Abstract
The nucleolus is the site of ribosome synthesis in the nucleus, whose integrity is essential. Epigenetic mechanisms are thought to regulate the activity of the ribosomal RNA (rRNA) gene copies, which are part of the nucleolus. Here we show that human cells lacking DNA methyltransferase 1 (Dnmt1), but not Dnmt33b, have a loss of DNA methylation and an increase in the acetylation level of lysine 16 histone H4 at the rRNA genes. Interestingly, we observed that SirT1, a NAD+-dependent histone deacetylase with a preference for lysine 16 H4, interacts with Dnmt1; and SirT1 recruitment to the rRNA genes is abrogated in Dnmt1 knockout cells. The DNA methylation and chromatin changes at ribosomal DNA observed are associated with a structurally disorganized nucleolus, which is fragmented into small nuclear masses. Prominent nucleolar proteins, such as Fibrillarin and Ki-67, and the rRNA genes are scattered throughout the nucleus in Dnmt1 deficient cells. These findings suggest a role for Dnmt1 as an epigenetic caretaker for the maintenance of nucleolar structure.
INTRODUCTION
Transcription of ribosomal RNA (rRNA) genes generates pre-rRNAs that are subsequently cleaved and processed into 28S, 18S and 5.8S rRNAs. These rRNAs are thereafter packaged with ribosomal proteins to form the large and small subunits of ribosomes. There are about 400 copies of the human rRNA gene in a diploid cell. These copies are arranged in clusters of tandem repeats, known as Nucleolar Organizer Regions (NORs), distributed on the short arms of human acrocentric chromosomes (1). Transcriptional activation of NORs by RNA polymerase I (Pol I) results in the formation of a membrane-free, well-differentiated intranuclear compartment, known as the nucleolus.
The rate of rRNA gene transcription directly reflects the cellular demand of protein production. It is well known, however, that even in growing cells only a subset of rRNA genes is active (2–5). In yeast, active and inactive rRNA genes are intermingled and randomly distributed in the NOR (5). Conversely, in higher eukaryotes there is a general belief that active and silent rRNA genes are clustered, thus forming active and silent NORs (5). In both cases, the precise mechanism underlying the switching on and off of each particular rRNA gene has remained particularly elusive. The direct involvement of chromatin structure in this switching process was established several years ago (6,7), and recent evidence indicates that epigenetic mechanisms, combining both DNA methylation and histone modifications, are also important in the regulation of rRNA gene activity (3,5).
In mouse cells, DNA methylation of a single CpG in the rDNA promoter has been shown to repress Pol I transcription (8). Moreover, the nucleolar remodeling complex (NoRC), an ATP-dependent chromatin remodeling complex that regulates rRNA gene silencing at the promoter level, and particularly the PHD finger/bromodomain of TIP5, the large subunit of the complex, interacts with the DNA methyltransferases (Dnmt) 1 and with the histone deacetylase HDAC1 (9,10). These observations are consistent with the idea that active and inactive rRNA genes have different chromatin conformations (2,3,5).
To elucidate whether the DNA methylation machinery has a critical role in the maintenance of rDNA chromatin conformation and nucleolar architecture, we have focused the present work in human cells disrupted at the Dnmt1 gene (11). We have observed that DNMT1 knockout cells, but not wild-type or DNMT3b knockouts, present a severe DNA hypomethylation at ribosomal DNA associated with a loss of recruitment of the class III histone deacetylase SirT1 and an increase in the acetylation levels of lysine 16 of histone H4 at these genomic loci. Furthermore, these chromatin changes in the ribosomal DNA compartment are associated with a fragmentation of the nucleolar structure.
MATERIALS AND METHODS
Cell culture
Human HCT116 colon cancer cells and HCT116 somatic knockouts lacking Dnmt1 (11) or Dnmt3B (12) were grown in McCoy's 5A medium supplemented with 10% fetal bovine serum (Invitrogene). Knockout cells were also supplemented with 0.1 mg/ml hygromycin B (Sigma).
DNA methylation analysis
The DNA methylation status at the promoter, and 5'-end regions of the 18S and 28S components of ribosomal DNA was analyzed by bisulfite genomic sequencing as described previously (13). Both strands were sequenced and 12 clones were analyzed for each sequence. For the promoter region, an additional DNA methylation analysis using the methyl-sensitive enzyme Hpa II was also used. PCR conditions and primer sequences are available upon request.
Northern blot
Cellular RNA was prepared from 10-cm plates of logarithmically growing cells using the Qiagen RNeasy Kit as specified by the manufacturer. Five micrograms of total cellular RNA was separated by electrophoresis on 1% agarose–MOPS gels and blotted in 20× SSC onto a Hybond N+ membrane overnight. The filter was washed with 2× SSC and then dried. The RNA was UV-cross-linked (Stratalinker, Stratagene). The filter was prehybridized for 1 hr at 68°C and hybridized overnight with a 32P-labeled riboprobe that hybridizes to nucleotides 1–155 of human pre-rDNA. Radioactive signals were visualized with a PhosphoImager and quantified with the Aida program.
