesting features. For example, codon 755 of the PB1 ORF specifies an invariant arginine (Table 2a). However, the codon CGG is relatively rare as it is used for <9% of arginine residues in the virus genome. Thus in over 300 genes, isolated from viruses with 10 different HA subtypes across a time span of 72 years, this normally rare codon is absolutely invariant (Table 2a). Given the high mutation rate of influenza and that five other arginine codons are one or two nucleotide substitutions away, this is suggestive of a high degree of constraint independent of protein sequence. Consistent with this, five of the upstream 12 codons are similarly invariant while another four show very low (MPD < 0.05) variation (Figure 1B). In contrast, the majority of codons in the influenza virus genome showed no strong conservation. For example, position 427 of PB2, which also encodes an invariant arginine residue, uses all six possible codons (Table 2b). The full datasets for each gene are available online (S2).
Table 2.
Examples of low and high MPD codon scores associated with invariant arginine (R) amino acid (AA) residues are tabulated
| Codon | AA | Obsa | Expb |
|---|---|---|---|
| (a) PB1 ORF, position 755 | |||
| CGU | R | 0 | 6.0 |
| CGC | R | 0 | 18.7 |
| CGA | R | 0 | 22.7 |
| CGG | R | 351 | 36.9 |
| AGA | R | 0 | 170.3 |
| AGG | R | 0 | 96.4 |
| MPDc: | 0 | 0.93 | |
| Normd: | 0 | ||
| (b) PB2 ORF, position 427 | |||
| CGU | R | 1 | 7.8 |
| CGC | R | 2 | 23.1 |
| CGA | R | 192 | 33.5 |
| CGG | R | 81 | 26.7 |
| AGA | R | 87 | 185.4 |
| AGG | R | 6 | 92.4 |
| MPD | 0.75 | 0.91 | |
| Norm: | 0.82 | ||
aObserved frequency of codons at specified position.
bHypothetical predicted occurence of codons based on overall codon usage data for the segment.
cCalculated mean pairwise difference (MPD) of the codon distribution.
dNorm = observed MPD/expected MPD.
Sources of high sequence constraint in coding regions
Some regions of high RNA sequence constraint could be explained by overlapping ORFs. The 5′-end of the PB1 gene contains a short internal ORF (PB1-F2) that is poorly conserved (29,30), but probably explains the low MPD scores between residues 32 and 50 of PB1 (Figure 1B). Similarly, the low variabilities of codon 168 onwards of NS1 (Figure 1H) are likely to result at least partly from the overlapping NS2/NEP ORF (22). The short overlap between the M1 ORF and exon 2 of M2 (23) was not associated with particularly low MPD scores although three of the four codons immediately 5′ to the M2 ORF (positions 235–238) are highly conserved (Figure 1G). This likely reflects the presence of previously characterized cis-acting sequences in the viral genome. The M2- and NS2- coding regions are accessed by splicing of the primary mRNAs and the low MPD scores of codons 8, 9, 236 and 238 of M1 and codons 10, 11, 167 and 168 of NS1 reflect the presence of splice donor and acceptor consensus motifs (22,23).
Some instances of RNA conservation may be explained by RNA sequences involved in modulating translation. The low MPD score of PB1 codon 31 perhaps reflects maintenance of an effective ribosomal initiation signal upstream of the initiating AUG codon for the PB1-F2 ORF (31). The AUG codons further downstream in the PB1-F2 ORF that have recently been proposed to direct expression of a shorter form of the protein (32) show only moderate conservation. However, the Kozak translation initiation consensus does not provide a good explanation for the general pattern of low MPD scores observed towards the 5′-coding end of most segments (Figure 1) as the consensus does not extend beyond the second translated codon (33,34). Similarly, the high frequency of low MPD scores observed towards the 3′-ends of the influenza virus ORFs is not easily explained through translational signals. In yeast, the identity of the penultimate codon has been shown to influence the efficiency of stop codon recognition (35), while bioinformatic analysis of higher eukaryotic ORFs has suggested a degree of non-randomness of the terminal tripeptide (36). However, there is no clear pattern of conservation evident in the influenza virus sequences in the final three codons of each gene and the low MPD scores extend 5′ to these sequences in all the ORFs analysed, even those without overlapping 3′-coding sequences (Figure 1). Another sequence motif that has been identified in the influenza virus genome as being important for translation of certain influenza virus mRNAs is the binding site for the cellular GRSF-1 protein (37,38). However, although the motif AGGGU occurs elsewhere in the viral ORFs as well as in the 5′-untranslated regions of NP and NS mRNAs, none of the occurrences in the coding regions are conserved.
