Abstract
Base J or β-d-glucosylhydroxymethyluracil is a DNA modification replacing a fraction of thymine in the nuclear DNA of kinetoplastid parasites and of Euglena. J is located in the telomeric sequences of Trypanosoma brucei and in other simple repeat DNA sequences. In addition, J was found in the inactive variant surface glycoprotein (VSG) expression sites, but not in the active expression site of T. brucei, suggesting that J could play a role in transcription silencing in T. brucei. We have now looked at the distribution of J in the genomes of other kinetoplastid parasites. First, we analyzed the DNA sequences immunoprecipitated with a J-antiserum in Leishmania major Friedlin. Second, we investigated the co-migration of J- and telomeric repeat-containing DNA sequences of various kinetoplastids using J-immunoblots and Southern blots of fragmented DNA. We find only ∼1% of J outside the telomeric repeat sequences of Leishmania sp. and Crithidia fasciculata, in contrast to the substantial fraction of non-telomeric J found in T. brucei, Trypanosoma equiperdum and Trypanoplasma borreli. Our results suggest that J is a telomeric base modification, recruited for other (unknown) functions in some kinetoplastids and Euglena.
INTRODUCTION
The bloodstream form of the African trypanosome Trypanosoma brucei, the causative agent of the human sleeping sickness, is covered by a variant surface glycoprotein (VSG) coat encoded by the VSG gene family. The VSG genes are expressed in telomeric polycistronic transcription units, named expression sites. Only one of the ∼20 expression sites is expressed at a time (1). In its mammalian host, T. brucei evades the host immune system by regularly replacing the VSG expressed, a process known as antigenic variation (see (2–6) for reviews). The presence of a DNA modification in the genome of T. brucei was first evoked when the telomeric VSG expression sites were mapped with restriction enzymes (7,8). Whereas the actively transcribed expression site was fully digested, the silent expression sites were only partially digested by some enzymes (7,8). This raised the possibility that silent sites contained a DNA modification and that this modification was involved in silencing (7,8). The DNA modification was later identified as β-d-glucosylhydroxymethyluracil, or base J (9,10). Base J is synthesized in two steps: a thymidine hydroxylase first hydroxylates thymidine in DNA leading to the formation of hydroxymethyl deoxyuridine (HMdU); then a glucosyltransferase converts HMdU into J by addition of a glucose moiety (11,12).
J has been found in the genomes of kinetoplastid parasites such as T. brucei, Trypanosoma cruzi, Crithidia fasciculata, Leishmania sp. and Trypanoplasma borreli (13). It is also present in the phagotrophic marine flagellate Diplonema (13) and in Euglena (14), two organisms distantly related to the kinetoplastids. Base J is located in the telomeric repeats of all kinetoplastid parasites analyzed (13). In the genome of T. brucei, J was also found in other DNA repetitive sequences such as the 50, 70 and 177 bp repeats, the 5S rRNA and the mini-exon (spliced leader) RNA genes (10,15). Van Leeuwen et al. (10) also located J in the inactive VSG expression sites, but not in the active site, consistent with the restriction digest findings of Bernards et al. and Pays et al. (7,8). We have now investigated the distribution of base J in more detail in Leishmania species and in the phylogenetically related species C. fasciculata, and T. cruzi, all of which lack the VSG expression sites and some of the DNA repetitive sequences containing J in T. brucei. Knowing the distribution of J in these species might shed some light on the function of J.
MATERIALS AND METHODS
J-immunoprecipitation
The immunoprecipitations of J-DNA-containing fragments were done as described in (13). Briefly, the genomic DNA was sonicated to ∼500 bp fragments, incubated with the J-antiserum in a TBSTE 1× buffer containing 10 mM Tris pH 8.0, 150 mM NaCl, 0.02% Tween-20, 2 mM EDTA, 0.1 mg tRNA per ml and 1 mg BSA per ml. The DNA was immunoprecipitated with immobilized rProteinA agarose beads (RepliGen). The immunoprecipitated DNA was washed four times and spotted on a nitrocellulose membrane along with a fraction of the input DNA. The membrane was hybridized with a variety of 32P end-labeled 30 bp DNA oligonucleotides or random-primed labeled PCR probes covering sequences of interest. The hybridization signals were quantified with a FLA-3000 apparatus (Fuji) using the Bas reader version 3.14 and the Tina version 2.09 softwares. The exact sequence of the DNA oligonucleotides used to make the probes are given in the Supplementary Data, Materials and Methods section.
