Abstract
In Drosophila, the Trithorax-group (trxG) and Polycomb-group (PcG) proteins interact with chromosomal elements, termed Cellular Memory Modules (CMMs). By modifying chromatin, this ensures a stable heritable maintenance of the transcriptional state of developmental regulators, like the homeotic genes, that is defined embryonically. We asked whether such CMMs could also control expression of genes involved in patterning imaginal discs during larval development. Our results demonstrate that expression of the hedgehog gene, once activated, is maintained by a CMM. In addition, our experiments indicate that the switching of such CMMs to an active state during larval stages, in contrast to embryonic stages, may require specific trans-activators. Our results suggest that the patterning of cells in particular developmental fields in the imaginal discs does not only rely on external cues from morphogens, but also depends on the previous history of the cells, as the control by CMMs ensures a preformatted gene expression pattern.
Keywords: cellular memory, Hedgehog, Polycomb, Trithorax, Drosophila
During Drosophila embryogenesis, the transcriptional state of homeotic genes is established in a spatially restricted pattern by a regulatory cascade involving the products of segmentation genes (Ingham and Martinez-Arias 1992). Later, the Trithorax-group (trxG) and Polycomb-group (PcG) proteins take over and maintain, respectively, the active and the silenced states of transcription (Francis and Kingston 2001). This ability of cells to remember and propagate their gene expression programs throughout the entire development was termed cellular memory. This basic developmental function has been conserved during evolution, as related mechanisms were identified in other model organisms (Goodrich et al. 1997; Deschamps et al. 1999).
It is thought that trxG and PcG proteins form multimeric complexes involved in modeling chromatin (Papoulas et al. 1998; Shao et al. 1999; Petruk et al. 2001). The enzymatic functions associated with the complexes could also be involved in setting heritable epigenetic marks on chromatin. PcG proteins have been found to bind to specific chromosomal elements, termed PcG-response elements (PREs; Zink et al. 1991; Simon et al. 1993). The silencing function of PcG proteins at PREs can be counteracted by trxG proteins binding in the vicinity, at trxG-response elements (TREs; Tillib et al. 1999). In the bithorax complex, the Fab7 element is needed for maintaining segment-specific expression of the homeotic Abdominal-B gene. A transgenic model system has been established showing that the silent state of the Fab7-PRE can be switched at embryogenesis to an activated state, allowing continuous transcription of a nearby reporter gene through many rounds of mitotic division and surprisingly also through meiosis (Cavalli and Paro 1998). Activity is dependent on the trxG proteins and is marked by hyperacetylated H4 (Cavalli and Paro 1998, 1999). The binding and interplay of PcG and trxG proteins at elements such as Fab7 ensure transcriptional memory, presumably by setting and maintaining epigenetic marks during DNA replication and mitosis. For this reason, the Fab7 element has been termed a Cellular Memory Module (CMM).
Although several PREs regulating developmentally important genes have been identified (en, ph, as well as from the bithorax and Antennapedia complexes; Zink et al. 1991; Simon et al. 1993; Fauvarque et al. 1995; Brown et al. 1998) and many more candidates exist, only a few PREs from the bithorax complex have been tested and characterized as CMMs (M. Prestel and R. Paro, unpubl.). It is not known whether the concept of epigenetic maintenance of gene expression states is restricted to genes involved in long-term decisions, such as the HOX/HOM genes (i.e., to restrict embryonic patterns) or may be a more general feature used at different times of development. In this respect, the recurring role of segmentation genes used also for tissue patterning may be a good example to test for such a function. Importantly, the knowledge of how selector genes and segmentation genes are transcriptionally regulated is of fundamental importance to understand how stem cells established at later stages of development can maintain their identity throughout the entire development.
The product of one of these segmentation genes, hedgehog (hh), known to be acting as a morphogen (Heemskerk and DiNardo 1994), is essential for many crucial developmental pathways involved in the regulation of growth and patterning in both invertebrate and vertebrate species. In humans, misactivation of the Hh pathways leads to congenital diseases (e.g., prosencephaly; Villavicencio et al. 2000), and is associated with many kinds of tumors and cancers such as basal cell carcinomas and primitive neuroectodermal tumors (Toftgard 2000; Taipale and Beachy 2001). In Drosophila, one of its roles is to pattern leg and wing imaginal discs through the activation of decapentaplegic and wingless expression (Basler and Struhl 1994). In these discs, hh is initially activated in the posterior (P) compartment by Engrailed (En; Tabata et al. 1992; Zecca et al. 1995), which plays the key role in specifying the posterior identity (Kornberg et al. 1985; Simmonds et al. 1995). In late third-instar wing discs, Hh induces expression of en in the anterior compartment in a thin stripe along the antero–posterior (A–P) boundary (Blair 1992; Strigini and Cohen 1997). Several mechanisms seem to prevent hh and en expression from spreading into the anterior (A) compartment. For example, Polyhomeotic (PH) probably directly or indirectly maintains the repression of hh in the anterior cells abutting the A–P boundary (Maschat et al. 1998), whereas Groucho represses both hh and en in anterior cells (de Celis and Ruiz-Gomez 1995; Apidianakis et al. 2001).
How cells building compartments can maintain their determined identity until the completion of development is still unclear. The trxG and PcG proteins are known to control en expression (Busturia and Morata 1988; Moazed and O'Farrell 1992; Breen et al. 1995; Brizuela and Kennison 1997; Strutt 1997; Maschat et al. 1998). Previous studies found indications that hh expression itself might also be regulated by the trxG and PcG proteins (Felsenfeld and Kennison 1995; Randsholt et al. 2000). In this paper, we present evidence that hh expression is, indeed, directly controlled by the action of trxG and PcG proteins. We characterize a 3.4-kb fragment situated upstream of the hh transcription start site that exhibits CMM activity, and we show that in wing imaginal disc, initial activation of hh expression by En can be inherited through mitosis to daughter cells, even after En has ceased to act. The maintenance of hh expression is not caused by any kind of positive feedback loop but is dependent on the trxG and PcG proteins. We conclude that, during development, hh transcription is controlled by a CMM. Therefore, CMM switching may be a mechanism widely used at any time during development to maintain transcriptional states of genes with diverse functions.
