Abstract
IL-2 gene expression in activated T-cells is initiated by chromatin remodeling at the IL-2 proximal promoter and conversion of a transcriptional repressor into a potent transcriptional activator. A purine-box regulator complex was purified from activated Jurkat T-cell nuclei based on sequence-specific DNA binding to the antigen receptor response element (ARRE)/nuclear factor of activated T-cells (NF-AT) target DNA sequence in the proximal IL-2 promoter. ARRE DNA-binding subunits were identified as NF90, NF45 and systemic lupus erythematosis autoantigens, Ku80 and Ku70. Monoclonal antibodies to Ku80, Ku70 and NF90 specifically inhibit constitutive and inducible ARRE DNA-binding activity in Jurkat T-cells. Ku80, Ku70 and NF90 bind specifically to the IL-2 gene promoter in vivo, as demonstrated by chromatin immunoprecipitation. Activation of Jurkat T-cells and mouse primary spleen cells induces binding of Ku80 and NF90 to the IL-2 promoter in vivo, and decreases binding of Ku70 to the IL-2 promoter in vivo, and these dynamic changes are inhibited by immunosuppressants cyclosporin A and triptolide. Dynamic changes in binding of Ku80, Ku70 and NF90 to the IL-2 proximal promoter in vivo correlate with chromatin remodeling and transcriptional initiation in activated T-cells.
INTRODUCTION
Expression of T-cell growth factor, IL-2, is tightly regulated. Activation of T-cells through the T-cell antigen and CD28 coreceptors signals up to a 100-fold increase in IL-2 mRNA expression within 6 hr followed by protein secretion (1). IL-2 gene expression in activated T-cells involves chromatin remodeling at the IL-2 proximal promoter that is closely linked to cooperative binding of transcription factors to the antigen receptor response element (ARRE)/nuclear factor of activated T-cells (NF-AT), NF-κB, AP-1 and Oct-1 sequences and intense transcriptional activation (2). The A-T-rich purine-box/ARRE/NF-AT target site serves a unique role in regulating IL-2 transcription: a specific transcriptional repressor preexisting in resting T-cells is converted during T-cell activation into a potent transcriptional activator, through mechanisms that involve melting of chromatin (2–4). Proteins are prebound to the distal ARRE/NF-AT site in the IL-2 promoter in the nucleus of resting Jurkat and EL-4 T-cells and T-cell activation triggers expansion of the footprint of this purine-box regulator complex (5,6).
We previously described the purification from stimulated Jurkat T-cells of an inducible nuclear purine-box regulator that binds specifically to the ARRE/NF-AT target DNA sequence in the IL-2 promoter (7,8). Subunits of this labile purine-box regulator, NF45 and NF90, are zinc-finger DNA- and RNA-binding proteins. NF90 contains two double-stranded RNA-binding domains (dsRBD) that bind structured RNAs including IL-2 (9,10). Antisera to NF90 and NF45 specifically inhibited ARRE/NF-AT DNA-binding and in vitro transcription (7). NF45 and NF90 regulate transcriptional activation of the IL-2 promoter and other genes (11–14), posttranscriptional mRNA stabilization and nuclear export of IL-2 and other genes (9,10) and translation (15,16). NF90 has been implicated in host antiviral responses and as a cellular cofactor involved in viral replication and translation (16–18). Targeted disruption of NF90 is associated with profound T-cell lymphocytopenia, severe impairment of IL-2 gene expression and ARRE/NF-AT transcriptional activation (Shi et al, J. Exp Med. in press).
Here we report that systemic lupus erythematosis autoantigens, Ku80 and Ku70, copurify with NF90 and NF45, and contribute to specific binding of the purine-box regulator to ARRE/NF-AT DNA sequences in vitro and to the IL-2 promoter in vivo (20,21). Ku80 and Ku70 are multifunctional nucleic acid binding proteins that interact with double- and single-stranded DNA and RNA (22). In complex with the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs), Ku80 and Ku70 participate in repair of double-stranded DNA breaks and in V(D)J recombination in lymphocytes (23,24). Ku80 and Ku70 bind specifically to purine-rich and A-T rich DNA sequences and regulate transcription (25–34), and mammalian DNA, HIV and HSV replication (29,35–39).
We present complementary experiments of protein purification, Electrophoretic mobility shift assays (EMSA)-antibody inhibition and chromatin immunoprecipitation that demonstrate specific binding of Ku80, Ku70 and NF90 to ARRE DNA-sequences in vitro and dynamic binding to the proximal IL-2 promoter in vivo.
MATERIALS AND METHODS
Cell culture and stimulation conditions
Jurkat T-cells were nonstimulated (NS) or stimulated for 4 hr with phorbol myristyl acetate (20 ng/ml) and ionomycin (2 µM), in the absence or presence of cyclosporin A (1000 ng/ml) or triptolide (200 or 1000 ng/ml, PG490, Pharmagenesis, Palo Alto, CA).
Electrophoretic mobility shift assays (EMSA)
Nuclear proteins (5 µg) were incubated with 32P-labeled oligonucleotide probe for 30 min at 4°C [25 mM HEPES pH 7.6, 50 mM KCl, 0.1 mM EDTA, 10% glycerol and 1.5 µg poly(dI-dC)]. The poly(dI-dC) was included as a nonspecific competitor and a source of free DNA ends. The amount of poly(dI-dC) was adjusted during the purification steps to reveal specific ARRE DNA-binding complexes in the presence of excess free probe. Protein–DNA complexes were separated on 4% nondenaturing acrylamide gels in 0.5× Tris–Borate EDTA. The wild-type ARRE-2 oligonucleotide probe is 5′ gatcGGAGGAAAAACTGTTTCATACAGAAGGCGT-3′ (–255 to –285 in the human IL-2 promoter), and the mutant ARRE-2 competitor oligonucleotide is 5′-gatcGAAAGGAGtAAAAAaTtTTTaATACAGAA-3′ (7). The NF-κB probe is 5′ agctAAAGAGGGACTTTCCCTAAA-3′. For EMSA-antibody inhibitions, 2 µl of ascites or 200–250 ng of purified mAB or polyclonal antisera were incubated with nuclear proteins for 30 min at 4°C before addition of radiolabeled probe and further incubation.
Antibodies
Anti-human Ku monoclonal antibodies used for EMSA were mAB162 (anti-Ku heterodimer), 111 (Ku80) and N3H10 (Ku70) (Becton–Dickinson and Neomarkers) (40). Other antibodies used for EMSA, Western and ChIP: Ku80 (Santa Cruz sc-1485), Ku70 (sc-1486), NF90 (mAB DRBP76 BD), NFATp (sc-1151), NFATc1 (mAB clone 7A6, sc-7294), NFATc2 (mAB clone G1-D10, sc-7295) and cPLA2 (sc-438).
