Abstract
The early cell divisions of Xenopus laevis and other metazoan embryos occur in the presence of constitutively high levels of the cell cycle regulator cyclin E1. Upon completion of the 12th cell division, a time at which many maternal proteins are downregulated by deadenylation and destabilization of their encoding mRNAs, maternal cyclin E1 protein is downregulated while its mRNA is polyadenylated and stable. We report here that stable polyadenylation of cyclin E1 mRNA requires three cis-acting elements in the 3′ untranslated region; the nuclear polyadenylation sequence, a contiguous cytoplasmic polyadenylation element and an upstream AU-rich element. ElrA, the Xenopus homolog of HuR and a member of the ELAV gene family binds the cyclin E1 3′UTR with high affinity. Deletion of these elements dramatically reduces the affinity of ElrA for the cyclin E1 3′UTR, abolishes polyadenylation and destabilizes the mRNA. Together, these findings provide compelling evidence that ElrA functions in polyadenylation and stabilization of cyclin E1 mRNA via binding these elements.
INTRODUCTION
The first 12 cleavage divisions in Xenopus laevis embryos are rapid and occur in the absence of detectable RNA synthesis. At the end of these divisions (∼7 h post-fertilization, hpf), the midblastula transition (MBT) begins, characterized by the initiation of global zygotic transcription and slowing of the cell cycle (1,2). Prior to the MBT, synthesis and degradation of maternal cyclins, whose mRNAs are transcribed and stored in the oocyte, drive progression through the cell cycle. At the MBT, several maternal cyclins are no longer synthesized, resulting in remodeling of the embryonic cell cycle. Cyclin E1 protein is distinct from other maternal cyclins in that it is present at constitutively high levels during the first 12 cell cycles and is abruptly degraded at the MBT (3–5). How high levels of cyclin E1 are maintained and then abolished are largely unknown.
We have shown that translation of polyadenylated cyclin E1 mRNA contributes to constitutively high protein levels before the MBT (6,7). At the MBT, cyclin E1 mRNA remains polyadenylated and stable, despite the disappearance of cyclin E1 protein (3,5). This behavior is in distinct contrast to that of cyclins A1 and B2, who are synthesized and degraded during each cell cycle and disappear after the MBT due to rapid deadenylation and destabilization of their mRNAs (7,8). Their 3′ untranslated regions (3′UTRs) specify the distinct behavior of these maternal cyclin mRNAs (7). In addition, RNA gel mobility shift and UV-cross-link assays showed that a protein of ∼36 kDa from Xenopus embryo extracts bound the cyclin E1 3′UTR but not the 3′UTR of cyclin A1 or B1 (unpublished). The purpose of the current study is to identify this 36-kDa binding protein.
The cyclin E1 3′UTR contains several U- and AU-rich regions. These include an AU-rich element (ARE) of the sequence U5AU5AU6A2 at position 1684, which is 45-nt upstream of the nuclear polyadenylation sequence or NPS (A2UA3), and a U15 contiguous with the NPS, which fits the definition of a cytoplasmic polyadenylation element (CPE, 9). CPEs are U-rich sequences that specify polyadenylation in Xenopus oocytes (maturation specific) or embryos (embryonic specific) and require the NPS for activity.
The 60-kDa CPE-binding protein (CPEB) binds maturation-specific CPEs and is essential for cytoplasmic polyadenylation (10,11). The 36-kDa ElrA, a member of the ELAV family of RNA-binding proteins, has been shown to bind two mRNAs depending on the presence of embryonic-specific CPEs, but a function in polyadenylation has not been shown (12). The majority of CPEB is degraded at fertilization but ElrA is present at all times during Xenopus development (13). ElrA did not bind one maturation-specific CPE tested, that of histone B4 (12). HuR is the human homolog of ElrA and it binds AREs in the 3′UTRs of mRNAs to stabilize them (14). Since a 36-kDa protein uniquely bound the cyclin E1 3′UTR, we test here whether it is ElrA. We show that ElrA binds the 3′UTR of cyclin E1 mRNA in a manner depending on the ARE and CPE that function in polyadenylation and stabilization.
MATERIALS AND METHODS
Embryos and microinjections
Embryos were obtained and staged according to standard protocols and maintained in 0.1 × Marc's modified Ringer's medium at 23°C (15,16). For mRNA microinjections, two-cell embryos were injected with 27.6 nl of in vitro-transcribed capped mRNA in water (100 000 c.p.m./μl). Embryo samples were collected at the indicated time points. Total RNA from 5 embryos was extracted with TRIzol (Invitrogen). Embryo protein extracts were prepared as previously described (8).