Nuclear run-on analysis
Nuclear run-on was performed according to the protocol described by Ashe et al. (14). Cells were harvested by centrifugation, washed with PBS and resuspended in RSB (10 mM Tris pH 7.5, 10 mM NaCl, 3 mM MgCl2 3 mM) + 0.5% NP-40. After incubation on ice (5 min), nuclei were pelleted through a cushion of RSB + 0.5% NP-40 + 10% sucrose. Nuclei were resuspended in an equal volume of transcription buffer (40 mM Tris at pH 7.9, 0.3 M NaCl, 10 mM MgCl2, 40% glycerol, 2 mM DTT). RNA was labeled by the addition of ATP, GTP and CTP (250 µM each) plus 60 µCi[-32P]-UTP (800 Ci/mmol, Amersham). Transcription reactions were carried out at 30°C for 15 min and terminated by centrifugation for 30 s. Nuclei were resuspended in HSB (10 mM Tris at pH 7.5, 0.5 M NaCl, 10 mM MgCl2) containing 10 units of RNase-free DNase and incubated at 30°C for 5 min. The reactions were deproteinized by addition of 200 µg/ml of proteinase K, 0.2% SDS, at 37°C for 30 min. RNA was extracted with phenol/chloroform, ethanol precipitated and incubated in DNase buffer (10 mM Tris at pH 7.5, 10 mM MgCl2) containing 10 units of DNase for 30 min at 37°C. RNA was again extracted, ethanol precipitated and resuspended in water. After partial RNA hydrolysis by 0.2 M NaOH on ice for 5 min, the RNA was neutralized with 0.1 M Tris/0.1 M HCl. Hybridization of the labeled RNA to a prehybridized filter was performed overnight at 42°C in 50% formamide, 6× SSPE, 5× Denhardt's solution, 0.1% SDS and 100 µg/ml of tRNA. Probes were generated for 200 bp fragments comprising the ribosomal gene promoter, 5′-NTS, 18S, 28s and tubulin. Filters were made with a slot blot (Bio-Rad) with 5 µg of ssDNA applied to each slot. Filters were washed in 1× SSPE/0.1% SDS at room temperature for 20 min and in 0.1× SSPE/0.1% SDS at 65°C for 20 min. Signals were quantitated with a Molecular Dynamics PhosphorImager.
Chromatin immunoprecipitation (ChIP)
To investigate the modification status of histones in repetitive sequences, standard ChIP assays were performed as previously described (13). Commercial antibodies anti-acetyl-K16 H4, anti-acetyl-K9 H3 and anti-dimethyl-K9 H3 were purchased from Upstate biotechnologies and anti-SIRT1 from Abcam. In all cases, chromatin was sheared to an average length of 0.25–1 kb for this analysis. PCR amplification was performed in 25 μl with specific primers for each of the analyzed sequences. For each set of primers, the sensitivity of PCR amplification was evaluated on serial dilutions of total DNA collected after sonication (input fraction). Primers and conditions for each sequence are available upon request.
Quantitative RT-PCR
Of total cellular RNA, 2.5 μg was reverse-transcribed using MMLV-RT (Invitrogen). The amounts of different RNA species were estimated using the TaqMan method in an ABIPrism 7000 Sequence Detection System (Applied Biosystems) using primer specific for the 47S (from +13 to +130) and 28S (from +8077 to +8276) rDNA sequences. Oligonucleotide sequences are available upon request.
Quantification of histone H4 acetylation by high-performance capillary electrophoresis (HPCE)
The degree of histone H4 acetylation was measured as described previously (13). Briefly, histones were extracted by acid extraction with the addition of 0.25 M HCl followed by acetone precipitation, fractionated by reverse-phase HPLC (Beckman–Coulter) on a Delta-Pak C18 column (Waters) and eluted with an acetonitrile gradient (20–60%) in 0.3% trifluoroacetic acid using an HPLC gradient system.
RNA interference of SIRT1
For SirT1 knockdown, we performed RNA interference experiments using three different siRNAs validated in human cells (Qiagen; Cat. Nos.: SI00098434, SI00098441 and SI00098448). A validated nonsilencing siRNA (Cat. No.: 10227281) was used as negative control. A mixture of SirT1 siRNAs, 30 nM each, and control siRNA, 90 nM, were transfected in HCT116 cells using Oligofectamine (Gibco) as indicated by the manufacturer. Transfected cells were harvested after 72 hr and analyzed.