Protein purification
Jurkat T-cells were stimulated for 12 hr with 20 ng/ml PMA + 2 µM ionomycin. Nuclear proteins were extracted from chromatin with 0.3 M ammonium sulfate for 30 min at 4°C and the supernatants were precipitated with 0.2 g/ml ammonium sulfate. The protein pellets were resuspended in buffer C25 (25 mM HEPES, pH 7.6, 25 mM KCl, 0.1 mM EDTA, 1 mM DTT, 10% glycerol) and dialyzed against two changes of C25. Nuclear proteins (300 mg) were purified using a diethylaminoethyl column (Whatman DE-52, 20 × 2.2 cm2) eluted with a linear gradient of 25–400 mM KCl. Complex I eluted around 200 mM KCl, and these fractions were pooled, dialyzed against C25 and chromatographed on a carboxymethyl–agarose column eluted with a gradient of KCl (Pharmacia, 14 × 2.2 cm2). Complex I eluted around 180 mM KCl. Fractions were pooled and dialyzed against C25 and loaded onto an octylamine–agarose column (Pharmacia, 5 × 0.8 cm2) and eluted stepwise with buffer C containing 0.1, 0.2, 0.25, 0.3 and 0.4 M KCl. Complex I eluted maximally at 0.2 M KCl, and complexes II and III eluted maximally at 0.4 M KCl. Octylamine fraction 0.2 containing complex I (pool A) was adjusted to 50 mM KCl and coupled to a mutant or wild-type ARRE oligonucleotide affinity column (200 µl volume) (7). The affinity columns were washed with buffer C50 and eluted stepwise with three column volumes each of buffer C containing 0.1, 0.2, 0.3 or 0.4 M KCl. Complex I eluted at 0.2 M KCl with better recovery from the mutant compared to the ARRE affinity column. Complexes II and III present in octylamine 0.4 M fraction (pool B) were diluted to 50 mM KCl and applied to a wild-type ARRE affinity column. Complex II/III peak activity eluted maximally from the ARRE column at 0.4 M KCl.
Coimmunoprecipitation studies
NF45 IgG was covalently coupled to protein-A agarose using bis(sulfosuccinimidyl)suberate (BS3, Pierce). Jurkat T-cells (1–2 × 107 cells/condition) were either nonstimulated, stimulated with PMA + ionomycin or stimulated in the presence of CsA (1000 ng/ml) or triptolide (1000 ng/ml), and then nuclear extracts were prepared. Immunoprecipitations were performed by incubating 150 mg of Jurkat T-cell nuclear proteins with 50 µl of protein A beads (control) or anti-NF45-protein A beads for 2 hr at 4°C, and then washing 4× with 200 µl of intermediate stringency buffer (50 mM Tris–Cl pH 7.5, 250 mM NaCl, 1% NP-40, 0.1% SDS). The specific immunoprecipitates were fractionated by SDS–PAGE (8% separating gel), and proteins were transferred to nitrocellulose using a semidry blotter (BioRad). Primary antibodies for Western immunoblotting were goat αKu70 and αKu80 (Santa Cruz), mouse αKu80 (mAB 111), rabbit αNF45 and αNF90 and rabbit αDPK1.
Chromatin immunoprecipitation
Jurkat T-cells or C57BL/6 mouse primary spleen cells were nonstimulated or stimulated for 4 hr with PMA + ionomycin, in the absence or presence of CsA (1000 ng/ml) or triptolide (200 ng/ml), then formaldehyde crosslinking and chromatin immunoprecipitation (ChIP) was performed as described (36). PCR primers were designed to amplify the human IL-2 promoter: F 5′-GAGTTACTTTTGTATCCCCACCCCC (–317 to –292 in the IL-2 promoter), R 5′-CCTGTACATTGTGGCAGGAGTTGAGG (+33 to 58); IL-2 intron 3: F 5′-GCTTAAGAGGATACAGAACACTGCAACAG, R 5′-CCCTACCCCATCATAGTATCAATGCAGGTG; origin negative control myc1: F 5′-TTCTCAACCTCAGCACTGGTGACA, R 5′-GACTTTGCTGTTTGCTGTCAGGCT; myc11 origin of replication: F 5′-TATCTACACTAACATCCCACGCTCTG, myc11: R 5′-CATCCTTGTCCTGTGAGTATAAATCATCG. Mouse PCR primers were: IL-2 proximal promoter (270 bp): F 5′-CTGCCACCTAAGTGTGGGCTAACCCGACC, R 5′-GCATGCTGTACATGCCTGCAGGACTTGAGG; IL-2 intron 3 (424 bp): F 5′-CCACAATGTGGGTGGGTCACTGCAATTGAAC, R 5′-GTTAGGCCACTCTAGTGAGCTCTTCTGGC; adenosine deaminase origin of DNA replication (268 bp): F 5′-CTGAGACTATCCTCCAGGTCTTCTAATGGGG, R 5′-GGATGACCCTTTCATGGCTGCCTATGACCAACAG. PCR amplifications used a three-step protocol with 59°C annealing temperature and 35 cycles.
RESULTS
Purine-box regulator complexes contain ARRE DNA-binding subunits NF90, NF45, Ku80 and Ku70
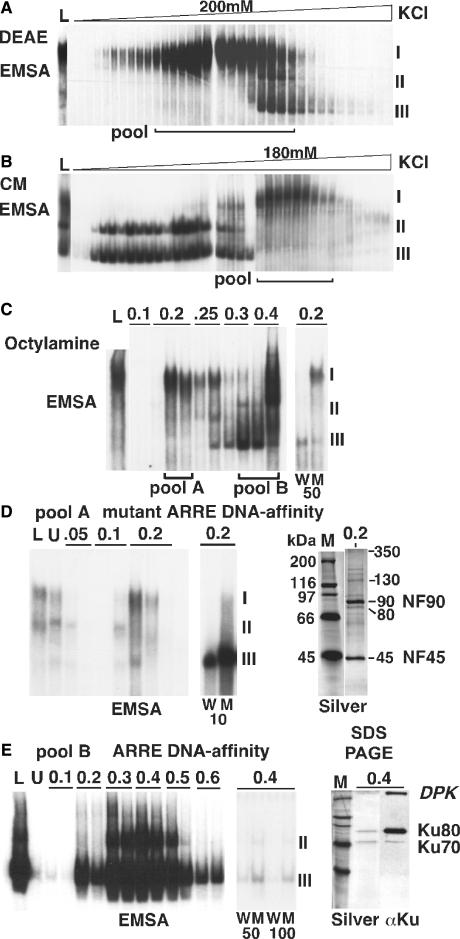
Activated Jurkat T-cells contain an inducible nuclear purine-box regulator ARRE/NF-AT DNA-binding complex (Figures 1 and 2) (7,8,20). Purine-box regulator complex I migrates slowly as an elongated complex in EMSA and exhibits highly specific ARRE DNA-binding activity. At all stages of purification complex I converted into faster migrating complexes II and III that exhibited tighter bands in EMSA (Figure 1) and this observation led us to infer that all three complexes might share common ARRE DNA-binding subunits. The conversion of complex I into complexes II and III was promoted by exposure to salt concentrations above 0.4 M and to DNA. We developed a purification strategy for complex I that minimized exposure to high salt and DNA, which involved sequential diethylaminoethyl (Figure 1A), carboxymethyl (Figure 1B) and octylamine (Figure 1C) ion-exchange columns followed by DNA-affinity chromatography (Figure 1D and E). Complex I was substantially enriched in the octylamine elution 0.2 M KCl/pool A (Figure 1C). Oligonucleotide competition experiments demonstrated greater inhibition of complex I binding using the wild-type compared to the mutant ARRE sequence (Figure 1C, lanes W versus M). Fifty nanograms of wild-type ARRE oligonucleotide completely inhibited complex I binding and promoted conversion of residual complex I into complex III. In contrast, 50 ng of mutant ARRE oligonucleotide only partially inhibited complex I, and did not promote conversion of complex I into III. This result suggested a moderate affinity of complex I for the mutant ARRE sequence. We successfully employed an immobilized mutant ARRE affinity column for gentle purification of complex I using elution with low concentrations of salt. We loaded octylamine-eluted pool A containing exclusively complex I (Figure 1C) onto a mutant ARRE DNA-affinity column (Figure 1D) and recovered maximal complex I activity at 0.2 M KCl, coeluting with substantial amounts of complex III, and associated with proteins of 45, 80, 90, 130 and 350 kDa (Figure 1D, right). This result suggested that affinity-purified complex I could convert into complex III, and that complexes I and III might share common ARRE DNA-binding subunits. Ten nanograms of wild-type ARRE oligonucleotide completely inhibited complex I and promoted conversion of complex I into complex III (Figure 1D, lane W). In contrast, 10 ng of mutant ARRE oligonucleotide was less potent than wild type in inhibiting complex I formation (hence the residual smear), and less potent in inhibiting complex III (Figure 1D, lane M). These EMSA competitions show that complexes I and III each exhibit greater affinity for the wild-type ARRE compared to the mutant ARRE oligonucleotide. We previously characterized the 45 and 90 kDa proteins as NF45 and NF90 (8).