J-immunoblot
The detection of J-containing fragments on Southern blots was done as detailed in (13). Briefly, ∼250–500 ng of DNA was digested with various frequently cutting restriction enzymes, the DNA fragments were size-fractionated in a 0.7–0.8% agarose gel, in TBE 1× (89 mM Tris, 89 mM boric acid, 2 mM EDTA). DNA was transferred to a nitrocellulose membrane by Southern blotting (16). The membrane was blocked overnight in 5% skim milk in TBST buffer (10 mM Tris pH 8.0, 150 mM NaCl, 0.02% Tween-20). The membrane was incubated with the J-antiserum (10) for 2 h at a 1/3000 dilution in 5% skim milk in TBST and the membrane was washed for over one hour with TBST. The J-containing fragments were detected by using a goat anti-rabbit immunoglobulin HRP conjugate (Biosource) at a 1/3000 dilution in 5% skim milk in TBST. The membrane was incubated and washed for over one hour, followed by detection by enhanced chemiluminescence and autoradiography. The membrane was afterwards hybridized with a 32P end-labeled telomeric oligonucleotide (TTAGGG)5 probe in a formamide-based buffer at 42°C overnight and washed four times with 6× SSC (900 mM NaCl, 90 mM Na citrate), 0.1% SDS.
Quantitation of the amount of J found outside the telomeres in Leishmania, Crithidia and Euglena
Approximately 10 μg of genomic DNA digested with frequently cutting restriction enzymes was serially diluted by a factor 2 in order to see when the signal given by the J-antiserum is linear. The DNA fragments were size-fractionated in an agarose gel and blotted as described in the J-immunoblot section. The signals obtained with the J-immunoblots were compared with those obtained with the telomeric Southern blot. The portions of the blot reacting with the J-antiserum, but not with the telomeric probe were boxed. The optical density value in the boxed area was then divided with the optical density value obtained in the whole lane, after correction for the background. Only values in the linear range were used to quantify the amount of J found outside the telomeric repeats. The percentage of J and telomeric repeats co-migrating in the lower fraction of the gel in Euglena was obtained via similar procedures. The signals obtained from the J-immunoblots were detected with the SuperSignal Enhanced Chemiluminescence kit from Pierce, using the imaging system FluorChem v2.0 from Alpha Innotech Corporation. The signals obtained from the telomeric Southern blots were quantitated with a FLA-3000 apparatus (Fuji) using the Bas reader version 3.14 and Tina version 2.09 softwares.
RESULTS
A search for J in DNA sequences outside the telomeric repeat by J-immunoprecipitation in Leishmania
Little is known about the location of base J in kinetoplastid parasites other than T. brucei (10,15) besides that it is found in telomeric repeats (13). In order to get more information, we looked at the distribution of J in the genome of Leishmania. Using an antiserum against J-DNA (10), we immunoprecipitated J-containing DNA sequences of L. major Friedlin (the reference strain for the Leishmania genome project (17)) and hybridized them with DNA sequences of interest. Since J is found in several repetitive DNA sequences of T. brucei, we included most of the known repetitive DNA sequences of L. major Friedlin, such as the telomeric repeats, the subtelomeric repeats LST-RA, LST-RB, LST-RC, LST-RE, LST-RI and LST-RJ (18) and the 63 bp repeat found inside the ribosomal locus (19). We also looked for the presence of J in repetitive genes like the mini-exon, the rRNA18S and the rRNA24S as well as in the α-tubulin gene cluster and in the single copy gene JBP1, which encodes the J-binding protein 1 (20). The results of these experiments, are presented in Table 1. Substantial immunoprecipitation was only observed for the telomeric repeats.
Table 1.
Distribution of J in various DNA sequences of L. major Friedin as determined by anti-J immunoprecipitation
| % IPa | ± | n | |
|---|---|---|---|
| Telomeric repeats | 7.7 | 4.8 | 5 |
| LST-RA | 0.2 | 0.2 | 5 |
| LST-RB | 0.1 | 0.0 | 2 |
| LST-RC | 0.1 | 0.0 | 2 |
| LST-RE | 0.0 | 0.0 | 3 |
| LST-RI | 0.1 | 0.0 | 2 |
| LST-RJ | 0.2 | 0.3 | 5 |
| α-tubulin | 0.1 | 0.0 | 2 |
| 63 bp repeat | 0.0 | 0.0 | 2 |
| rRNA 18S | 0.1 | 0.1 | 2 |
| rRNA 24S | 0.0 | 0.0 | 2 |
| JBP1 | 0.0 | 0.0 | 2 |
| Mini-exon | 0.1 | 0.1 | 3 |
aThe percentage of immunoprecipitation (%IP) was calculated by dividing the hybridization signals in the immnunoprecipitated fraction by the total input.
