Abstract
Tumour-derived p53 mutants are thought to have acquired ‘gain-of-function’ properties that contribute to oncogenicity. We have tested the hypothesis that p53 mutants suppress p53-target gene expression, leading to enhanced cellular growth. Silencing of mutant p53 expression in several human cell lines was found to lead to the upregulation of wild-type p53-target genes such as p21, gadd45, PERP and PTEN. The expression of these genes was also suppressed in H1299-based isogenic cell lines expressing various hot-spot p53 mutants, and silencing of mutant p53, but not TAp73, abrogated the suppression. Consistently, these hot-spot p53 mutants were able to suppress a variety of p53-target gene promoters. Analysis using the proto-type p21 promoter construct indicated that the p53-binding sites are dispensable for mutant p53-mediated suppression. However, treatment with the histone deacetylase inhibitor trichostatin-A resulted in relief of mutant p53-mediated suppression, suggesting that mutant p53 may induce hypo-acetylation of target gene promoters leading to the suppressive effects. Finally, we show that stable down-regulation of mutant p53 expression resulted in reduced cellular colony growth in human cancer cells, which was found to be due to the induction of apoptosis. Together, the results demonstrate another mechanism through which p53 mutants could promote cellular growth.
INTRODUCTION
The p53 suppressor gene is mutated in ∼50% of all human cancers (1–3). Mutations in p53 have been shown to abrogate its cardinal functions in promoting apoptosis, cell-cycle arrest and DNA repair, thereby leading to cancer development and progression (1,2). Activation of p53, which is a transcription factor, results in the transactivation of many target genes that regulate these biological processes. Thus, abrogation of DNA-binding function of p53 results in amelioration of p53-dependent transcription, and hence, target genes required for the efficient execution of the biological processes are not activated. The significance of the DNA-binding property of p53 in regulating many of its biological functions is highlighted by the large percentage (∼90%) of mutations found in DNA-binding domain (DBD) of p53 in human cancers (1,3). Of these, there are several hot-spot residues such as R175, G245, R248, R249, R273 and R282 that are more prone to mutations than others (1,3). R248 and R273 are DNA-contact mutants and R175, G245, R249 and R282 are conformational mutants (4), and like most other mutations found in the DBD, all of them have compromised DNA-binding activity (1,5).
Mutated p53 is often overexpressed in tumour cells (1,2,6), due to their inability to effectively activate MDM2, which negatively regulates p53 abundance (7,8). Whether the accumulated p53 in tumour cells have any specific functions supporting cellular growth has been intensively researched recently. There is accumulating evidence that mutant p53 may not only have lost the tumour-suppressive functions but may have also acquired additional pro-oncogenic properties (6,8), leading to the concept that mutant p53 may have acquired novel oncogenic ‘gain-of-function’ activities (6). In this respect, several biochemical and biological functions of mutant p53 that are independent of wild-type p53's activities were described. It was shown that mutant p53 could transactivate oncogenic targets such as c-myc (9), anti-apoptotic gene BAG-1 (10), growth-promoting genes as asparagine synthetase and hTERT (11,12) and the multi-drug resistance gp180 protein (MDR1) (13). At the same time, not much is known if mutant p53 has any negative, inhibitory role on common p53-target gene expression. In this respect, only one report by Zalcenstein et al. demonstrated that the p53 mutant R175H could down-regulate the expression from the p53-dependent CD95(FAS/APO-1) promoter (14). However, whether such a phenomenon is universal and affects the status of other wild-type p53-target genes is unclear.
Recent findings have also suggested that activation of mutant p53 by small molecules such as PRIMA-1 could restore sequence-specific DNA binding and the active conformation to mutant p53 proteins in vitro and in vivo in living cells, leading to anti-tumour effects, which was dependent on the presence of mutant p53 (15,16). Although PRIMA-1 reconstituted the wild-type function of mutant p53, it cannot be excluded that mutant p53 may have been involved in the suppression of p53-target gene expression, and modulation of such an activity could also be one reason for the activation of its tumour-suppressive functions. There is insufficient evidence to propose a mechanism by which mutant p53 could have acquired a ‘gain-of-function’ by down-regulating the classical p53-target gene expression.
We thus asked whether mutant p53 would be able to modulate the expression of classical wild-type p53 responsive target genes. We report here that down-regulation of mutant p53 expression in several human cancer cell lines harbouring mutant p53 by siRNA-mediated silencing induced the expression of p53-dependent apoptotic and repair genes. Consistently, several p53-target genes were found to be down-regulated in isogenic cell lines stably expressing the various hot-spot p53 mutants, which also correlated with down-regulation of p53-target gene promoter activity by various mutant p53 expression in transient transfection assays. Down-regulation of target gene activity was found not to be dependent on the presence of p53-binding sites, but was markedly reduced in the presence of histone deacetylase (HDAC) inhibitor trichostatin-A (TSA), suggesting that mutant p53-mediated p53-target gene suppression is at least in part due to hypo-acetylation of histones. Finally, silencing of p53 expression in human cancer cells resulted in reduction in cellular colony formation, indicating that mutant p53 expression indeed could support cellular growth. Thus, our finding suggests a novel function of mutant p53 that can contribute to cancer progression.
MATERIALS AND METHODS
Cell culture and plasmids
The p53 null H1299 human lung cancer cell line and the 13 derivate isogenic cell lines expressing vector (pCDNA) or the six hot-spot mutations (R175H, G245S, R248W, R249S, R273H and R282 W) either as an arginine or a proline polymorphic variant at codon 72 has been established in the laboratory and described previously (5). H1299 cells stably expressing the temperature-sensitive p53 mutant either as an arginine or proline polymorphic form at codon 72 has been previously established in the laboratory and has been described (17). At 37°C, these cells express p53 in a mutant conformation, and are hence functionally inactive. Upon temperature shift to 32°C, the p53 adopts a wild-type conformation and is active (17).
Mutant p53-expressing CNE-2 nasopharyngeal carcinoma cells, T47D breast carcinoma cells and HUH-7 hepatoma cells are gift from Dr Hui K M. The human pancreatic cancer cell line PaCL4 has a missense alteration in one allele of p300 gene and the second allele is inactivated, as determined in an analysis to identify cell lines and tumours with p300 mutations (gift from Dr Levrero M) (18,19). All cell lines were cultured in DMEM containing 10% bovine fetal serum. All H1299-based isogenic cell lines were also kept on G418 (800 μg/ml).
All p53 mutant and wild-type p53 plasmids were generated and have been described (5,17). P73DD, which inhibits TAp73 function, has been described (20). The following p53-target gene promoter-reporter constructs were used in this study: p21-luciferase (luc), gadd45-luc, p53AIP-1-luc, Bax-luc, Igfbp3-luc, p53R2-luc, cyclinG1-luc and hTert-luc, and have been described (5,20). p21-promoter deletion constructs 0-luc, 2-luc and 4-luc were gift from Dr Wafik el-Deiry, and have been described (21).
