Abstract
Specific binding of HIV-1 viral protein NCp7 to a unique 35-base RNA stem-loop SL1 is critical for formation and packaging of the genomic RNA dimer found within HIV-1 virions. NCp7 binding stimulates refolding of SL1 from a metastable kissing dimer (KD) into thermodynamically stable linear dimer (LD). Using UV melting, gel electrophoresis and heteronuclear NMR, we investigated effects of various site-specific mutations within the full-length SL1 on temperature- or NCp7-induced refolding in vitro. Refolding involved intramolecular melting of SL1 stems but not dissociation of the intermolecular KD interface. Refolding required only two NCp7 molecules per KD but was limited by the amount of NCp7 present, implying that the protein does not catalytically promote refolding. Efficient refolding depended strictly on the presence and, to a lesser degree, on sequence of a highly conserved G-rich internal loop that normally limits thermal stability of the SL1 stem. Adding two base pairs to the lower stem created a hyperstable SL1 mutant that failed to refold, even when bound by NCp7 at high stoichiometries. NMR analysis of these kinetically trapped mutant RNA–protein complexes indicated that NCp7 initiates refolding by dissociating base pairs in the upper stem of SL1. This study illuminates structural transitions critical for HIV-1 assembly and replication.
INTRODUCTION
Human immunodeficiency virus (HIV-1), as with all retroviruses, carries two copies of its single-stranded RNA genome in the form of a noncovalent dimer inside each virion. This dimeric RNA genome is involved in strand transfers during reverse transcription, recombination and packaging [reviewed in (1)]. Formation of the dimer is a poorly understood process involving at least two distinct steps. Initially, genomic RNA dimerizes inside the host cell, after which it is packaged into viral particles via interaction with the Gag polyprotein. Inside the new viral particles, Gag is proteolytically processed, and one proteolytic product, the nucleocapsid protein (NCp7) directs further changes (maturation) of the viral RNA dimer, making it more compact and thermostable (2). The initial RNA–RNA interactions have been mapped to a so-called dimer initiation sequence, corresponding to a 35-nucleotide (nt) stem-loop, termed SL1, which is also part of the HIV-1 RNA packaging signal in the 5′-untranslated region (3–5). SL1 carries a palindromic 6-nt sequence in its apical loop, which allows it to spontaneously dimerize in vitro in the absence of any proteins via a loop interaction. The initially formed kissing-loop dimer (KD) is metastable; it is converted in vitro into a mature linear dimer (LD) by NCp7 (6) in a process believed to mimic the maturation of HIV-1 viral RNA in vivo. NCp7 stimulates the refolding as a RNA chaperone (7–9), by destabilizing intramolecular base pairing in KD and therefore lowering the energy barrier for the conversion to the thermodynamically more stable LD conformation. NCp7 also displays chaperone activity in other stages of the HIV-1 life cycle, such as strand transfer during reverse transcription [reviewed in (8)], and in various in vitro systems [e.g. (10)]. Both the initial RNA–RNA interactions and NCp7-mediated refolding of RNA dimer are essential steps required for dimerization, efficient packaging, viral maturation and, as a consequence, optimal viral infectivity.
Several groups have extensively investigated the role of NCp7 in dimer maturation, including optimal RNA–protein stoichiometry, and kinetics of conformational rearrangement induced by NCp7 in vitro [reviewed in (1)]. Many of these studies have been conducted with a truncated 23-nt version of SL1, which lacked the internal loop and the lower part of the stem. In particular, it has been shown for the 23-nt SL1 constructs that both temperature-induced and NCp7-induced conversion of KD into LD occurs via rearrangement of strands in the helical stems without disruption of the loop–loop base pairing (11,12). However, it is not clear how this process is affected by the lower stem of SL1, including the G-rich internal loop, which is extremely conserved among HIV-1 isolates. A mere over-stabilization of the SL1 stems relative to the kissing loop helical structure may modify the order of a temperature-induced melting of the SL1 KD, with the kissing loop moiety melting first and stems melting next (13,14). Indeed, the G-rich internal loop may be a critical participant in the NCp7-driven refolding process. NCp7 has been shown to preferentially recognize unpaired purines via hydrophobic interactions with zinc finger residues in two other stem-loop RNA structures of the packaging signal, SL2 and SL3 (15,16). In SL1 RNA, there are unpaired purines flanking the 6-nt palindrome in the loop region as well as a purine-rich asymmetric internal loop (Figure 1); both sites have been shown to specifically interact with NCp7 (17).
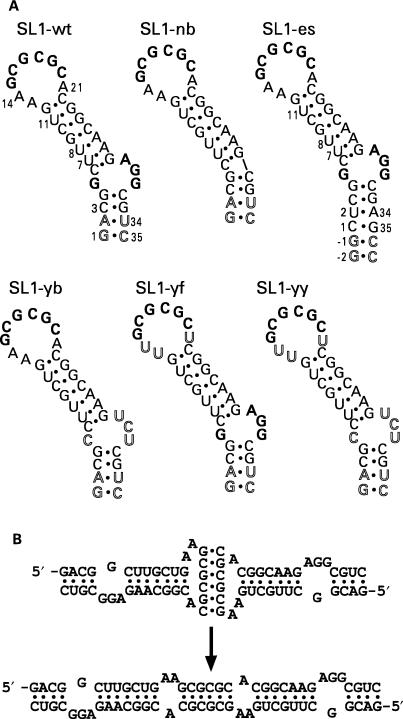
Figure 1.