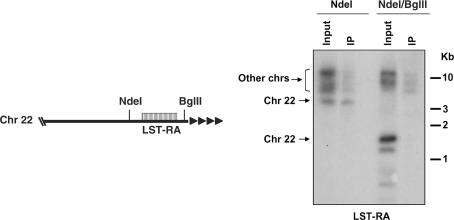
Two subtelomeric repeats, LST-RA and LST-RJ, were immunoprecipitated at a higher frequency than the other DNA sequences tested (Table 1). These subtelomeric repeats are directly adjacent to the telomeric repeats in some chromosomes (Figure 1) (18) and could have been pulled down because of their linkage to the telomeric repeats. We investigated this by immunoprecipitating, with the J-antiserum, genomic DNA digested with the restriction enzyme NdeI alone or in combination with BglII. Following a NdeI digest, the LST-RA repeats stay still linked to the telomeric repeats, whereas a NdeI-BglII digest separates the LST-RA from the telomeric repeats in the chromosome 22 of L. major Friedlin (Figure 1). Immunoprecipitated DNA fragments were size-fractionated by electrophoresis in an agarose gel, blotted and hybridized with a LST-RA probe. As shown in Figure 1, the LST-RA repeats were only immunoprecipitated when still associated with the telomeric repeats (i.e. following a NdeI digest) (Figure 1). The LST-RA repeats present in other chromosomes were probably still linked to the telomeric repeats following a NdeI-BglII double digest as they were immunoprecipitated by the J-antiserum (Figure 1). Similar data were obtained for LST-RJ in immunoprecipitation experiments where this subtelomeric repeat was dissociated from the telomeric repeats by restriction enzyme digestion (data not shown). We conclude from these data that the slightly higher percentages of immunoprecipitation seen for the LST-RA and the LST-RJ DNA repeats in Table 1 are due to the proximity of these repeats to the telomeric repeats of L. major and not to significant amounts of J in these subtelomeric sequences. The 0.1% immunoprecipitation of all other fragments is due to background immunoprecipitation observed with DNA without J (10,13,15).
Figure 1.
Determination of the presence of J in the LST-RA subtelomeric repeats of L. major Friedlin by J-immunoprecipitation of digested genomic DNA. The left panel shows the localization of the NdeI and BglII sites around the LST-RA repeats in chromosome 22 (17). The black and white rectangle depicts the LST-RA repeats, the black triangles represent the telomeric repeats. The right panel shows the result of the immunoprecipitation with the J-antiserum of NdeI or NdeI/BglII digested DNA. Immunoprecipitated fragments were size-fractionated by electrophoresis, blotted and hybridized with a DNA oligonucleotide containing the LST-RA sequence. Five percent of the amount of DNA used for the immunoprecipitation was loaded in the input lane. The bands at the top of the gel presumably represent LST-RA-containing DNA fragments from other chromosomes that are still linked to the telomeric repeats following a NdeI or a NdeI/BglII double digest. The multiplicity of shorter fragments hybridizing with the LST-RA probe in the input lane of the NdeI/BglII digest might be explained by the presence of a polymorphism in some of the LST-RA repeats in chromosome 22 in our Leishmania population or some of these fragments could come from other chromosomes.
J and telomeric repeat-containing DNA fragments of Leishmania co-migrate after electrophoresis in an agarose gel
In order to verify that the localization of J in Leishmania is mostly telomeric, we digested genomic DNA of L. major Friedlin with a combination of frequently cutting restriction enzymes (making sure that most subtelomeric LSTR repeats would be digested by these enzymes). The telomeric repeats are not digested by these enzymes, however. The digested DNA was size-fractionated by electrophoresis in an agarose gel, blotted and incubated with the J-antiserum followed by hybridization with a telomeric probe. Incubation of the membrane with the antiserum did not interfere with the subsequent hybridization of the telomeric probe (data not shown). Figure 2A shows that the fragments recognized by the J-antiserum co-migrated with telomeric repeat-containing fragments in Leishmania. Other sequences like the subtelomeric LST-RA repeats were not co-localizing with the J-containing fragments (Figure 2A). This confirms that J is not detectable outside the telomeric sequences of L. major Friedlin with our methods. As a control for this experiment, we tried to force the parasite to synthesize J in DNA sequences that are normally without J by growing L. major Friedlin on medium containing HMdU (hydroxymethyldeoxyuridine). This precursor of J is converted into HMdUTP, incorporated into the DNA and converted into J in T. brucei (11). Whereas in T. brucei J levels can be raised 20-fold with HMdU (11), feeding of Leishmania with HMdU resulted in a modest 1.5–2-fold increase in J levels (data not shown). Digestion of the DNA of HMdU fed Leishmania with frequently cutting restriction enzymes, followed by incubation with the J-antiserum and hybridization with a telomeric probe did not result in a J-DNA distribution different from that in Figure 2A (data not shown). Hence, we cannot show that non-telomeric Leishmania sequences can in principle be immunoprecipitated with J-antiserum, following HMdU feeding.
Figure 2.