3′UTR constructs and in vitro transcription
pGbA1 and pGbE1 were previously described (17). pT7E1pA was constructed by first generating the cyclin E1 3′UTR by amplifying nucleotides 1387–1784 (Genbank Accession Z13966) by PCR with primers E1-KF ggggtacccAGTGCTTTAACTCTG-TGC and E1-BR cgggatccAAAAA-AAACAGCTGTCTTCTAAACAGC. The 5′ and 3′ primers contained a KpnI and BamHI site (underlined), respectively. The resulting PCR product was digested with KpnI and BamHI and inserted between these sites in pGbORF/mosEDEN, which deleted the EDEN sequence. pT7E1 was derived from the previously described pGbE1 (7) by digesting with EcoRI and XbaI to excise the globin ORF and recircularizing the plasmid. pT7E1 was used to create cyclin E1 3′UTR deletion mutants. pT7E1Δ1575–1784 was constructed by deleting the AvaI/EcoRV fragment and then recircularizing the plasmid. pT7E1Δ1387–1571 was constructed by deleting the XbaI/AvaI fragment from pT7E1 and recircularizing the plasmid. Smaller deletions were made using the Stratagene QuikChange mutagenesis protocol using primers (sense strand primers listed): pT7E1Δ1749–1754, ATCTATCTTTTTTTTTTTTTTTGATGCTGTTTAGA AGACAGC; pT7E1ΔCPE (Δ729-1754), GAATGCTGC TACATATCGATGCT-GTTTAGAAG; pT7E1ΔARE/CPE/NPS (Δ1684-1754), CTGGCATGAGTGTTGCCG ATGCTGTTTAGAAGACAGC; pT7E1ΔARE+/CPE (Δ1648-1748), CAGATGAGCTAATCAAAGTGCAAT AAAGATGCTGTTTAGAAG; pT7E1ΔARE (Δ1684-1715), GAATCTGGCATGAGTGTTGCCTGCTGCTA CATATCTATC. For in vitro transcription, pT7E1Δ1575-1784 was linearized with Pvu II. All other plasmids were linearized with EcoRV. 32P-labeled, capped mRNAs were synthesized using T7 polymerase according to the manufacturer's recommendations (Promega).
GST-ElrA purification
GST-ElrA was purified using glutathione-sepharose beads (GS) following the manufacturer's protocol (Pharmacia). Briefly, a 300 ml culture of E. coli dH5α, transformed with pGEX-ElrA, was induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 3 h at room temperature. Bacteria were pelleted at 4000 × g, resuspended in lysis buffer, incubated on ice for 30 min and then briefly sonicated. Lysed bacteria were centrifuged and GST-ElrA was purified by incubation of the supernatant with GS-beads. The amount of GST-ElrA obtained was quantitated by Bradford assay. pGEX-ElrA was previously described (12).
Gel shifts, UV-cross-link assays and determination of Kd
Gel shifts were done according to Westmark et al. (18). Briefly, 250 ng of bacterially expressed GST-ElrA or protein from one-fifth of an embryo was incubated with the indicated 32P-labeled RNA (4 fmol) in gel shift buffer (15 mM HEPES pH 8, 10% glycerol, 10 mM KCl, 1 mM DTT, 1 unit/μl RNasin and 200 ng tRNA) in a total volume of 10 μl for 15 min at room temperature. Here, 20 U of RNase T1 was added, and samples digested for 20 min at 37°C. Samples were resolved on 6% native polyacrylamide gels. Gels were fixed, dried and visualized by phosphorimaging. For gel shift competition assays, unlabeled competitor RNA (cyclin A1 3′UTR, Genbank Accession P18606) was added along with the radiolabeled RNA in the specified amounts. For Kd determinations, gel shift assays were performed as described above except that 1 fmol of 32P-labeled RNA was incubated with the indicated concentration of GST-ElrA for 15 min and then analyzed on native polyacrylamide gels (RNase T1 was not used). Free and bound RNA were quantified using ImageQuant (Molecular Dynamics). Free RNA was defined as the position where the RNA migrated in the absence of protein. UV-cross-link assays were done similarly to gel shift assays except that following treatment with RNase T1, samples were UV-cross-linked for 5 min on ice using a Stratalinker 1800 (Stratagene) and resolved on a denaturing 10% SDS-PAG.