(A) Nucleotide sequences of the six RNA constructs used in this study. SL1-wt is a 35-nt construct with the wild-type sequence of the Lai isolate, but with the two bottom base pairs reversed. Constructs SL1-yb and SL1-yf have pyrimidines replacing the internal loop purines or the flanking adenines, respectively, while SL1-yy has pyridine replacement in both the internal loop and flanking regions. The construct SL1-nb has no internal loop, and SL1-es is extra stabilized by an additional two GC base pairs in the bottom stem. The palindromic 6-nt sequence and the G-rich internal loop are shown in bold; non-wild-type residues are shown in outline. (B) Kissing (top) and linear (bottom) dimers of SL1-wt.
In this article, we show for the full-length 35-nt SL1 RNA that the KD-to-LD conversion occurs via transfer of strands in the stem portions of SL1 without breaking the loop–loop interactions of the 6-nt palindromic part of SL1. We also investigate the role of the native unpaired purines in the NCp7-induced KD-to-LD refolding. In addition, we have been successful in trapping an intermediate conformation in the refolding process and have demonstrated via NMR that the initial destabilization occurs in the upper stem of SL1, between the KD interface and the G-rich internal loop.
MATERIALS AND METHODS
RNA synthesis
A series of RNA oligonucleotides, wild-type SL1 and its mutants (Figure 1), were prepared by run-off transcription in vitro using T7 RNA polymerase from synthetic DNA templates, as previously described (18); most were 35 nt long. Each RNA construct was purified by 20% polyacrylamide gel electrophoresis (PAGE) followed by electroelution. RNAs were dialyzed against 10 mM EDTA, then 1.0 mM EDTA and finally against sterile water for 48 h. Each RNA sample was quantified by ultraviolet (UV) spectrophotometry and stored at −20°C. For refolding experiments by heteronuclear NMR, an SL1 construct (SL1-es) having two extra GC base pairs in the bottom of the stem (Figure 1) was synthesized similarly but with all four uridines isotopically labeled with 13C- and 15N-enriched rUTP (Silantes, Germany).
NCp7 expression and purification
The full-length 55 amino acid HIV-1 NCp7 was expressed and purified as described previously (15). The plasmid used for expressing NCp7 was a generous gift from Professor Michael Summers. After purifying NCp7 on a G50 size-exclusion column, protein was concentrated using a Centricon (3 K cutoff) and stored in sodium acetate buffer, pH 6.4 containing 0.1 mM ZnCl2. The integrity and purity of NCp7 were checked by SDS-PAGE and mass spectrometry.
RNA sample preparation and dimerization assay
In each case of RNA sample preparation, a 5-μM RNA aliquot was heated to 90°C for 3 min and chilled in ice water for 3 min. While on ice water, a 2× dimerization buffer (20 mM sodium phosphate pH 6.4, 100 mM NaCl and 0.2 mM MgCl2) of equal volume was added to the sample. This gave a final strand concentration of RNA of 2.5 μM. As a final step, samples were incubated either at ambient temperature for folding RNA into the KD or at 55°C for an LD of SL1 RNA. The existence of desired RNA dimer was checked by running dimer samples on 10% native PAGE in 1× TBE (Tris-Borate EDTA) or 1× TBM (Tris-Borate-Magnesium) buffer. The 10× TBE buffer contained 89 mM Tris, 89 mM boric acid and 2 mM EDTA; in 10× TMB, EDTA was replaced with 2 mM MgCl2. The KD form requires magnesium to be stable in the gel, and it dissociates into monomers when exposed to EDTA-containing TBE buffer. The RNAs folded into the KD form showed bands for a dimeric species in TBM and monomer bands in TBE buffer. On the other hand, RNAs incubated at 55°C and folded into the LD did not require magnesium for dimeric stability, hence showing a dimer band both in TBE and TBM buffers. Dependence of KD stability on the presence of magnesium has been previously reported by several groups (19–21).
NCp7-mediated maturation of SL1 RNA ‘kissing dimer’
Electrophoretically characterized KD samples of all SL1 RNAs, i.e. wild type and mutants, were mixed with NCp7. In each case, four aliquots of 2.5 μM KD RNA were prepared in duplicate. NCp7 was added to each aliquot to achieve final RNA monomer-to-protein molar ratios of 2:0, 2:2, 2:4 and 2:8. These complexes were incubated at ambient temperature for either 2 h or overnight. NCp7-mediated dimer maturation occurred during this step. After incubation, NCp7 was stripped of RNA by phenol–chloroform extraction. RNA was then ethanol precipitated and resuspended in 1× dimerization buffer after dialyzing with the same buffer to bring RNA back to native solution conditions. Finally, native PAGE and UV absorbance melting experiments were performed on each sample to characterize the RNA dimeric form(s) that existed after NCp7 incubation. Most experiments were carried out in duplicate.
UV melting experiments
All UV experiments were carried out on a CARY 3E UV–visible spectrophotometer equipped with a temperature controller. RNA dimer samples were heated over a temperature range of 15–90°C with heating rates of 0.5–1.0°C per min, 0.1°C resolution step size; the absorbance was measured at 275 nm (A275). Each experiment entailed both heating and cooling. Data were processed using polynomial curve fittings and thermal derivatives of melting curves were calculated using KaleidaGraph software (version 3.6.4).
Native gel electrophoresis
PAGE was performed on each RNA sample after NCp7 protein extraction, as described above. Native PAGE experiments were carried out using a 10% bisacrylamide ratio of matrix at 4°C in either TBE or TBM buffer. All gels were pre-equilibrated at 100 V for 2 h prior to sample loading. A native loading dye with 10% glycerol was used in each case. Gels were run for 2–4 h or until the bromophenol blue ran to the bottom. Gels were then stained with ethidium bromide and visualized under UV light.