Small-interfering (si) RNA against p53 (gac ucc agu ggu aau cua), p73 (ggc aau aau cuc ucg cag u) or scrambled, control siRNA (uuc ucc gaa cgu guc acg u) were obtained (synthesized by Xeragon) and used for gene silencing experiments.
Transfections and luciferase assays
Here, 2 × 105 cells (in 6-well dishes) were used in transfection experiments with LIPOFECTAMINE PLUS-Reagent, as per manufacturer's protocols (Stratagene), or by the calcium phosphate method. Both methods gave similar results. The transcriptional activity of various p53 constructs were determined by luciferase assays in human H1299 cells and PaCL4 cells by transfecting the various promoter constructs (0.5 μg) with empty vector, wild-type p53 (arginine or proline) or the various p53 mutant constructs (0.1 μg) and β-galactosidase construct (0.1 μg). Luciferase activity was determined by chemiluminance and normalized against β-galactosidase activity 24 h after transfections, as described (20). In experiments where TSA (Upstate Biotech.) was added, cells were pre-treated with TSA (100 ng/ml) for 6 h before transfection. TSA was present during transfection and luciferase activity was analysed 18 h after transfection. All experiments were done at least thrice independently and data represents mean from one of the experiments.
For silencing gene expression, the indicated siRNA (5 μg) were transfected into cells using TransMessenger Transfection Reagent as per manufacturer's instruction (Qiagen). Cells were harvested for 48 h after transfection and used for RNA analysis.
Colony formation assay
HUH-7 and T47D cells were co-transfected with 1 μg pCDNA and 10 μg of pSuper-based plasmid containing oligonucleotide sequences for silencing p53 [as described in ref. (22)] or control, scrambled sequences, with Effectene transfection reagent (Qiagen). Further controls included cells without any transfection. Twenty-four hours after transfection, medium was changed and cells were selected on medium containing G418 (500 μg/ml) for 14 days. Colonies were colored with crystal violet solution (MERCK).
RNA extraction and RT-PCR
Total RNA was prepared from cells using TRIZOL Reagent (Invitrogen) as per manufacturer's instructions. Total RNA (1–5 μg) was converted into single-strand cDNA using Superscript II (Invitrogen) as per manufacturer's instructions. Semi-quantitative RT-PCR analysis was performed under the following conditions: TAp73: 94°C–50 s, 72°C–1 min, 54°C–50 s for 31 cycles (expected size 350 bp); gapdh: 94°C–35 s, 72°C–70 s, 54°C–50 s for 23 cycles (500 bp); gadd45: 94°C–35 s, 72°C–40 s, 53°C–35 s for 30 cycles (454 bp); p53: 94°C–30 s, 72°C–90 s, 60°C–30 s for 30 cycles (587 bp); p21: 94°C–50 s, 72°C–60 s, 54°C–50 s for 30 cycles (331 bp); PTEN: 95°C–30 s, 72°C–75 s, 55°C–30 s for 31 cycles (1153 bp); PERP: 94°C–50 s, 72°C–50 s, 52°C–50 s for 34 cycles (500 bp); c-jun: 95°C–30 s, 72°C–30 s, 64°C–30 s for 31 cycles (280 bp); pig3: 95°C–30 s, 70°C–50 s, 57°C–30 s for 30 cycles (349 bp); p53R2: 94°C–30 s, 70°C–70 s, 54°C–40 s for 30 cycles (1000 bp). All reactions were preceded by heating at 94°C for 3 min, and ended with heating at 72°C for 10 min. Primers used in this study are as follows: TAp73 for: 5′-TCT GGA ACC AGA CAG CAC CT; rev: 5′-GTG CTG GAC TGC TGG AAA GT; gapdh for: 5′-ACC CCT TCA TTG ACC TCA AC; rev: 5′–CAG CGC CAG TAG AGG CAG; gadd45 for: 5′-TTG GAG GAA TTC TCG GCT GGA GAG CAG AGG; rev: 5′CCA TTG ATC CAT GTA ACT TTC CCG GCA; p53 for: 5′-AGG AGC CGC AGT CAG ATC; rev: 5′- ACT CGG ATA AGA TGC TGA G; p21 for: 5′-CGA CTG TGA TGC GCT AAT GG; rev: 5′CCG TTT TCG ACC CTG AGA; PTEN for: 5′-GCCATCATCAAAGAGATCGTT; rev: 5′GGATCAGAGTCAGTGG; PERP for: 5′-TACTCAGCGCCATCGCCTTC; rev: 5′-TGTGTAGAAGTACCTGGGCTTG; c-jun for: 5′-ATG CCC TCA ACG CCT CGT TCC TCC; rev: 5′-CTG CTC GTC GGT CAC GTT CTT CTT GGG; pig3 for:5′-GTG CAC TTT GAC AAG CCG GGA GGA; rev: 5′-CAG CCT GGG TCA GGG TCA ATC CCT; p53R2 for: 5′-ATGGGCGACCCGGAAAGGCCGGAA;rev: 5′-TCTGCATCCAAGGTGAAGACGTT.
Immunoblot analysis
Cell lysates were prepared as described (23). Protein lysates were analysed by immunoblotting using anti-p53 (clone DO-1), gadd45, PTEN and p21 (Santa Cruz) and anti-Actin (Sigma) antibodies, as described (23). Detection was performed using enhanced chemiluminescent reagent (Amersham).
Cell death analysis
Cell death was determined by analysing the population of cells with a sub-G1 DNA content, by staining with propidium iodide (PI). Briefly, cells were harvested 3–5 days after siRNA transfection, washed once in PBS and fixed in 70% ethanol overnight before being stained with PI for analysis of the DNA-content. All analyses were performed by flow cytometry in duplicates and twice independently.