Immunoprecipitation
Five embryos injected with radiolabeled cyclin E1 3′UTR at the two cell stage (between 1.5 and 2 hpf) were collected at 2 and 6 h post-injection (4 and 8 hpf). Protein extracts were prepared from embryos as previously described (8), diluted to a final volume of 500 μl in IP buffer (15 mM HEPES pH 8, 10% glycerol, 10 mM KCl, 1 mM DTT, 200 ng tRNA, 320 U RNasin) and incubated 15 min at room temperature. Samples were then treated with 400 U of RNase T1 (20 μl of 20 U/μl) for 30 min at 37°C and UV-cross-linked for 5 min on ice. Either anti-HuR (raised against the human protein, Santa Cruz, 3A2), anti-ElrA (raised against the Xenopus protein, Drs Peter Good and Michael Sheets) or pre-immune serum was added and samples were incubated at 4°C for 1 h. Next, 15 μl of protein G plus agarose (Santa Cruz) was added and samples were rotated at 4°C for 1 h. Four washes were performed with IP wash buffer (15 mM HEPES pH 8, 10% glycerol, 10 mM KCl, 1 mM DTT, 200 ng tRNA and 1% NP-40). Protein G beads were resuspended in SDS reducing buffer, heated to 95°C for 5 min, and supernatants resolved on a 10% SDS-PAG. The gel was fixed, dried and visualized by phosphorimaging.
Poly(A) tail analysis
Total RNA was extracted from five embryos injected with radiolabeled, capped RNAs and resolved by electrophoresis on 6% polyacrylamide/8 M urea gels as described previously (19). Gels were fixed, dried and visualized by phosphorimaging.
RESULTS
ElrA binds to the 3′UTR of cyclin E1 mRNA
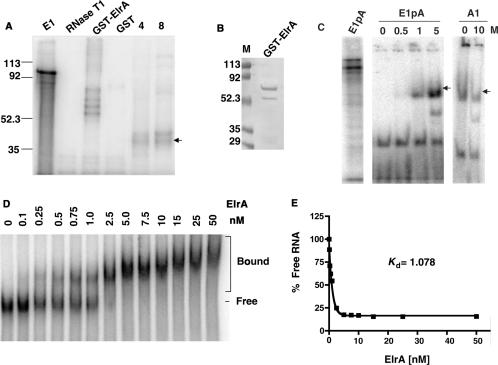
UV-cross-link assays with protein extracts of embryos either before (4 h or stage 7) or after (8 h or stage 9) the MBT showed that the 36-kDa protein bound both before and after the MBT (Figure 1A, lanes labeled 4 and 8). Since ElrA is also present both before and after the MBT, we asked if it can bind the cyclin E1 3′UTR. To determine if ElrA can bind to the cyclin E1 3′UTR, we performed UV-cross-link assays with GST-ElrA and a radiolabeled mRNA consisting of only the cyclin E1 3′UTR (E1). When recombinant GST-ElrA was prepared, it eluted as a doublet, with the upper band ∼62 kDa and the faster migrating band at ∼55 kDa (Figure 1B). This faster migrating band is most likely a C-terminal cleavage product of the full-length GST-ElrA that lacks RRM3. The UV-cross-link assay shown in Figure 1A demonstrates that GST-ElrA bound the cyclin E1 3′UTR. In contrast, GST did not bind suggesting that ElrA specifically targets the 3′UTR.
Figure 1.
GST-ElrA binds the 3′UTR of cyclin E1 mRNA. (A) UV-cross-link assay: radiolabeled cyclin E1 3′UTR (E1) was incubated with GST-ElrA, GST or 4 and 8 h embryo extracts. E1 lane is the RNA without added protein or RNase T1. RNase T1 lane is E1 digested with this enzyme in the absence of added protein. Molecular weight markers are indicated to the left of the phosphorimage in kDa. The arrow indicates a 36-kDa protein that binds in embryo extracts. n = 5. (B) Comassie blue stained gel of purified GST-ElrA. Molecular weight markers are indicated to the left of the gel in kDa. (C) Gel shift competition assay: GST-ElrA was incubated with radiolabeled cyclin E1 3′UTR containing a poly(A)65 tail (E1pA) in the presence of increasing molar excess (M) of either unlabeled E1pA or an unlabeled competitor RNA (A1, cyclin A1 3′UTR). E1pA lane is the RNA alone without added protein or RNase T1. Arrow indicates ElrA bound mRNA. Samples were electrophoresed on native gels and analyzed by phosphorimaging; the digested unbound RNA running at the bottom of the gel is not shown. Experiment was also performed in the absence of a poly(A)65 tail with identical results. n = 3. (D) Binding assay to determine the dissociation constant of GST-ElrA for the cyclin E1 3′UTR. Radiolabeled cyclin E1 3′UTR (1 fmol) was incubated with GST-ElrA over a range of 0–50 nM. Reactions were electrophoresed on native gels and analyzed by phosphorimaging. The bracket shows bound RNA and the line shows free RNA (n = 4). The results of the binding assay were quantitated to determine the dissociation constant (Kd,) shown in panel (E).