Results
hedgehog transcription is directly controlled by PcG and trxG proteins
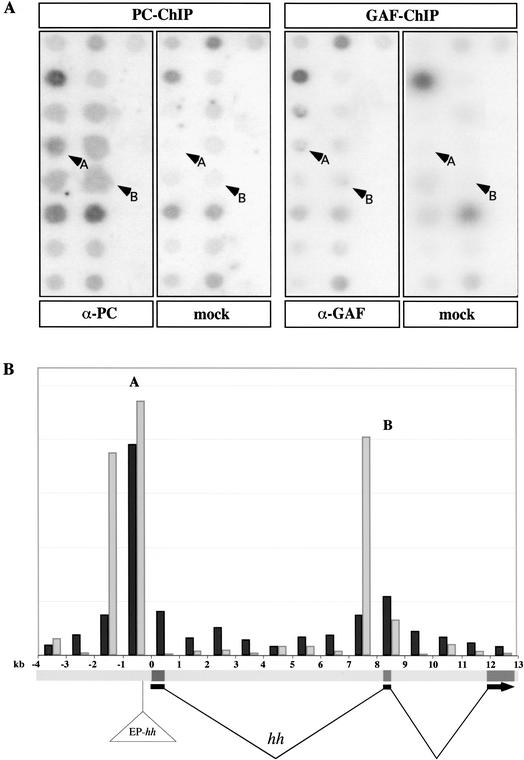
The immunoprecipitation technique using cross-linked chromatin (XChIP) allows the mapping of in vivo DNA target sites of chromatin proteins (Strutt and Paro 2000). Because one Polycomb (PC, a member of the PcG) binding site on polytene chromosomes coincides with the cytological position of hh at 94E, we decided to apply this method to ask whether there are PC and GAGA factor (GAF/Trl, a member of the trxG) binding sites in the hh genomic region. These two factors had previously been found to be hallmarks of CMMs (Strutt et al. 1997), and the GAF has been shown to be associated with some PcG complexes and necessary for the silencing function of PREs (Horard et al. 2000; Busturia et al. 2001). Initially we hybridized the immunoprecipitated material to a genomic stretch of 45 kb encompassing the hh gene (data not shown). This led us to identify PC/GAF-binding sites in regions close to the transcription unit. To further fine-map the location of the PC/GAF-binding sites we subdivided the region around the hh gene into 1-kb-sized PCR fragments (from 4 kb upstream of the hh transcription start site according to the transcript CG4637 from Flybase, to 13.4 kb downstream to the end of the gene; see Fig. 1). Slot-blot hybridizations of immunoprecipitated material (Fig. 1A) revealed two main sites where PC and GAF are strongly enriched (Fig. 1B). The first site (A) is located in a region between 0.07 and 1.06 kb upstream of the transcription start site, whereas the second binding site (B) is found in a region spanning the second exon of the hh gene and spreading about 0.4 kb on both sides of the exon. On both sites we observe a substantial overlap between PC- and GAF-binding sites. The presence of this particular arrangement of PC- and GAF-binding sites in the hh genomic region suggests that these PcG and trxG proteins directly control hh expression.
Figure 1.
Binding of PC and GAF to the hh genomic region in embryos. (A) Slot-blot hybridization. Chromatin from Drosophila wild-type embryos was either mock-immunoprecipitated or immunoprecipitated with anti-PC or anti-GAGA antibodies. Then 1-kb PCR fragments from the hh genomic region were blotted on a nylon membrane, and the immunopurified DNA was radiolabeled and used as a probe for hybridization (arrows A and B show the signals corresponding to the strongest enrichment compared to mock). (B) The graph depicts the relative enrichments of immunopurified DNA compared with mock (PC enrichment is shown in black, GAF enrichment in gray). The protein distribution shows two main peaks of PC- and GAF-binding sites. One peak is situated upstream of the transcription start site (peak A), whereas the second one spans the second exon and spreads into the neighboring introns (peak B). The transposon EP-hh is inserted 364 bp upstream of the hh transcription start site.
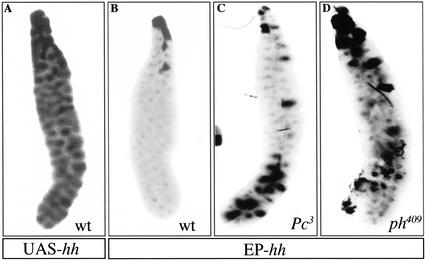
To investigate this at the functional level, we assessed the accessibility of the hh promoter region to a trans-activating factor. It is known that a PRE placed in the vicinity of an Upstream Activating Sequence (UAS) is able to counteract GAL4 binding, preventing expression of the reporter gene (Zink and Paro 1995; Fitzgerald and Bender 2001). We took advantage of the availability of an EP line possessing a UAS site close to the endogenous hh transcription start site (Rørth et al. 1998) to test whether the hh-PREs could inhibit the activation of transcription induced by GAL4. The EP3521 line (termed here EP-hh) possesses an EP transposon containing several UAS sites, and is inserted in the hh promoter region (−0.36 kb, see Fig. 1B). The endogenous hh gene is not transcribed in salivary glands. By using an hs-GAL4 line, which is known to be leaky at 25°C, weak expression of GAL4 in larval salivary glands is observed. When hs-GAL4 is crossed to a line containing UAS-hh integrated randomly in the genome, in situ stainings reveal that at 25°C, by the action of GAL4, the hh mRNA is present in high amounts in all the salivary gland cells (Fig. 2A). However, when hs-GAL4 is crossed to the EP-hh line, in which the UAS sites are juxtaposed to the presumptive PRE, hh transcription was observed in only a very few cells situated mainly at the base of the glands (Fig. 2B). We reasoned, because in most cells transcription is inhibited, that the PcG proteins binding the PREs in the vicinity of the hh promoter block the accessibility of GAL4 to the UAS sites. Accordingly, reducing the amount of some of the PcG proteins in the cells by repeating the experiment with flies heterozygous for the Pc3 allele (Fig. 2C) or with males hemizygous for the ph409 allele (Fig. 2D) induces partial derepression of transcription of the endogenous hh gene in a substantial number of gland cells. These results indicate that the repression observed in most of the salivary gland cells in the EP line is caused by the action of the PcG proteins through their binding to the identified PREs. As such, these experiments demonstrate that the transcription of hh is directly repressed by the PcG proteins.
Figure 2.
The PcG proteins repress transcription of the hh gene in salivary glands. At 25°C, the hs-GAL4 driver is leaky in salivary glands. It can activate transcription of a UAS-hh reporter construct (A). However, when using the EP-hh line (in which an EP element is inserted near the endogenous hh promoter) in the same conditions, hh transcription is observed in a very few cells only (B). Repeating the same experiment in flies heterozygous mutant for Pc3 (C) or ph409 (D) shows that hh transcription becomes derepressed in more cells in the salivary glands.