Immunological methods
The following antibodies were used: rabbit polyclonal antibodies raised against Fibrillarin (Abcam) and Ki-67 (Santa Cruz Biotech), acetyl lysine 9 histone H3, acetyl lysine 16 histone H4 and SirT1 (Upstate) and mouse monoclonal against UBF (Santa Cruz Biotech.) and SirT1 (Abcam). Secondary antibodies included anti-mouse and anti-rabbit IgG horseradish peroxidase conjugates (Amersham Biosciences) and anti-mouse and anti-rabbit IgG Cy3 and Cy2 conjugates (Jackson Immunoresearch). Immunolocalization, immunoblot and immunoprecipitation and chromatin immunoprecipitation (ChIP) assays were performed essentially as described (13).
Fluorescence in situ hybridization (FISH)
For 2D analysis, cells were swollen in 0.5% trisodium citrate/0.25% KCl before fixation in methanol acetic acid (MAA; 3:1 v/v) using standard procedures. To preserve the 3D nuclear structure, cells grown on slides were washed 3× in PBS before permeabilization in CSK buffer for 5 min on ice. A plasmid probe for human rDNA was labeled by nick translation with either biotin-16-dUTP or digoxigenin-11-dUTP. A biotin-labeled paint for the p-arm of chromosome 9 was purchased from Cambio. One hundred and fifty nanograms of the chromosome paint and 10–30 µg CotI DNA (Gibco) or 200-ng-labeled rDNA probe and 3 µg Cot 1 DNA, were used per slide. FISH was performed as described previously (15,16).
AgNOR staining
Subconfluent cells grown on coverslips were fixed with absolute methanol for 6 min and subjected to staining with a drop of 50% silver nitrate in distilled water adjusted to pH 3 with formic acid, at 80°C for 2–3 min. Slides were washed in distilled water, air-dried and mounted in DePeX. The degree of dispersion of nucleolar components was assessed using ImageJ 1.32j software by (i) measuring the distance between the two most distant Ag-NOR units within nuclei (Nu), normalizing this value to the greatest diameter of the corresponding nucleus (N) and (ii) recording the number of Ag-NOR granules in each nucleus after conversion to binary images at a fixed threshold of 40 (on a 0–255 black–white scale). At least 50 and 100 cells were analyzed in each case, respectively.
Electron microscopy
Cells were grown to subconfluence in F-25 flasks, washed in 0.1 M sodium cacodylate, pH 6.9 and fixed in a solution of 2.5% glutaraldehyde in 0.1 M sodium cacodylate for 3 hr at room temperature (RT). After washing and dehydration in ethanol, samples were embedded in Epon 812. Ultrathin sections were contrasted by using the uranyl–EDTA-lead technique and examined at 80 kV under a JEOL 1010 electron microscope.
RESULTS
Loss of Dnmt1 induces rDNA gene hypomethylation without changes in the transcription rate
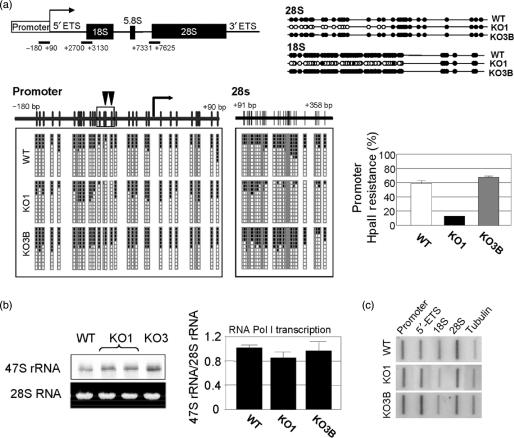
The somatic knockout cells of Dnmt1 (KO1) undergo loss of DNA methylation in DNA repeats, such as juxtacentromeric satellites, but KO1 cells do not show significant hypomethylation at the promoter CpG islands of tumor suppressor genes (11,12). We wondered how the loss of Dnmt1 could impact the methylation status of ribosomal DNA that it is distributed in tandem repeats. Although we and others have briefly studied this initial question (13,17), we have now specifically addressed this issue in a morphological and chromatin context. We first analyzed the DNA methylation status by bisulfite genomic sequencing in three regions of the rRNA gene: the proximal promoter including the transcription start site and the 5′-end sequences of the 18S and 28S regions (Figure 1a). The promoter, 18S and 28S regions of a fraction of the rRNA genes in WT cells had CpG methylation. In contrast, the 18S and 28S regions had undergone CpG hypomethylation in KO1 cells compared to the knockouts of Dnmt3b (KO3B) or to parental WT cells (Figure 1a). Interestingly, the analysis of the CpG methylation content of the rDNA promoter region showed that only a cluster of four CpG dinucleotides (positions –60, –68, –70 and –76), especially two of them (–60 and –68) were specifically demethylated in KO1 cells (Figure 1a). These observations indicate that loss of Dnmt1 in human cells affects the methylation status of specific regions of the rRNA gene repeat.
Figure 1.