To test if recombinant ElrA binds any AU-rich sequence or specifically binds the cyclin E1 3′UTR, we performed a gel shift competition assay using the cyclin A1 3′UTR as competitor (Figure 1C). The cyclin A1 3′UTR contains a maturation-specific CPE contiguous with the NPS and two AREs of the consensus sequence U2AU3AU2. GST-ElrA was incubated with rabiolabeled cyclin E1 3′UTR either with or without a poly(A)65 tail (E1pA or E1) in the presence of increasing amounts of competitor; either unlabeled E1pA, E1, or unlabeled cyclin A1 3′UTR (A1). Results were the same in the presence or the absence of the poly(A) tail, so are shown for only E1pA. While a one molar excess of unlabeled E1pA effectively competed off the majority of bound 32P-labeled E1pA, a ten molar excess of unlabeled cyclin A1 3′UTR (A1), was unable to compete for ElrA binding. This data shows that ElrA bound specifically to the cyclin E1 3′UTR and in agreement with prior HuR studies (20,21), its binding did not require a poly(A) tail, suggesting that ElrA can bind prior to polyadenylation.
We next performed native gel shifts to determine the affinity of ElrA for the cyclin E1 3′UTR (Figure 1D). 32P-labeled cyclin E1 3′UTR was incubated with increasing amounts of GST-ElrA over a range of 0–50 nM. Complex formation began with 0.1 nM of GST-ElrA and steadily increased, with complete binding occurring with 5 nM. ElrA bound to the 3′UTR of cyclin E1 with an apparent Kd of 1 nM (Figure 1E). Overall, these data show that the cyclin E1 3′UTR is bound by ElrA with high affinity in vitro.
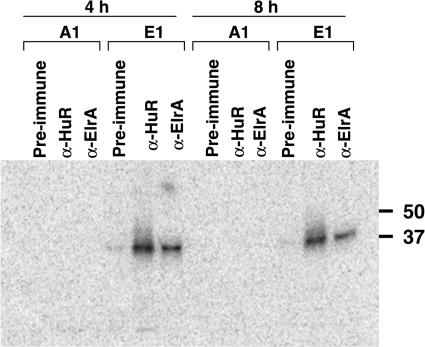
To ask if endogenous ElrA binds the cyclin E1 3′UTR, we immunoprecipitated protein–RNA complexes with antibody raised against human HuR or Xenopus ElrA (13). Radiolabeled cyclin E1 or cyclin A1 3′UTR was microinjected into 2-h (2 cell) embryos and protein extracts were prepared at 4 or 8 h. Protein extracts were UV-cross-linked and then immunoprecipitated with anti-HuR, anti-ElrA or pre-immune serum. Both antibodies immunoprecipitated RNA–protein complexes with the cyclin E1 but not the cyclin A1 3′UTR (Figure 2), showing that endogenous ElrA associates specifically with the 3′UTR of cyclin E1 both before and after the MBT.
Figure 2.
Endogenous ElrA binds the 3′UTR of cyclin E1 mRNA. Radiolabeled cyclin E1 3′UTR or cyclin A1 3′UTR was microinjected into two cell-embryos between 1.5 and 2 h post-fertilization (hpf) and time points were taken at 4 and 8 h. Protein extracts were prepared, treated with RNase T1, and UV-cross-linked to preserve RNA–protein complexes. Cross-linked embryo extracts were immunoprecipitated with either anti-HuR, anti-ElrA or pre-immune serum as described in the materials and methods section. Phosphorimages of the immunoprecipitated RNA–protein complexes are shown. Molecular weight markers are indicated to the right in kDa. n = 3.
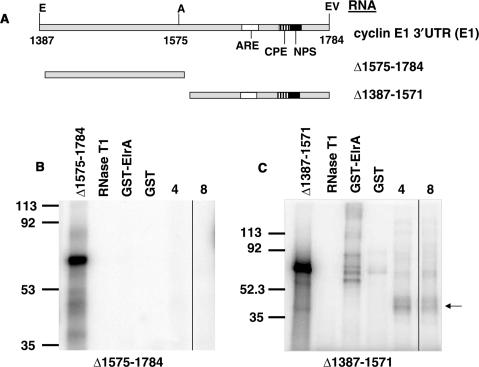
ElrA binds the distal part of the cyclin E1 3′UTR
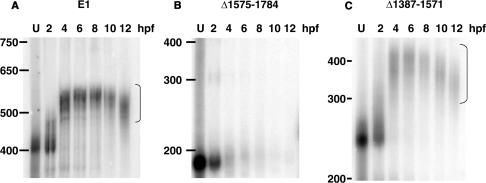
Since the distal part of the cyclin E1 3′UTR contains an ARE and a potential CPE, we next asked if ElrA binds this region. The cyclin E1 3′UTR sequence with these regions indicated is shown in Supplementary Figure S1. We deleted either the distal (▵1575–1784) or the proximal (▵1387–1571) part of the cyclin E1 3′UTR (Figure 3A) and assessed the effect on GST-ElrA binding by UV-cross-link analyses (Figure 3B and C). ElrA did not bind the proximal part of the cyclin E1 3′UTR (Figure 3B). In contrast, ElrA bound the distal part equally as well as it bound the full-length 3′UTR (compare Figure 3C and 1A). Similarly, the distal part of the cyclin E1 3′UTR bound the 36-kDa protein similar to the full-length 3′UTR (Figure 3C, lanes labeled 4 and 8), whereas there were no detectable proteins bound to the proximal part (Figure 3B).