A fragment of the upstream regulatory region of hedgehog exhibits a CMM activity
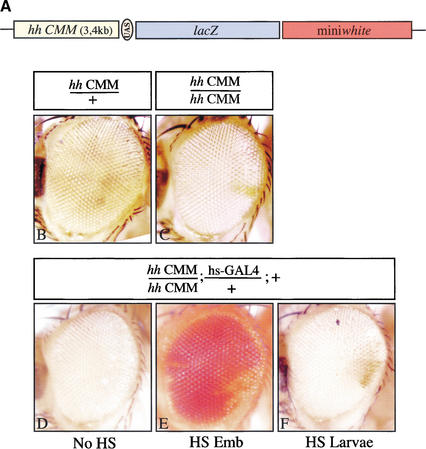
Having shown that the hh gene is controlled by the PcG proteins, we were interested to see whether the mapped PC/GAF-binding sites could function as CMMs. We produced transgenic flies using the vector that allows us to test for the maintenance of the reporter gene expression through cell divisions (Cavalli and Paro 1998). A 3.4-kb fragment, starting from position −3760 to −402 bp upstream of the hh transcription start site (according to transcript CG4637 from Flybase), and containing the PRE identified in the hh promoter region (peak A, Fig. 1), was linked to a GAL4/UAS-inducible lacZ gene (UAS-lacZ) and miniwhite as a reporter and transformation marker (Fig. 3A). Most of the lines obtained (15/22) exhibit pairing-sensitive silencing, a phenomenon often associated with PREs, when homozygous for the construct, indicated by the variegated expression of miniwhite in the eyes (Fig. 3B,C; Fauvarque and Dura 1993; Kassis 1994; Zink and Paro 1995). A short GAL4 pulse produced in these flies during embryogenesis by activation of the hs-GAL4 driver leads to homogeneous expression of the lacZ gene in the entire embryo (data not shown). When these embryos are transferred back to 21°C and are allowed to develop to adulthood, >90% of the offspring of the two lines tested displayed partial or homogeneous miniwhite derepression in the eyes (Fig. 3D,E). These results show that the upstream 3.4-kb fragment is able to maintain the initial state of transcription of the reporter gene throughout development and therefore exhibits CMM properties.
Figure 3.
A fragment of the upstream regulatory region of hedgehog exhibits a CMM activity. A 3.4-kb fragment, termed hh CMM, containing the PRE identified in the hh promoter region, was cloned into the pUZ transformation vector (A), and transgenic flies were generated. Flies heterozygous for the transgene show reduced miniwhite expression (B). This is even more pronounced in flies homozygous for the transgene depicting pairing-dependent silencing of miniwhite (C). Transgenic flies, homozygous for the hh CMM construct and containing the hs-GAL4 driver raised at 21°C have repressed miniwhite expression (D). However, when submitted to an embryonic GAL4 pulse and raised afterward at 21°C until adulthood, the activation of the reporter genes is maintained until adult stages, and flies exhibit red eye color (E). When a GAL4 pulse is given during larval stages, the activation of the reporter genes is not maintained throughout development, and miniwhite stays repressed in the eyes (F).
During imaginal disc development, hedgehog expression can be inherited through cell divisions independently of the initial trans-activator
Having shown that the hh gene is controlled by PcG proteins and that a DNA fragment upstream of the hh transcription start site can function as a CMM in a transgenic assay, we wanted to test whether the hh gene itself, in its original chromatin environment, is regulated by CMM activity during imaginal disc development, when cells undergo a high number of divisions. It is known that all wing pouch cells are progenies of the cells determined at the dorso–ventral (D–V) boundary at early larval stages (Klein 2001). We hypothesized that if the transcription of a gene possessing a CMM is activated in cells during early larval development at the D–V boundary, then transcription should be inherited to daughter cells after mitosis, resulting in expression of the gene in all wing pouch cells.
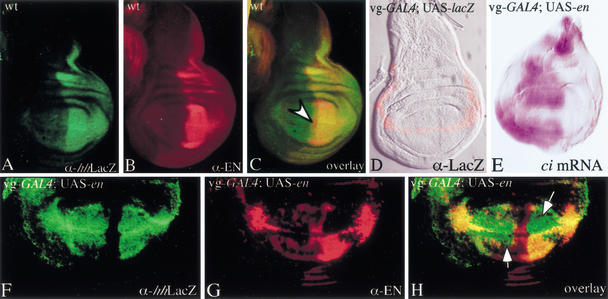
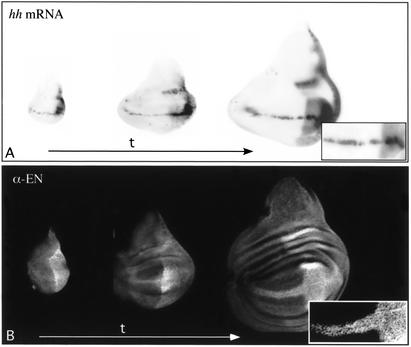
During embryonic and larval development, En induces transcription of hh in the posterior compartment of leg and wing imaginal discs, where the two factors substantially colocalize (Fig. 4A–C; Tabata et al. 1992; Guillen et al. 1995; Zecca et al. 1995). Even though it is not presently clear whether En directly activates hh expression, this regulatory feature gives us a tool to test for CMM activity at the hh gene. We expressed UAS-en at the D–V boundary using a vestigial-GAL4 driver (vg-GAL4; Simmonds at al. 1995). This transgene combination allows expression of GAL4 in a thin stripe (1 or 2 cells thick) along the D–V boundary during wing disc development (Fig. 4D). Double stainings of such late third-instar wing discs reveal that, surprisingly, En does not only induce a thin stripe of hh–lacZ expression (reflecting the hh expression pattern in the P30 enhancer trap line) in cells along the D–V boundary as expected, but also in all the posterior and anterior wing pouch cells (except in a stripe along the A–P boundary; Fig. 4F). Strong UAS-en expression is detected in cells at the D–V boundary and lower levels of En in some regions of the anterior wing pouch (Fig. 4G). The repression of the endogenous en observed in some parts of the posterior compartment is explained by the fact that high levels of En could cause repression of the endogenous en in the P compartment (Guillen et al. 1995). Strikingly, the overlay of Hh–LacZ and En stainings clearly reveals large domains, in both anterior and posterior wing pouch, with strong hh expression in the absence of En, suggesting that the transcription of hh in these cells becomes independent of En (Fig. 4H). Furthermore, it is known that En represses cubitus interruptus (ci) expression (Eaton and Kornberg 1990; Schwartz et al. 1995), and it has been shown that clones of A cells lacking Ci express low levels of Hh protein (Methot and Basler 1999). To check whether the activation of hh in the wing pouch cells is caused by the repression of ci expression by En, ci expression was examined in vg-GAL4; UAS-en wing imaginal disc. The stainings revealed that ci repression by En is restricted to the cells at the D–V boundary only (Fig. 4E), indicating that hh expression in the wing pouch cells of the A compartment is not caused by a down-regulation of ci. These observations suggest that hh expression is activated by En at the D–V boundary in early larval development, and is inherited, even in the absence of the initial trans-activator (En), through mitosis in the cells forming, in later stages, the wing pouch.
Figure 4.