RESULTS
Silencing of p53 expression results in upregulation of p53-target genes in several human cancer cell lines expressing mutant p53
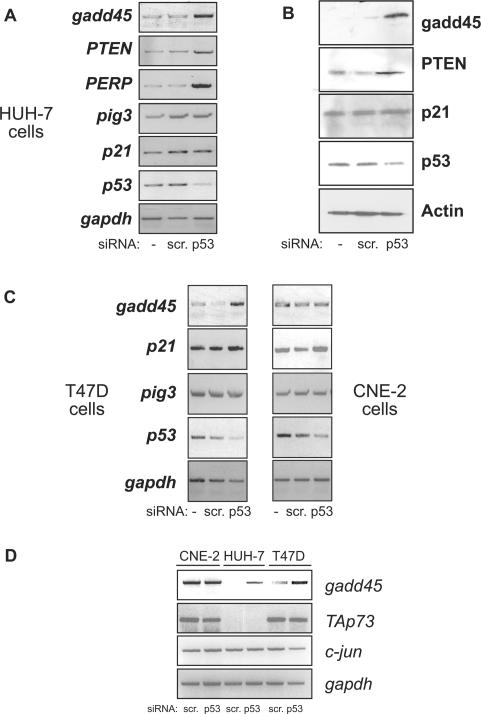
We first tested the hypothesis that silencing of mutant p53 expression will affect the expression of endogenous p53-target genes. To this end, we utilized three human cancer cell lines, namely the HUH-7 hepatoma cell line which harbours the A220G mutation; the T47D breast cancer cell line which carries the C194T mutation and the CNE-2 nasopahryngeal carcinoma cell line that harbours the R280T mutation (24–26). p53 mRNA levels were reduced upon siRNA-mediated silencing using p53-specific siRNA and not in control scrambled siRNA-transfected or untransfected cells in all cases (Figure 1A–C). Concomitant to reduction in p53 levels, the levels of gadd45, PTEN and PERP were upregulated in HUH-7 cells (Figure 1A). The upregulation of PTEN and gadd45 was also confirmed at the protein level by immunoblotting (Figure 1B). However, the expression levels of other p53-target genes such as p21 and pig3 were unaltered upon p53 silencing in HUH-7 cells (Figure 1A and B). This variable upregulation of p53-target genes was also noticed in the other two cell lines. p53 silencing resulted in upregulation of gadd45 and to some extent of p21 in T47D cells, and only marginal upregulation of p21 in CNE-2 cells (Figure 1C). The levels of pig3 did not change in both cells lines whereas gadd45 remained unchanged in CNE-2 cells (Figure 1C). This effect of increase in expression of various genes was restricted to p53-target genes as expression of unrelated genes such as TAp73 and c-jun were unaltered by silencing of p53 (Figure 1D). It was interesting to note that TAp73 expression was naturally silenced in HUH-7 cells, and together with the absence of any change in TAp73 levels in the other two cell lines, the data exclude the possibility that the increase in the expression of p53-target genes upon p53 silencing was due to relief of mutant p53-mediated suppression of p73 function.
Figure 1.
Silencing of mutant p53 expression results in upregulation of p53-target gene expression. (A and B) Mutant p53 harbouring HUH-7 hepatoma cells (A220G) were untransfected (−) or transfected with either control scrambled siRNA or p53-specific siRNA. Cells were collected 48 h later for mRNA analysis of the indicated target genes by reverse-transcriptase (RT) PCR reaction (A) or by immunoblotting with the specific antibodies (B). (C) RT-PCR target gene analysis was similarly performed in T47D breast cancer cells (C194T) (left panel) and CNE-2 nasopharyngeal carcinoma cells (R280T) (right panel). (D) mRNA analysis shows direct comparison of p53-regulated gadd45 gene expression as well as p53-independent TAp73 and c-Jun expression levels in all three cell lines.
H1299-based isogenic cell lines expressing hot-spot p53 mutants express reduced levels of p53-target genes, whose expression can be induced by silencing mutant p53 expression
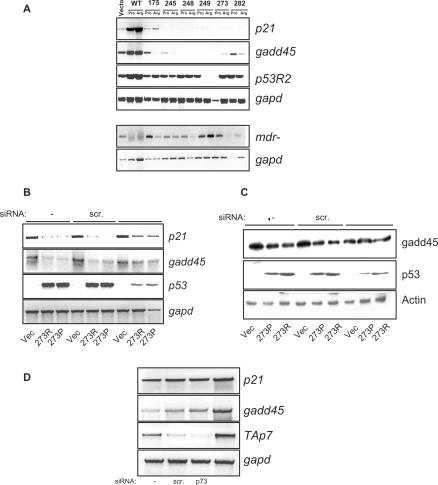
The results obtained from the human cancer cell lines expressing endogenous mutant p53 indicated that mutant p53 may be actively silencing p53-target gene expression. To further investigate this possibility, we analysed the expression status of several p53-target genes such as p21, gadd45 and p53R2, in a panel of H1299-based isogenic cell lines stably expressing the six hot-spot p53 mutants, which has been generated in our previous study (5). The isogenic cell lines express approximately equal amounts of the six commonly found human cancer-derived hot-spot mutations such as R175H, G245S, R248W, R249S, R273H and R282W, in either of the polymorphic forms of the common polymorphism at codon 72, i.e. arginine or proline (R or P) (5). H1299 cells expressing empty expression pcDNA vector or temperature-sensitive mutant p53 in either the R or P forms and cultured at 32°C, during which the p53-target genes are activated (19), were used as positive controls. Using these cell lines, we noticed that the p53-target gene expression was markedly reduced in almost all mutant-p53-expressing cell lines compared to the vector-expressing cell line, whereas expression of active wild-type p53 resulted in induction of these target genes (Figure 2A). The reduction was more pronounced with respect to p21 and gadd45 levels, whereas the reduction of p53R2 levels was less pronounced. Although there were variations in the levels of p53R2 expression between cells expressing either the R or P form of various p53 mutants, all the cell lines had reduced p53R2 levels compared to wild-type p53-expressing cells (Figure 2A), indicating that the effect may be general and attributable to mutant p53. The levels of p53 were almost similar in all cell lines, excluding any possibility that p53 levels may influence the expression of the target genes (Figure 2B, and data not shown). Moreover, expression of the multi-drug resistance gene-1 (mdr-1) was not suppressed but elevated in cell lines expressing the p53 mutants (Figure 2A, lower panel). These data suggested that mutant p53 expression indeed results in the suppression of p53-target gene expression.
Figure 2.
Mutant p53 expression leads to suppression of p53-target gene expression, which is relieved upon silencing of mutant p53 expression. (A) H1299-based isogeneic cell lines stably expressing the six hot-spot p53 mutants (i.e. 175, 245, 248, 249, 273 and 282) either in the proline (Pro) or arginine (Arg) codon 72 polymorphic form were analysed for steady-state p53-target gene expression levels, as indicated, by reverse-transcriptase PCR reaction. H1299-cells stably expressing the temperature-sensitive p53 mutants, which adopt a wild-type conformation at 32°C, were grown at 32°C and used as positive controls for p53-target gene activation (WT-Pro and Arg). Levels of mdr-1 were also evaluated. (B and C) H1299-cells expressing p53 mutant R273H in the proline (P) or arginine (R) codon 72 polymorphic form were transfected with either control scrambled siRNA or p53-specific siRNA or left untransfected (−). Cells were collected 48 h later for mRNA analysis of the indicated target genes by RT-PCR reaction (B) or by immunoblot analysis (C). (D) Vector-expressing H1299 cells were transfected with control scrambled siRNA, p73-specific siRNA, p73DD cDNA or left untransfected (−), and the levels of target genes were analysed by RT-PCR reaction.