The general pattern of low sequence diversity exhibited at most segment termini is best explained by reference to known or suspected packaging signals. The terminal conserved regions in the PB2, PB1, PA and NP genes correspond well to the minimal sequences found in DI RNAs or in efficiently packaged engineered vRNAs (Figure 1A–C, E) (11,12,16–18,20,39,40). Similarly, for segment 6, our MPD data are consistent with the minimal sequences within the 5′ and 3′-ends of the NA ORF necessary for efficient packaging (13). In particular, the sharp drop-off in packaging efficiency noted when codon 7 of NA was deleted matches well with its low MPD score (0.07) in our analysis. The low sequence diversity exhibited by the 3′-end of the HA ORF (Figure 1D) also corresponds to the sequences identified as important for packaging of segment 4 (14). The average size of the conserved regions [∼30 codons/90 nt (Figure S1I)] is also consistent with the existing experimental data. Thus the location of the majority of the clusters of low-diversity codons is consistent with the hypothesis that cis-acting segment-specific packaging signals are evolutionarily conserved sequences.
Potential novel features in the virus genome
An interesting feature of our analysis concerns the areas of high sequence conservation that do not correlate with any previously identified RNA motifs or with putative terminally located packaging signals. The most prominent of these lies around codons 200–250 of PA but others are visible around codons 50 and 70 of M1 and possibly around codon 350 of PB2 (Figure 1A, C, G). In the case of the P genes, experiments aimed at mapping packaging signals in these segments started out with the assumption that internal sequences were not involved (17–19) and therefore do not provide a test of this function. However, known DI RNAs do not generally include these internal regions (39,40), arguing against a role in segment packaging. In neither case is there an obvious out of frame AUG codon that could initiate translation of a reasonable length alternative ORF (as with PB1-F2 for instance) and in any case, it is not clear how such an ORF could be accessed for translation in the PA or PB2 genes as we are unaware of any reports of mRNA splicing or IRES function in these segments.
No experimental information is available yet on the packaging of segment 7. Nevertheless, by analogy with other segments, we predict that the low-diversity sequences at the 5′-end of the M1 and 3′-end of the M2 ORFs will prove to be important. However, the patches of sequence conservation centred around codons 50 and 70 are substantially further into the gene than any other segment packaging signal so far mapped or inferred (Figure 1) so it is possible these sequences have other embedded function(s). One study has identified a fourth alternatively spliced mRNA formed by some strains of influenza A virus (41), but the splice donor used to form this RNA is only partially conserved with the critical GU bases 3′ to the cleavage site (in codon 41 of M1) forming a minority population in our database. Furthermore, the highly conserved codons in M1 centred around positions 50 and 70 would lie in the intron of this transcript, making this putative mRNA an unlikely explanation for the conservation. Further work is therefore required to identify the cause and function of these areas of RNA conservation.