Southern blot and J-immunoblot of genomic DNA of L. major Friedlin, T. brucei and E. gracilis digested with frequently cutting restriction enzymes. (A) Genomic DNA of L. major Friedlin was digested with the enzymes AluI, BsrGI, BstUI, CfoI, HaeII, HpaII, Sau3AI and TaqI and size-fractionated by electrophoresis in an agarose gel and blotted on a nylon membrane. The blot was incubated with the J-antiserum (α-J) and after analysis, hybridized with telomeric (telo) and LST-RA radioactively labeled DNA oligonucleotide probes. (B) Southern blot and J-immunoblot of genomic DNA of T. brucei, L. major, and E. gracilis. DNA of T. brucei was digested with AluI, AvaII, CfoI, HinfI, RsaI, SspI. DNA of Euglena and L. major was digested with the enzymes listed in A. The combination of restriction enzymes was optimized to digest the greatest variety of DNA repetitive sequences. The left panel shows the result after incubation with the J-antiserum (α-J). The right panel shows the result of the hybridization with the telomeric probe (telo).
As an additional control, we repeated the experiment shown in Figure 2A using genomic DNA of T. brucei and of Euglena gracilis, in which the bulk of J is located outside the telomeres (10,14,15). Figure 2B shows that a substantial portion of the J-DNA fragments present in the lower part of the gel did not hybridize with the telomeric probe in T. brucei. In E. gracilis, most of the J-containing fragments were found in the lower part of the gel, whereas the telomeric repeat-containing fragments remained predominantly in the upper part of the gel (Figure 2B). This confirms that J is mostly located outside the telomeric repeats in Euglena, as proposed by Dooijes et al. (14).
Location of J in DNA fragments of other kinetoplastid parasites
We extended our analysis of J location to other Leishmania species and kinetoplastid parasites using a similar approach to the one presented in Figure 2. J and telomeric repeat-containing fragments also co-migrated in DNA from Leishmania tarentolae, the amastigote and promastigote forms of Leishmania donovani and in DNA from C. fasciculata and T. cruzi, but not in DNA from the fish parasite T. borreli and from the horse parasite Trypanosoma equiperdum (Figure 3). As the telomeric fragments from C. fasciculata run fast through the gel (Figure 3), the possibility remained that the telomeric fraction also contained non-telomeric fragments with J. To test this possibility, we digested the DNA with a more limited set of restriction enzymes. Figure 4A shows that the fragments obtained with these digests still show strict co-migration of J-DNA with telomeric repeats in contrast to the results obtained for identical digests of T. brucei DNA (Figure 4B). We conclude from these results that in C. fasciculata the bulk of J is also located in the telomeric repeats, but that these repeats are much shorter than in the stocks of Leishmania that we have analyzed (see Supplementary Data, Results and Discussion).
Figure 3.
Southern blot and J-immunoblot of genomic DNA of various kinetoplastid parasites digested with frequently cutting restriction enzymes. DNA was digested with the restriction enzymes AluI, AvaII, CfoI, HinfI, RsaI, SspI, size-fractionated and blotted as described in Figure 2. The left panel shows the result after incubation with the J-antiserum (α-J). The right panel shows the result of the hybridization with the telomeric probe (telo).
Figure 4.
Southern blots and J-immunoblots of genomic DNA of C. fasciculata and T. brucei (bloodstream form) digested with a variety of frequently cutting restriction enzymes. The DNA was treated as in Figure 2. The blot was incubated with the J-antisera (α-J) followed by hybridization with a telomeric probe (telo). (A) Southern blot of genomic DNA of C. fasciculata. (B) Southern blot of genomic DNA of T. brucei (bloodstream form). The bands migrating at the top of the lanes 1, 2 and 4 that strongly react with the J-antisera are probably due to the 50 bp repeats as these are digested by RsaI (which was used in the lanes 3, 5, 6 and 7). Lanes 1. AluI, HpaII, BsrGI, HaeII; 2. AluI, HpaII, Sau3AI, TaqI; 3. AluI, HpaII, CfoI, RsaI; 4. BsrGI, HaeII, Sau3AI, Taq I; 5. BsrGI, HaeII, CfoI, RsaI; 6. Sau3AI, RsaI, CfoI, TaqI; 7. AluI, HpaII, BsrGI, HaeII, Sau3AI, RsaI, CfoI, TaqI. ND stands for not digested.
Determination of the sensitivity of the J-immunoblots
To determine how much J outside the telomeres we can detect with our methodology, we did a J-immunoblot with digested genomic DNA of L. major mixed in with a serial dilution of digested genomic DNA of Euglena (Figure 5). Euglena and Leishmania contains similar amounts of J [see (14,21)], but the Euglena non-telomeric J-containing DNA fragments are found in the lower portion of the agarose gel (see Figures 2B and 3). This mimics well what one would expect for J found outside the telomeres in Leishmania. In this mixing experiment, we detected a signal in our J-immunoblots until 60 ng of Euglena DNA (Figure 5). This corresponds to 12–25% of the DNA we normally use for a J-immunoblot (between 250 and 500 ng). As the J-immunoblot and telomeric Southern blot of Figure 3 gave superimposable patterns for Leishmania, Crithidia and T. cruzi, we conclude that there must be less than 12–25% of J outside the telomeric repeats in these parasites.