Figure 1.
Purification of purine-box regulator complexes reveals ARRE–DNA-binding subunits NF90, NF45, Ku80 and Ku70. EMSA were used to monitor enrichment of ARRE–DNA-binding complexes from nuclear extracts of Jurkat T-cells stimulated with PMA + Ionomycin. (A) Diethylaminoethyl (DEAE) elution. Fractions enriched in complex I (and small amounts of complexes II and III) were pooled and dialyzed. (B) Carboxymethyl (CM) elution. Note the substantial conversion of complex I into complexes II and III. Fractions containing complex I only were pooled and dialyzed. (C) Octylamine elutions with steps of KCl. Right, oligonucleotide competitions demonstrate greater inhibition of complex I in elution 0.2 with 50 ng of wild type (W) compared to mutant (M) ARRE, yet 50 ng of M does produce substantial inhibition of complex I. (D) DNA-affinity elution. Octylamine elutions 0.2 M KCl/pool A, enriched in complex I only, were diluted and loaded onto a mutant ARRE–DNA-affinity column and eluted with the indicated steps of KCl. Note the presence of complexes I and III in eluted fraction 0.2, associated with principal protein subunits at 45, 80, 90, 130 and 350 kDa shown by SDS–PAGE with silver staining. Ten nanograms of wild-type (W) ARRE oligonucleotide inhibits complex I and promotes conversion to complex III more potently than mutant (M) oligonucleotide. (E) DNA-affinity elution. Octylamine elutions enriched in complexes II and III, pool B, were diluted and loaded onto a wild-type ARRE–DNA-affinity column, and maximal DNA-binding activity was eluted at 0.4 M KCl, associated with protein subunits at 70, 80 and 350 kDa shown by SDS–PAGE with silver staining. Internal tryptic peptide sequences of the 70 and 80 kDa proteins identified them as Ku70 and Ku80. Western immunoblot with human JM anti-Ku sera (right) confirms the identity of Ku70 and Ku80 (lane 4), and suggests the slowest migrating protein to be DNA-dependent protein kinase (DPK) catalytic subunit.
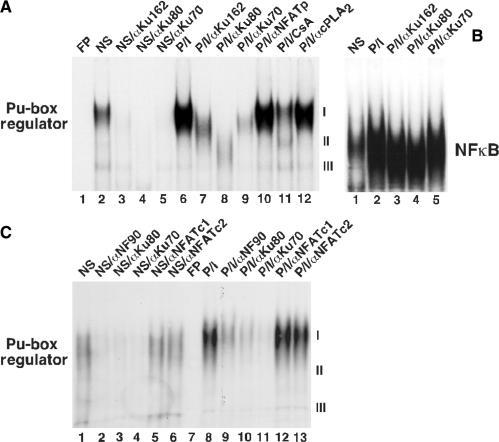
Figure 2.
Purine-box regulator in T-cell nuclear extracts contains ARRE–DNA-binding subunits Ku80, Ku70 and NF90. EMSA of nuclear proteins from NS and PMA + ionomycin (P/I)-stimulated Jurkat T-cells. (A) Monoclonal antibodies against Ku were preincubated with nuclear proteins before addition of 32P-labeled oligonucleotide probe: 162 recognizes the native Ku70/Ku80 heterodimer, 111 recognizes Ku80 and N3H10 recognizes Ku70. Polyclonal antibodies against NF-ATp or cytoplasmic phospholipase A2 (cPLA2) did not inhibit binding of purine-box regulator complex, while cyclosporin A (CsA) caused partial inhibition. (B) Ku monoclonal antibodies did not inhibit induced NF-κB DNA-binding. (C) Monoclonal antibodies to NF90 (mAB anti-DRBP76), Ku80 and Ku70, but not NFATc1 or NFATc2, specifically inhibited purine-box regulator ARRE–DNA-binding activity in nuclear extracts of nonstimulated and P/I-stimulated Jurkat T-cells.
We purified complexes II and III in octylamine elution 0.4 M KCl/pool B (Figure 1C) using a wild-type ARRE DNA-affinity column (Figure 1E) and determined that maximal activity eluted at 0.4 M KCl. ARRE sequence-specific binding was demonstrated by the greater inhibition of complexes II and III with 50 and 100 ng of wild type compared to mutant competitor oligonucleotides (Figure 1E, lanes W versus M). Complexes II and III were associated with proteins of 70, 80 and 350 kDa (Figure 1E, right). Internal tryptic peptide sequence information allowed us to identify these proteins as the systemic lupus erythematosis autoantigens, Ku70 and Ku80 (data not shown). Human anti-Ku serum JM reacted specifically with the purified 70 and 80 kDa proteins in a Western immunoblot (Figure 1E, right, lane αKu). We interpret the 350 kDa immunoreactive protein to represent DNA-dependent protein kinase catalytic subunit (DPKcs), known to interact with Ku70 and Ku80 (21).