NMR experiments for thermal melting profiles of kissing and linear dimers
All NMR experiments were performed on Varian INOVA 600 MHz NMR spectrometers. For 2D 15N-HSQC experiments (22), a spectrometer equipped with a cold probe was used. Using the jump-and-return pulse sequence, 1D NMR profiles of imino resonances were collected at various temperatures between 10 and 60°C for KD and LD samples of SL1-wt RNA. In both cases, a 300-μM RNA dimer sample was used in a Shigemi NMR tube with a diameter of 8 mm. Data were processed using vnmr (Varian) and NMRpipe (23) software with a Gaussian window function.
RESULTS
SL1 constructs
We synthesized five 35-nt-long RNAs, and one extra-stabilized RNA of 39 nt; these six RNA samples represented the wild-type sequence of SL1 and desired modifications as described in Figure 1. The RNA construct SL1-wt corresponds to the wild-type sequence of SL1 RNA with the two bottom base pairs reversed for optimal transcription yield. Four other constructs were designed to investigate the role of unpaired purines in the KD-to-LD conversion: SL1-yb (i.e. pyrimidine bulge) mutant carries a pyrimidine-rich internal loop in place of the wild-type purine-rich internal loop. SL1-yf (pyrimidine flanks) has flanking adenines replaced with uracils, and a double mutant, SL1-yy, has pyrimidines substituted for both flanking adenines and internal loop purine residues. SL1-nb (no bulge) carries a continuous stem without an asymmetric internal loop. Finally, to investigate the effect of overall stem stability, we extended the lower stem by two extra GC base pairs in the SL1-es construct (i.e. extra stable or extended stem). The orientation of residues 1, 2, 34 and 35 in SL1-es is the same as in wild-type HIV-1 sequences, unlike our other five constructs.
Characterization of dimer formation by native PAGE
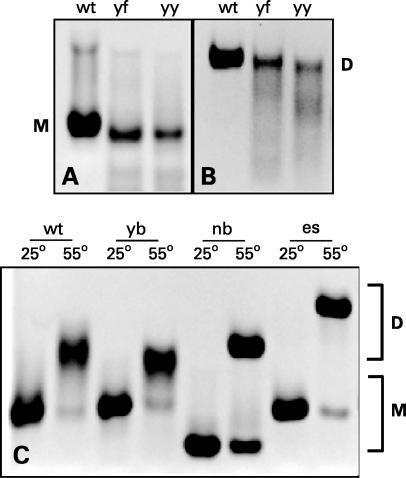
The KD form of SL1 dissociates into monomers during migration through the pores of polyacrylamide gels in TBE unless magnesium is present in the running buffer (20). On the other hand, the LD form runs as a dimer during PAGE even in the absence of magnesium; this can be used as an assay to distinguish the two dimeric forms (21). All SL1 constructs studied here formed KD (Figure 2A and B and not shown). However, SL1-yf and SL1-yy with flanking adenines mutated to pyrimidines appear to form less stable KDs, because some smearing was observed between the monomer and dimer positions in the magnesium-containing gels (Figure 2B); smearing implies a dynamic equilibrium between dimer and monomer in the gel.
Figure 2.
Electrophoretic behavior of various SL1 constructs; ‘M’ and ‘D’ indicate the position of monomers and dimers, respectively. SL1 RNA was run in native 10% polyacrylamide gels at 100 V in either TBE or TBM buffer. (A) Kissing dimers (KDs) of SL1 in TBE buffer. (B) KDs of SL1 in TBM buffer. (C) KDs of SL1 constructs were incubated for 1 h at either 25 or 55°C as indicated and separated in PAGE run in TBE buffer. Only linear dimers of SL1 migrate as dimers in TBE buffer.
Incubation at elevated temperature converts KD into LD (21,24). Incubation of SL1 constructs at 55°C for 2 h converted them into stable LD forms, which ran as dimers in the magnesium-free gels (Figure 2C and not shown); the second smaller band running at the monomer position corresponds to the portion of the SL1, which has not been converted and remained in the KD form. This unconverted portion was especially significant for SL1-nb and SL1-es, for which the stems were over-stabilized either by deleting the internal loop or by adding two extra base pairs.
Temperature-induced KD-to-LD conversion studied by UV and NMR
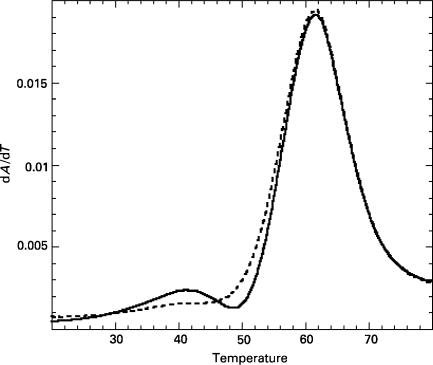
We recorded UV thermal melting profiles for 3-μM samples of KD and LD forms of our SL1 RNA constructs. LD forms of all six constructs melted cooperatively with a single transition, at 62°C in the case of SL1-wt (Figure 3). In contrast, KD forms exhibited a biphasic melting transition when the temperature was slowly increased from 15 to 90°C. In the case of SL1-wt, the first transition occurred at 41°C, followed by the main melting transition at 62°C. The first transition at 41°C was no longer observed when UV absorbance was recorded during the subsequent slow cooling of the sample from 90 to 15°C or during the next heating, indicating that KD converted into LD during the slow annealing. The presence of LD forms in these samples was confirmed by running native PAGE in TBE and TBM buffers.
Figure 3.
UV thermal melting profiles of kissing (solid curve) and linear (dashed curve) dimers of SL1-wt (3 μM RNA in the 1× dimerization buffer): Kissing dimer exhibits a biphasic melting curve with transitions at 41 and 62°C, while the linear dimer melts with a single transition at 62°C.