Effect of synonymous mutations on segment packaging
As an experimental test of the validity of our analysis, we examined the effect of single codon synonymous changes within the terminal-coding regions on segment packaging. First, mutations were introduced into the 3′-coding region of PB2. The experimental system used an engineered minigenome version of segment 1 containing the GFP gene as a quantifiable marker. Mutations were introduced into the cDNA clone PB2(159)GFP(166) which expresses a vRNA analogue encoding (in the negative sense) GFP flanked by the indicated number of nucleotides from the 3′- and 5′-termini respectively of the segment as prior analysis has shown that this RNA is efficiently packaged into virus particles (16). The construct PB2(159)GFP(34) was used as a negative control, as the loss of all coding regions from the 3′-end of the PB2 ORF severely reduces packaging of the vRNA (16). Cells were transfected with the vRNA expression plasmids, superinfected with influenza virus A/WSN/33 and infection allowed to proceed. The resulting progenies were then used to infect a second set of cells and the proportion of infected cells that expressed GFP (indicating successful packaging of the recombinant vRNA segment) quantified by flow cytometry. This system measures the ability of the PB2 minigenomes to compete with the wild-type PB2 segment for packaging, which is (very likely) directly related to the minigenome packaging efficiency. As expected (16), the ‘wild type’ PB2(159)GFP(166) vRNA was incorporated into virus particles as nearly 30% of secondarily infected cells expressed GFP (Table 3). However, single or double nucleotide synonymous changes in any of the highly conserved PB2 codons 744, 745, 748, 751 or 757 reduced transmission of the segment between 20- and 50-fold (Table 3). Strikingly, these effects were almost as severe as deletion of the entire 3′-coding region of PB2 [Table 3; PB2(159)GFP(34)]. In contrast, synonymous mutation of the relatively poorly conserved codons 731 or 737 resulted in only a small decrease (<2-fold) in apparent vRNA packaging efficiency (Table 3). Decreased transmission of the vRNA reporter molecule could result from reduced packaging efficiency or from a lowered replication efficiency in the primary cells that reduced the amount of material available for incorporation into virions. To distinguish between these possibilities, we measured the replication efficiency of the vRNAs in the absence of virus budding. Cells were transfected with the vRNA expression constructs along with plasmids that express PB1, PB2, PA and NP, the minimum viral proteins required to support transcription and replication of the viral genome (3,42). After incubation, accumulation of the vRNA molecules was quantified by primer extension (28). None of the mutations introduced into highly conserved codons significantly reduced vRNA accumulation, although curiously, mutation of the non-conserved codon 731 did affect vRNA levels (Table 3). We therefore conclude that the analysis of codon conservation successfully identified single nucleotide changes that significantly affect segment packaging.
Table 3.
Effect of synonymous mutations in the PB2-coding region on segment 1 packaging
| vRNAa | Mutation | MPD score | Packaging levels (% of GFP-expressing cells)b | vRNA expression levels (% of PB2(159) GFP(166))c |
|---|---|---|---|---|
| PB2(159) GFP(34) | n/a | n/a | 0.4 ± 0.1 | 130 ± 10 |
| PB2(159) GFP(166) | n/a | n/a | 27.5 ± 7.3 | 100 |
| mut 731 | GUG -> GUC | 0.360 | 18.5 ± 1.6 | 20 ± 0 |
| mut 737 | CGG -> CGC | 0.539 | 17.9 ± 4.4 | 35 ± 5 |
| mut 744a | CUU -> CUA | 0 | 0.8 ± 0.2 | 80 ± 20 |
| mut 744b | CUU -> UUA | 0 | 0.5 ± 0.1 | 65 ± 5 |
| mut 745 | ACU -> ACA | 0 | 1.2 ± 0.3 | 55 ± 5 |
| mut 748 | CAG -> CAA | 0 | 1.1 ± 0.3 | 75 ± 25 |
| mut 751 | ACC -> ACG | 0.008 | 0.5 ± 0.3 | 110 ± 10 |
| mut 757 | GCC -> GCG | 0 | 1.1 ± 0.4 | 70 ± 10 |
aPB2(159)GFP(166) denotes vRNAs containing PB2 codons 1–44 and 716–759 (synonymous mutations introduced as indicated), while PB2(159)GFP(34) refers to a vRNA lacking all 3′-PB2-coding regions.
bMean ± standard deviation of four measurements from two independent clones.
cMean ± range from two independent clones as determined by primer-extension analysis. n/a, not applicable.
Next, we tested similar mutations in the 3′-coding region of NA, which we identified previously as critical for the propagation of viruses harbouring dicistronic NA segments (43). In this case, the engineered vRNA segments contained the CAT gene as an assayable marker. Synonymous mutations were introduced into the CAT38 construct which contains NA codons 456–469, while the CAT35 construct lacking all 3′-NA coding sequences was used as a low-packaging efficiency control (43). When the single absolutely conserved codon (468) in this region of NA was mutated, packaging was severely impaired, while mutations on two codons with high MPDs (461 and 464) had less effect (Table 4). Mutation of codons with intermediate levels of conservation led to variable packaging efficiencies, with some alterations having no apparent effect (e.g. codon 467) but the majority of changes decreased segment 6 incorporation (Table 4). None of the mutations decreased expression levels of the vRNA segments by >∼2-fold (Table 4). The overall poorer correlation between MPD score and experimentally measured effects on packaging when NA codons were mutated compared to PB2 (Tables 3 and 4) probably reflects the lower statistical significance of the conservation in segment 6 (Materials and methods section) and may be improvable by better sequence alignment of a larger dataset. Nevertheless, overall these data (Tables 3 and 4) confirm the ability of our analysis to identify functionally important RNA conservation and highlight the surprising finding that single nucleotide changes can have a dramatic effect on segment packaging.