DNA methylation analyses at the rRNA genes in Dnmt1 deficient cells and rDNA transcription rate. (a) Schematic representation of the rRNA gene, depicting three regions along the gene repeat: the proximal promoter, including the transcription start site, and the initial sections of the 18S and 28S regions, subjected to bisulfite genomic sequencing. Methylated and nonmethylated CpG positions are represented as black and white circles, respectively. Twelve different clones are shown. For the promoter, a cluster of four CpG dinucleotides are specifically demethylated in KO1 cells. Left, quantification of the resistance to methylation-dependent HpaII digestion in the described rDNA promoter region. (b) Left, northern-blot analysis of the expression of the precursor 47S rRNA transcript in duplicate samples. The expression of the 28S RNA was included as an internal loading control. Right, RT-PCR quantitative analysis of the expression of the 47S precursor with respect to the 28S rRNA as a measure of RNA Pol I transcription. (c) Run-on experiments of transcriptional activity in four regions of the rRNA gene. The tubulin promoter was included as control for normalization purposes.
The above results prompted us to investigate the activity of the rRNA gene using initially two approaches: a global analysis of the expression of the precursor 47S pre-rRNA by northern blot (Figure 1b) and a quantitative measure of the expression of the 47S precursor relative to the expression of the 28S RNA by quantitative RT-PCR (Figure 1b). Similar levels of 47S pre-RNA transcripts were present in all three cell lines using both methods (Figure 1b). We have further analyzed the transcription of the rRNA gene by nuclear run-on experiments. Using this approach, we found again that no significant differences were detected in the rDNA transcription activity of wild-type HCT116 in comparison with Dnmt1 or Dnmt3b knockout cells (Figure 1c). These data suggest that Dnmt1 deficiency directly affects the methylation of the rRNA gene but has no significant effect on the rDNA transcription rate. Our results on rDNA transcription are different from those presented by other group using the same cell system (17). This discrepancy can be explained by the methodologies and genomic regions used to analyze rDNA transcription.
Loss of Dnmt1 induces an increased AcK16H4 mark at rDNA gene mediated by the loss of SirT1 recruitment
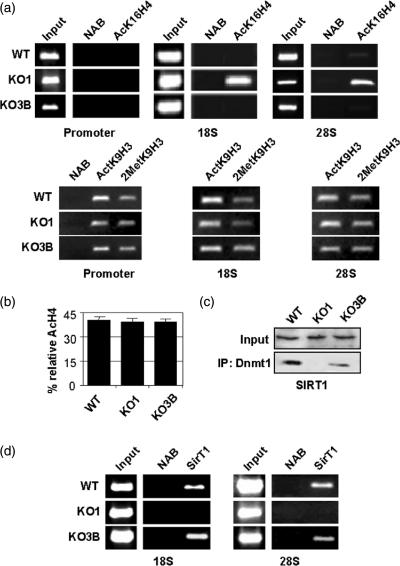
Chromatin conformation is directly affected by DNA methylation and by the biochemical modification pattern of histone tails (18,19). It is well-established that active or relaxed chromatin conformations are associated with acetylation of several lysine residues of histones H3 and H4 (18,19). Formation of long-term silenced or compacted chromatin is thought to be achieved by methylation of deacetylated lysines as well as methylated CpG residues in the DNA (20). Here, we have used ChIP assays to analyze the presence of one methylated residue, lysine 9 histone H3 (MetK9H3) and of two acetylated lysine residues, lysine 9 histone H3 (AcK9H3) and lysine 16 histone H4 (AcK16H4) at the three rDNA regions analyzed by DNA methylation. We have found equivalent amounts of dimethyl-MetK9H3 and AcK9H3 residues at the promoter, 18S and 28S rDNA regions gene in WT, KO1 and KO3B cells (Figure 2a). Strikingly, the AcK16H4 residue was strongly enriched in the 18S and 28S regions in KO1 cells compared to WT and KO3B cells (Figure 2a), but not in the promoter region (Figure 2a). However, despite the increase in the AcK16H4 content in rRNA genes, no significant differences were found between the overall degree of acetylation of histone H4 in KO1 cells and WT or KO3B cells (Figure 2b), indicating local effects in the acetylation pattern of histone tails after Dnmt1 depletion.
Figure 2.
Dnmt1 deficiency results in increased AcK16H4 and loss of SirT1 recruitment to rDNA gene regions. (a) ChIP analysis of the distribution of acetylated lysine 16 histone H4 (AcK16H4) (upper panels), acetylated lysine 9 histone H3 (AcK9H3) and di-methylated lysine 9 histone H3 (MetK9H3) (lower panels) in three regions along the rRNA gene (Promoter, 18S and 28S) showing a specific increase of AcK16H4 in the 18S and 28S regions in KO1 cells. For each protein, results are representative of at least two experiments. Input and nonantibody (NAB) lanes are also shown. (b) HPCE quantification of the overall content of acetylated histone H4 in showing no significant differences between cell lines. (c) Immunoprecipitation assay of human Dnmt1 showing its association with human sirtuin1 (SirT1), that it is lost in KO1 cells. (d) ChIP analysis of the distribution of human SirT1 in 18S and 28S regions of the rDNA genes showing loss of SirT1 occupancy in Dnmt1 deficient cells.