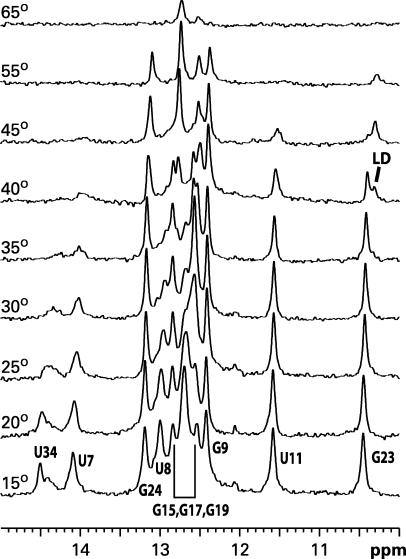
The nature of the temperature-induced transition at 41°C can be identified by monitoring the imino proton NMR signals of SL1-wt (Figure 4). The G23 imino proton, which forms part of the wobble GU pair in the upper stem, resonates at slightly different frequencies in the two structural isomers of SL1: at ∼10.45 p.p.m. in KD (25) and at 10.36–10.39 p.p.m. (depending on temperature and salt conditions) in LD (26). The lower trace in Figure 4 shows the imino proton spectrum of the KD form of SL1-wt at 15°C. As temperature increased and base pairs destabilized, the resonances gradually broadened reflecting faster exchange with water; the resonances completely disappeared above 65°C (62°C is the midpoint of melting as monitored by UV absorbance). However, at an intermediate temperature, 40°C, the G23 resonance split and displayed two separate peaks corresponding to the resonances of both KD and LD. At higher temperatures, only one signal from LD remained. The most plausible explanation for this observation is that the stems melt at this temperature (midpoint 41°C as monitored by UV) and refold into the more stable LD conformation. The GC pairs in the palindromic interface of the dimer (imino protons G15, G17, G19) remained intact throughout the refolding process and were the last to melt as temperature was increased. The lower portion of the stem (U34) appears to melt just before the refolding event and not to form again in the refolded LD above 40°C; alternatively, it forms but the base pairs are not stable enough for the imino protons to be observed due to rapid proton exchange with water with or without intermediate exchange between different conformations. The final temperature-driven dissociation of strands at 62°C occurs from this refolded LD form. A similar melting behavior has been reported previously for a shorter 23-nt SL1 construct lacking the G-rich internal loop and the lower part of the stem (11).
Figure 4.
Temperature dependence of imino proton resonances in 1D NMR spectra. Here, 300 μM SL1-wt RNA was folded into the kissing dimer (KD) form and gradually heated in the NMR spectrometer. At 40°C, the G23 imino proton split into two species corresponding to the original kissing dimer and newly emerging linear dimer.
NCp7-induced KD-to-LD conversion studied by native PAGE
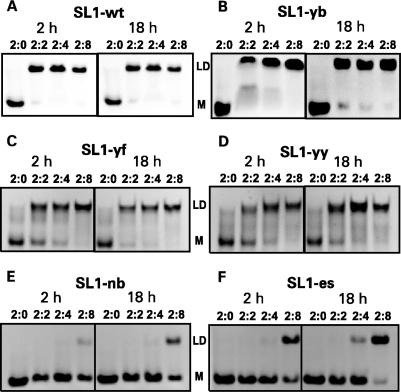
We incubated NCp7 with the KD forms of various SL1 constructs at ambient temperature and with RNA strand-to-protein ratios of 2:2, 2:4 and 2:8. As a control, KD samples without any NCp7 were also included in each set. Incubations were carried out for 2 h and overnight (18 h). After the incubation, RNA was extracted from the complexes as described in the Materials and methods section. Prior to further experiments, we established that the RNA extraction step did not affect the RNA folding. Extracted RNA was run in native PAGE in TBE buffer; under these conditions, any SL1 RNA that refolded into LD migrates as a dimer but any RNA that remained in KD form dissociates into a monomer (vide supra). The results shown in Figure 5 depict the efficiency of NCp7-mediated dimer maturation for each SL1 construct.
Figure 5.
Kissing dimers (KDs) of all six SL1 constructs were incubated at ambient temperature for 2 h (left panels) or 18 h (right panels) with NCp7 in the RNA strand-to-protein ratios of 2:0, 2:2, 2:4 and 2:8. RNA was ethanol-precipitated after phenol–chloroform extraction, dialyzed in the 1× dimerization buffer and separated in native PAGE in the TBE buffer. ‘LD’ indicates the position of the dimer bands, which correspond to the linear dimers at these conditions. KDs dissociate to monomers during PAGE (shown by ‘M’). Panels (A–F) show results for SL1-wt, SL1-yb, SL1-yf, SL1-yy, SL1-nb and SL1-es, respectively.
After incubation of the wild-type SL1-wt with NCp7, complete dimer conversion was achieved within 2 h when two protein molecules were present per dimer of RNA as indicated by the presence of the LD band (Figure 5A). Increasing the amount of NCp7 or the incubation time produced no further changes, indicating that the reaction was already completed within 2 h with the 2:2 ratio. To estimate the minimum time required for complete refolding of SL1-wt under these conditions, we varied the incubation time from 30 s to 1 h for the 2:2 ratio complex: complete dimer conversion was achieved within 1 min of incubation (data not shown).