Figure 5.
Determination of the sensitivity of the J-immunoblots. J-immunoblot containing 250 ng of DNA of L. major Friedlin digested with AluI, RsaI, Sau3AI and mixed with 2-fold serially diluted AluI, RsaI, Sau3AI digested DNA of Euglena (starting with 250 ng of DNA). The DNA was treated as described in Figure 2.
Percentage of J outside the telomeric repeats in Leishmania and Crithidia
To determine more rigorously whether there is any J outside the telomeres in Leishmania and Crithidia, we increased the sensitivity of our assays, by raising the amount of digested genomic DNA on our blots to 5–10 μg, about 20-fold the concentration used in Figures 2, 3 and 5. With these high amounts of DNA, we detected J-containing DNA fragments that did not hybridize with the telomeric probe in L. tarentolae, L. major and C. fasciculata (Figure 6). We used serial dilutions of the DNA to quantitate the signals in Figure 6 as described in the Materials and methods section. By this approach, we found ∼1% of J is in non-telomeric J-containing DNA sequences both in L. tarentolae and in Crithidia. In L. major, the majority of the bands recognized by the J-antiserum hybridized with the telomeric probe, with the exception of a band of ∼10 kb (Figure 6B). In the light of the data obtained with L. tarentolae and as the non-telomeric J-containing DNA fragments are more evident in L. tarentolae than in the L. major J-immunoblots, we assume that the proportion of J found outside the telomeric repeats for L. major is ∼1% at most. Our attempts to isolate the non-telomeric J-containing DNA fragments in L. tarentolae by J-immunoprecipitation of gel extracted DNA fragments have been unsuccessful so far.
Figure 6.
Identification of non-telomeric J-containing DNA fragments in Leishmania and Crithidia. (A) Approximately 10 μg of AluI, Sau3AI digested DNA of L. tarentolae, (B) AluI, RsaI, Sau3AI digested DNA of L. major Friedlin and (C) BsrGI, CfoI, RsaI, HaeII digested DNA of C. fasciculata were serially diluted by a factor two and treated as described in Figure 2. The portions of the J-immunoblot containing non-telomeric J-containing DNA sequences used for the quantitation of the amount of J found outside the telomeres are depicted by the asterik symbol. The enzymes used for Leishmania permit an optimal visualization of the non-telomeric J-containing DNA fragments.
DISCUSSION
Base J was discovered in T. brucei as a DNA modification present in silenced VSG expression sites and absent in active ones, hence, its historical connection to gene silencing. Subsequently, van Leeuwen et al. showed that J is present in all simple sequence DNA repeats of T. brucei, roughly 50% being located in the telomeric repeats (10,15,22). J is also present in the telomeric repeats of other kinetoplastid parasites like Leishmania, Crithidia and T. cruzi, which lack most DNA sequences containing J in T. brucei (13). We now investigated whether J is found outside the telomeric repeats in these kinetoplastid parasites and we show that over 98% of J is found in the telomeric repeats in Leishmania and Crithidia. This suggests that J cannot be involved in gene silencing in these organisms by spreading in subtelomeric regions, as J is mostly a modification of telomeric repeat sequences. Obviously, this conclusion is as good as the methods used, which have limited sensitivity.
Sensitivity of the J-immunoprecipitations
Using the J-antiserum, we immunoprecipitated ∼7–8% of the telomeric fragments of L. major (Table 1), whereas van Leeuwen et al. reported immunoprecipitating 22% of the telomeric fragments in L. donovani (13). This could be due to a difference in the amount of J in the telomeric repeats of L. donovani and L. major or to experimental variability as seen before with this antiserum (15), which is now over ten years old (10).
The background in our J-immunoprecipitations is 0.1% (Table 1). Model experiments by van Leeuwen et al. have shown that 0.6% of DNA fragments containing one J base per 943 bp are still precipitated by the J-antiserum, i.e. well above the background observed here (10). If there is any J in the DNA sequences we tested in Leishmania, it must be less than 0.1 mole percent (i.e. 1 J per ∼943 bp). We do not think that this is a significant amount, as the J-antiserum immunoprecipitated more than 2% of the various DNA repeats tested in T. brucei (10,11,15).