Purine-box regulator complex in unfractionated nuclear extracts contains ARRE DNA-binding subunits Ku80, Ku70 and NF90
We proceeded to test whether antibodies against Ku80, Ku70 or NF90 affected the purine-box regulator/ARRE DNA-binding complex in nuclear extracts prepared from resting and activated Jurkat T-cells (Figure 2). In nonstimulated Jurkat T-cells, a purine-box regulator complex can be detected in the nucleus (Figure 2A, lane 2), and T-cell activation with PMA + ionomycin (P/I) strongly induces the DNA-binding affinity of this complex (Figure 2A, lane 6 versus 2). Activation in the presence of CsA inhibits the induction of ARRE–DNA-binding activity down to the NS level (Figure 2, lane 11 versus 6). Monoclonal antibodies against the Ku heterodimer, individual Ku80 or Ku70 subunits (40), each produced nearly complete inhibition of both the constitutive and inducible purine-box regulator complex (Figure 2A, lanes 3–5 versus 2 and 7–9 versus 6). We observed no significant inhibition of the purine-box regulator with polyclonal antibodies against NF-ATp/NFATc2 or unrelated cytoplasmic phospholipase A2 (Figure 2A, lanes 10 and 12). The absence of a supershift may be a consequence of the labile nature of the purine-box regulator, and its dissociation by subunit-specific antibodies. As a control for specificity, the mAbs to Ku caused no inhibition of the induced NF-κB DNA-binding complex (Figure 2B, lanes 3–5 versus 2). A mAB to NF90 (anti-DRBP76) also inhibited the purine-box regulator complex I in nonstimulated and stimulated T-cells comparably to polyclonal anti-Ku80 and anti-Ku70 (Figure 2C, lanes 2–4 versus 1 and 9–11 versus 8). Under our experimental conditions (that used 0.3 M ammonium sulfate to extract nuclear proteins), mAbs against nuclear factor of activated T-cells, NFATc1 and NFATc2 (characterized to bind ARRE sequences in vitro following 0.42 M KCl nuclear extractions) did not affect the purine-box regulator complex I in nonstimulated or stimulated T-cells (Figure 2C, lanes 5 and 6 versus 1 and 12 and 13 versus 8). These EMSA-antibody inhibition experiments confirm the results of our purification (Figure 1) and demonstrate that Ku80, Ku70 and NF90 are specific ARRE DNA-binding subunits of the purine-box regulator in nonstimulated and stimulated T-cell nuclear extracts.
Interactions between Ku, DNA-PK, NF45 and NF90 are weakened during T-cell activation
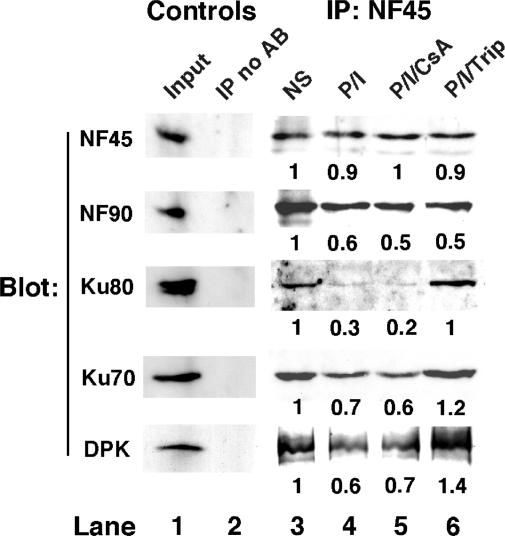
We characterized T-cell activation-induced changes in purine-box regulator subunit interactions by coimmunoprecipitation (Figure 3). Jurkat T-cells were either nonstimulated, stimulated with PMA + ionomycin for 4 hr or stimulated in the presence of immunosuppressants CsA or triptolide (41). NF45, NF90, Ku80, Ku70 and DNA-PKcs were readily detected by immunoblotting in nuclear extract from nonstimulated T-cells (Figure 3, lane 1), but were not detected in protein A precipitates performed in the absence of NF45 antibody (Figure 3, lane 2). Extracted nuclear proteins were immunoprecipitated with immobilized antibody against NF45 and the immunoprecipitates (IP) were washed at low, moderate or high stringencies and analyzed by immunoblotting. Using moderate stringency washing conditions of the NF45 IPs, immunoblotting demonstrated coimmunoprecipitation of NF90, Ku80, Ku70 and DNA-PKcs in nuclear extracts from NS T-cells (Figure 3, lane 3). T-cell stimulation with PMA + ionomycin (P/I) was consistently associated with coordinate decreases in the associations of Ku80, Ku70 and DNA-PK with NF45 (Figure 3, lane 4 versus 3). T-cells stimulated in the presence of CsA (P/I/CsA) also showed a decrease in the associations of Ku and DNA-PK with NF45 as in stimulated cells (Figure 3, lane 5 versus 3), whereas T-cells stimulated in the presence of the more potent immunosuppressant, triptolide (P/I/Trip), showed a pattern of association of Ku and DNA-PK with NF45 most similar to that in nonstimulated cells (Figure 3, lane 6 versus 3). Under high stringency washing conditions we observed that Ku70 and Ku80 coimmunoprecipitated with NF45 in nuclear extracts from nonstimulated T-cells, but Ku proteins were washed away from NF45 in extracts from stimulated T-cells (data not shown). These results support and complement our protein purification results (Figure 1) and establish that NF45, NF90, Ku70, Ku80 and DNA-PKcs interact dynamically in the nucleus of T-cells. Our results are supported by a recent report that used tandem affinity purification in HEK293 cells to demonstrate specific interactions between Ku70, Ku80 and NF90/ILF3 (42).
Figure 3.
Association of NF45 and NF90 with Ku80, Ku70 and DNA-dependent protein kinase is altered during T-cell activation. Jurkat T-cells were either NS, stimulated with PMA + ionomycin (P/I) for 4 hr or stimulated in the presence of CsA (P/I/CsA, 1000 ng/ml) or triptolide (P/I/Trip, 1000 ng/ml). Extracted nuclear proteins (lane 1) were co-immunoprecipitated with protein A beads (control, lane 2) or immobilized antibody against NF45 (lanes 3–6) and the immunoprecipitates (IP) were washed at moderate stringency and analyzed by Western immunoblotting. The observation of decreased association of Ku70 and Ku80 with NF45 in activated T-cells was reproduced more than 6×.