Pyrimidine substitutions in the G-rich internal loop had very little, if any, effect: the KD-to-LD conversion of SL1-yb was practically as effective as that of SL1-wt during a 2-h incubation at the 2:2 ratio (Figure 5B). On the other hand, pyrimidine substitutions for adenines flanking the central palindrome (in SL1-yf and SL1-yy) had a more noticeable effect: the conversion was clearly not complete after 2 h of incubation at the 2:2 ratio. Interestingly, for these three mutants, increasing the incubation time or increasing the amount of NCp7 led to a more complete conversion (Figure 5C and D). The situation was dramatically different for SL1 constructs with over-stabilized stems, SL1-nb (deleted internal loop) and SL1-es (two extra base pairs). NCp7-mediated refolding of these latter two mutants was markedly inefficient, especially for SL1-nb (Figure 5E and F). At least eight NCp7 molecules per RNA dimer were required to convert SL1-nb into LD after 18 h of incubation, and even then, the conversion was only partial. The SL1-es RNA converted a little more readily, but it still required a higher protein-to-RNA ratio than the wild-type sequence.
A trapped folding intermediate of SL1-es studied by NMR
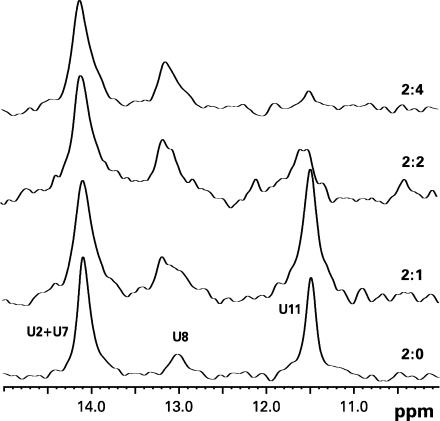
The evidence of kinetically trapped intermediate species of SL1-es–NCp7 complexes (Figure 5F) presented us with an opportunity to investigate the refolding process by NMR. We monitored changes in imino proton and nitrogen resonances by acquiring 15N-HSQC (heteronuclear singlequantum correlation) NMR spectra for the complex of NCp7 with SL1-es, which incorporated 13C,15N-labeled nucleosides at the four native uridines in the stem. The 15N-HSQC is an NMR experiment that correlates covalently connected nitrogen and proton nuclei. There are four uridines in SL1-es: U7, U8, U11 are located in the upper stem with U11 being part of a wobble GU pair, and U2 is located in lower stem below the internal loop. All four of these uridines correspond to true wild-type SL1 (see Figure 1); the first three have the same position (and numbers) in SL1-es and SL1-wt, and U2 in SL1-es is located opposite to U34 in SL1-wt, since our SL1-wt sequence has that base pair inverted for optimal transcription efficiency (Figure 1).
15N-HSQC spectra were acquired at 15°C as NCp7 was incrementally added to the 150-μM U-labeled SL1-es KD sample. Aliquots of highly concentrated NCp7 were successively added to achieve RNA strand-to-protein ratios of 2:0, 2:1, 2:2 and 2:4 in the NMR tube. Each data point represents an incubation of ∼3 h before the next aliquot. For clarity, the data are presented as the first 1D slices of HSQC, which show resonances of imino protons directly attached to labeled nitrogen nuclei (Figure 6); positions of the 15N resonances did not change during the NCp7 titration (not shown). The lower trace in Figure 6 corresponds to the free KD form of SL1-es; only imino protons from the four labeled uridines contribute to the observed signal. The strongest signal comes from U11 at 11.46 p.p.m., which is a part of the wobble pair (see also Figure 4). The imino proton of U8 resonates at 13.2 p.p.m., and imino protons of U7 and U2 overlap at 14.2 p.p.m. (in SL1-wt, imino protons from U7 and U34 are well resolved; Figure 4). Signals from both U7 and U8 are relatively weak, probably due to conformational properties of the UU:AA stretch in RNA; they also have relatively small intensity in the LD form of SL1-wt (26).
Figure 6.
First slices of 15N-HSQC NMR experiments on complexes of U{15N}-labeled SL1-es kissing dimer and NCp7 in the 2:0, 2:1, 2:2 and 2:4 RNA strand-to-protein ratios at 15°C. Only signals from imino protons of uridines are observed in this experiment.
The effect of increasing the amount of NCp7 on the SL1-es structure can be monitored by imino proton signals of labeled uridines. When only one protein molecule was present per RNA dimer, the effect was relatively small; only the line shape of U8 from the upper stem was somewhat affected, indicating a dynamic process at this site. The effect was much more significant on the U11 residue when greater amounts of NCp7 were present. The U11 signal dramatically broadened in the 2:2 complex and completely disappeared with four protein molecules per RNA dimer (i.e. two NCp7 molecules per RNA strand), indicating complete destabilization of the wobble GU pair from the upper stem. Because the imino resonances of U2 and U7 overlapped, we could not monitor them individually. However, the cumulative effect on both residues was negligible. It is clear that the primary effect of NCp7 was on the upper stem at these conditions; there was no evidence that the structure or stability of the lower stem, which is over-stabilized in SL1-es by two extra base pairs, was affected at all. Although the U8 residue appeared to be the first one affected at sub-stoichiometric protein amounts, the most dramatic effect was on the wobble GU pair located close to the dimeric interface of the KD form of SL1.
No evidence of NCp7 turnover during the refolding process
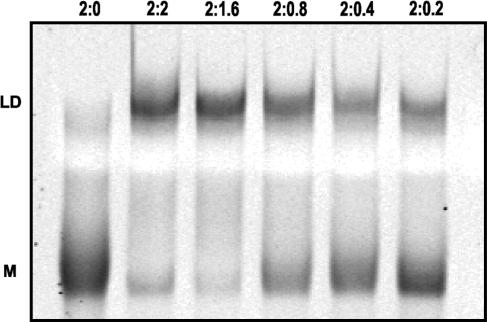
We carried out overnight incubations of SL1-wt KD with sub-stoichiometric amounts of NCp7 in RNA strand-to-protein ratios of 2:0.2, 2:0.4, 2:0.8, 2:1.6 and 2:2. With two NCp7 molecules per RNA dimer, as expected, dimer maturation was complete. If NCp7 behaved as an enzyme, then samples with lower protein content would be expected to display an increasing extent of dimer maturation over time. However, even after overnight incubation, the amount of RNA converted to LD remained strictly proportional to the amount of NCp7 present in the reaction (Figure 7). These results imply that once NCp7 transforms KD into LD, the protein remains bound to the mature LD RNA and does not jump to another KD molecule.