Table 4.
Effect of synonymous mutations in the NA-coding region on segment 6 packaging
| vRNAa | Mutation | MPD score | Relative packaging efficiency (on a 0.1–100 scale)b | vRNA expression levels (% of CAT35)b |
|---|---|---|---|---|
| CAT35 | n/a | n/a | 0.1 | 100 |
| CAT38 | n/a | n/a | 100 | 111 ± 5 |
| mut 461 | GCU -> GCA | 0.364 | 15.0 ± 4.5 | 94 ± 19 |
| mut 464 | CCG -> CCC | 0.393 | 9.5 ± 1.5 | 44 ± 9 |
| mut 463a | UUG -> UUA | 0.098 | 0.5 ± 0.1 | 103 ± 17 |
| mut 463b | UUG -> CUG | 0.098 | 11.9 ± 4.3 | 44 ± 7 |
| mut 466 | ACC -> ACG | 0.065 | 35.5 ± 1.0 | 55 ± 7 |
| mut 467 | AAU -> AAA | 0.047 | 64.2 ± 6.7 | 91 ± 12 |
| mut 468 | GAC -> GAU | 0 | 2.4 ± 0.4 | 333 ± 91 |
| mut 469 | AAG -> AAA | 0.061 | 6.6 ± 1.2 | 235 ± 43 |
aCAT38 denotes vRNAs containing NA codons 456–469 required for efficient packaging, while CAT35 refers to a vRNA lacking all 3′-NA-coding region. Synonymous mutations were introduced into the CAT38 vRNA as indicated.
bMean ± standard deviation of four measurements from two independent clones as determined by CAT ELISA. n/a, not applicable.
Are certain packaging signals virus strain specific?
For segment 8, the conserved sequences found at the end of the NS2 ORF agree with experimental data (15). Experimentally however, a more important contribution came from the sequences corresponding to codons 2–9 of NS1 as synonymous mutations here reduced packaging efficiency by 3–4-fold (15). Surprisingly, none of the wobble bases of these codons were conserved at the 95% level in our dataset and the lowest MPD score was 0.21 (Figure 2A). NS genes fall into two lineages that diverged a century ago (44,45), so we analysed the A and B alleles separately (Table 1). Significantly, the individual gene families displayed sequence conservation within the region identified as crucial for efficient packaging. In allele A, codons 5 and 7 showed very low MPD scores and codon 6 was also relatively invariable, while in NS1(B), codons 3–5 and 7 were conserved (Figure 2A). Furthermore, the nature of the conservation differed. Within the sequences coding for amino acid 5–8, 7 of the 12 conserved nucleotides differ between alleles A and B (Figure 2A). Sequences further into the coding region of NS1 (at least up to codon 30) also contribute to packaging of the segment (15) but here too the patterns of conservation was largely different between the two alleles (S2). The non-coding region immediately upstream of the NS1-coding region is also necessary but not sufficient to direct efficient packaging of segment 8 (15) and these sequences are conserved between the two NS alleles. Nevertheless, the differing nature of the conserved sequences within the crucial region of the NS1-coding sequence raises the possibility that the packaging signal differs significantly between the two lineages of NS genes.
Figure 2.
Varying patterns of RNA conservation in the NS1 and HA ORFs. (A) Consensus nucleotide and amino acid (single letter code) sequences of the first 11 codons of the overall NS1 dataset and the A and B lineages are shown along with the codon MPD scores. Residues not conserved at the 95% level are shown by asterisks. Highly conserved triplets (MPD value <0.05) are shown in red. Conserved residues that differ between lineages are highlighted. Underlining denotes the splice donor sequence. (B) Consensus sequences for the C-terminal 27 codons of HA subtypes 1, 3 and 5, encompassing the packaging signal for H1. MPD scores are not shown for clarity but conserved triplets are color-coded red (MPD < 0.05) or blue (MPD < 0.1) and highlighted. Homology between the H5 sequence and conserved nucleotides in H1 or H3 sequences are indicated by lines.