Ku80, Ku70 and NF90 bind specifically and dynamically to the IL-2 gene promoter in vivo, as demonstrated by chromatin immunoprecipitation
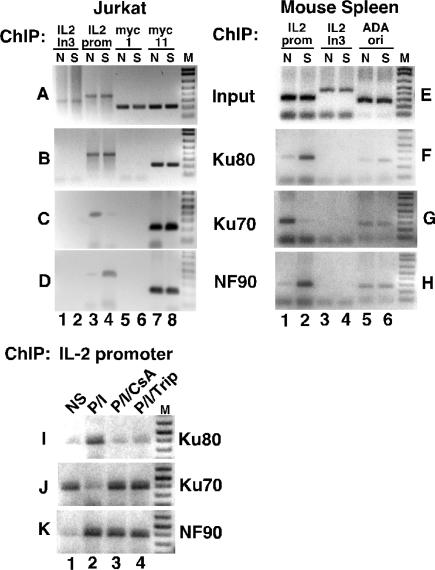
We characterized interactions of Ku80, Ku70 and NF90 with the IL-2 promoter in resting and activated T-cells in vivo, using formaldehyde crosslinking and chromatin immunoprecipitation (ChIP) experiments (Figure 4). For our ChIP studies, we designed PCR primers to interrogate the presence of the proximal IL-2 gene promoter that contains the ARRE/NF-AT DNA-binding sequence, and a region within intron 3 of the IL-2 gene, 4 kb removed, as a control. The myc11 origin of DNA replication sequence near the myc promoter, known to bind Ku80 and Ku70, was used as a positive control, whereas the myc1 sequence, 5.9 kb removed, served as a negative control (36,37). Input chromatin prepared from nonstimulated and stimulated Jurkat T-cells resulted in PCR amplification products using combinations of oligonucleotide primers for the IL-2 intron 3, IL-2 gene promoter, myc1 and myc11 sequences (Figure 4A). Ku80 ChIP using mAB 111 showed no precipitation of IL-2 intron 3 (Figure 4B, lanes 1 and 2), but nearly equivalent precipitation of IL-2 promoter DNA from both nonstimulated and stimulated T-cells (Figure 4B, lanes 3 and 4). We observed identical Ku80 ChIP results using a polyclonal antibody (data not shown). Our Ku70 ChIP experiment showed no binding to IL-2 intron 3 (Figure 4C, lanes 1 and 2), and specific binding of Ku70 to the IL-2 promoter in nonstimulated T-cells (Figure 4C, lane 3). Remarkably, T-cell stimulation caused a substantial decrease in Ku70 binding to the IL-2 promoter (Figure 4C, lane 4). As expected, the Ku80 and Ku70 ChIP experiments showed that the myc1 sequence was negative, while the myc11 origin sequence was positive (Figure 4B and C, lanes 5–8). NF90 ChIP using mAB DRBP76 showed no binding to IL-2 intron 3 (Figure 4D, lanes 1 and 2) and weak binding to the IL-2 promoter in nonstimulated T-cells (Figure 4D, lane 3). Notably, T-cell stimulation was associated with a substantial increase in NF90 binding to the IL-2 promoter (Figure 4D, lane 4 versus 3). Additionally, NF90 bound specifically to the myc11 origin sequence (Figure 4D, lanes 5–8). This is the first evidence that NF90 binds specifically to A-T-rich mammalian origins of DNA replication. The changes in DNA-binding of Ku80, Ku70 and NF90 induced by T-cell stimulation occurred specifically at the IL-2 gene promoter, and did not occur at the myc11 origin (Figure 4B–D, lanes 7 and 8).
Figure 4.
Ku80, Ku70 and NF90 bind specifically and dynamically to the IL-2 promoter in vivo. Jurkat T-cells or mouse primary spleen cells were NS or stimulated for 4 hr with PMA + ionomycin (S) then nuclear proteins were crosslinked to chromatin in vivo with 1% formaldehyde. (A) Sheared and restricted chromatin was used as template for PCR amplifications (35 cycles) of IL-2 intron 3 (negative control), IL-2 proximal promoter, myc1 (negative control) and myc11 (origin of DNA replication, positive control for Ku binding) sequences. (B) Ku80 chromatin immunoprecipitation (ChIP) was performed using mAB 111. (C) Ku70 ChIP was performed using polyclonal antibody. (D) NF90 ChIP was performed using mAB DRBP76. (E) Input chromatin from mouse spleen cells was used as template for amplification of IL-2 proximal promoter, IL-2 intron 3 (negative control) and adenosine deaminase origin of DNA replication (positive control for Ku binding). (F) Ku80 ChIP. (G) Ku70 ChIP. (H) NF90 ChIP. (I–K) Jurkat T-cells were NS or stimulated for 4 hr with PMA + ionomycin (P/I) or stimulated in the presence of immunosuppressants cyclosporin A (P/I/CsA) or triptolide (P/I/Trip), then IL-2 promoter ChIP was performed, using specific antibodies against Ku80, Ku70 and NF90 and 30 cycles of PCR amplification.
We performed similar ChIP experiments on primary mouse spleen cells that were nonstimulated or stimulated for 4 hr with PMA + ionomycin (Figure 4E–H). Ku70 and Ku80 bind to the adenosine deaminase (ADA) origin of replication in vivo, and therefore we used this amplicon as a positive control for Ku ChIP experiments in mouse cells (37). Ku80 ChIP shows weak binding to the IL-2 promoter in nonstimulated cells (Figure 4F, lane 1) that is induced upon stimulation (Figure 4F, lane 2 versus 1). Ku70 ChIP shows substantial binding to the IL-2 promoter in nonstimulated cells that decreases upon stimulation (Figure 4G, lanes 1 and 2). NF90 ChIP shows induced binding to the IL-2 promoter upon stimulation (Figure 4H, lane 2 versus 1). Ku80, Ku70 and NF90 showed no binding to IL-2 intron 3 (Figure 4F–H, lanes 3 and 4), and no stimulation-induced changes in binding to the ADA origin (Figure 4F–H, lanes 5 and 6).
Immunosuppressants cyclosporin A and triptolide inhibit dynamic changes in IL-2 promoter binding by Ku80 and Ku70
Cyclosporin A and triptolide inhibit IL-2 gene expression at the levels of chromatin remodeling (43) and transcriptional activation (41). We previously showed that triptolide is more potent than CsA in its inhibition of induced ARRE DNA-binding activity, and operates through different mechanisms as demonstrated by its ability to inhibit CsA-resistant T-cell activation stimulated by PMA + CD28 (41). We used ChIP to characterize how CsA and triptolide affected IL-2 chromatin remodeling in Jurkat T-cells as reflected by dynamic changes in IL-2 proximal promoter binding by Ku80, Ku70 and NF90 (Figure 4I–K). In these experiments we observed that P/I stimulation induced binding of Ku80 to the IL-2 promoter (Figure 4I, lane 2 versus 1), and CsA and triptolide each inhibited this induction (Figure 4I, lanes 3, 4 versus 2). Reciprocally, stimulation with P/I decreased binding of Ku70 to the IL-2 promoter (Figure 4J, lane 2 versus 1), and CsA and triptolide each inhibited this decrease (Figure 4J, lanes 3, 4 versus 2). Stimulation with P/I-induced binding of NF90 to the IL-2 promoter (Figure 4K, lane 2 versus 1), and CsA and triptolide did not diminish this induction (Figure 4K, lanes 3, 4 versus 2). Thus, T-cell immunosuppressants that inhibit IL-2 transcriptional activation (41) apparently operate through mechanisms that inhibit dynamic changes in DNA-binding of Ku80 and Ku70 to the IL-2 proximal promoter in vivo.
DISCUSSION
In this study we present complementary experiments of native protein purification, EMSA-antibody inhibition and chromatin immunoprecipitation that together demonstrate specific and dynamic binding of purine-box regulator subunits, Ku80, Ku70 and NF90 to the ARRE DNA sequence in vitro and to the IL-2 proximal promoter in vivo. These dynamic changes in binding of Ku80, Ku70 and NF90 to the IL-2 proximal promoter correlate temporally with induced IL-2 chromatin remodeling and transcriptional initiation in activated T-cells.