We thus analysed the effects of silencing mutant p53 expression in some of these cell lines. To this end, cell lines expressing p53 mutants R273 (in the arginine or proline form) were utilized. p53 expression was silenced by siRNA-mediated silencing, which resulted in an increase in p21 and gadd45 levels in the 273 mutant p53-expressing cell lines, but not in vector-expressing cell lines (Figure 2B). Similar to the RT-PCR data, immunoblot analysis indicated that the levels of gadd45 were reduced in mutant p53 cells compared to vector-expressing cells that were untransfected or transfected with scrambled siRNA (Figure 2C). By contrast, the levels of gadd45 were almost comparable between vector-expressing cells and those expressing mutant p53 in which p53 expression was silenced (Figure 2C), indicating that expression of mutant p53 indeed resulted in the suppression of p53-target gene expression.
As mutant p53 was shown to inactivate TAp73 activity (27,28), the observed decrease in p53-target gene expression in cells expressing mutant p53 could be due to inactivation of p73 function. To investigate this possibility, we silenced the expression of TAp73 in H1299 cells (control vector-expressing cells) by p73-specific siRNA (Figure 2D). Transfection of scrambled siRNA resulted in marginal increase in gadd45 levels (Figure 2D). However, silencing of p73 expression did not lead to a decrease in the levels of p21 and gadd45 in these cells (Figure 2D). In addition, we expressed the dominant negative p73DD protein, which is able to inhibit TAp73 activity by binding to it (29). Expression of p73DD did not also cause a reduction of p21 and gadd45 levels, confirming that decrease in TAp73 levels or activity are not causal to lower levels of p53-target gene expression in mutant p53-expressing cell lines. Moreover, these results suggest that down-regulation of p53-target gene expression may be a direct consequence of mutant p53 expression.
Hot-spot p53 mutants down regulate promoter activity of p53-target genes
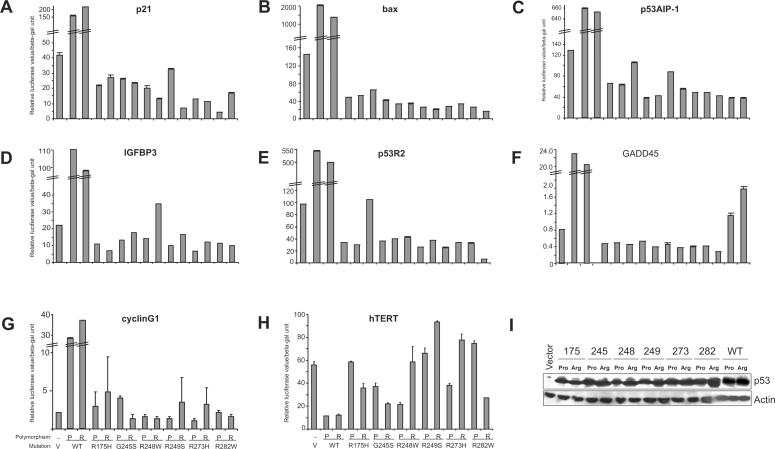
To understand the mechanism of down-regulation of p53-target genes by mutant p53, we performed transient co-transfection experiments to analyse the effects of various hot-spot p53 mutant constructs on p53-target gene promoter activity, in H1299 cells. We utilized seven p53-dependent promoters, including p21, GADD45, p53AIP-1, BAX, IGFBP3, cyclinG1 and p53R2 (Figure 3A–G). Expression of wild-type p53 in either the proline or arginine polymorphic variant resulted in massive activation of all promoters (Figure 3A–G). By contrast, expression of the six hot-spot p53 mutants in either of the polymorphic forms generally resulted in the suppression of promoter activity throughout, compared to vector (Figure 3A–G), consistent with the findings obtained with endogenous levels of target genes. The extent of suppression was dependent on the kind of mutation and the promoters used in the experiments. It should be noted that although there were variations in the levels of suppression of target gene promoters between cells expressing either the R or P form of various p53 mutants, expression of all forms of mutants led to a decrease in target gene promoter activity (Figure 3A–G), indicating that the effect may be general and attributable to mutant p53. We have excluded the possibility that the differences in p53 levels are causal to the differences in promoter activity, as all mutant p53 constructs were expressed to about equal extents (Figure 3I, showing representative results). Moreover, this suppressive effect was not unspecific as it was not observed with the hTERT promoter, which is generally suppressed by wild-type p53 (20). Although wild-type p53 suppressed hTERT promoter activity, most mutant p53 did not (Figure 3H). Thus, the data indicate that various hot-spot p53 mutants have the ability to suppress the expression from p53-target gene promoters.
Figure 3.
Mutant p53 expression results in down-regulation of p53-target gene promoters. (A–H) H1299 cells were transiently transfected with plasmids expressing the empty vector (pCDNA) (V), wild-type (WT) or the p53 mutant constructs either in a Proline (P) or Arginine (R) form, together with the various reporter plasmids expressing the firefly luciferase gene under the transcriptional control of the indicated gene promoters. Luciferase activity was analysed 24 h post-transfection. Transfections were carried out in triplicates and at least three independent times and the standard deviations are indicated. Wild-type p53 expression led to massive induction of the p53-target gene promoters, and hence, the graphs are depicted to highlight the down-regulation by mutant p53. (I) Representative western blot analysis was performed using half the samples from the transfections to determine the expression status of p53 in all cases. Actin levels show loading control.
Presence of p53-binding site on p21 promoter is not required for down-regulation by mutant p53
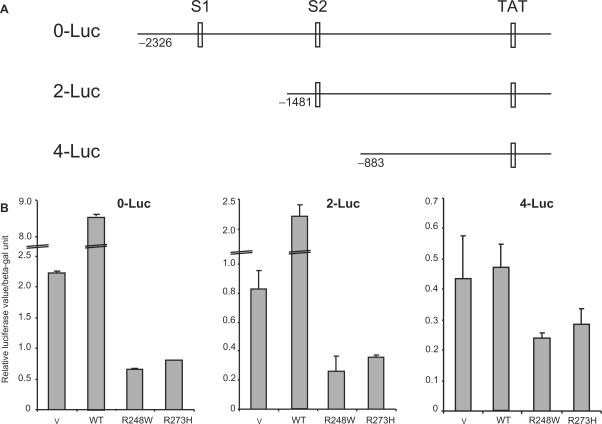
It was interesting to note that all the mutants used in this study are DNA-binding mutants and hence, do not have the property to bind DNA in a sequence-specific manner (1). However, the results suggest that the suppression by mutant p53 is specific to p53-target gene promoters. Hence, we investigated if the p53-binding sites on the promoters are required for the suppression. To this end, we utilized the proto-type promoter constructs from the p21 promoter having (0-Luc) or lacking one or both of the p53- binding sites (2-Luc or 4-Luc, respectively) (Figure 4A) (21). Lack of one p53-binding site did not affect the ability of wild-type p53 to activate the promoter or the ability of representative p53 mutants (R248 and R273) to suppress it (Figure 4B, middle panel). However, although deletion of both p53-binding sites abrogated wild-type p53's ability to activate the promoter, the mutant p53 were still able to suppress this activity (Figure 4B, rightmost panel). Thus, the results suggest that the presence of p53-binding sites are not absolutely essential for mutant p53 to down regulate p53-target gene promoters.