The fraction of J in the telomeric repeats in Leishmania versus other organisms
The difference between J location in T. brucei and Leishmania is readily visible by comparison of the patterns obtained between our J-immunoblots and telomeric Southern blots (see Figures 2–4 and 6). Whereas the patterns are comparable with Leishmania, the J-immunoblots of T. brucei give various prominent bands that do not strongly react with the telomeric probe (see Figures 2–4). We think that these bands correspond to repetitive DNA sequences not digested to their monomeric units by the restriction enzymes used. Indeed all simple sequence repeats in T. brucei are known to be imperfect.
We did not determine the percentage of J found outside the telomeric repeats in T.cruzi, but a fraction of J is apparently found in the subtelomeric regions in this parasite (Robert Sabatini, personal communication). We assume that this fraction is less than 12–25% of J, as we would otherwise have detected it in our J-immunoblot assays. With Euglena DNA, we find that 95% of the total J signal co-migrated with only 7% of the total telomeric signal (data not shown). We do not see why J would be concentrated in this small proportion of telomeric fragments and we therefore conclude that the bulk of J is located outside the telomeric repeats, confirming the conclusion of experiments with intact cells using co-immunofluorescence/in situ hybridization (14).
Sensitivity of the J-immunoblots/telomeric Southern blot approach
By using large amounts of DNA fragments in Southern blots we detected some non-telomeric J-containing DNA sequences in Leishmania and Crithidia corresponding to ∼1% of the J in the genome. This method has limitations, as the J-containing sequences located outside the telomeres that we can detect are dependent on the restriction enzymes used. For instance, we could have missed J-containing repetitive DNA sequences not cut by the restriction enzymes used. Such sequences run among the telomeric fragments during electrophoresis and would not have been detected in Leishmania. In the present study, we used a variety of restriction enzymes, which cut the majority of the DNA repetitive sequences found in L. major Friedlin. We are therefore confident that we are not underestimating the amount of J found outside the telomeric repeats because of the set of restriction enzymes used.
The sensitivity of the J-immunoblots and telomeric Southern blots were rather similar and differed depending on the size of the DNA fragments analyzed. Longer DNA fragments still gave a signal on both blots after a 1000-fold dilution, whereas the signal of the shorter fragments was lost when they were diluted 30-fold (Figure 6). Presumably, longer fragments contain more J residues and telomeric repeats than the shorter ones. We conclude from these values that our approach was sensitive enough to detect 3% of non-telomeric J-containing DNA fragments.
Regulation of the J levels and J location in Leishmania
Although growth of L. major Friedlin or L. tarentolae on medium with HMdU resulted in a 1.5–2-fold increase in J levels in DNA, we were unable to show the formation of J outside the telomeric repeats. If the newly synthesized J were randomly distributed in the genome, we would have detected it by our J-immunoblots, which we calculated to be sensitive enough to detect 12–25% of J outside the telomeric repeats. Our speculative conclusion is that the majority of the newly synthesized J following HMdU feeding must be present in the telomeric repeats in Leishmania. This could be due to the preferential incorporation of HMdU in telomeric sequences, to preferential excision of HMdU from non-telomeric sequences or to the incomplete conversion of HMdU to J by the glucosyltransferase outside the telomeres of Leishmania. We have no evidence for any of these possibilities. Our inability to get detectable amounts of J into non-telomeric sequences of Leishmania contrasts with the situation in T. brucei where HMdU feeding results in its random incorporation in the genome followed by conversion to J (11). Whereas in T. brucei J levels can be increased or decreased by 20-fold without any major effect on cell viability (11,23), J levels cannot be substantially changed in Leishmania following HMdU or BrdU (a nucleoside analog that non-specifically inhibits J synthesis (11)) feeding. So far, we have only managed to modify J levels by at most 2–4-fold in Leishmania by feeding these nucleoside analogs (Genest,P.A. and Borst,P., unpublished data). It is therefore possible that J levels and location are strictly regulated in Leishmania and that HMdU feeding only results in an increase in J levels in the DNA sequences that already contain J.
Non-telomeric J-containing DNA sequences in Leishmania and Crithidia
As we were unable to isolate the non-telomeric J-containing DNA sequences of L. tarentolae, the identity of these sequences remains unknown. Remarkably, non-telomeric J-containing DNA sequence were much more apparent in L. tarentolae than in L. major following digestion with AluI and Sau3AI (see Figure 6). Possibly the non-telomeric J-containing repetitive DNA sequences of L. tarentolae are more effectively fragmented by these restriction enzymes than those of L. major. It is also possible that J is present in subtelomeric repeats of L. tarentolae, but not in L. major (see Table 1 and Figure 1), as these sequences differ between Leishmania species (24) and could therefore be differently modified. In L. major, a band of ∼10 kb was recognized by the J-antiserum and not by the telomeric probe following digestion with AluI and Sau3AI, (Figure 6B) and in Crithidia, we identified non-telomeric J-containing DNA fragments that were bigger than the telomeric repeat-containing fragments (Figure 6C). We think that these fragments must be of repetitive nature since they were not digested by the frequently cutting restriction enzymes used. Interestingly, a band running at the migration front strongly hybridized with the telomeric probe, but was not recognized by the J-antiserum in L. major (Figure 6B). We do not know the location of these telomeric repeats in the genome.