These results might suggest that Dnmt1 could be required for the recruitment of a histone deacetylase with preference for H4-K16 to the 18S and 28S regions of the rRNA gene. The yeast sirtuin 2 (Sir2) protein is a NAD+-dependent histone deacetylase of nucleolar localization (21). In humans, there are at least seven Sir2-like proteins (SirT1-7). Interestingly, depletion of SirT1 in human cells causes hyperacetylation of H4-K16 and H3-K9 (22) and in mouse appears to regulate rDNA transcription, in part, by deacetylation of TAFI68 (23). Using immunoprecipitation assays, we found that SirT1 was associated with human Dnmt1, and that this association was obviously lost in KO1 cells, but not in KO3B cells (Figure 2c). Interestingly, ChIP assays showed that SirT1 can bind the rDNA 18S and 28S regions, and that this binding was lost in KO1 cells (Figure 2d). These results suggest that Dnmt1 exerts a key role in the recruitment of SirT1 to the nucleolar compartment.
Dnmt1 deficient cells have a structurally disorganized nucleolus
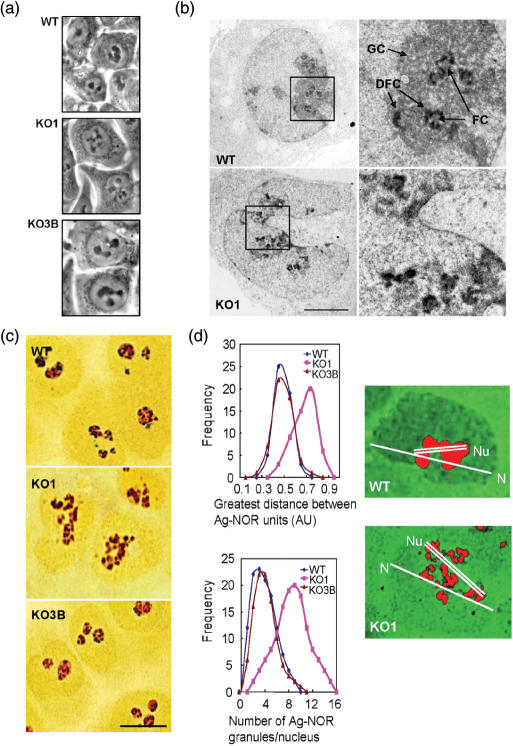
These DNA methylation and histone modification changes in ribosomal DNA prompted us to characterize in detail the morphology of the nucleolar compartment in this cell system. Under phase-contrast microscopy, living WT and KO3B cells presented between one and three large, spherical intranuclear masses corresponding to nucleoli (Figure 3a). In contrast, the nucleolar masses appeared distorted and fragmented in several small spots in KO1 cells (Figure 3a). A fine ultrastructural analysis revealed that the subcellular organization of the nucleolus was also altered in cells lacking Dnmt1. After uranyl–EDTA-staining, the Dense Fibrillar Component (DFC), the site of Pol I transcription and of maturation of pre-rRNA transcripts (2), was observed in WT and KO3B cells as strong electron-dense ring-like structures surrounding the Fibrillar Centers (FC), which are depots of rRNA genes (Figure 3b). This figure was altered in KO1 cells, in which we observed fragmented and dispersed DFCs (Figure 3b). Most evidently, the Granular Component (GC), the site of pre-ribosomal particle assembly (2), observed as a compact material surrounding the DFCs in WT and KO3B cells, was clearly diminished and disorganized in KO1 cells (Figure 3b). Selective light microscope staining of Ag-NOR proteins with silver nitrate clearly revealed the pattern of nucleolar disorganization in KO1 cells (Figure 3c). The number of Ag-NOR granules and the degree of dispersion (greatest intergranular distance) were very similar for WT and KO3B cells, whereas most KO1 cells had a more dispersed nucleolar pattern and more Ag-NOR granules (Figure 3d).
Figure 3.
Structural disorganization of the nucleolar compartment in human cells lacking Dnmt1. (a) Phase-contrast images of living cells showing the pattern of nucleolar organization in WT and KO3B cells, and the distortion of this pattern in KO1 cells, showing the fragmentation of the nucleolar compartment into several small spots. (b) Ultrastructural analysis of the nucleolar compartment after uranyl–EDTA-lead staining of ribonucleoproteins, showing the classical pattern of nucleolar organization in Fibrillar Centers (FC), Dense Fibrillar Component (DFC) and Granular Components (GC) in WT cells (similar in KO3B cells) and the distortion of this pattern in KO1 cells. Images in right panel are enlarged views of the region delimited by the squares in the left panel. Bar: 5 μm. (c) Selective Ag-NOR staining showing the morphological changes of the nucleolus in KO1 cells. Bar: 10 μm. (d) Left, degree of dispersion of nucleolar components in WT, KO1 and KO3B cells, measured as the increase of both the distance between Ag-NOR granules (upper) and the number of Ag-NOR granules per nucleus (lower). Right, example of pseudocolor images obtained from Ag-NOR-stained WT and KO1 cells, depicting the two cellular parameters, nuclear (N) and nucleolar (Nu) distances, used to calculate the degree of dispersion of nucleolar components.