Figure 4.
Presence of p53 binding sites on p21 promoter is not required for down-regulation by mutant p53. (A) Schematic of p21 promoter indicating the position of the p53 binding sites (S1 and S2) as described (21). (B) Luciferase reporter assays were preformed by transfecting vector, wild-type p53, or the indicated p53 mutant cDNA constructs into H1299 cells as described, with the various p21 promoter constructs. Transfections were carried out in triplicates and at least three independent times and the standard deviations are indicated.
Treatment with Trichostatin A relieves mutant p53-mediated suppression of p53-target genes
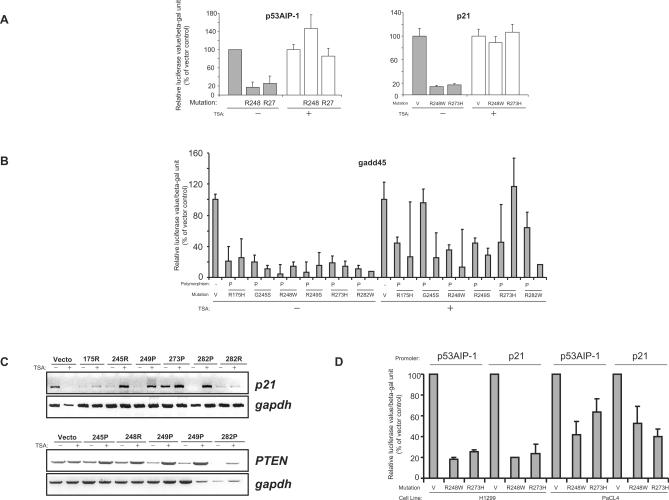
One possible mechanism of gene suppression is hypo-acetylation of histones by histone deacetylases (HDAC), and wild-type p53 has been shown to suppress several gene promoters by recruitment of HDACs (30). We therefore explored the possibility that p53 mutants exerted their suppressive effects on p53-target genes by affecting the chromatin acetylation status. To this end, we first utilized the HDAC inhibitor, Trichostatin A (TSA) (31), and evaluated its effect on the ability of mutant p53 (R248 and R273) to suppress the p53AIP-1and p21 promoters. Treatment with TSA resulted in a massive induction of the promoter activity even in the presence of the p53 mutants (Figure 5A), compared to TSA-untreated cells where there was a consistent suppression by the mutants (Figure 5A). There was also a slight increase in the promoter activity of vector-transfected cells treated with TSA (Figure 5A). However, the effect of increase in promoter activity was much more pronounced when mutant p53 was transfected, suggesting that TSA-mediated relief of suppression was at least partially dependent on the presence of mutant p53. This was further confirmed when we evaluated the effects of all six hot-spot mutants (in both the proline or arginine polymorphic variations) on the gadd45 promoter. Treatment with TSA again resulted in the relief of suppression of promoter activity, though to varying extents depending on the p53 mutants used, as well as the polymorphic status (Figure 5B).
Figure 5.
Treatment with Trichostatin A relieves mutant p53-mediated suppression of p53-target genes, which is reduced in cells lacking p300. (A and B) Luciferase reporter assays were performed as described above using the indicated p53 constructs in the presence (+) or absence (−) of trichostatin-A (TSA). Cells were pre-treated with TSA (100 ng/ml) for 6 h before transfection and TSA was present during transfection and luciferase activity was analysed 18 h after transfection. Promoter activity was determined using the p53AIP-1, p21 (A) and the gadd45 promoter constructs (B) The level of luciferase activity in mutant p53-transfected cells are shown as the percentage activity compared to the vector-transfected cells, which was set at 100%. (C) H1299-based isogenic cells lines stably expressing the indicated mutant p53 in either the proline (P) or arginine (R) codon 72 polymorphic form were treated in the presence (+) or absence (−) of TSA as described for 18 h and the levels of p21 and PTEN mRNA were analysed by reverse-transcriptase PCR reaction. (D) p53-target gene promoter activity was determined using the p53AIP-1 or p21 promoter constructs in p300 deficient (pancreatic cancer cell line PaCL4) or proficient (lung adenocarcinoma H1299) cells.
To evaluate if treatment with TSA will affect the levels of endogenous p53-target genes in mutant p53-expressing cells, we analysed the levels of both p21 and PTEN in several H1299-based isogenic cells lines expressing various mutant p53 in either the proline or arginine polymorphic forms. Treatment with TSA resulted in a significant increase in the levels of p21 in most of the cell lines expressing mutant p53 (Figure 5C, top panel). This was in contrast to vector-expressing cells in which TSA caused a down- regulation of p21 levels (Figure 5C, top panel). Similarly, expression of PTEN was elevated upon TSA treatment in all mutant p53-expressing cells lines, albeit to different extents (Figure 5C, lower panel). However, the increase of PTEN levels in vector-expressing cells were marginal and not of the magnitude as observed with mutant p53-expressing cells (Figure 5C, lower panel). It should be noted that cells expressing p53 mutants in either the proline or arginine polymorphic forms were responsive to TSA treatment, though to varying extents, suggesting that the polymorphism may not be crucial in mutant p53's ability to suppress target gene expression. Moreover, consistent with the promoter activity, different p53 mutants had varying abilities to suppress target gene expression, which upon TSA treatment, resulted in upregulation of target genes to varying extents. Together, the data suggest that mutant p53-mediated p53-target gene suppression is at least in part due to hypo-acetylation of histones.
p300 is a transcriptional co-activator that is involved in regulating acetylation of proteins (32). To confirm the role of histone deacetylation in the down-regulation of promoters by mutant p53, we used the human pancreatic PaCL4 cell line in which p300 is deleted, and exhibit reduced histone acetylation activity (18). Two p53 mutants (R248 and R273) were used to evaluate their ability to suppress p53AIP-1 and p21 promoter activity, in both H1299 and PaCL4 cells. Whereas the mutant p53 consistently suppressed promoter activity to a significant extent in H1299 cells, this effect was reproducibly less pronounced in PaCL4 cells (Figure 5D). These data thus support the hypothesis that mutant p53 probably down-regulates p53-target gene promoters by decreasing the acetylation status.