Figure 7.
Native PAGE experiments on SL1-wt RNA kissing dimer (KD) in the presence of sub-stoichiometric amounts of NCp7, run in the TBE buffer. KD of SL1-wt was incubated with NCp7 at ambient temperature for 18 h in the RNA strand-to-protein ratio of 2:0, 2:2, 2:1.6, 2:0.8, 2:0.4 and 2:0.2, as indicated. The upper band in each lane (‘LD’) corresponds to mature linear dimer, while the lower monomer band (‘M’) emanates from the residual KD that dissociates during PAGE in the TBE buffer. Increasing intensity of the monomer band and simultaneous decrease in the dimer band across the lanes correlates with the amount of NCp7 present in each complex.
DISCUSSION
The first thermal transition, melting of stems, occurs at 41°C in the KD form of the full-length SL1 (Figure 3). This occurs at a temperature ∼20°C higher than in the truncated 23-nt SL1 constructs (11), but in either case this transition occurs >20°C below the temperature required to dissociate the KD complex into monomers. This substantial difference in the stabilities of SL1 stems compared to the more stable kissing loop interface is apparently important for optimal NCp7-induced refolding. It is estimated from the single-molecule stretching-induced melting experiments that at saturating amounts of NCp7, this chaperone protein can destabilize each base pair in a double-stranded DNA duplex by ∼1 kcal/mol (27). This is consistent with the general chaperone properties of NCp7 (7,8) in that destabilization lowers energy barriers between metastable kinetically trapped intermediates and thermodynamically stable conformations. Although caution must be used when extrapolating thermodynamic data from duplex DNA to SL1 RNA, it is clear nevertheless that there is an upper limit on the thermal stability of the stems, above which NCp7 will be hindered in its ability to facilitate the KD-to-LD conversion. Although the relative stability of stems is the same in KD and LD, the energy barrier for the conversion depends on this stability. Indeed, the conversion is dramatically impeded in the over-stabilized constructs, SL1-nb and SL1-es, at room temperature and one-to-one RNA-protein ratio (Figure 5). A larger protein:RNA stoichiometry was necessary to convert these constructs to LD, and still the conversion was not complete under the conditions employed. It is interesting that in the case of double-stranded DNA, the degree of helix destabilization also depends on the binding stoichiometry of NCp7 (28).
The G-rich internal loop appears to be an important destabilizing element in SL1, and consequently critical to the refolding process. In SL1-es, the upper and lower stems have seven and six base pairs, respectively; this mutant is converted to LD by NCp7 more readily than SL1-nb, where the deletion of the internal loop makes the stem 11 base pairs long (Figure 1). The first thermal transition occurs in SL1-es is at 48°C (not shown); it is interesting that incubating the KD form of DIS39, an RNA construct similar to SL1-es, at 37°C (rather than at room temperature as in our study) reportedly induces nearly complete conversion to LD even at a one-to-one RNA-protein ratio (21).
Mutating all residues in the G-rich internal loop to pyrimidines has very little effect on the ability of NCp7 to promote the KD-to-LD conversion (Figure 5B). It appears that the exact sequence of the internal loop is not important for the dimer maturation in vitro as long as the overall stability of SL1 stem is not too high. However, the SL1 G-rich internal loop sequence is extremely conserved in HIV-1 sequences. Among >300 HIV-1 isolates for which the SL1 stem-loop was completely sequenced (29), only two have sequence variations in the internal loop: one has G-AAG and another has G-GGG instead of the prevalent G-AGG (see Supplementary Data). This high degree of conservation suggests that this internal loop sequence has roles other than destabilization of the SL1 stem. One such role is in packaging of genomic RNA into virions: mutating all residues of the internal loop to uridines (in the context of a different sequence of the upper stem of SL1) reduces the packaging efficiency to 18% of the wild-type level, and reduces the overall infectivity by 1.1 log units in vivo (30). Deleting the internal loop altogether had a similar effect: the packaging and overall infectivity are reduced to 14% and by 1.0 log units, respectively (30). It has also been shown that mutations in the SL1 internal loop impair dimerization of genomic RNA in vivo, decreasing the amount of dimeric form while increasing the amount of monomeric viral RNA found inside viral particles (31). Deletions AGG31 or G5 or substitution G5C (numbering as in Figure 1) decreased the fraction of dimeric form to 62, 62 and 75%, respectively, relative to that in wild-type viruses. The AGG31 deletion and G5C substitution are likely to increase the stability of the SL1 stem: the former because a rather large internal loop is substituted with a 1-nt bulge, and the latter because a GC base pair is substituted for the GA mismatch in the wild-type sequence (26,32–35). However, it is unlikely that the G5 deletion would increase the stability of the stem, because the resulting 3-nt bulge is still large and furthermore lacks a somewhat stabilizing GA base pair. All observations together suggest that there may be differences in the dimerization processes in vivo and in vitro. One possible explanation for the divergent effects of mutations in the internal loop may be stabilization of an alternative global secondary structure that occludes the palindromic sequence in SL1 and thus prevents formation of initial KD. This possibility seems attractive in view of the notion that the HIV-1 leader RNA can adopt two alternative structures that have distinctly different biological functions in packaging and gene expression (36,37). This can also explain the remarkably strict conservation of sequences in the lower stem of SL1 (see Supplementary Data), and the finding that many mutations are detrimental for dimerization of genomic RNA in viral particles, even when such mutations preserve base pairing in the lower stem (31). However, in the case of the G5 deletion, such a possibility was excluded by Shen et al. (31) based on secondary structure predictions using the mfold algorithm (38).