UAS-en expressed at the D–V boundary induces expression of hh in most of the wing pouch cells. All discs are shown dorsal side up, with anterior to the left. In wild-type third instar wing imaginal disc, hh–lacZ (A) and en (B) are expressed in the posterior compartment. However, in late discs, Hh induces an extension of en expression into the anterior compartment (C, arrowhead). The vg-GAL4 driver induces expression of the UAS-lacZ reporter gene at the D–V boundary in wing imaginal discs (D). When UAS-en is misexpressed in a stripe along the D–V boundary using the vg-GAL4 driver, ci is only repressed at the D–V boundary by En (E). However, En is able to activate hh–lacZ expression in most of the wing pouch cells (anterior and posterior) at a constant high level (F), whereas strong UAS-en expression is detected at the D–V boundary and lower levels of EN in some regions of the wing pouch (G). The overlay (H) of hh–lacZ and en expression domains shows large regions in the wing pouch where hh–lacZ is expressed in the complete absence of En (arrows), indicating that at this stage hh expression is maintained independently of En.
hedgehog inheritance of expression in the wing imaginal disc is not caused by a positive feedback loop
hh inheritance of transcription to daughter cells could be explained alternatively by the existence of a positive feedback loop allowing continuous maintenance of hh expression. This positive feedback loop would be activated once hh is expressed, either by autoactivation or cross-activation with another factor, like En, for instance. To investigate this possibility, we misexpressed hh along the D–V boundary, using the vg-GAL4 driver and a UAS-hh transgene. Although UAS-hh is continuously strongly expressed at the D–V boundary from the second instar larval stage, in situ stainings do not reveal any inheritance of hh transcription to daughter cells, because the presence of hh mRNA is always restricted to a thin row of cells at the D–V boundary, even in late third-instar wing discs (Fig. 5A). This result demonstrates that the previously observed inheritance of hh expression in wing pouch cells of vg-GAL4; UAS-en flies is not caused by autoactivation by Hh itself nor by any positive feedback loop.
Figure 5.
Misexpression of UAS-hh at the D–V boundary induces en expression but does not activate transcription of the endogenous hh gene. The figure shows wing imaginal discs from second instar larvae to late third instar larvae. UAS-hh is strongly misexpressed at the D–V boundary by the vg-GAL4 driver, starting when the D–V boundary is established (A), but is not maintained in the progenitor cells in the wing pouch. en expression gets progressively activated at the D–V boundary in late larval development (B). The magnifications of the D–V boundary (inserts) show that in late third-instar wing imaginal disc, Hh induces en expression non-cell-autonomously. The en expression domain is broader (7 cells thick) than that of hh (2 cells thick), indicating that in late larval wing pouch cells, En is not able to activate hh expression.
Furthermore, antibody stainings in such discs display a progressive activation of en expression along the D–V boundary during development. In late third-instar larvae, a strong En signal is observed, testifying to the functional activity of the protein produced by UAS-hh. Higher magnification shows that in these discs, Hh is able to induce en expression non-cell-autonomously in a stripe of ∼7 rows of cells (Fig. 5B). However, the fact that at this stage, hh expression is only limited to a stripe of 2 rows of cells indicates that En is no longer able to induce transcription of the endogenous hh gene, in contrast with early larval stages. It implies that the low levels of En protein observed in some of the anterior wing pouch cells of vg-GAL4; UAS-en third-instar larvae (Fig. 4G) is most probably caused by a late activation of en transcription by Hh. In addition, hh expression in these cells cannot be due to activation by low or undetectable levels of En protein, because we have now shown that even strong doses of En do not activate hh transcription in this region at this stage of development.
The maintenance of the transcriptional state of hedgehog through cell division depends on PcG and trxG proteins
When UAS-en is misexpressed at the D–V boundary in a wild-type genetic background using vg-GAL4 (Fig. 6A), it induces hh expression in most of the cells of the wing pouch except in a stripe along the A–P boundary where hh seems to be repressed. Whereas UAS-en is strongly misexpressed at the D–V boundary, the endogenous en gene is weakly misactivated in some cells of the anterior wing pouch (Fig. 6B).
Figure 6.
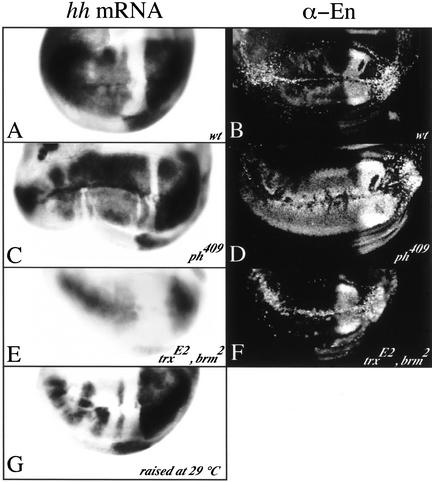
The PcG and trxG proteins control the inheritance of hh expression in the wing pouch cells. UAS-en is misexpressed using the vg-GAL4 driver in all wing discs shown. In a wild-type background, a high level of hh mRNA is detected in most of the wing pouch cells except in a stripe at the A–P boundary (A); en is expressed strongly at the D–V boundary and more weakly in some region of the wing pouch (B). In a ph409 mutant background, hh (C) and en (D) are more strongly derepressed than in wild-type flies. The stripe where hh was not expressed in a wild-type background is reduced, indicating a dependence on PH-regulation. In double heterozygous mutants for trxE2 and brm2, hh expression is activated at the D–V but is not maintained through cell divisions and progressively fades away (E). en is strongly expressed at the D–V boundary but not in the other wing pouch cells (F). For embryos raised at 29°C until the start of the second instar larval stage, hh transcription is ectopically activated in only a few clones in the wing pouch (G), indicating that, at this temperature, the Pc-repression of the hh gene is stronger and transcription is more difficult to be switched on. However, once switched on, it is inherited through cell divisions, in contrast to the trxG mutants.
Repeating the same experiment in a genetic background hemizygous mutant for an hypomorphic allele of polyhomeotic (ph409) leads to a broader domain of expression of hh (Fig. 6C). Remarkably, the region along the A–P boundary seems to be less refractory to activation of hh transcription, given that the territory of the repressed domain is reduced. Endogenous en is itself overexpressed in the anterior compartment (Fig. 6D). This is consistent with the previous findings demonstrating that its expression can be derepressed in a PcG gene mutant background (Busturia and Morata 1988; Moazed and O'Farrell 1992; Randsholt et al. 2000). In our case in the anterior wing pouch cells, the activation of en transcription by Hh is probably more efficient than in a wild-type background because en cannot be correctly silenced by PH.