The packaging signal in segment 4 has been shown to extend into the 3′-end of the HA-coding region (14). However, this was determined for the H1 serotype and the 16 HA subtypes are not well conserved at the amino acid level within this region (Skehel, 2004; alignment hosted at: http://www.flu.lanl.gov/review/HAalignment.html). We therefore extended our analysis to other subtypes of HA. H3 sequences were more conserved overall than H1 (Table 1) but similar to H1, this was particularly marked towards the 3′-end of the ORF. However, when the location of the conserved codons in this crucial 3′-region was compared between H1 and H3 sequences, the pattern was very different, showing almost no superposition (Figure 2B). The differences between the H1 and H3 codons that show evidence of selective pressure against synonymous changes supports the hypothesis that strain-specific differences between packaging signals exist. We next examined H5 sequences as the possibility of the highly pathogenic avian H5N1 influenza currently circulating in the Far East and elsewhere reassorting with human-adapted H1N1 or H3N2 viruses is particularly significant. Unfortunately, MPD analysis of the H5 HA ORF was not informative as the available sequences are virtually all from the recent outbreaks and show insufficient diversity to be useful (data not shown). The nucleotide sequence of recent H5 genes within the region corresponding to the H1 packaging signal is more closely related to the H1 gene than the H3 (68% identical compared to 43%; Figure 2B). This difference is maintained when only those triplets that are conserved at the wobble position are considered but nevertheless, the H5 gene shows a substantial number of differences from both (Figure 2B). Given that single nucleotide mutations in the PB2 and NA packaging signals can substantially reduce incorporation of the segment (Tables 3 and 4), we speculate that efficient inclusion of the H5 gene into an otherwise human-adapted virus may require significant adaptation of the packaging signal.
CONCLUSION
Although our measure of synonymous variation is largely constrained to mapping conservation of every third nucleotide, it offers higher resolution than the deletion analyses that provide the majority of the experimental data on segment incorporation. Our dataset (S2) offers sufficient detail to suggest suitable positions and synonyms to be explored by reverse genetic experiments aimed at elucidating the functions of the conserved RNA sequences (Tables 3 and 4). Analysing the growth characteristics of recombinant viruses containing mutations at the conserved positions will provide the ultimate test of the bioinformatics analysis and this work is in progress. We also expect the dataset will be useful in the design of experiments whose aim is to generate functionally important mutations only in the viral proteins. Though current consensus is that segment-specific packaging signals exist in influenza, there is little information on how these signals operate. In the absence of evidence for a protein factor able to identify eight separate and unique sequences, the most attractive hypothesis is that the RNA packaging signals interact with each other, so that each segment has preference for packing with a given other segment or two in such a way to make it likely that a complete set of segments is packaged (13,18). This model is complicated by the fact that vRNA is wrapped around NP to form a complex RNP structure in which on average, 24 nt of RNA are associated with each NP monomer (2). Base pairing (not necessarily only Watson–Crick type) between segments is still theoretically possible, perhaps mediated or initiated by short hairpin-loops protruding away from the RNP in a manner perhaps analogous to dimerization of retroviral genomes (46). However, it is difficult to analyse RNAs for complementarity where the bases involved are discontinuous, and we have not been able to identify any convincing relationships between the conserved regions on different segments. One might expect the extended nature of the packaging signals coupled with an ordered RNP structure would result in detectable periodicity in the spacing of conserved residues. However, despite the tendency of the human eye to see patterns in data such as Figures 1 and S1, several approaches including discrete Fourier and wavelet-style methods and the pairwise distance distributions shown in Figure S1 have failed to detect any significant periodicity in the data (data not shown). Any periodicity might be masked by flexibility in the RNP structure, or alternatively, even if RNP interactions are primarily mediated by RNA–RNA interactions, the participation of other specific helping or adaptor factors cannot be ruled out. It is interesting that mutations in NS1 and PA that inhibit particle formation without apparently affecting vRNA synthesis or trafficking have been identified (47,48).