J function in the various kinetoplastid parasites according to its genomic location
The most plausible interpretation of our results is that the bulk of J is restricted to telomeres in Leishmania and Crithidia. These kinetoplastids are more related to each other than the ones that do have significant amounts of extra-telomeric J, as shown by the phylogenetic tree in Figure 7. We propose that J may have arisen at first in a common ancestor of the organisms shown in Figure 7 to help in the maintenance of the unique character of telomeric sequences. In some evolutionary branches such as the African trypanosomes and Euglena, J may have been recruited for functions in other repetitive sequences or for gene silencing, but in Leishmania and Crithidia it has remained mainly telomeric. In Leishmania, this telomeric function of J appears to be essential, as disruption of the gene for the J-binding protein 1, a protein specifically binding to J-DNA (20) is lethal (25). We are currently trying to use this finding to determine why Leishmania telomeres need J.
Figure 7.
Phlyogenetic tree of kinetoplastid parasites and E. gracilis adapted from (26–28). The percentage of J in the telomeric repeats that we determined for the different species is indicated.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR online.
ACKNOWLEDGEMENTS
We thank Dr Saara Vainio, Dr Robert Sabatini, Dr Stephen M. Beverley, Dr Peter J. Myler, Dr Gloria Rudenko, Dr Fred van Leeuwen, Dr Bas van Steensel, Dr Hein te Riele and Dr Jos Jonkers for critical reading of this manuscript. We are indebted to Dr Marc Ouellette and Gaetan Roy for the kind gift of L. donovani genomic DNA samples and to Dr William Martin and Dr Meike Hoffmeister for the E. gracilis genomic DNA samples. We thank Dr Wendy Gibson for her help with the phylogeny of the kinetoplastids. This work was funded by a grant from the Netherlands Organization for Scientific Research and Chemical Sciences (NWO-CW) to P.B. The authors have no financial interest conflicting with this article. Funding to pay the Open Access publication charge was provided by the Netherlands Cancer Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chaves I, Rudenko G, Dirks-Mulder A, Cross M, Borst P. Control of variant surface glycoprotein gene-expression sites in Trypanosoma brucei. EMBO J. 1999;18:4846–4855. doi: 10.1093/emboj/18.17.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borst P, Ulbert S. Control of VSG gene expression sites. Mol. Biochem. Parasitol. 2001;114:17–27. doi: 10.1016/s0166-6851(01)00243-2. [DOI] [PubMed] [Google Scholar]
- 3.Pays E, Vanhamme L, Perez-Morga D. Antigenic variation in Trypanosoma brucei: facts, challenges and mysteries. Curr. Opin. Microbiol. 2004;7:369–374. doi: 10.1016/j.mib.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Vanhamme L, Pays E, McCulloch R, Barry JD. An update on antigenic variation in African trypanosomes. Trends Parasitol. 2001;17:338–343. doi: 10.1016/s1471-4922(01)01922-5. [DOI] [PubMed] [Google Scholar]
- 5.Cross GA. Antigenic variation in trypanosomes: secrets surface slowly. Bioessays. 1996;18:283–291. doi: 10.1002/bies.950180406. [DOI] [PubMed] [Google Scholar]
- 6.Barry JD, McCulloch R. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 2001;49:1–70. doi: 10.1016/s0065-308x(01)49037-3. [DOI] [PubMed] [Google Scholar]
- 7.Bernards A, Harten-Loosbroek N, Borst P. Modification of telomeric DNA in Trypanosoma brucei; a role in antigenic variation? Nucleic Acids Res. 1984;12:4153–4170. doi: 10.1093/nar/12.10.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pays E, Delauw MF, Laurent M, Steinert M. Possible DNA modification in GC dinucleotides of Trypanosoma brucei telomeric sequences; relationship with antigen gene transcription. Nucleic Acids Res. 1984;12:5235–5247. doi: 10.1093/nar/12.13.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gommers-Ampt JH, van Leeuwen F, de Beer AL, Vliegenthart JF, Dizdaroglu M, Kowalak JA, Crain PF, Borst P. beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75:1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- 10.van Leeuwen F, Wijsman ER, Kieft R, van der Marel GA, van Boom JH, Borst P. Localization of the modified base J in telomeric VSG gene expression sites of Trypanosoma brucei. Genes Dev. 1997;11:3232–3241. doi: 10.1101/gad.11.23.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Leeuwen F, Kieft R, Cross M, Borst P. Biosynthesis and function of the modified DNA base beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol. Cell. Biol. 1998;18:5643–5651. doi: 10.1128/mcb.18.10.