Substituting uracils for the unpaired adenines flanking the kissing loop interface also had only a moderate effect on dimer maturation in vitro; the rate of NC-7-induced conversion was slightly attenuated in SL1-yf and SL1-yy (Figure 5C and D). These flanking residues are less strongly conserved among HIV-1 strains than are those of the internal loop. The conformations of the flanking adenine residues also appear to be quite variable in NMR solution (25,34,39) and X-ray crystal [reviewed in (40)] structures of SL1 constructs; in fact, these residues may be flexible in solution. Nevertheless, flanking regions composed entirely of pyrimidines have never been observed in nature, and purines, especially adenines, are strongly predominant in the 5′-flanking region (see Supplementary Data). It is possible that purines are important for stabilizing the initial kissing interaction (25); KD forms of SL1-yf and SL1-yy appeared to be less stable than their purine-containing counterparts during PAGE (Figure 2B). It is interesting that the all-flanking-U mutation studied in the context of a longer 216-nt RNA construct did not show any dimers at all during PAGE (41). It is likely that the mechanical force imposed on SL1 dimers during migration through the gel pores is stronger for longer RNA molecules.
In this study, we have established, using the over-stabilized SL1-es construct, that the upper stem of the SL1 hairpin is the region predominantly affected by NCp7 binding (Figure 6). Other investigators have shown that there are two high-affinity binding sites of NCp7 on each SL1 hairpin: one associated with the KD interface and the other with the G-rich internal loop (17). However, NCp7 efficiently converts SL1-wt into LD in the 2:2 ratio in vitro (Figure 5), i.e. with one protein per each SL1 hairpin. It is likely that there is an equilibrium entailing two bound states of NCp7 under such conditions, because the binding constants of the two are comparable (17). However, NCp7 does not appear to dissociate from SL1-wt after the conversion to LD, because no turnover was observed at sub-stoichiometric ratios (Figure 7). A similar observation has been reported previously with dimer maturation in a heterologous system, which included the HIV-1 NCp7 and a synthetic 345-nt fragment of Harvey sarcoma virus leader RNA. In that model system, the conversion of an initial, metastable RNA dimer into its thermodynamically stable form occurred rapidly when the concentration of NCp7 was greater than some critical value, but it did not go to completion even after 10 h of incubation with one protein molecule per 50 nt (42). Similar dependence on a critical threshold protein concentration has also been observed in other reactions involving NCp7 chaperone activity [reviewed in (8)]. Although interpretations are complicated for RNA molecules that contain multiple NCp7-binding sites (17), there are only two such sites on SL1-wt. It is likely that the G-rich internal loop is responsible for the slow dissociation rate from LD; this might account for the observation that NCp7 acts catalytically in refolding a truncated SL1 construct that lacked both the internal loop and the lower stem (12). Perhaps NCp7 is able to slide rapidly off the blunt end of an RNA stem, and the internal loop in the wild-type sequence prevents such a dissociation mechanism. Further studies will be needed to determine the biological significance, if any, of the slow dissociation kinetics of NCp7 from the LD form of SL1.
In conclusion, we demonstrated for the full-length SL1 that refolding of KD into LD occurs without dissociation of the KD interface. This was expected and consistent with the data for shorter SL1 constructs (11,12). The KD-to-LD conversion of SL1-wt requires at least two NCp7 molecules per RNA dimer. This refolding is not catalytic for the full-length SL1, in contrast to shorter SL1 constructs devoid of the G-rich internal loop and the lower stem (12). The presence of the conserved internal loop is critical for the NCp7-induced refolding. However, the exact sequence of the internal loop appears to be less important in in vitro experiments. Over-stabilization of the lower stem led to a kinetically trapped KD form of SL1-es; adding NCp7 to this form destabilizes the upper stem but fails to completely dissociate it.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institute of Health grants AI36636 and AI46967. Funding to pay the Open Access publication charge was provided by AI46967.
Conflict of interest statement. None declared.