The same experiment repeated in a genetic background now doubly heterozygote for the trxG genes trithorax (trxE2) and brahma (brm2) consistently shows that hh expression is activated at the D–V boundary, but can hardly be maintained through cell divisions in the anterior compartment, because in in situ staining the Hh signal progressively fades away from the D–V boundary (Fig. 6E). As expected, in such a case, en expression in the anterior compartment is restricted to the D–V boundary, because Hh might not be present in a sufficient amount to activate transcription of the endogenous en gene in the subsequent wing pouch cells (Fig. 6F).
Furthermore, it is known that PcG-mediated silencing is enhanced at higher temperature (Fauvarque and Dura 1993), and this hyperrepressed state can be inherited through cell divisions (Cavalli and Paro 1998). Based on these observations, we reasoned that raising embryos at 28°C instead of 18°C would make the Pc-mediated silencing more difficult to derepress, and influence the activation of hh transcription by En. vg-GAL4; UAS-en embryos were allowed to develop at 28°C until the beginning of second instar larvae, when the D–V boundary is established in wing discs and UAS-en is expressed there. As expected, stainings on third instar imaginal discs reveal ectopic clones of wing pouch cells expressing hh (Fig. 6G). However, the frequency of cells expressing hh is lower than in discs of larvae grown at 18°C, indicating that the Pc-mediated silencing was harder to erase at 28°C. Nevertheless, in contrast with trxG mutant flies, once the transcription has initially been activated in this case, it is maintained in the subsequent daughter cells as suggested by the presence of clones spreading from the D–V midline to the limits of the wing pouch.
These experiments demonstrate that once initiated by En, the maintenance of the transcriptional state of hh to the daughter cells can be attributed to the action of the PcG and trxG proteins. We conclude that the CMM activity of the hh upstream region we have described in the transgenic assay is also efficient when considered in its natural chromatin environment and is responsible for the inheritance of the initial transcriptional state of hh from the initiation to the completion of the wing pouch development.
The switching of a CMM during larval stages may require specific trans-activating factors
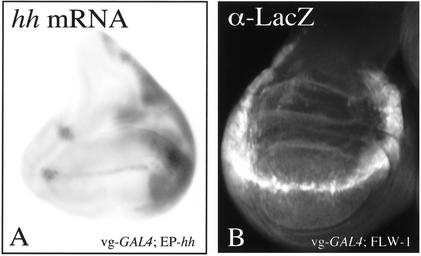
We previously reported that in the GAL4/UAS system, a GAL4 pulse, when provided in larval stages, was only able to transiently activate transcription of the reporter gene, but no heritable switching of the Fab7-CMM was observed because transcription was lost as soon as the trans-activator (GAL4) was down-regulated (Cavalli and Paro 1998). These observations led to the hypothesis that Pc-mediated silencing might be more stable in larval stages than in embryonic stages, and CMMs cannot be switched to mitotically heritable activity at these later stages. Consistent with these data, we have noticed that the upstream 3.4-kb fragment showing a CMM activity could not be switched to an active state through a GAL4 pulse produced during larval stages as demonstrated by the lack of miniwhite derepression in the eyes of the adult flies (Fig. 3G).
However, in contrast to these experiments, we have now shown that the endogenous hh CMM can be switched to an active state in larval wing pouch cells upon an En pulse. The switch occurs in second instar larval stages, when the D–V boundary is established through the action of the Notch pathway (Kim et al. 1996; Klein 2001) and GAL4 expressed by the vg driver. At this moment, en misexpression induces a switch of the endogenous hh CMM at the D–V boundary to an active state, leading to maintenance of hh transcription in all wing pouch cells. We wanted to test whether GAL4 is also able to directly switch the endogenous hh CMM, in its natural chromatin environment, in larval stages or whether this feature is restricted to specific trans-activators like En. To perform this experiment, we used the previously described line containing an EP-element inserted into the hh promoter region (EP-hh). By inducing GAL4 in the cells it is possible to activate expression of the endogenous hh gene. We postulated that, by promoting transcription of the endogenous hh gene, the hh CMM may be switched to an active state in wing pouch cells. As observed on in situ preparations of late third-instar discs, endogenous hh transcription is activated by GAL4 at the D–V boundary, but is not maintained through cell division in wing pouch cells (Fig. 7A). In comparison, also the well-characterized Fab7-CMM is itself not switched to the active state after GAL4 induction at the D–V boundary because expression of the reporter gene is not maintained in daughter wing pouch cells (Fig. 7B). We conclude that the GAL4 trans-activator is not able to switch a CMM in larval stages, although this can be carried out by the action of a gene-specific trans-activator, alone or more likely in association with other factors.
Figure 7.
The GAL4 trans-activator is not able to switch a CMM when expressed during larval stages. hh is transcribed at the D–V boundary using the EP-hh line in combination with the vg-GAL4 driver (A). However, transcription is not maintained in the daughter cells of the wing pouch. Similarly, expression of lacZ is not maintained when FLW-1 flies (Cavalli and Paro 1998) are crossed with vg-GAL4 flies (B). This indicates that at this stage the Fab7-CMM cannot be switched to the active state by GAL4.
Discussion
Initially, CMMs were found to maintain the embryonically defined expression of selector genes encoding the HOX/HOM factors, used to established long-term cellular identities. However, CMMs appear to be also used to freeze developmental decisions taken at later stages. Indeed, the expression pattern of hh is subject to substantial changes over time, depending on the morphogenetic field needed to be patterned. Yet, the finding that hh expression, once activated, is also maintained by CMM mechanisms suggests that this type of control through chromatin-based epigenetic features is much more widespread and influenced by external signals. Our results indicate that CMMs, if controlled by the correct trans-activator, can be switched and maintained in the active state at any time during development.
Developmental relevance of the presence of CMMs at the hedgehog gene and other segmentation genes
Very little is known about how the gene expression pattern of cells building compartments in imaginal discs is inherited through cell divisions. Except for some homeotic genes, it is generally assumed that auto- and cross-regulations allow selector and segmentation gene expression to be maintained until the adult stage. However, here we show that at least in the case of hh, a cellular memory system can take over to carry out the maintenance. It had already been proposed that trxG proteins might be needed to allow a proper inheritance of En expression in the cells of the posterior compartment (Breen et al. 1995). It was also suggested that a positive feedback loop between en and hh could achieve their own maintenance (de Celis and Ruiz-Gomez 1995). Our results indicate that this does not seem to be the case because the windows of time in which En can activate hh and Hh can activate en seem not to overlap over the entire wing development. During embryogenesis and early larval development (at least until the D–V boundary is established in wing disc), En is able to activate hh. We have shown that this competence disappears later, in particular in third instar larvae, when even high amounts of En cannot activate hh transcription in at least the anterior compartment of the disc. On the other hand, Hh seems to acquire the competence to activate en transcription in late larval stages. These results are consistent with the fact that in late larval stages, the Hh gradient is able to induce a stripe of en expression at the A–P boundary, whereas En does not in turn induce hh expression in this domain (Blair 1992; Strigini and Cohen 1997). Thus, because no feedback loop seems to exist, the data suggest that the hh CMM has a role in maintaining hh expression in the posterior domain during late stages of development.