While Ku and DNA-PKcs are accepted to bind to free ends of double-stranded (ds) DNA and promote DNA repair (23,24), their functions as sequence-specific DNA-binding proteins and transcriptional regulators remain more ambiguous (44). An important recent study showed that ds DNA breaks are formed during estrogen induction of the pS2 promoter by enzymatic activity of DNA topoisomerase IIβ, and these newly formed ds DNA breaks are repaired by Ku80, Ku70, DNA-PKcs and poly(ADP)ribose polymerase recruited specifically to the pS2 promoter (34). The authors proposed that transient ds DNA break formation and DNA repair might facilitate rapid chromatin unwinding linked to transcriptional activation.
Ku is an abundant nuclear protein, and its affinity for free-DNA-ends creates the potential for artifacts in EMSA experiments as a consequence of Ku binding to the ends of linear oligonucleotide probes. In this study, we were careful to employ sufficient amounts of unlabeled competitor poly(dI-dC) as a source of free-DNA-ends to minimize this nonspecific binding and reveal sequence-specific binding of Ku to the radiolabeled ARRE oligonucleotide. Sequence-specific binding of Ku to internal A-T-rich target sequences similar to ARRE has been demonstrated using closed DNA minicircles or plasmids that lack free ends (28,35).
Chromatin immunoprecipitation (ChIP) allows characterization of interactions between nuclear proteins and their cognate DNA sequences in the native chromatin context. Sequence-specific binding of Ku70 and Ku80 to A-T-rich origins of DNA replication in vivo was demonstrated using ChIP (36,37). Rosenfeld and colleagues performed ChIP experiments and demonstrated specific recruitment of Ku80, Ku70 and DNA-PK to the pS2, Dio1, MMP12, PSA and RARβ promoters upon induction in vivo, and that activation of these promoters was regulated through transient dsDNA break formation and DNA repair (34).
T-cell activation is recognized to induce IL-2 chromatin remodeling and transcriptional activation (2). The identities of the chromatin remodeling proteins that bind to and regulate the IL-2 proximal promoter in vivo are not entirely clarified. We isolated and characterized NF90, NF45, Ku80 and Ku70 as specific ARRE–DNA-binding proteins in vitro, using EMSA to monitor our biochemical purification under nondenaturing conditions (7,8) (Figure 1). We now extend these findings and are the first to demonstrate that Ku80, Ku70 and NF90 bind dynamically to the chromatin at the IL-2 proximal promoter in vivo, using ChIP experiments. T-cell stimulation induces binding of Ku80 and NF90 to the IL-2 promoter, and reciprocally decreases binding of Ku70 to the IL-2 promoter in vivo. The time course of dynamic changes in binding of Ku80, Ku70 and NF90 to the IL-2 promoter correlates with induced IL-2 chromatin remodeling in activated T-cells, assayed by micrococcal nuclease sensitivity (45). The dynamic binding of Ku80, Ku70 and NF90 to the IL-2 promoter during T-cell activation is specific, as we observed no stimulation-induced changes in the specific binding of these proteins to myc11 or ADA origins of DNA replication (46).
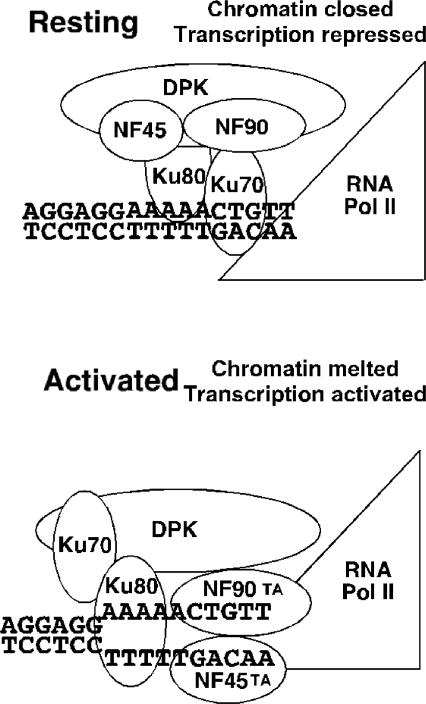
From our experiments, we propose a model for regulation of endogenous IL-2 gene expression (Figure 5). In nonstimulated T-cells the purine-box regulator complex is present in the nucleus and contains subunits Ku80, Ku70, NF90, NF45 and DNA-PKcs. Antibodies against Ku80, Ku70 or NF90 disrupt EMSA complex I in vitro in stimulated and nonstimulated T-cell nuclear extracts, suggesting that these ARRE–DNA–proteins are physically and functionally associated. ChIP experiments demonstrate substantial binding of Ku70 to the IL-2 promoter in nonstimulated T-cells in vivo. T-cell activation induces conformational changes in the purine-box regulator and IL-2 chromatin remodeling associated with decreased binding of Ku70 and increased binding of Ku80 and NF90 to the IL-2 promoter in vivo. A proposed reorganization of subunits during T-cell activation that could alter the interactions of Ku70, Ku80 and NF90 with IL-2 promoter sequences is shown. Whether Ku70 might completely dissociate from the IL-2 promoter and the purine-box regulator complex during T-cell activation in vivo remains unknown at this time.
Figure 5.
Model for dynamic binding of Ku80, Ku70, NF90, NF45 and DNA-PKcs to the IL-2 proximal promoter in vivo in resting and activated T-cells. (A) In resting T-cells, Ku70 and Ku80 bind to the ARRE sequence in the IL-2 proximal promoter and contribute to a closed chromatin conformation and transcriptional repression. (B) T-cell activation induces chromatin remodeling associated with decreased binding of Ku70 to the IL-2 promoter and increased binding of NF90 and Ku80 to the IL-2 promoter. Associations between NF45/NF90 heterodimer and Ku70. Ku80 and DNA-PKcs are weakened. IL-2 chromatin remodeling is linked to the conversion of the purine-box regulator from a transcriptional repressor into a potent transcriptional activator, indicated by new interactions of transactivation (TA) domains on NF90 and NF45 with RNA Pol II.
We propose that increased association of NF90 and NF45 with the chromatin at the IL-2 promoter is associated with transcriptional activation, indicated by transactivation (TA) domains of these proteins interacting with RNA polymerase II. Antisera against NF45 and NF90 specifically inhibited ARRE–DNA-binding as well as basal and stimulated in vitro transcription (7). We recently showed that stable overexpression of NF45 in Jurkat T-cells conferred a 100-fold-specific increase in ARRE/NF-AT luciferase transcriptional activation (11). Additionally, we have observed that stable transgenic expression of NF90 in Jurkat T-cells is associated with 70–80-fold-specific increase in ARRE/NF-AT luciferase transcriptional activation (Shi et al, J. Exp Med. in press). Conversely, we have determined that targeted disruption of NF90 in mice is associated with profound impairment of IL-2 gene expression and ARRE/NF-AT luciferase transcriptional activation in activated T-cells (Shi et al, J. Exp Med. in press).