Figure 3.
ElrA binds the distal portion of the cyclin E1 3′UTR. (A) Schematic of 3′UTR deletions. E1 is the full-length 3′UTR of cyclin E1 mRNA with nucleotides (numbers), restriction sites (E-EcoRI, A-AvaI, EV-EcoRV) and the relative positions of the cis-elements indicated (ARE, AU-rich element; CPE, cytoplasmic polyadenylation element; NPS, nuclear polyadenylation sequence). ▵1575-1784 has the distal part of the 3′UTR deleted while Δ1387-1571 has the proximal part deleted. Phosphorimages of UV-cross-link assays of GST-ElrA with (B) ▵1575-1784 or (C) Δ1387-1571 are shown. The indicated radiolabeled RNA was incubated with GST-ElrA, GST or 4- and 8-h embryo extracts. RNase T1 lane is the RNA digested with this enzyme in the absence of added protein. Molecular weight markers are indicated to the left in kDa. The arrow indicates a 36-kDa protein that binds in embryo extracts. n = 3.
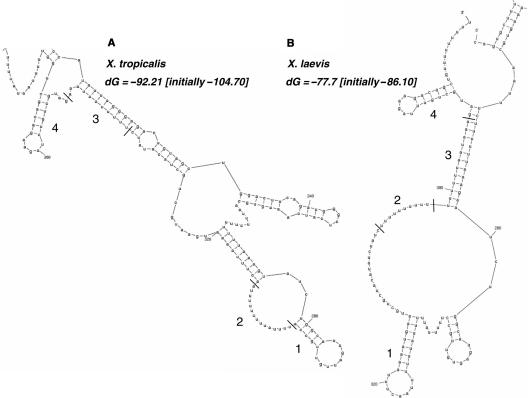
Consistent with these results, the ARE is predicted by mfold analysis (22) to participate in a stem loop similar to one forming an HuR target site (Figure 4B, region 1; (23)). The U15 CPE fits the consensus sequence for an embryonic CPE that has been shown to bind ElrA (12) and also conforms to a nine-nucleotide consensus motif (NNUUNNUUU) reported as a single-stranded binding site for HuR (Figure 4B, region 2; (24)). Similar structures or sequences are also present in the distal part of the 3′UTR of Xenopus tropicalis cyclin E1 mRNA (Figure 4A, regions 1 and 2). The X. tropicalis structure (Figure 4A, region 1) is more GC rich, but similarly contains an 8-nt loop. The remaining six uridines of the X. laevis CPE and the NPS form a stem with upstream sequence (Figure 4B, region 3), as does the equivalent sequence in X. tropicalis, a five uridine stretch followed by the NPS (Figure 4A, region 3). Lastly, a region of sequence similarity following the NPS forms a stem loop in both species (Figure 4A and B, region 4).
Figure 4.
Cyclin E1 3′UTR structure as predicted by the mfold algorithm (22). The entire 3′UTR of either X. tropicalis (A, Genbank accession NM_001016328) or X. laevis (B, Genbank accession Z13966) was folded using the mfold algorithm. The most stable predicted structure is shown, along with its free-energy value (dG) as compared to the initial free energy before folding. Only the distal part of the 3′UTR is shown due to space limitations (X. laevis nt 1641–1784, X. tropicalis nt 1617–1861). Large numbers show similar sequences and/or structures between the species as explained in the text. Small numbers refer to nucleotides with the first nucleotide of the 3′UTR set as 1.