We have noticed the existence of a domain along the A–P boundary that seems to be refractory to a switch of the hh CMM to an active state (see Figs. 4, 6). Interestingly, it appears that in this region Groucho and PH contribute to a strong repression system preventing hh expression from being activated in the anterior compartment in wild-type flies (de Celis and Ruiz-Gomez 1995; Maschat et al. 1998; Apidianakis et al. 2001). Thus, these proteins may counteract a stable switch of the CMM to an active state. Consistent with this result is the reduction of the thickness of this refractory domain in flies mutant for ph (Fig. 6).
It has been reported that large clones lacking en/inv expression in the posterior compartment of wing discs show reduced or no Hh protein, although this was not a universal feature of small clones (Sanicola et al. 1995; Tabata et al. 1995). Apparently, in this situation the loss of en/inv in the cells, especially when induced early in development, might cause a substantial reprogramming of the gene expression pattern leading to repression of hh, perhaps owing to the appearance of new repressors. In this case, the initially activated CMM would not be able to overcome the repression.
From our results, it is likely that CMMs have major direct roles in the inheritance of the expression of hh in the development of wing imaginal discs (we could also imagine that the well-defined en-PRE could also act as a CMM). Furthermore, hh and its vertebrate homologs are expressed in many other tissues during development, in which its activation and/or maintenance are independent of En and not yet elucidated (i.e., eye, gut, lung; Bitgood and McMahon 1995; Hoch and Pankratz 1996; Strutt and Mlodzik 1996; Warburton et al. 2000). Further studies will help us to understand how the hh CMM may be involved in regulating the gene in different tissues.
Dynamic CMM states during development
The finding that genes necessary to pattern imaginal discs can be regulated by CMMs is in disagreement with models in which the elaboration of pattern in multicellular fields is solely based on information conferred by the local concentration of secreted signaling molecules (morphogen model). In addition to this, we propose that the establishment of a specific gene expression program in cells at various developmental stages depends on both the information conferred by the morphogens surrounding the cell and its history. Thus, a cell fate will be specified by the transcriptional activation or repression of new genes, as a result of surrounding information, as well as by the maintenance of old transcriptional states established earlier and inherited by CMMs through the action of the PcG and trxG proteins. It has already been suggested that the gene optomotor-blind could be regulated by a cellular memory mechanism in imaginal discs (Lecuit et al. 1996), although it was not directly demonstrated which mechanism could allow inheritance of transcription.
It is important to note that the state of activation of a CMM does not have to be established, once and for all, during embryogenesis, but can be modified or stably switched later in development. This may be especially true for genes patterning imaginal discs for which the expression pattern is established during larval development in contrast to homeotic genes defining the A–P axis during embryogenesis. However, it seems that general trans-activating factors like GAL4, which are able to establish the active state of a CMM during embryogenesis, are not able to modify or switch the CMM state later in development, suggesting that the chromatin state of a CMM is more difficult to reprogram at late developmental stages. During larval stages, many cell divisions have been accomplished and cells are getting more and more restricted in their determination state. The chromatin could then be in a “mature” conformation stable enough to transmit a previously established transcriptional state despite the potentially contradictory actions of other transcription factors found simultaneously in the nucleus. Nevertheless, other transcription factors such as En (in the case where En directly activates hh) seem to be able, alone or by recruiting cofactors, to stably switch a CMM from a repressed to an active state during larval stages. At these stages, the switching of CMMs could require specific factors to set epigenetic marks. It could be envisaged that the En complex is able to attract some kind of chromatin-remodeling machinery that would have the potency to erase the memory and leave the chromatin competent to be reprogrammed.
In this way, it seems that the cell memory system is a complex and dynamic process during development, in which the role of CMMs is to heritably maintain a previously established transcriptional state until new specific patterning cues are able to redirect the epigenetic marks of the CMMs. However, this makes it also quite clear that during the establishment of a morphogenetic field, besides the local specifying signaling events, the previous history of a determining gene should be taken into account.
Materials and methods
DNA vectors and cloning strategy
The 3.4-kb fragment upstream of the transcription start site was amplified by PCR. aattaaccctcactaaagggagagcggccgcCGTTTT TAGTTTGCTGCCTGCATT was used as the upper primer, and taatacgactcactatagggagactagtACACTATCGCCTCGAGTT CATTCC as the lower primer (where the capital letters denote the sequence homologous to the genomic hh upstream region). Thereby, new restriction sites were created at both ends. The PCR product was digested with the NotI and SpeI restriction enzymes, and the resulting fragment was cloned via the NotI and SpeI sites into the pUZ vector (Lyko et al. 1997).
Fly strains and handling
Flies were maintained on standard culture medium at 18°C, except when stated otherwise. Embryos of the strain w1118 were used as a host for generating the transgenic lines. In a modified version of the GAL47-1 (Brand et al. 1994), the hsGAL4 construct was inserted into the CyO chromosomes, and the miniwhite marker gene was mutated with EMS. This allows the hs-GAL4 driver to be followed during crossings [gift from M. Prestel (Zentrum für Molekulare Biologie, University of Heidelberg, Germany)]. ph409 is a hypomorphic viable mutation, and Pc3 is considered to be a strong antimorph mutant. trxE2 and brm2 are two amorphic mutations, recombined on the same chromosome. The vg-GAL4 line expresses GAL4 in a thin stripe at the dorso–ventral boundary of wing imaginal discs (Simmonds et al. 1995). The hs-GAL4 line is able to produce a high amount of GAL4 protein upon heat shock. However, at 25°C it is known to be leaky in salivary glands, as low amounts of GAL4 are produced. The EP3521 line, termed here EP-hh (Rørth et al. 1998), possesses an EP element inserted upstream of the hh gene. Upon GAL4 induction, a functional Hh protein is expressed (Rørth et al. 1998). The FLW-1 line possesses the Fab7-CMM controlling expression of the reporter genes lacZ and miniwhite (Cavalli and Paro 1998). UAS-en (Guillen et al. 1995; Tabata et al. 1995) and UAS-hh (Fietz et al. 1995) are lines able to express functional En and Hh protein, respectively, upon a GAL4 pulse. The hh–lacZ line P30 (Lee et al. 1992), in which lacZ expression reflects expression of the endogenous hh gene, was used for immunostaining. For the heat-shock experiments, in order to produce a short pulse of GAL4 protein in the embryos, flies were allowed to lay overnight on apple juice agar plates at 21°C, and embryos (4–16 h old) were heat-shocked at 37°C in a waterbath for 55 min. Second instar larvae were heat-shocked in small vials incubated in a waterbath at 37°C for 1 h.