Knock-down of mutant p53 expression results in suppression of cellular colony formation due to increased apoptosis
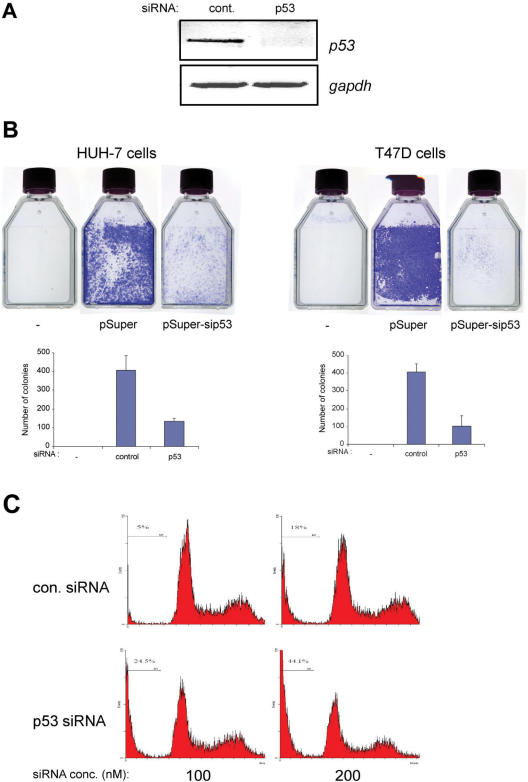
Since silencing of mutant p53 expression led to the induction of p53-target genes involved in growth arrest and apoptosis, we evaluated if knock-down of endogenous mutant p53 expression would also affect cellular growth. Hence, we silenced p53 expression in HUH-7 cells which harbours the A220G mutation in p53 and in T47D cells that carry the C194T mutation, and where silencing led to increased expression of p53-target genes (Figure 1A–C). Plasmid-based scrambled control siRNA and p53-specific siRNA were transfected and cells were selected for ∼2 weeks to monitor cellular colony formation, which reflects the long-term propensity of cells to grow. Parallel cultures were evaluated for specific and effective silencing of p53 expression (Figure 6A, shown for HUH-7). Whereas scrambled siRNA did not affect the growth of both cell lines, p53-specific siRNA expression resulted in markedly reduced number of colonies in both cases (Figure 6B, upper panel showing representative results). Absence of any transfection and subsequent selection resulted in no surviving colonies, confirming the efficacy of the selection procedure. Enumeration of cellular colonies indicated that there was a significant decrease in the number of colonies upon silencing of p53 expression (HUH-7––control versus p53 siRNA: 407 versus 133; P = 0.0073 and T47D—control versus p53 siRNA: 406 versus 103; P = 0.0133).
Figure 6.
Silencing of p53 expression results in reduced colony formation and elevated cell death. (A and B) p53 expression was silenced as described in HUH-7 and T47D cells using pSuper-based control or p53-specific siRNA, or were left untransfected. Cells were selected for two weeks on G418. Parallel cultures were used for semi-quantitative RT-PCR analysis of p53 expression status (representative results shown for HUH-7 cells) (A) and colonies were visualized by staining with crystal violet solution (B, upper panel). All experiments were performed at least thrice independently and representative results are shown. Colonies were counted both manually and using colony count software. Similar results were obtained using both methods. Data are presented as the mean ± SD error of the mean. (C) Cell death was analysed by measuring the sub-G1 DNA content reflective of apoptotic cells, by flow cytometry. HUH-7 cells were transfected with scrambled or p53 siRNA (100 or 200 nM) and the percentage cell death was determined 5 days after transfection. Representative results are shown.
We finally evaluated if the reduction in colony formation upon silencing p53 expression was due to elevated apoptosis. Comparison of apoptotic cells with a sub-G1 DNA content indicated that there was a significant increase when p53 siRNA was transfected compared to scrambled siRNA (Figure 6C for HUH-7 cells: % sub-G1 cells —scr. siRNA versus p53 siRNA: for 100 nM—5.0 versus 24.5; for 200 nM: 18.3 versus 44.1). Together, the data suggest that suppression of mutant p53 expression inhibits cellular growth probably via induction of apoptosis.
DISCUSSION
The results presented here demonstrate that expression of mutant p53, which does not have the ability to transactivate p53-target gene expression, results in down-regulation of various p53-target genes in several cell lines. Silencing of mutant p53 expression led to relief of this suppression and thus, activation of various p53-target genes. Cellular colony formation was reduced upon silencing of mutant p53 expression due to increased cell death, correlating with upregulation of p53-target gene expression. Thus, the down-regulation of p53-target genes by mutant p53 appears to be a general phenomenon, and hence, may constitute one mechanism for the gain of function of mutant p53.
It is intriguing that not much attention has been paid to this phenomenon, which is quite obvious upon closer analysis of results of promoter-reporter assays. Such assays have been traditionally used and most researchers have concluded from these that mutant p53 do not have the ability to activate the promoters of p53-target genes. However, the down-regulation has somewhat gone unnoticed by many, as it is not striking at first glance. However, careful analysis of a recently published article indicated that expression of various hot-spot p53 mutants indeed resulted in down-regulation of the p21 promoter [Supplementary Data of ref. (33)—Table 2]. Moreover, consistent with an earlier report that mutant p53 could suppress CD95 promoter activity, the data presented here strongly argue that suppression of p53-target gene expression could be a general mechanism by which mutant p53 could support oncogenesis.
The upregulation of p53-target genes upon silencing of mutant p53 expression was not unspecific, as expression of other genes such as c-jun, TAp73 and mdr-1 was not affected. Furthermore, p53 mutants were found to activate mdr-1 gene expression and the hTERT promoter activity as has been previously reported (12,13). Moreover, this phenomenon of suppression required the presence of mutant p53, as cells devoid of p53 did not show changes in the levels of target genes, and cellular colony growth was not affected upon p53 silencing in these cells, indicating that the expression of mutant p53 was essential. However, there are several variations that we observed in our studies: the extent to which the various p53-target genes were upregulated upon p53 silencing and the magnitude of target gene suppression by the proline or arginine polymorphic variants of mutant p53. For example, most of the genes analysed were upregulated in HUH-7 cells, whereas not all of them were upregulated in CNE-2 cells, thereby reflecting the cell-to-cell variation. It could be also that different p53 mutants have differential capacity to suppress target genes. Besides, although all the mutants had the ability to suppress gene expression, there were variations between the arginine and proline polymorphic forms, similar to that observed with wild-type p53 forms, which have also been shown to differentially regulate target gene expression (17). However, we were unable to observe a general pattern if either of the polymorphic forms could better suppress target gene expression than the other. The cause of this variation, which was also apparent in experiments in which cells were treated with TSA, is not clear and warrants further investigation. Nonetheless, the phenomenon of mutant p53-mediated suppression of p53-target genes appears to be consistent in most mutant p53-containing cell lines examined.