Transcriptional regulation by Ku and DNA-PKcs at specific promoters has been previously reported, with suggestions of opposing roles for Ku70 and Ku80 (25–34). For example, Ku and DNA-PKcs mediated inhibition of RNA Pol II transcription at the hsp70 promoter (27) and at the NRE1 site in the mouse mammary tumor virus promoter (28). Inhibitory effects of Ku on transcription correlated with the level of expression of Ku70 protein: (i) transgenic overexpression of Ku70 protein in rat fibroblasts suppressed the induction of hsp70 in response to heat shock (47) and (ii) in mouse lymphosarcoma cells, heat shock was associated with a rapid disappearance of Ku70 protein (48). A transcriptional activating role for Ku80 was suggested by (i) the report that transfection of a Ku80 expression plasmid into Ku80-deficient MCF-7 breast cancer cells activated transcription of a human glucocorticoid receptor reporter gene (26), (ii) the result that rat fibroblasts which overexpressed Ku80 showed enhanced induction of hsp70 in response to heat shock (47) and (iii) in vitro transcription studies that demonstrated specific enhancement of hsp70 promoter transcriptional initiation associated with expression of Ku80 cDNA in xrs-6 Ku80-deficient cells (49).
Our results suggest novel and important roles for Ku80, Ku70, NF90 and NF45 in the regulation of IL-2 chromatin remodeling and gene expression in activated T-cells. Future investigations should elucidate how Ku80 and Ku70 may serve reciprocal roles in regulating IL-2 chromatin remodeling and gene expression in activated T-cells.
ACKNOWLEDGEMENTS
We thank Mark Krasnow for encouragement. Funding to pay the Open Access publication charge was provided by NIH grants R01-AI39624 and R01-HL62588, and a gift from the Donald E. and Delia B. Baxter Foundation to PNK.
Conflict of interest statement. None declared.
REFERENCES
- 1.June CH, Ledbetter JA, Lindsten T, Thompson CB. Evidence for the involvement of three distinct signals in the induction of IL-2 gene expression in human T lymphocytes. J. Immunol. 1989;143:153–161. [PubMed] [Google Scholar]
- 2.Garrity PA, Chen D, Rothenberg EV, Wold BJ. Interleukin-2 transcription is regulated in vivo at the level of coordinated binding of both constitutive and regulated factors. Mol. Cell Biol. 1994;14:2159–2169. doi: 10.1128/mcb.14.3.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mouzaki A, Weil R, Muster L, Rungger D. Silencing and trans-activation of the mouse IL-2 gene in Xenopus oocytes by proteins from resting and mitogen-induced primary T-lymphocytes. EMBO J. 1991;10:1399–1406. doi: 10.1002/j.1460-2075.1991.tb07660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mouzaki A, Rungger D. Properties of transcription factors regulating interleukin-2 gene transcription through the NFAT binding site in untreated or drug-treated naive and memory T-helper cells. Blood. 1994;84:2612–2621. [PubMed] [Google Scholar]
- 5.Shaw JP, Utz PJ, Durand DB, Toole JJ, Emmel EA, Crabtree GR. Identification of a putative regulator of early T cell activation genes. Science. 1988;241:202–205. [PubMed] [Google Scholar]
- 6.Randak C, Brabletz T, Hergenrother M, Sobotta I, Serfling E. Cyclosporin A suppresses the expression of the interleukin 2 gene by inhibiting the binding of lymphocyte-specific factors to the IL-2 enhancer. EMBO J. 1990;9:2529–2536. doi: 10.1002/j.1460-2075.1990.tb07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corthesy B, Kao PN. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J. Biol. Chem. 1994;269:20682–20690. [PubMed] [Google Scholar]
- 8.Kao PN, Chen L, Brock G, Ng J, Kenny J, Smith AJ, Corthesy B. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J. Biol. Chem. 1994;269:20691–20699. [PubMed] [Google Scholar]
- 9.Shim J, Lim H, J RY, Karin M. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol. Cell. 2002;10:1331–1344. doi: 10.1016/s1097-2765(02)00730-x. [DOI] [PubMed] [Google Scholar]
- 10.Shi L, Zhao G, Qiu D, Godfrey WR, Vogel H, Rando TA, Hu H, Kao PN. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J. Biol. Chem. 2005;280:18981–18989. doi: 10.1074/jbc.M411034200. [DOI] [PubMed] [Google Scholar]
- 11.Zhao G, Shi L, Qiu D, Hu H, Kao PN. NF45/ILF2 tissue expression, promoter analysis, and interleukin-2 transactivating function. Exp. Cell Res. 2005;305:312–323. doi: 10.1016/j.yexcr.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 12.Reichman TW, Muniz LC, Mathews MB. The RNA binding protein nuclear factor 90 functions as both a positive and negative regulator of gene expression in mammalian cells. Mol. Cell Biol. 2002;22:343–356. doi: 10.1128/MCB.22.1.343-356.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saunders LR, Perkins DJ, Balachandran S, Michaels R, Ford R, Mayeda A, Barber GN. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 2001;276:32300–32312. doi: 10.1074/jbc.M104207200. [DOI] [PubMed] [Google Scholar]
- 14.Nie Y, Ding L, Kao PN, Braun R, Yang JH. ADAR1 interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Mol. Cell Biol. 2005;25:6956–6963. doi: 10.1128/MCB.25.16.6956-6963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu YH, Grabowski GA. Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid beta-glucosidase and other mRNAs. Mol. Genet. Metab. 1999;68:441–454. doi: 10.1006/mgme.1999.2934. [DOI] [PubMed] [Google Scholar]
- 16.Langland JO, Kao PN, Jacobs BL. Nuclear factor-90 of activated T-cells: A double-stranded RNA-binding protein and substrate for the double-stranded RNA-dependent protein kinase, PKR. Biochemistry. 1999;38:6361–6368. doi: 10.1021/bi982410u. [DOI] [PubMed] [Google Scholar]
- 17.Liao HJ, Kobayashi R, Mathews MB. Activities of adenovirus virus-associated RNAs: purification and characterization of RNA binding proteins. Proc. Natl. Acad. Sci. USA. 1998;95:8514–8519. doi: 10.1073/pnas.95.15.8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isken O, Grassmann CW, Sarisky RT, Kann M, Zhang S, Grosse F, Kao PN, Behrens SE. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 2003;22:5655–5665. doi: 10.1093/emboj/cdg562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi L, Godfrey WR, Lin J, Zhao G, Kao PN. NF90 regulates inducible IL-2 gene expression in T-cells. J. Exp Med. doi: 10.1084/jem.20052078. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoki Y, Zhao G, Qiu D, Shi L, Kao PN. CsA-sensitive purine-box transcriptional regulator in bronchial epithelial cells contains NF45, NF90, and Ku. Am. J. Physiol. 1998;275:L1164–1172. doi: 10.1152/ajplung.1998.275.6.L1164. [DOI] [PubMed] [Google Scholar]