In situ hybridization
In situ hybridization to imaginal discs was performed according to the protocol of T. Wolff (2000). The hh mRNA probe was DIG-labeled using the hh cDNA cloned in pBluescript as template. The hybridization signal was detected using an anti-DIG-alkaline-phosphatase antibody.
Chromatin immunoprecipitation and dot blot analysis
The chromatin immunoprecipitation was performed following a standard procedure described in Strutt and Paro (2000). For the dot-blot, 14 primer pairs were designed for the elaboration of 1-kb-sized PCR fragments covering the hh genomic region. PCRs were performed using genomic DNA as template. After blotting the PCR products on nylon membranes, the immunoprecipitated and the mock DNA were radiolabeled, and the membranes were probed individually with their respective labeled DNA. Filters were exposed overnight to a Phosphorimager screen, and scanned. Signals were quantified by using NIH image software (version 1.62). For each dot, the intensity of the signal was quantified and the background was subtracted from it. Then, relative enrichment of the immunoprecipitated material was calculated by dividing the intensity of the signal obtained for the PC and GAF chromatin immunoprecipitations with the ones obtained for their respective mocks.
Acknowledgments
We thank N. Randsholt and S. Cohen for flies, P. Lawrence for flies and anti-EN antibodies, and I. Guerrero for the hh cDNA. We are also grateful to B. Koch and M. Hild for providing chromatin preparations, and L. Ringrose for helpful comments on the manuscript. The research of R.P. is supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL paro@sun0.urz.uni-heidelberg.de; FAX 49-6221-545891.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.242702.
References
- Apidianakis Y, Grbavec D, Stifani S, Delidakis C. Groucho mediates a Ci-independent mechanism of hedgehog repression in the anterior wing pouch. Development. 2001;128:4361–4370. doi: 10.1242/dev.128.21.4361. [DOI] [PubMed] [Google Scholar]
- Basler K, Struhl G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature. 1994;368:208–214. doi: 10.1038/368208a0. [DOI] [PubMed] [Google Scholar]
- Bitgood MJ, McMahon AP. hedgehog and Bmp genes are coexpressed at many diverse sites of cell–cell interaction in the mouse embryo. Dev Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- Blair SS. engrailed expression in the anterior lineage compartment of the developing wing blade of Drosophila. Development. 1992;115:21–33. doi: 10.1242/dev.115.1.21. [DOI] [PubMed] [Google Scholar]
- Brand AH, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. Methods Cell Biol. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- Breen TR, Chinwalla V, Harte PJ. Trithorax is required to maintain engrailed expression in a subset of engrailed-expressing cells. Mech Dev. 1995;52:89–98. doi: 10.1016/0925-4773(95)00393-f. [DOI] [PubMed] [Google Scholar]
- Brizuela BJ, Kennison JA. The Drosophila homeotic gene moira regulates expression of engrailed and HOM genes in imaginal tissues. Mech Dev. 1997;65:209–220. doi: 10.1016/s0925-4773(97)00081-6. [DOI] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- Busturia A, Morata G. Ectopic expression of homeotic genes caused by the elimination of the Polycomb gene in Drosophila imaginal epidermis. Development. 1988;104:713–720. doi: 10.1242/dev.104.4.713. [DOI] [PubMed] [Google Scholar]
- Busturia A, Lloyd A, Bejarano F, Zavortink M, Xin H, Sakonju S. The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development. 2001;128:2163–2173. doi: 10.1242/dev.128.11.2163. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- ————— Epigenetic inheritance of active chromatin after removal of the main transactivator. Science. 1999;286:955–958. doi: 10.1126/science.286.5441.955. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Ruiz-Gomez M. groucho and hedgehog regulate engrailed expression in the anterior compartment of the Drosophila wing. Development. 1995;121:3467–3476. doi: 10.1242/dev.121.10.3467. [DOI] [PubMed] [Google Scholar]
- Deschamps J, van den Akker E, Forlani S, De Graaff W, Oosterveen T, Roelen B, Roelfsema J. Initiation, establishment and maintenance of Hox gene expression patterns in the mouse. Int J Dev Biol. 1999;43:635–650. [PubMed] [Google Scholar]
- Eaton S, Kornberg TB. Repression of ci-D in posterior compartments of Drosophila by engrailed. Genes & Dev. 1990;4:1068–1077. doi: 10.1101/gad.4.6.1068. [DOI] [PubMed] [Google Scholar]
- Fauvarque MO, Dura JM. Polyhomeotic regulatory sequences induce developmental regulator-dependent variegation and targeted P-element in Drosophila. Genes & Dev. 1993;7:1508–1520. doi: 10.1101/gad.7.8.1508. [DOI] [PubMed] [Google Scholar]
- Fauvarque MO, Zuber V, Dura JM. Regulation of polyhomeotic transcription may involve local changes in chromatin activity in Drosophila. Mech Dev. 1995;52:343–355. doi: 10.1016/0925-4773(95)00412-t. [DOI] [PubMed] [Google Scholar]
- Felsenfeld AL, Kennison JA. Positional signaling by hedgehog in Drosophila imaginal disc development. Development. 1995;121:1–10. doi: 10.1242/dev.121.1.1. [DOI] [PubMed] [Google Scholar]
- Fietz MJ, Jacinto A, Taylor AM, Alexandre C, Ingham PW. Secretion of the amino-terminal fragment of the hedgehog protein is necessary and sufficient for hedgehog signalling in Drosophila. Curr Biol. 1995;5:643–650. doi: 10.1016/s0960-9822(95)00129-1. [DOI] [PubMed] [Google Scholar]
- Fitzgerald DP, Bender W. Polycomb group repression reduces DNA accessibility. Mol Cell Biol. 2001;21:6585–6597. doi: 10.1128/MCB.21.19.6585-6597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE. Mechanisms of transcriptional memory. Nat Rev Mol Cell Biol. 2001;2:409–421. doi: 10.1038/35073039. [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature. 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- Guillen I, Mullor JL, Capdevila J, Sanchez-Herrero E, Morata G, Guerrero I. The function of engrailed and the specification of Drosophila wing pattern. Development. 1995;121:3447–3456. doi: 10.1242/dev.121.10.3447. [DOI] [PubMed] [Google Scholar]
- Heemskerk J, DiNardo S. Drosophila hedgehog acts as a morphogen in cellular patterning. Cell. 1994;76:449–460. doi: 10.1016/0092-8674(94)90110-4. [DOI] [PubMed] [Google Scholar]
- Hoch M, Pankratz MJ. Control of gut development by fork head and cell signaling molecules in Drosophila. Mech Dev. 1996;58:3–14. doi: 10.1016/s0925-4773(96)00541-2. [DOI] [PubMed] [Google Scholar]