REFERENCES
- 1.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: an inseparable pair. Nat. Rev. Microbiol. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 2.Fu W, Gorelick RJ, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paillart JC, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 5.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 6.Muriaux D, De Rocquigny H, Roques B-P, Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop–loop complex into a stable dimer. J. Biol. Chem. 1996;271:33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 7.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 8.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 9.Darlix JL, Lopez-Lastra M, Mely Y, Roques B. Nucleocapsid protein chaperoning of nucleic acids at the heart of HIV structure, assembly and cDNA synthesis. In: Kuiken C, Foley B, Freed E, Hahn B, Marx P, McCutchan F, Mellors JW, Wolinsky S, Korber B, editors. HIV Sequence Compendium 2002. Los Alamos, NM: Los Alamos National Laboratory; 2003. pp. 69–88. [Google Scholar]
- 10.Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl. Acad. Sci. USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theilleux-Delalande V, Girard F, Huynh-Dinh T, Lancelot G, Paoletti J. The HIV-1(Lai) RNA dimerization. Thermodynamic parameters associated with the transition from the kissing complex to the extended dimer. Eur. J. Biochem. 2000;267:2711–2719. doi: 10.1046/j.1432-1327.2000.01292.x. [DOI] [PubMed] [Google Scholar]
- 12.Rist MJ, Marino JP. Mechanism of nucleocapsid protein catalyzed structural isomerization of the dimerization initiation site of HIV-1. Biochemistry. 2002;41:14762–14770. doi: 10.1021/bi0267240. [DOI] [PubMed] [Google Scholar]
- 13.Weixlbaumer A, Werner A, Flamm C, Westhof E, Schroeder R. Determination of thermodynamic parameters for HIV DIS type loop–loop kissing complexes. Nucleic Acids Res. 2004;32:5126–5133. doi: 10.1093/nar/gkh841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorenz C, Piganeau N, Schroeder R. Stabilities of HIV-1 DIS type RNA loop–loop interactions in vitro and in vivo. Nucleic Acids Res. 2006;34:334–342. doi: 10.1093/nar/gkj435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 16.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 17.Shubsda MF, Paoletti AC, Hudson BS, Borer PN. Affinities of packaging domain loops in HIV-1 RNA for the nucleocapsid protein. Biochemistry. 2002;41:5276–5282. doi: 10.1021/bi016045+. [DOI] [PubMed] [Google Scholar]
- 18.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. In: Dahlberg JE, Abelson JN, editors. RNA Processing Part A: General Methods Meth. Enzym. Vol. 180. New York: Academic Press; 1989. pp. 51–62. [DOI] [PubMed] [Google Scholar]
- 19.Marquet R, Baudin F, Gabus C, Darlix JL, Mougel M, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 1991;19:2349–2357. doi: 10.1093/nar/19.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laughrea M, Jette L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi KI, Baba S, Chattopadhyay P, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. Structural requirement for the two-step dimerization of human immunodeficiency virus type 1 genome. RNA. 2000;6:96–102. doi: 10.1017/s1355838200991635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kay LE, Keifer P, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 1992;114:10663–10665. [Google Scholar]
- 23.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 24.Baba S, Takahashi K, Nomura Y, Noguchi S, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. Conformational change of dimerization initiation site of HIV-1 genomic RNA by NCp7 or heat treatment. Nucleic Acids Res. Suppl. 2001;1:155–156. doi: 10.1093/nass/1.1.155. [DOI] [PubMed] [Google Scholar]
- 25.Mujeeb A, Clever JL, Billeci TM, James TL, Parslow TG. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat. Struct. Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 26.Ulyanov NB, Mujeeb A, Du Z, Tonelli M, Parslow TG, James TL. NMR structure of the full-length linear dimer of stem-loop-1 RNA in the HIV-1 dimer initiation site. J. Biol. Chem. 2006;281:16168–16177. doi: 10.1074/jbc.M601711200. [DOI] [PubMed] [Google Scholar]
- 27.Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, et al. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34:593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein's nucleic acid chaperone function. J. Mol. Biol. 2006;363:867–877. doi: 10.1016/j.jmb.2006.08.070. [DOI] [PubMed] [Google Scholar]
- 29.Leitner T, Foley B, Hahn B, Marx P, McCutchan F, Mellors JW, Wolinsky S, Korber B. HIV Sequence compendium 2005. Los Alamos, NM: Ios Alamos National Laboratory; 2005. [Google Scholar]
- 30.Clever JL, Parslow TG. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen N, Jette L, Wainberg MA, Laughrea M. Role of stem B, loop B, and nucleotides next to the primer binding site and the kissing-loop domain in human immunodeficiency virus type 1 replication and genomic-RNA dimerization. J. Virol. 2001;75:10543–10549. doi: 10.1128/JVI.75.21.10543-10549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence DC, Stover CC, Noznitsky J, Wu Z, Summers MF. Structure of the intact stem and bulge of HIV-1 Psi-RNA stem-loop SL1. J. Mol. Biol. 2003;326:529–542. doi: 10.1016/s0022-2836(02)01305-0. [DOI] [PubMed] [Google Scholar]
- 33.Yuan Y, Kerwood DJ, Paoletti AC, Shubsda MF, Borer PN. Stem of SL1 RNA in HIV-1: structure and nucleocapsid protein binding for a 1 × 3 internal loop. Biochemistry. 2003;42:5259–5269. doi: 10.1021/bi034084a. [DOI] [PubMed] [Google Scholar]
- 34.Baba S, Takahashi K, Noguchi S, Takaku H, Koyanagi Y, Yamamoto N, Kawai G. Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers. J. Biochem. (Tokyo) 2005;138:583–592. doi: 10.1093/jb/mvi158. [DOI] [PubMed] [Google Scholar]
- 35.Greatorex J, Gallego J, Varani G, Lever A. Structure and stability of wild-type and mutant RNA internal loops from the SL-1 domain of the HIV-1 packaging signal. J. Mol. Biol. 2002;322:543–557. doi: 10.1016/s0022-2836(02)00776-3. [DOI] [PubMed] [Google Scholar]
- 36.Ooms M, Huthoff H, Russell R, Liang C, Berkhout B. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 2004;78:10814–10819. doi: 10.1128/JVI.78.19.10814-10819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbink TE, Ooms M, Haasnoot PC, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44:9058–9066. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 38.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 39.Kieken F, Paquet F, Brule F, Paoletti J, Lancelot G. A new NMR solution structure of the SL1 HIV-1Lai loop–loop dimer. Nucleic Acids Res. 2006;34:343–352. doi: 10.1093/nar/gkj427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ennifar E, Dumas P. Polymorphism of bulged-out residues in HIV-1 RNA DIS kissing complex and structure comparison with solution studies. J. Mol. Biol. 2006;356:771–782. doi: 10.1016/j.jmb.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 41.Clever JL, Wong ML, Parslow TG. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng YX, Copeland TD, Henderson LE, Gorelick RJ, Bosche WJ, Levin JG, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.