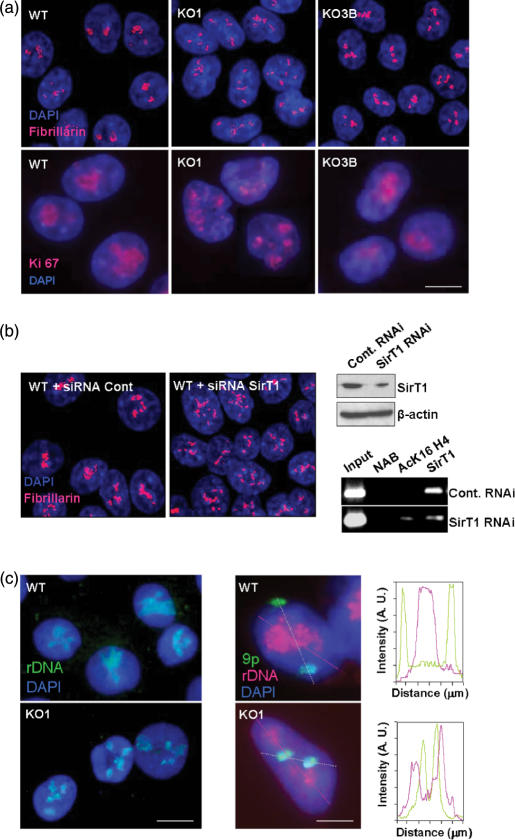
We next established the cellular distribution of Fibrillarin and Ki-67, proteins of prominent nucleolar localization. As expected, we observed that fibrillarin and Ki-67 were associated with large spherical intranuclear masses corresponding to the nucleolus in WT and KO3B cells (Figure 4a). Fibrillarin and Ki-67 were also associated with remnants of the nucleolar compartment in KO1 cells, which, in this case, exhibited the disorganization pattern of the nucleolus in several masses of reduced size (Figure 4a). Interestingly, because we have observed that SirT1 was associated with human Dnmt1 (Figure 2c), we wondered if the loss of SirT1 might cause a similar nucleolar phenotype. To test this hypothesis, we knocked-down by RNA interference experiments the SirT1 gene in HCT-116 wild-type cells (Figure 4b). ChIP analyses demonstrated that SirT1-RNAi-depleted cells presented a loss of Sirt1 occupancy and increased AcK16H4 in the described rDNA regions (Figure 4b). After 72 hr of specific knockdown targeting of SirT1, these HCT-116 cells exhibited a distorted pattern of the nucleolus in several masses of reduced size using immunolocalization of Fibrillarin, a prominent nucleolar marker (Figure 4b). The observed disorganization of the nucleolar compartment in SirT1-knocked-down cells is remarkably similar to the one we had observed in KO1 cells (Figure 4a). Thus, these results suggest that both Dnmt1 and SirT1 may act as natural epigenetic caretakers of the nucleolus structure.
Figure 4.
Immunolocalization of nucleolar proteins and FISH analyses of rRNA genes in Dnmt1 deficient cells. (a) Fibrillarin and Ki-67 immunolocalization (red) and DAPI-stained nuclei (blue). In all cell lines, fibrillarin and Ki-67 were found in close association with nucleolar masses, showing the fragmentation of the nucleolar compartment in several masses of reduced size in KO1 cells. (b) Left, RNA interference knockdown of SirT1 in wild-type HCT-116 cells induces a disorganization of the nucleolus detected by immunolocalization of the prominent nucleolar protein Fibrillarin. Right, SirT1 interference is demonstrated by western blot (top) and an illustrative ChIP analyses of 18S rRNA region demonstrates loss of Sirt1 occupancy and increased AcK16H4 in the SirT1 depleted cells (down). (c) Left panels: fluorescence-microscopy images of a 2D-FISH analysis showing the distribution of rRNA genes (green) in DAPI-stained nuclei (blue). rRNA genes were found to be grouped in one to three large, definite clusters in WT cells. In contrast, rRNA genes were dispersed in several clusters scattered throughout the nucleus in KO1 cells. Middle panels: fluorescence-microscopy images of a 3D-FISH analysis showing the distribution of rRNA genes (red) and the p-arm of the chromosome 9 (green) in DAPI-stained nuclei (blue). Right panels represent the densitometric analysis of red and green light intensity through the lines depicted in the middle panel.