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulbert S, Cross M, Boorstein RJ, Teebor GW, Borst P. Expression of the human DNA glycosylase hSMUG1 in Trypanosoma brucei causes DNA damage and interferes with J biosynthesis. Nucleic Acids Res. 2002;30:3919–3926. doi: 10.1093/nar/gkf533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Leeuwen F, Taylor MC, Mondragon A, Moreau H, Gibson W, Kieft R, Borst P. beta-D-glucosyl-hydroxymethyluracil is a conserved DNA modification in kinetoplastid protozoans and is abundant in their telomeres. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2366–2371. doi: 10.1073/pnas.95.5.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dooijes D, Chaves I, Kieft R, Dirks-Mulder A, Martin W, Borst P. Base J originally found in kinetoplastida is also a minor constituent of nuclear DNA of Euglena gracilis. Nucleic Acids Res. 2000;28:3017–3021. doi: 10.1093/nar/28.16.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Leeuwen F, Kieft R, Cross M, Borst P. Tandemly repeated DNA is a target for the partial replacement of thymine by beta-D-glucosyl-hydroxymethyluracil in Trypanosoma brucei. Mol. Biochem. Parasitol. 2000;109:133–145. doi: 10.1016/s0166-6851(00)00247-4. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Russell DW. Molecular Cloning. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Pess; 2001. [Google Scholar]
- 17.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, et al. The genome of the kinetoplastid parasite. Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunkin SM, Kiser P, Myler PJ, Stuart K. The size difference between Leishmania major Friedlin chromosome one homologues is localized to sub-telomeric repeats at one chromosomal end. Mol. Biochem. Parasitol. 2000;109:1–15. doi: 10.1016/s0166-6851(00)00215-2. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Calvillo S, Sunkin SM, Yan S, Fox M, Stuart K, Myler PJ. Genomic organization and functional characterization of the Leishmania major Friedlin ribosomal RNA gene locus. Mol. Biochem. Parasitol. 2001;116:147–157. doi: 10.1016/s0166-6851(01)00310-3. [DOI] [PubMed] [Google Scholar]
- 20.Cross M, Kieft R, Sabatini R, Wilm M, de Kort M, van der Marel GA, van Boom JH, van Leeuwen F, et al. The modified base J is the target for a novel DNA-binding protein in kinetoplastid protozoans. EMBO J. 1999;18:6573–6581. doi: 10.1093/emboj/18.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toaldo CB, Kieft R, Dirks-Mulder A, Sabatini R, van Luenen HG, Borst P. A minor fraction of base J in kinetoplastid nuclear DNA is bound by the J-binding protein 1. Mol. Biochem. Parasitol. 2005;143:111–115. doi: 10.1016/j.molbiopara.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Leeuwen F, Wijsman ER, Kuyl-Yeheskiely E, van der Marel GA, van Boom JH, Borst P. The telomeric GGGTTA repeats of Trypanosoma brucei contain the hypermodified base J in both strands. Nucleic Acids Res. 1996;24:2476–2482. doi: 10.1093/nar/24.13.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cross M, Kieft R, Sabatini R, Dirks-Mulder A, Chaves I, Borst P. J-binding protein increases the level and retention of the unusual base J in trypanosome DNA. Mol. Microbiol. 2002;46:37–47. doi: 10.1046/j.1365-2958.2002.03144.x. [DOI] [PubMed] [Google Scholar]
- 24.Fu G, Barker DC. Characterisation of Leishmania telomeres reveals unusual telomeric repeats and conserved telomere-associated sequence. Nucleic Acids Res. 1998;26:2161–2167. doi: 10.1093/nar/26.9.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genest PA, ter Riet B, Dumas C, Papadopoulou B, van Luenen HG, Borst P. Formation of linear inverted repeat amplicons following targeting of an essential gene in Leishmania. Nucleic Acids Res. 2005;33:1699–1709. doi: 10.1093/nar/gki304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson AG, Stevens JR, Lukes J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Hamilton PB, Stevens JR, Gaunt MW, Gidley J, Gibson WC. Trypanosomes are monophyletic: evidence from genes for glyceraldehyde phosphate dehydrogenase and small subunit ribosomal RNA. Int. J. Parasitol. 2004;34:1393–1404. doi: 10.1016/j.ijpara.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 28.Yurchenko V, Lukes J, Xu X, Maslov DA. An integrated morphological and molecular approach to a new species description in the Trypanosomatidae: the case of Leptomonas podlipaevi n. sp., a parasite of Boisea rubrolineata (Hemiptera: Rhopalidae) J. Eukaryot. Microbiol. 2006;53:103–111. doi: 10.1111/j.1550-7408.2005.00078.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.