While our work was in progress, two recent publications reported that suppression of mutant p53 expression could indeed lead to inhibition of cellular growth. Bossi et al. utilized several human cancer cell lines harbouring mutant p53 and found that silencing of mutant p53 expression led to a reduction in tumour malignancy (34). The other report indicated that depletion of mutant p53 expression resulted in the activation of p21, bax and PIG3 promoter activity and led to cellular cytotoxicity (35). In addition, similar suppression of p53 expression in H1299 cells stably expressing the mutant R175H p53 was also shown to decrease cellular growth (36). Together, these data support our findings and suggest that suppression of mutant p53 expression may be one important way to relief the mutant p53-mediated down-regulation of p53-target gene expression, and hence, could lead to inhibition of cellular growth.
It is at present unclear whether mutant p53-mediated suppression of p53-target gene expression is direct. As mutant p53 was shown to inactivate TAp73 activity (27,28), the observed decrease in p53-target gene expression in cells expressing mutant p53 could be due to inactivation of p73 function. However, we have excluded this possibility as siRNA-mediated silencing of TAp73 expression and inactivation of TAp73 activity by expression of the dominant negative p73DD protein did not lead to a decrease of expression of p53-target genes, further indicating that down-regulation of p53-target gene expression is a direct consequence of mutant p53 expression and not due to p73 inactivation. Wild-type p53 has been shown to activate target genes by binding to p53-specific binding sequences found in the target gene promoters (21). However, investigations with the prototype p21 promoter indicated that both the p53-binding sites are dispensable for mutant p53-mediated suppression. It is thus not clear how mutant p53 is able to recognize target gene promoters specifically. Because all the p53 mutants used in this study have lost their sequence-specific DNA-binding ability (data not shown), the findings suggest that mutant p53 may still bind to p53-target gene promoters at different positions, independent of their DBD, or affect gene regulation by other means. Support for this hypothesis comes from several studies. First, it was shown that mutant p53 may promote gene amplification by activating topoisomerase I, leading to genomic instability (37). Next, Zalcenstein et al. demonstrated that mutant R175H p53 was able to bind to CD95 promoter sequences in the region distinct from the p53-binding sites where wild-type p53 binds (14). Consistently, Gohler et al. suggested that mutant p53 binds selectively and with high affinity to non-B DNA, which was found to be dependent on the stereo-specific configuration of the DNA, and not on DNA sequence (38). It is not clear at present if there is a consensus sequence or specific configuration in all the p53-target gene promoters to which mutant p53 could bind, and further investigations are required to elucidate this possibility.
Nonetheless, the mechanism of mutant p53-mediated suppression of target gene expression appears to partly depend on HDAC activity. Treatment of cells with the HDAC inhibitor TSA resulted in relief of suppression that was dependent on the presence of mutant p53. It was reported that TSA could also initiate p53-like functions by inducing a variety of p53-inducible genes in a p53-independent manner (31). Thus, it is possible that both mutant p53-dependent relief of down-regulation and p53-independent transactivation mechanisms may work simultaneously to activate these target genes. However, comparison of promoter activity upon TSA treatment in vector and mutant p53 expressing cells indicated that release from suppression was much stronger in presence of mutant p53, thereby excluding non-specific p53-independent effects as a major causal mechanism at work. Recent data by Blagosklonny et al. who had utilized another HDAC inhibitor, FR901228, also support our findings (35). Treatment with FR901228 resulted in the induction of mRNA of p53-target genes, and consequent cytotoxicity, specifically in cells expressing mutant p53 (35). Furthermore, our studies demonstrated that mutant p53-mediated suppression was less pronounced in tumour cells lacking p300, the transcriptional co-activator protein that plays a central role in co-ordinating and integrating multiple signal-dependent events (32). However, we were unable to observe a synergistic increase in target gene suppression by co-transfection of p300 with mutant p53 (data not shown). Thus, p300 may be required but not sufficient for mutant p53-dependent down-regulation of p53-target gene expression, which occurs at least in part by decreasing the acetylation status.
Taken together, the results presented here propose another mechanism through which mutant p53 may contribute to unregulated cellular growth and oncogenesis. Together with the recent reports demonstrating mutant p53's ability to support cellular viability, the presence of the overexpressed mutant p53 in tumour cells may be a good therapeutic target for clinical intervention. Future studies should thus be aimed at efficiently suppressing mutant p53 expression in human tumours in in vivo models to evaluate the efficacy of ‘anti-mutant p53 therapy’.
ACKNOWLEDGEMENTS
We thank Drs Hui KM, Levrero M and El-Deiry W for providing plasmids and cell lines. F. Vikhanskaya is a visiting scientist from the Institute of Cytology RAS, St. Peterburg, Russia.We thank the National Medical Research Council, Singapore, for their generous funding and support to K.S. Funding to pay the Open Access publication charge was provided by National Medical Research Council, Singapore.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat. Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Olivier M, Hussain SP, Caron de Fromentel C, Hainaut P, Harris CC. TP53 mutation spectra and load: a tool for generating hypotheses on the etiology of cancer. IARC Sci. Publ. 2004;157:247–270. [PubMed] [Google Scholar]
- 4.Roemer K. Mutant p53: gain-of-function oncoproteins and wild-type p53 inactivators. Biol. Chem. 1999;380:879–887. doi: 10.1515/BC.1999.108. [DOI] [PubMed] [Google Scholar]
- 5.Vikhanskaya F, Siddique MM, Lee MK, Broggini M, Sabapathy K. Evaluation of the combined effect of p53 codon 72 polymorphism and hotspot mutations in response to anticancer drugs. Clin. Cancer Res. 2005;11:4348–4356. doi: 10.1158/1078-0432.CCR-04-1547. [DOI] [PubMed] [Google Scholar]
- 6.Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 2000;60:6788–6793. [PubMed] [Google Scholar]