The distal region of the cyclin E1 3′UTR specifies polyadenylation
Previously we showed that when placed downstream of a heterologous ORF and 5′UTR (that of globin) the cyclin E1 3′UTR conferred stable polyadenylation on a chimeric mRNA. We asked if ElrA binding might play a role in this polyadenylation. We tested this possibility, using in vitro synthesized, m7G-capped reporter RNAs consisting only of the full-length cyclin E1 3′UTR (E1) or deletion mutants Δ1387–1571 and Δ1575–1784, to ask if the distal region of the 3′UTR that binds ElrA directs polyadenylation. E1 and Δ1387–1571 became polyadenylated soon after injection into fertilized embryos, as evidenced by a decrease in gel mobility (Figure 5A and C). In contrast, Δ1575–1784 was not polyadenylated and was unstable, as the gel mobility did not change and the signal decreased rapidly after injection (Figure 5B).
Figure 5.
The distal portion of the 3′UTR of cyclin E1 mRNA specifies polyadenylation. Radiolabeled cyclin E1 3′UTR (A) or 3′UTR deletions (B) ▵1575-1784 or (C) Δ1387-1571 were injected into two cell-embryos. Total RNA was extracted at the indicated hour post-fertilization (hpf), electrophoresed on a denaturing gel and analyzed by phosphorimaging. U is RNA before injection. Nucleotide markers are indicated on the left. Bracket indicates polyadenylated RNA. The 2-hpf sample was taken 5 min post-injection. n = 5.
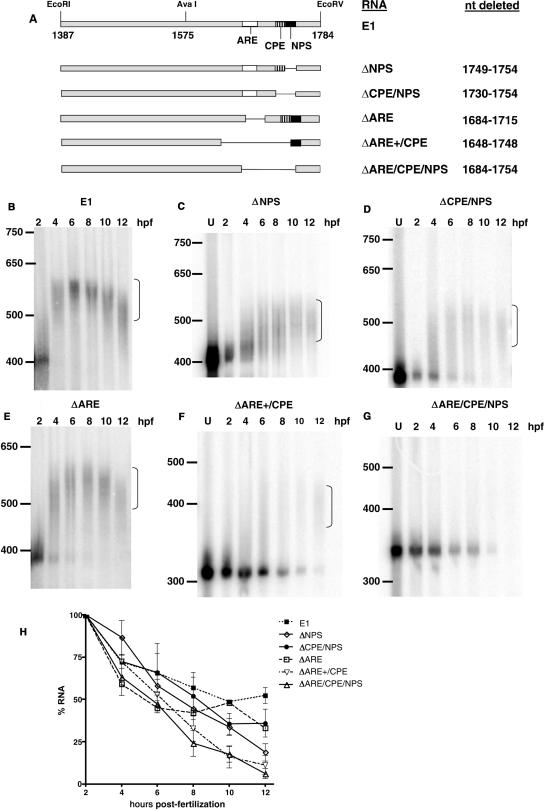
To identify which region of the distal 3′UTR was responsible for polyadenylation, we created several smaller deletions within the context of the complete 3′UTR (Figure 6A). The NPS was first deleted as it has been reported to be required for CPE-mediated polyadenylation (25,26). Surprisingly, polyadenylation occurred in the absence of the NPS but with slower kinetics and a shorter poly(A) tail (Figure 6C) when compared to the full-length 3′UTR (Figure 6B). Deletion of both the NPS and the contiguous CPE resulted in a polyadenylation pattern similar to deletion of the NPS alone, except that more of the RNA remained unadenylated for longer (Figure 6D). Polyadenylation in the absence of these elements indicated that additional cis-acting element(s) were present. Deletion of the upstream ARE very slightly affected polyadenylation, with some unadenylated RNA remaining at 4 and 6 hpf (Figure 6E). In contrast, a deletion beginning 36-nt upstream of the ARE and continuing through the CPE completely abolished polyadenylation and destabilized the RNA (Figure 6F). A more limited deletion of these 36 nt and the ARE (▵1648-1712) has an identical adenylation pattern (data not shown) suggesting that the upstream sequence, which is U-rich, is also important for polyadenylation and mRNA stability (see Figures 4B and S1, for the cyclin E1 3′UTR sequence). Similarly, deleting from the ARE through the NPS abolished polyadenylation and destabilized the RNA (Figure 6G). A summary of the effects of the deletions on mRNA stability over several experiments is shown in Figure 6H. All deletions decreased stability of the mRNA, with simultaneous deletion of the ARE, CPE and NPS having the most dramatic effect. Taken together, these data strongly indicate that the ARE, CPE and NPS are all required for the proper polyadenylation and stabilization of cyclin E1 mRNA.
Figure 6.