- 21.Ting NS, Kao PN, Chan DW, Lintott LG, Lees-Miller SP. DNA-dependent protein kinase interacts with antigen receptor response element binding proteins NF90 and NF45. J. Biol. Chem. 1998;273:2136–2145. doi: 10.1074/jbc.273.4.2136. [DOI] [PubMed] [Google Scholar]
- 22.Tuteja R, Tuteja N. Ku autoantigen: a multifunctional DNA-binding protein. Crit. Rev. Biochem. Mol. Biol. 2000;35:1–33. doi: 10.1080/10409230091169177. [DOI] [PubMed] [Google Scholar]
- 23.Lees-Miller SP. The DNA-dependent protein kinase, DNA-PK: 10 years and no ends in sight. Biochem. Cell Biol. 1996;74:503–512. doi: 10.1139/o96-054. [DOI] [PubMed] [Google Scholar]
- 24.Chu G. Double strand break repair. J. Biol. Chem. 1997;272:24097–24100. doi: 10.1074/jbc.272.39.24097. [DOI] [PubMed] [Google Scholar]
- 25.Roberts MR, Han Y, Fienberg A, Hunihan L, Ruddle FH. A DNA-binding activity, TRAC, specific for the TRA element of the transferrin receptor gene copurifies with the Ku autoantigen. Proc. Natl. Acad. Sci. USA. 1994;91:6354–6358. doi: 10.1073/pnas.91.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warriar N, Page N, Govindan MV. Expression of human glucocorticoid receptor gene and interaction of nuclear proteins with the transcriptional control element. J. Biol. Chem. 1996;271:18662–18671. doi: 10.1074/jbc.271.31.18662. [DOI] [PubMed] [Google Scholar]
- 27.Yang SH, Nussenzweig A, Li L, Kim D, Ouyang H, Burgman P, Li GC. Modulation of thermal induction of hsp70 expression by Ku autoantigen or its individual subunits. Mol. Cell Biol. 1996;16:3799–3806. doi: 10.1128/mcb.16.7.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giffin W, Torrance H, Rodda DJ, Prefontaine GG, Pope L, Hache RJ. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 29.Jeanson L, Mouscadet JF. Ku represses the HIV-1 transcription: identification of a putative Ku binding site homologous to the mouse mammary tumor virus NRE1 sequence in the HIV-1 long terminal repeat. J. Biol. Chem. 2002;277:4918–4924. doi: 10.1074/jbc.M110830200. [DOI] [PubMed] [Google Scholar]
- 30.Mo X, Dynan WS. Subnuclear localization of Ku protein: functional association with RNA polymerase II elongation sites. Mol. Cell Biol. 2002;22:8088–8099. doi: 10.1128/MCB.22.22.8088-8099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu P, LaVallee PA, Lin JJ, Hoidal JR. Characterization of proteins binding to E-box/Ku86 sites and function of Ku86 in transcriptional regulation of the human xanthine oxidoreductase gene. J. Biol. Chem. 2004;279:16057–16063. doi: 10.1074/jbc.M305856200. [DOI] [PubMed] [Google Scholar]
- 32.Lebrun P, Montminy MR, Van Obberghen E. Regulation of the pancreatic duodenal homeobox-1 protein by DNA-dependent protein kinase. J. Biol. Chem. 2005;280:38203–38210. doi: 10.1074/jbc.M504842200. [DOI] [PubMed] [Google Scholar]
- 33.Mayeur GL, Kung WJ, Martinez A, Izumiya C, Chen DJ, Kung HJ. Ku is a novel transcriptional recycling coactivator of the androgen receptor in prostate cancer cells. J. Biol. Chem. 2005;280:10827–10833. doi: 10.1074/jbc.M413336200. [DOI] [PubMed] [Google Scholar]
- 34.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz MT, Matheos D, Price GB, Zannis-Hadjopoulos M. OBA/Ku86: DNA binding specificity and involvement in mammalian DNA replication. Mol. Biol. Cell. 1999;10:567–580. doi: 10.1091/mbc.10.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novac O, Matheos D, Araujo FD, Price GB, Zannis-Hadjopoulos M. In vivo association of Ku with mammalian origins of DNA replication. Mol. Biol. Cell. 2001;12:3386–3401. doi: 10.1091/mbc.12.11.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sibani S, Price GB, Zannis-Hadjopoulos M. Decreased origin usage and initiation of DNA replication in haploinsufficient HCT116 Ku80+/− cells. J. Cell Sci. 2005;118:3247–3261. doi: 10.1242/jcs.02427. [DOI] [PubMed] [Google Scholar]
- 38.Waninger S, Kuhen K, Hu X, Chatterton JE, Wong-Staal F, Tang H. Identification of cellular cofactors for human immunodeficiency virus replication via a ribozyme-based genomics approach. J. Virol. 2004;78:12829–12837. doi: 10.1128/JVI.78.23.12829-12837.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor TJ, Knipe DM. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 2004;78:5856–5866. doi: 10.1128/JVI.78.11.5856-5866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reeves WH, Satoh M, Wang J, Chou CH, Ajmani AK. Systemic lupus erythematosus. Antibodies to DNA, DNA-binding proteins, and histones. Rheum. Dis. Clin. North Am. 1994;20:1–28. [PubMed] [Google Scholar]
- 41.Qiu D, Zhao G, Aoki Y, Shi L, Uyei A, Nazarian S, Ng JC, Kao PN. Immunosuppressant PG490 (triptolide) inhibits T-cell interleukin-2 expression at the level of purine-box/nuclear factor of activated T- cells and NF-kappaB transcriptional activation. J. Biol. Chem. 1999;274:13443–13450. doi: 10.1074/jbc.274.19.13443. [DOI] [PubMed] [Google Scholar]
- 42.Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Wang J, Woltring D, Gerondakis S, Shannon MF. Histone dynamics on the interleukin-2 gene in response to T-cell activation. Mol. Cell Biol. 2005;25:3209–3219. doi: 10.1128/MCB.25.8.3209-3219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson CW, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins [published erratum appears in Nature 1996 Nov 28;384(6607):384] Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 45.Rao S, Procko E, Shannon MF. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J. Immunol. 2001;167:4494–4503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- 46.Sibani S, Price GB, Zannis-Hadjopoulos M. Ku80 binds to human replication origins prior to the assembly of the ORC complex. Biochemistry. 2005;44:7885–7896. doi: 10.1021/bi047327n. [DOI] [PubMed] [Google Scholar]
- 47.Li GC, Yang SH, Kim D, Nussenzweig A, Ouyang H, Wei J, Burgman P, Li L. Suppression of heat-induced hsp70 expression by the 70-kDa subunit of the human Ku autoantigen. Proc. Natl. Acad. Sci. USA. 1995;92:4512–4516. doi: 10.1073/pnas.92.10.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghoshal K, Jacob ST. Heat shock selectively inhibits ribosomal RNA gene transcription and down-regulates E1BF/Ku in mouse lymphosarcoma cells. Biochem. J. 1996;317:689–695. doi: 10.1042/bj3170689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodard RL, Lee KJ, Huang J, Dynan WS. Distinct roles for Ku protein in transcriptional reinitiation and DNA repair. J. Biol. Chem. 2001;276:15423–15433. doi: 10.1074/jbc.M010752200. [DOI] [PubMed] [Google Scholar]