- 7.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 8.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 9.Frazier MW, He X, Wang J, Gu Z, Cleveland JL, Zambetti GP. Activation of c-myc gene expression by tumor-derived p53 mutants requires a discrete C-terminal domain. Mol. Cell. Biol. 1998;18:3735–3743. doi: 10.1128/mcb.18.7.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X, Pater A, Tang SC. Cloning and characterization of the human BAG-1 gene promoter: upregulation by tumor-derived p53 mutants. Oncogene. 1999;18:4546–4553. doi: 10.1038/sj.onc.1202843. [DOI] [PubMed] [Google Scholar]
- 11.Scian MJ, Stagliano KE, Deb D, Ellis MA, Carchman EH, Das A, Valerie K, Deb SP, Deb S. Tumor-derived p53 mutants induce oncogenesis by transactivating growth-promoting genes. Oncogene. 2004;23:4430–4443. doi: 10.1038/sj.onc.1207553. [DOI] [PubMed] [Google Scholar]
- 12.Xu D, Wang Q, Gruber A, Bjorkholm M, Chen Z, Zaid A, Selivanova G, Peterson C, Wiman K, et al. Downregulation of telomerase reverse transcriptase mRNA expression by wild type p53 in human tumor cells. Oncogene. 2000;19:5123–5133. doi: 10.1038/sj.onc.1203890. [DOI] [PubMed] [Google Scholar]
- 13.Sampath J, Sun D, Kidd VJ, Grenet J, Gandhi A, Shapiro LH, Wang Q, Zambetti GP, Schuetz D. Mutant p53 cooperates with ETS and selectively up-regulates human MDR1 not MRP1. J. Biol. Chem. 2001;276:39359–39367. doi: 10.1074/jbc.M103429200. [DOI] [PubMed] [Google Scholar]
- 14.Zalcenstein A, Stambolsky P, Weisz L, Muller M, Wallach D, Goncharov TM, Krammer PH, Rotter V, Oren M. Mutant p53 gain of function: repression of CD95(Fas/APO-1) gene expression by tumor-associated p53 mutants. Oncogene. 2003;22:5667–5676. doi: 10.1038/sj.onc.1206724. [DOI] [PubMed] [Google Scholar]
- 15.Bykov VJ, Issaeva N, Shilov A, Hulcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 16.Bykov VJ, Selivanova G, Wiman KG. Small molecules that reactivate mutant p53. Eur. J. Cancer. 2003;39:1828–1834. doi: 10.1016/s0959-8049(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 17.Siddique M, Sabapathy K. Trp53-dependent DNA-repair is affected by the codon 72 polymorphism. Oncogene. 2006;25:3489–3500. doi: 10.1038/sj.onc.1209405. [DOI] [PubMed] [Google Scholar]
- 18.Costanzo A, Merlo P, Pediconi N, Fulco M, Sartorelli V, Cole PA, Fontemaggi G, Fanciulli M, Schiltz L, et al. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell. 2002;9:175–186. doi: 10.1016/s1097-2765(02)00431-8. [DOI] [PubMed] [Google Scholar]
- 19.Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF, Daigo Y, Russell P, Wilson A, et al. Mutations truncating the EP300 acetylase in human cancers. Nat. Genet. 2000;24:300–303. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- 20.Toh WH, Kyo S, Sabapathy K. Relief of p53-mediated telomerase suppression by p73. J. Biol. Chem. 2005;280:17329–17338. doi: 10.1074/jbc.M500044200. [DOI] [PubMed] [Google Scholar]
- 21.el-Deiry WS, Tokino T, Waldman T, Oliner JD, Velculescu VE, Burrell M, Hill DE, Healy E, Rees JL, et al. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res. 1995;55:2910–2919. [PubMed] [Google Scholar]
- 22.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 23.Lee MK, Hande MP, Sabapathy K. Ectopic mTERT expression in mouse embryonic stem cells does not affect differentiation but confers resistance to differentiation- and stress-induced p53-dependent apoptosis. J. Cell Sci. 2005;118:819–829. doi: 10.1242/jcs.01673. [DOI] [PubMed] [Google Scholar]
- 24.Tan G, Heqing L, Jiangbo C, Ming J, Yanhong M, Xianghe L, Hong S, Li G. Apoptosis induced by low-dose paclitaxel is associated with p53 upregulation in nasopharyngeal carcinoma cells. Int. J. Cancer. 2002;97:168–172. doi: 10.1002/ijc.1591. [DOI] [PubMed] [Google Scholar]
- 25.Moon WS, Chang K, Tarnawski AS. Overexpression of metastatic tumor antigen 1 in hepatocellular carcinoma: Relationship to vascular invasion and estrogen receptor-alpha. Hum. Pathol. 2004;35:424–429. doi: 10.1016/j.humpath.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Strano S, Munarriz E, Rossi M, Cristofanelli B, Shaul Y, Castognoli L, Levine AJ, Sacchi A, Cesareni G, et al. Physical and functional interaction between p53 mutants and different isoforms of p73. J. Biol. Chem. 2000;275:29503–29512. doi: 10.1074/jbc.M003360200. [DOI] [PubMed] [Google Scholar]
- 27.Marin MC, Jost CA, Brooks LA, Irwin MS, O'Nions J, Tidy JA, James N, McGregor JM, Harwood CA, et al. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat. Genet. 2000;25:47–54. doi: 10.1038/75586. [DOI] [PubMed] [Google Scholar]
- 28.Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, Yulug I, Merlano M, Numico G, et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003;3:387–402. doi: 10.1016/s1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 29.Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 30.Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, George DL. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirose T, Sowa Y, Takahashi S, Saito S, Yasuda C, Shindo N, Furuichi K, Sakai T. p53-independent induction of Gadd45 by histone deacetylase inhibitor: coordinate regulation by transcription factors Oct-1 and NF-Y. Oncogene. 2003;22:7762–7773. doi: 10.1038/sj.onc.1207091. [DOI] [PubMed] [Google Scholar]
- 32.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell. Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 33.Kakudo Y, Shibata H, Otsuka K, Kato S, Ishioka C. Lack of correlation between p53-dependent transcriptional activity and the ability to induce apoptosis among 179 mutant p53s. Cancer Res. 2005;65:2108–2114. doi: 10.1158/0008-5472.CAN-04-2935. [DOI] [PubMed] [Google Scholar]
- 34.Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, Sacchi A. Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene. 2006;25:304–319. doi: 10.1038/sj.onc.1209026. [DOI] [PubMed] [Google Scholar]
- 35.Blagosklonny MV, Trostel S, Kayastha G, Demidenko ZN, Vassilev LT, Romanova LY, Bates S, Fojo T. Depletion of mutant p53 and cytotoxicity of histone deacetylase inhibitors. Cancer Res. 2005;65:7386–7392. doi: 10.1158/0008-5472.CAN-04-3433. [DOI] [PubMed] [Google Scholar]
- 36.Scian MJ, Stagliano KE, Ellis MA, Hassan S, Bowman M, Miles MF, Deb SP, Deb S. Modulation of gene expression by tumor-derived p53 mutants. Cancer Res. 2004;64:7447–7454. doi: 10.1158/0008-5472.CAN-04-1568. [DOI] [PubMed] [Google Scholar]
- 37.El-Hizawi S, Lagowski JP, Kulesz-Martin M, Albor A. Induction of gene amplification as a gain-of-function phenotype of mutant p53 proteins. Cancer Res. 2002;62:3264–3270. [PubMed] [Google Scholar]
- 38.Gohler T, Jager S, Warnecke G, Yasuda H, Kim E, Deppert W. Mutant p53 proteins bind DNA in a DNA structure-selective mode. Nucleic Acids Res. 2005;33:1087–1100. doi: 10.1093/nar/gki252. [DOI] [PMC free article] [PubMed] [Google Scholar]