Splitting the influenza sequence dataset according to NS clades or HA subtypes suggested that the packaging signals may not be universal. It is possible that evolutionary separation of virus strains has allowed signals to evolve separately within lineages. This accords with experimental work observing that some reassortants are hard to make in the laboratory, and others are apparently attenuated, including the NS1 B allele in the background of an allele A virus (49) as well as several avian HA subtypes (including H5) in the background of a human H3 segment 7 (50,51). Many studies have concluded that the loss of fitness of such reassortants results from incompatibilities between viral gene products. We propose that mismatches between specific packaging signals can also reduce virus viability. Given the relatively large size and likely discontinuity of the packaging signals so far mapped, it is unlikely that such incompatibility would totally exclude reassortment. However, even a modest reduction in virus fitness that is viable in the laboratory is likely to reduce successful transmission in the wild and thus potentially make the viral reproductive ratio less than one (R0 < 1), preventing large outbreaks (52). At least two pandemics have resulted from reassortant viruses (1,4). It is now of utmost urgency to fully understand the factors that govern virus reassortment, and in particular to assess how much adaptation of the H5 HA gene might be required to allow its efficient packaging into an otherwise human-adapted virus.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online.
ACKNOWLEDGEMENTS
We thank Sylvie Gerbaud for contributing to the experimental work and Sylvie van der Werf for helpful discussion. This work was supported by grants from the Royal Society to J.R.G. Wellcome Trust (no. 073126) and BBSRC (no. S18874) to P.D. and the MRC (nos. G9901213 and G0300009) to L.T. and P.D. E.D.S.A. was supported by a fellowship from the French Ministère de la Recherche et de la Technologie. J.C.vK. is supported by an MRC Capacity Building Studentship. J.R.G. thanks the Department of Zoology, University of Cambridge for support during this work. Funding to pay the Open Access publication charge was provided by Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Webby RJ, Webster RG. Are we ready for pandemic influenza? Science. 2003;302:1519–1522. doi: 10.1126/science.1090350. [DOI] [PubMed] [Google Scholar]
- 2.Portela A, Digard P. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 2002;83:723–734. doi: 10.1099/0022-1317-83-4-723. [DOI] [PubMed] [Google Scholar]
- 3.Elton D, Digard P, Tiley L, Ortin J. Influenza Virology; Current Topics. Wymondham: Caister Academic Press; 2006. pp. 1–36. [Google Scholar]
- 4.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luytjes W, Krystal M, Enami M, Pavin JD, Palese P. Amplification, expression, and packaging of foreign gene by influenza virus. Cell. 1989;59:1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- 6.Enami M, Sharma G, Benham C, Palese P. An influenza virus containing nine different RNA segments. Virology. 1991;185:291–298. doi: 10.1016/0042-6822(91)90776-8. [DOI] [PubMed] [Google Scholar]
- 7.Bancroft CT, Parslow TG. Evidence for segment-nonspecific packaging of the influenza a virus genome. J. Virol. 2002;76:7133–7139. doi: 10.1128/JVI.76.14.7133-7139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noda T, Sagara H, Yen A, Takada A, Kida H, Cheng RH, Kawaoka Y. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439:490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- 9.Duhaut SD, McCauley JW. Defective RNAs inhibit the assembly of influenza virus genome segments in a segment-specific manner. Virology. 1996;216:326–337. doi: 10.1006/viro.1996.0068. [DOI] [PubMed] [Google Scholar]
- 10.Odagiri T, Tashiro M. Segment-specific noncoding sequences of the influenza virus genome RNA are involved in the specific competition between defective interfering RNA and its progenitor RNA segment at the virion assembly step. J. Virol. 1997;71:2138–2145. doi: 10.1128/jvi.71.3.2138-2145.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duhaut S, Dimmock NJ. Approximately 150 nucleotides from the 5′ end of an influenza A segment 1 defective virion RNA are needed for genome stability during passage of defective virus in infected cells. Virology. 2000;275:278–285. doi: 10.1006/viro.2000.0502. [DOI] [PubMed] [Google Scholar]