Dnmt1 deficient cells have an altered rRNA gene location in the nucleolus
We next investigated the nuclear distribution of the rRNA gene repeats by FISH. We found that the rRNA genes were grouped in one to three large, definite clusters in WT cells (Figure 4c), closely corresponding to the nucleolar masses observed under phase-contrast microscopy (Figure 3a) and the immunolocalization pattern of Fibrillarin and Ki-67 (Figure 4a). In sharp contrast, rRNA genes were scattered throughout the nucleus in several clusters of reduced size in KO1 cells (Figure 4c), also resembling the spatial disorganization pattern of the nucleolar compartment observed in KO1 cells after phase contrast-microscopy (Figure 3a) or Fibrillarin and Ki-67 immunolocalization (Figure 4a). We also analyzed the nuclear distribution of rRNA genes with respect to a definite chromosome territory, the p-arm of chromosome 9 (9p), in cells whose 3D architecture was preserved. In most WT and KO3B cells, rRNA genes occupy a large and definite territory in the center of the cell, where both p9 territories are associated with the nuclear membrane and frequently adopt a radial arrangement (Figure 4c). In KO1 cells, rRNA genes are scattered throughout the nuclear volume (Figure 4c). In addition, in many KO1 cells p9 territories had lost the radial arrangement or were detached from the nuclear membrane, and were intermingled with rRNA genes (Figure 4c), suggesting that alteration of the nucleolar architecture may affect the location of other nuclear territories.
DISCUSSION
Here we have presented an example of the close relationship between epigenetic modifications of the DNA molecule and large-scale subcellular phenotypes that define the architecture of a nuclear territory. We have found that human cancer cells lacking Dnmt1 have a substantial disorganization of the nucleolar compartment that is associated with the specific loss of CpG methylation in the rRNA gene repeat. This effect can be considered specific to Dnmt1, since depletion of a related DNA methyltransferase (Dnmt3B) in the same genetic background did not yield the same phenotype. Indeed, recent characterization of the nuclear proteome indicates that Dnmt1, but not Dnmt3B, is present in the nucleolus (24).
Human cells lacking Dnmt1 showed extensive hypomethylation of CpG residues in the 5′-end sequences of the 28S and 18S regions of the rRNA gene. Coincident with this demethylation pattern, an increase in the acetylation of the lysine 16 residue of histone H4 was observed in these regions. Similar associations of DNA methylation and histone acetylation patterns have been described in other genomic regions and DNA methyltransferases are frequently found in protein complexes containing histone deacetylases (20), supporting the notion that Dnmt1 is required for the recruitment to regions inside the rRNA gene of a histone deacetylase using K16H4 as substrate. Here we describe for the first time that Dnmt1 can associate with SirT1, and the binding of SirT1 with the rRNA gene is abrogated in cells lacking Dnmt1. Consistent with this, in yeast, the mechanism responsible for rRNA silencing involves the Sir2 protein, a member of the group of Silent Information Regulatory (Sir) proteins (21). Sir2 binds to Net1, which specifically associates with rRNA genes and recruits Sir2 to the nucleolus (25). In net1▵ cells Nop1, the yeast homologue of vertebrate fibrillarin, redistributes over the entire nucleus, suggesting that the integrity of the nucleolus as a compartment has been lost. Notably, Sir2 is primarily located in the nucleolar compartment in the yeast. In humans, SirT7 is the only member of the sirtuin family which has been found stably associated with the nucleolus (26). The steady-state distribution of SirT1 is essentially nucleoplasmic, showing a very faint nucleolar location [(26) and data not shown]. This observation might imply that a transit nucleoplasmic/nucleolar shuttling is required for SirT1 in order to act in the nucleolus. The idea of a rapid nucleolar transit of SirT1 is consistent with reports in which the nucleolus is described as a dynamic compartment showing a rapid entry/exit rate of different factors (24,27,28).
In several organisms the nucleoli maintain a tight association with the heterochromatic regions from several chromosomes, including those that are devoid of a NOR, e.g. centromeres and the heterochromatic sequences proximal and distal to the NOR on the p-arms of human acrocentric chromosomes (1). We have previously shown that lack of Dnmt1 in human cancer cells results in the extensive loss of heterochromatinization of large genomic regions containing Satellite 2 repeats (11,13). The Dnmt1-dependent loss of heterochromatinization and acquisition of a euchromatic conformation was accompanied by the loss of DNA methylation and a shift in the histone H3 biochemical pattern (13). This suggests that Dnmt1 can exert its large-scale maintenance function in definite nuclear territories. We propose that, in a similar way to the Dnmt1-dependent pattern of centromeric heterochromatin, containing Satellite 2 repeats, a Dnmt1-dependent mechanism of maintenance of the structural organization also operates in the nucleolar compartment, where a large number of genomic repeats, the rRNA genes, are contained.
Therefore, the proposed action of Dnmt1 in maintaining the nucleolar compartment architecture represents another role of DNA methylation in cell biology, since the nucleolus is not only the site for ribosome assembly, but also for many other functions, such as aging, regulation of oncogene and tumor-suppressor activities, modulation of telomerase function and modification of small RNAs (29,30).