- Horard B, Tatout C, Poux S, Pirrotta V. Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol Cell Biol. 2000;20:3187–3197. doi: 10.1128/mcb.20.9.3187-3197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Martinez-Arias A. Boundaries and fields in early embryos. Cell. 1992;68:221–235. doi: 10.1016/0092-8674(92)90467-q. [DOI] [PubMed] [Google Scholar]
- Kassis JA. Unusual properties of regulatory DNA from the Drosophila engrailed gene: Three “pairing-sensitive” sites within a 1.6-kb region. Genetics. 1994;136:1025–1038. doi: 10.1093/genetics/136.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Klein T. Wing disc development in the fly: The early stages. Curr Opin Genet Dev. 2001;11:470–475. doi: 10.1016/s0959-437x(00)00219-7. [DOI] [PubMed] [Google Scholar]
- Kornberg T, Siden I, O'Farrell P, Simon M. The engrailed locus of Drosophila: In situ localization of transcripts reveals compartment-specific expression. Cell. 1985;40:45–53. doi: 10.1016/0092-8674(85)90307-1. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Brook WJ, Ng M, Calleja M, Sun H, Cohen SM. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature. 1996;381:387–393. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- Lee JJ, von Kessler DP, Parks S, Beachy PA. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell. 1992;71:33–50. doi: 10.1016/0092-8674(92)90264-d. [DOI] [PubMed] [Google Scholar]
- Lyko F, Brenton JD, Surani MA, Paro R. An imprinting element from the mouse H19 locus functions as a silencer in Drosophila. Nat Genet. 1997;16:171–173. doi: 10.1038/ng0697-171. [DOI] [PubMed] [Google Scholar]
- Maschat F, Serrano N, Randsholt NB, Geraud G. engrailed and polyhomeotic interactions are required to maintain the A/P boundary of the Drosophila developing wing. Development. 1998;125:2771–2780. doi: 10.1242/dev.125.15.2771. [DOI] [PubMed] [Google Scholar]
- Methot N, Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- Moazed D, O'Farrell PH. Maintenance of the engrailed expression pattern by Polycomb group genes in Drosophila. Development. 1992;116:805–810. doi: 10.1242/dev.116.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoulas O, Beek SJ, Moseley SL, McCallum CM, Sarte M, Shearn A, Tamkun JW. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- Randsholt NB, Maschat F, Santamaria P. polyhomeotic controls engrailed expression and the hedgehog signaling pathway in imaginal discs. Mech Dev. 2000;95:89–99. doi: 10.1016/s0925-4773(00)00342-7. [DOI] [PubMed] [Google Scholar]
- Rørth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, Weigmann K, Milan M, Benes V, Ansorge W, et al. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- Sanicola M, Sekelsky J, Elson S, Gelbart WM. Drawing a stripe in Drosophila imaginal disks: Negative regulation of decapentaplegic and patched expression by engrailed. Genetics. 1995;139:745–756. doi: 10.1093/genetics/139.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C, Locke J, Nishida C, Kornberg TB. Analysis of cubitus interruptus regulation in Drosophila embryos and imaginal disks. Development. 1995;121:1625–1635. doi: 10.1242/dev.121.6.1625. [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Simmonds AJ, Brook WJ, Cohen SM, Bell JB. Distinguishable functions for engrailed and invected in anterior–posterior patterning in the Drosophila wing. Nature. 1995;376:424–427. doi: 10.1038/376424a0. [DOI] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W, Shimell MJ, O'Connor M. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev Biol. 1993;158:131–144. doi: 10.1006/dbio.1993.1174. [DOI] [PubMed] [Google Scholar]
- Strigini M, Cohen SM. A Hedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development. 1997;124:4697–4705. doi: 10.1242/dev.124.22.4697. [DOI] [PubMed] [Google Scholar]
- Strutt DI, Mlodzik M. The regulation of hedgehog and decapentaplegic during Drosophila eye imaginal disc development. Mech Dev. 1996;58:39–50. doi: 10.1016/s0925-4773(96)00555-2. [DOI] [PubMed] [Google Scholar]
- Strutt H. “In vivo mapping of Polycomb and trithorax group proteins in chromatin of Drosophila melanogaster.” PhD thesis. UK: Open University (Great-Britain); 1997. [Google Scholar]
- Strutt H, Paro R. Mapping DNA target sites of chromatin proteins in vivo by formaldehyde crosslinking. Methods Mol Biol. 2000;119:455–467. doi: 10.1385/1-59259-681-9:455. [DOI] [PubMed] [Google Scholar]
- Strutt H, Cavalli G, Paro R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T, Eaton S, Kornberg TB. The Drosophila hedgehog gene is expressed specifically in posterior compartment cells and is a target of engrailed regulation. Genes & Dev. 1992;6:2635–2645. doi: 10.1101/gad.6.12b.2635. [DOI] [PubMed] [Google Scholar]
- Tabata T, Schwartz C, Gustavson E, Ali Z, Kornberg TB. Creating a Drosophila wing de novo, the role of engrailed, and the compartment border hypothesis. Development. 1995;121:3359–3369. doi: 10.1242/dev.121.10.3359. [DOI] [PubMed] [Google Scholar]
- Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- Tillib S, Petruk S, Sedkov Y, Kuzin A, Fujioka M, Goto T, Mazo A. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol Cell Biol. 1999;19:5189–5202. doi: 10.1128/mcb.19.7.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toftgard R. Hedgehog signalling in cancer. Cell Mol Life Sci. 2000;57:1720–1731. doi: 10.1007/PL00000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villavicencio EH, Walterhouse DO, Iannaccone PM. The sonic hedgehog–patched–gli pathway in human development and disease. Am J Hum Genet. 2000;67:1047–1054. doi: 10.1016/s0002-9297(07)62934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D, Schwarz M, Tefft D, Flores-Delgado G, Anderson KD, Cardoso WV. The molecular basis of lung morphogenesis. Mech Dev. 2000;92:55–81. doi: 10.1016/s0925-4773(99)00325-1. [DOI] [PubMed] [Google Scholar]
- Wolff T. Histological techniques for the Drosophila eye. Part I: Larva and pupa. In: Sullivan W, Ashburner M, Hawley W, editors. Drosophila Protocols. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 216–220. [Google Scholar]
- Zecca M, Basler K, Struhl G. Sequential organizing activities of engrailed, hedgehog and decapentaplegic in the Drosophila wing. Development. 1995;121:2265–2278. doi: 10.1242/dev.121.8.2265. [DOI] [PubMed] [Google Scholar]
- Zink D, Paro R. Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J. 1995;14:5660–5671. doi: 10.1002/j.1460-2075.1995.tb00253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink B, Engstrom Y, Gehring WJ, Paro R. Direct interaction of the Polycomb protein with Antennapedia regulatory sequences in polytene chromosomes of Drosophila melanogaster. EMBO J. 1991;10:153–162. doi: 10.1002/j.1460-2075.1991.tb07931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]