- 12.Duhaut SD, Dimmock NJ. Defective segment 1 RNAs that interfere with production of infectious influenza A virus require at least 150 nucleotides of 5′ sequence: evidence from a plasmid-driven system. J. Gen. Virol. 2002;83:403–411. doi: 10.1099/0022-1317-83-2-403. [DOI] [PubMed] [Google Scholar]
- 13.Fujii Y, Goto H, Watanabe T, Yoshida T, Kawaoka Y. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2002–2007. doi: 10.1073/pnas.0437772100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe T, Watanabe S, Noda T, Fujii Y, Kawaoka Y. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J. Virol. 2003;77:10575–10583. doi: 10.1128/JVI.77.19.10575-10583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujii K, Fujii Y, Noda T, Muramoto Y, Watanabe T, Takada A, Goto H, Horimoto T, Kawaoka Y. Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J. Virol. 2005;79:3766–3774. doi: 10.1128/JVI.79.6.3766-3774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dos Santos Afonso E, Escriou N, Leclercq I, van der Werf S, Naffakh N. The generation of recombinant influenza A viruses expressing a PB2 fusion protein requires the conservation of a packaging signal overlapping the coding and noncoding regions at the 5′ end of the PB2 segment. Virology. 2005;341:34–46. doi: 10.1016/j.virol.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 17.Liang Y, Hong Y, Parslow TG. cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 2005;79:10348–10355. doi: 10.1128/JVI.79.16.10348-10355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muramoto Y, Takada A, Fujii K, Noda T, Iwatsuki-Horimoto K, Watanabe S, Horimoto T, Kida H, Kawaoka Y. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J. Virol. 2006;80:2318–2325. doi: 10.1128/JVI.80.5.2318-2325.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Wit E, Spronken MI, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Evidence for specific packaging of the influenza A virus genome from conditionally defective virus particles lacking a polymerase gene. Vaccine. 2006;24:6647–6650. doi: 10.1016/j.vaccine.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Ozawa M, Fujii K, Muramoto Y, Yamada S, Yamayoshi S, Takada A, Goto H, Horimoto T, Kawaoka Y. Contributions of two nuclear localization signals of influenza a virus nucleoprotein to viral replication. J. Virol. 2007;81:30–41. doi: 10.1128/JVI.01434-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macken C, Lu H, Goodman J, Boykin L. The value of a database in surveillance and vaccine selection. In: Osterhaus A, Cox N, Hampson AW, editors. Options for the Control of Influenza IV. Amsterdam: Elsevier Science; 2001. pp. 103–106. [Google Scholar]
- 22.Lamb RA, Lai CJ. Sequence of interrupted and uninterrupted mRNAs and cloned DNA coding for the two overlapping nonstructural proteins of influenza virus. Cell. 1980;21:475–485. doi: 10.1016/0092-8674(80)90484-5. [DOI] [PubMed] [Google Scholar]
- 23.Lamb RA, Lai CJ, Choppin PW. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc. Natl. Acad. Sci. U.S.A. 1981;78:4170–4174. doi: 10.1073/pnas.78.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 25.Jenkins GM, Holmes EC. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res. 2003;92:1–7. doi: 10.1016/s0168-1702(02)00309-x. [DOI] [PubMed] [Google Scholar]
- 26.Zhou T, Gu W, Ma J, Sun X, Lu Z. Analysis of synonymous codon usage in H5N1 virus and other influenza A viruses. Biosystems. 2005;81:77–86. doi: 10.1016/j.biosystems.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 28.Mullin AE, Dalton RM, Amorim MJ, Elton D, Digard P. Increased amounts of the influenza virus nucleoprotein do not promote higher levels of viral genome replication. J. Gen. Virol. 2004;85:3689–3698. doi: 10.1099/vir.0.80518-0. [DOI] [PubMed] [Google Scholar]
- 29.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O'Neill R, Schickli J, et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 30.Chen GW, Yang CC, Tsao KC, Huang CG, Lee LA, Yang WZ, Huang YL, Lin TY, Shih SR. Influenza A virus PB1-F2 gene in recent Taiwanese isolates. Emerg. Infect. Dis. 2004;10:630–636. doi: 10.3201/eid1004.030412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 32.Zamarin D, Ortigoza MB, Palese P. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J. Virol. 2006;80:7976–7983. doi: 10.1128/JVI.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grunert S, Jackson RJ. The immediate downstream codon strongly influences the efficiency of utilization of eukaryotic translation initiation codons. EMBO J. 1994;13:3618–3630. doi: 10.1002/j.1460-2075.1994.tb06669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997;16:2482–2492. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mottagui-Tabar S, Tuite MF, Isaksson LA. The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem. 1998;257:249–254. doi: 10.1046/j.1432-1327.1998.2570249.x. [DOI] [PubMed] [Google Scholar]