ACKNOWLEDGEMENTS
Supported by FP6 through funding by the Epigenome Network of Excellence under contract LSHG-CT-2004—503433, the Heatlh (FIS01-04) and Education and Science (I + D + I MCYT08-03 and Consolider MEC09-05) Departments of the Spanish Government and the Spanish Association Against Cancer (AECC). J.E. was a recipient of an EMBO short-term Fellowship. Funding to pay the Open Access publication charge was provided by Consolider MEC09-05.
Conflict of interest statement. None declared.
REFERENCES
- 1.Henderson AS, Warburton D, Atwood KC. Location of ribosomal DNA in the human chromosome complement. Proc. Natl. Acad. Sci. USA. 1972;69:3394–3398. doi: 10.1073/pnas.69.11.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmo-Fonseca M, Mendes-Soares L, Campos I. To be or not to be in the nucleolus. Nat. Cell Biol. 2000;2:E107–E112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- 3.Grummt I, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 2003;4:641–649. doi: 10.1038/nrm1171. [DOI] [PubMed] [Google Scholar]
- 4.McStay B. Nucleolar dominance: a model for rRNA gene silencing. Genes Dev. 2006;20:1207–1214. doi: 10.1101/gad.1436906. [DOI] [PubMed] [Google Scholar]
- 5.Santoro R. The silence of the ribosomal RNA genes. Cell. Mol. Life. Sci. 2005;62:2067–2079. doi: 10.1007/s00018-005-5110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson WF, Flavell RB. DNase I sensitivity of ribosomal RNA genes in chromatin and nucleolar dominance in wheat. J. Mol. Biol. 1988;204:535–548. doi: 10.1016/0022-2836(88)90353-1. [DOI] [PubMed] [Google Scholar]
- 7.Conconi A, Widmer RM, Koller T, Sogo JM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 8.Santoro R, Grummt I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell. 2001;8:719–725. doi: 10.1016/s1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- 9.Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 2002;32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y, Grummt I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr. Biol. 2005;15:1434–1438. doi: 10.1016/j.cub.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 11.Rhee I, Jair K-W, Chiu Yen R-W, Lengauer C, Herman JG, Kinzler KW, Vogelstein B, Baylin SB, Schuebel KE. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 12.Rhee I, Bachman KE, Ho Park B, Jair K-W, Chiu-Yen R-W, Schuebel KE, Cui H, Feinberg AP, Lengauer C, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 13.Espada J, Ballestar E, Fraga MF, Villar-Garea A, Juarranz A, Stockert JC, Robertson KD, Fuks F, Esteller M. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J. Biol. Chem. 2004;279:37175–37184. doi: 10.1074/jbc.M404842200. [DOI] [PubMed] [Google Scholar]
- 14.Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahy NL, Perry PE, Gilchrist S, Baldock RA, Bickmore WA. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J. Cell Biol. 2002;157:579–589. doi: 10.1083/jcb.200111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majumder S, Ghoshal K, Datta J, Smith DS, Bai S, Jacob ST. Role of DNA methyltransferases in regulation of human ribosomal RNA gene transcription. J. Biol. Chem. 2006;281:22062–22072. doi: 10.1074/jbc.M601155200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 19.Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- 20.Vermaak D, Ahmad K, Henikoff S. Maintenance of chromatin states: an open-and-shut case. Curr. Opin. Cell Biol. 2004;15:266–274. doi: 10.1016/s0955-0674(03)00043-7. [DOI] [PubMed] [Google Scholar]
- 21.Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy BK, Grunstein M, Gasser SM. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 23.Muth V, Nadaud S, Grummt I, Voit R. Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 2001;20:1253–1362. doi: 10.1093/emboj/20.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen JS, Lam YW, Leung AKL, Ong S, Lyon CE, Lamond AL, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 25.Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 26.Mischita E, Park JY, Burneskis JM, Barret JC, Horikawam I. Evolutionary conserved and non conserved localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;20:1075–1080. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. A kinetic framework for a mammalian RNA polymerase In Vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- 28.Mekhail K, Khacho M, Carrigan A, Hache RRJ, Gunaratnan L, Lee S. Regulation of ubiquitin ligase dynamics by the nucleolus. J. Cell Biol. 2005;170:733–744. doi: 10.1083/jcb.200506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez-Verdun D, Roussel P, Gébrane-Younès J. Emerging concepts of nucleolar assembly. J. Cell Sci. 2002;115:2265–2270. doi: 10.1242/jcs.115.11.2265. [DOI] [PubMed] [Google Scholar]
- 30.Olson MO, Dundr M. The moving parts of the nucleolus. Histochem. Cell Biol. 2005;123:203–216. doi: 10.1007/s00418-005-0754-9. [DOI] [PubMed] [Google Scholar]