Polyadenylation requires a minimum of three cis-elements in the 3′UTR. (A) Schematic of 3′UTR deletion mutants used in panels (B–G). The names of the RNAs and the nucleotides deleted are indicated on the right. E1 is the full-length cyclin E1 3′UTR as described in Figure 3. (B–G) show polyadenylation assays for the 3′UTR deletion mutants indicated performed as described in Figure 3. U is RNA before injection. Nucleotide markers are indicated on the left. Bracket indicates polyadenylated RNA. The 2-hpf sample was taken 5-min post-injection. n = 2 for panel E; n = 3 for panels C, D and F; n = 4 for panels A and G. (H) Graph showing the percent of indicated RNA remaining (+/− SEM) at each timepoint averaged over all experiments. Quantitation was performed with Imagequant (Molecular Dynamics).
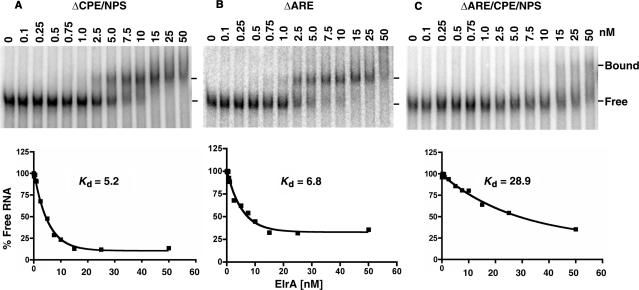
We next determined how deletion of these regions of the 3′UTR influenced ElrA binding. Deletion of both the CPE and NPS increased the Kd of ElrA for the cyclin E1 3′UTR 5-fold from 1 to 5 nM (Figure 7A), while a similar 7-fold increase was seen when either the ARE (Figure 7B) or NPS (data not shown) was deleted alone. When the ARE, CPE and the NPS were deleted the Kd increased to 29 nM, approximately another 5-fold (Figure 7C). These results indicate that ElrA binds similarly to the CPE/NPS and the ARE. Efficient binding requires all of these regions. The dramatic reduction in both polyadenylation and ElrA binding by deletion of these sequences provides compelling evidence that ElrA influences both the polyadenylation and stability of the cyclin E1 mRNA.
Figure 7.
Deletion of the 3′UTR cis-elements disrupts ElrA binding. Here, 1 fmol of the indicated radiolabeled RNA was incubated with GST-ElrA over a range of 0–50 nM. Reactions were electrophoresed on native gels, phosphorimaged (top panels) and the results quantitated (bottom panels). Bound and free RNA are indicated by the lines on the right. n = 2 for panels (A and B), n = 3 for panel (C).
DISCUSSION
These studies demonstrate that ElrA binds to the 3′UTR of cyclin E1 mRNA and that the region required for its binding functions in the polyadenylation and stabilization of the mRNA. ELAV was initially reported as an RNA binding protein whose presence was critical for neuronal development in Drosophila (27,28). Additional members of the ELAV family of proteins have been identified in several species and demonstrate a high degree of conservation (13,29). ELAV members play a role in post-transcriptional regulation with functions that include mRNA splicing, localization and stabilization (30–34). Our data implicate a function for ElrA in both cytoplasmic polyadenylation and mRNA stabilization.
It was previously reported that ElrA bound to two different maternal mRNAs in X. laevis embryos, Cl2 and the activin receptor mRNAs, in a manner dependent on the presence of their embryonic-specific CPEs (12). In contrast, ElrA did not bind to the histone B4 mRNA that contains a maturation-specific CPE. The embryonic CPE consensus sequence is oligo U12–27 and it functions at a range of 10–271 nt upstream of the NPS, while the maturation CPE consensus is U5AU and it functions from 100-nt upstream, contiguous with, or even downstream of the NPS (35). We report here, for the first time, that ElrA binds a maturation CPE simultaneously with an ARE in the cyclin E1 3′UTR. The CPE (U15A2U) in the cyclin E1 3′UTR meets both the location and sequence requirements for a maturation-type CPE. Contrarily, the cyclin A1 3′UTR does not bind ElrA (Figures 1B and 2) despite containing 2 AREs (U2AU3AU2) and a maturation-type CPE that functions in both oocytes and embryos (7,36). The cyclin A1 CPE sequence is identical to that of histone B4 (U5A2U) (12). It is likely that the additional 10 uridines in the cyclin E1 CPE contribute to stable ElrA binding. The ARE in the cyclin E1 3′UTR loosely meets the requirements for an embryonic CPE, located 45-nt upstream of the NPS and having the sequence U5AU5AU6. In comparison, the Cl2 embryonic CPE (U16GU4GU4) is 34-nt upstream of the NPS, while the activin receptor CPE is two U14 sequences separated by 42-nt and 164-nt upstream of the NPS. Regardless of the label given to them, ElrA binding to the cyclin E1 3′UTR requires the presence of these two U-rich regions that function in mRNA polyadenylation and stabilization.