- 36.Gatto GJ, Jr, Berg JM. Nonrandom tripeptide sequence distributions at protein carboxyl termini. Genome Res. 2003;13:617–623. doi: 10.1101/gr.667603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park YW, Wilusz J, Katze MG. Regulation of eukaryotic protein synthesis: selective influenza viral mRNA translation is mediated by the cellular RNA-binding protein GRSF-1. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6694–6699. doi: 10.1073/pnas.96.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kash JC, Cunningham DM, Smit MW, Park Y, Fritz D, Wilusz J, Katze MG. Selective translation of eukaryotic mRNAs: functional molecular analysis of GRSF-1, a positive regulator of influenza virus protein synthesis. J. Virol. 2002;76:10417–10426. doi: 10.1128/JVI.76.20.10417-10426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noble S, Dimmock NJ. Characterization of putative defective interfering (DI) A/WSN RNAs isolated from the lungs of mice protected from an otherwise lethal respiratory infection with influenza virus A/WSN (H1N1): a subset of the inoculum DI RNAs. Virology. 1995;210:9–19. doi: 10.1006/viro.1995.1312. [DOI] [PubMed] [Google Scholar]
- 40.Jennings PA, Finch JT, Winter G, Robertson JS. Does the higher order structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza viral RNA? Cell. 1983;34:619–627. doi: 10.1016/0092-8674(83)90394-x. [DOI] [PubMed] [Google Scholar]
- 41.Shih SR, Suen PC, Chen YS, Chang SC. A novel spliced transcript of influenza A/WSN/33 virus. Virus Genes. 1998;17:179–183. doi: 10.1023/a:1008024909222. [DOI] [PubMed] [Google Scholar]
- 42.Huang TS, Palese P, Krystal M. Determination of influenza virus proteins required for genome replication. J. Virol. 1990;64:5669–5673. doi: 10.1128/jvi.64.11.5669-5673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vieira Machado A, Naffakh N, Gerbaud S, van der Werf S, Escriou N. Recombinant influenza A viruses harboring optimized dicistronic NA segment with an extended native 5′ terminal sequence: induction of heterospecific B and T cell responses in mice. Virology. 2006;345:73–87. doi: 10.1016/j.virol.2005.09.050. [DOI] [PubMed] [Google Scholar]
- 44.Baez M, Zazra JJ, Elliott RM, Young JF, Palese P. Nucleotide sequence of the influenza A/duck/Alberta/60/76 virus NS RNA: conservation of the NS1/NS2 overlapping gene structure in a divergent influenza virus RNA segment. Virology. 1981;113:397–402. doi: 10.1016/0042-6822(81)90166-5. [DOI] [PubMed] [Google Scholar]
- 45.Kawaoka Y, Gorman OT, Ito T, Wells K, Donis RO, Castrucci MR, Donatelli I, Webster RG. Influence of host species on the evolution of the nonstructural (NS) gene of influenza A viruses. Virus Res. 1998;55:143–156. doi: 10.1016/s0168-1702(98)00038-0. [DOI] [PubMed] [Google Scholar]
- 46.Greatorex J. The retroviral RNA dimer linkage: different structures may reflect different roles. Retrovirology. 2004;1:22. doi: 10.1186/1742-4690-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garaigorta U, Falcon AM, Ortin J. Genetic analysis of influenza virus NS1 gene: a temperature-sensitive mutant shows defective formation of virus particles. J. Virol. 2005;79:15246–15257. doi: 10.1128/JVI.79.24.15246-15257.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Regan JF, Liang Y, Parslow TG. Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA. J. Virol. 2006;80:252–261. doi: 10.1128/JVI.80.1.252-261.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Treanor JJ, Snyder MH, London WT, Murphy BR. The B allele of the NS gene of avian influenza viruses, but not the A allele, attenuates a human influenza A virus for squirrel monkeys. Virology. 1989;171:1–9. doi: 10.1016/0042-6822(89)90504-7. [DOI] [PubMed] [Google Scholar]
- 50.Scholtissek C, Stech J, Krauss S, Webster RG. Cooperation between the hemagglutinin of avian viruses and the matrix protein of human influenza A viruses. J. Virol. 2002;76:1781–1786. doi: 10.1128/JVI.76.4.1781-1786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maines TR, Chen LM, Matsuoka Y, Chen H, Rowe T, Ortin J, Falcon A, Nguyen TH, Mai le Q, et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kermack WO, McKendrick AG. Contributions to the mathematical theory of epidemics – I. 1927. Bull. Math. Biol. 1991;53:33–55. doi: 10.1007/BF02464423. [DOI] [PubMed] [Google Scholar]