The 3′UTR of cyclin E1 directed polyadenylation independent of translation, as an RNA consisting only of the 3′UTR or its distal end was polyadenylated identically to the endogenous mRNA or a chimeric mRNA containing the cyclin E1 3′UTR (this article and 7). Of particular interest was the observation that deletion of the NPS, either alone or in association with the potential CPE, slowed but did not abolish polyadenylation. In polyadenylation directed by maturation-type CPEs, the NPS serves as a binding site for CPSF and its associated polyadenylation factors, including poly(A) polymerase. Our data show that a sequence in the cyclin E1 3′UTR directs polyadenylation independently of the NPS and CPE, rare for all known cytoplasmic polyadenylation reactions. Deletion of the ARE alone did not disrupt polyadenylation, but additional deletion of the upstream 36 nt did. An example of non-CPE directed polyadenylation is the cytosine-rich sequence important for polyadenylation of protein phosphatase 2A (37), but the sequence upstream of the ARE in the cyclin E1 3′UTR is not cytosine-rich. It is possible that the upstream region is needed to stabilize ElrA binding to the ARE or for binding of other proteins that may interact with ElrA. The combined action of these elements is most likely responsible for the continued polyadenylation and stability of the cyclin E1 mRNA following the MBT, when many other maternal mRNAs are deadenylated and destabilized. Interestingly, mfold analysis shows that in the most stable predicted structure, these upstream nucleotides form a stem by base pairing with the NPS (Figure 4B). These sequences and/or structures are roughly conserved in X. tropicalis cyclin E1 3′UTR (Figure 4A), suggesting that function is also conserved. We are currently testing this hypothesis.
ElrA binding these elements may either support the assembly of the cytoplasmic polyadenylation machinery or maintain polyadenylation of the cyclin E1 mRNA once it is attained. ElrA associates with the cyclin E1 3′UTR independently of its adenylation state, consistent with either of these potential roles. ElrA has been shown to protect the body of deadenylated mRNA from degradation in vitro, in association with the Xenopus cold-inducible RNA-BP (xCIRP) (38). As the cyclin E1 mRNA remains stably polyadenylated past the MBT, it is obviously not protecting the deadenylated mRNA body from degradation. Consistent with a role for ElrA in regulating cyclin E1 mRNA, we have recently shown that its human homolog, HuR, binds to and stabilizes the mRNA encoding human cyclin E1 (39). The sequences/structures that HuR binds in the human cyclin E1 3′UTR are currently being mapped.
Despite ElrA association and maintenance of polyadenylation past the MBT, cyclin E1 mRNA is translated effectively only during oocyte maturation and the first two cell cycles following fertilization (3). Cyclin E1 translation drops dramatically following the second cell cycle (6) followed by the terminal disappearance of the protein at the MBT (3,4,8). Overexpression of cyclin E1 before the MBT results in embryonic death (40). These studies suggest that tight regulation of the translational efficiency of cyclin E1 is critical for embryogenesis. How is cyclin E1 translationally repressed in early embryos? A translational repressor may bind the cyclin E1 mRNA following translation during cell cycles 1 and 2. The binding of both ElrA and a translational repressor would explain how cyclin E1 remains polyadenylated and is translationally repressed following the second cell cycle. Alternatively, ElrA may be involved in both polyadenylation and translational repression of cyclin E1. HuR has been shown to function in translational repression (41) and the ELAV family member ElrB binds to and translationally represses the maternal Vg1 mRNA during Xenopus oocyte maturation (42). ElrA is present in oocytes presenting the possibility that it associates with cyclin E1 mRNA during oocyte maturation to facilitate polyadenylation and/or translation. One scenario is that following fertilization, ElrA either associates with different protein partners or is post-translationally modified changing its function to a repressor of translation. We are at present investigating this possibility.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by Public Health Service grant CA095898–01A1 from the National Cancer Institute to R.S.H. We thank Therese Mitchell for technical assistance and Dr Michael Sheets (University of Wisconsin) and Dr Peter Good (NIH) for supplying ElrA constructs and antibodies. We thank Michael Sheets, Yann Audic and Al Klingelhutz for critical reading of the manuscript. Funding to pay the Open Access publication charge was provided by the NCI grant CA095898-01A1.
Conflict of interest statement. None declared.
REFERENCES
- 1.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 2.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 3.Rempel RE, Sleight SB, Maller JL. Maternal Xenopus Cdk2-cyclin E complexes function during meiotic and early embryonic cell cycles that lack a G1 phase. J. Biol. Chem. 1995;270:6843–6855. doi: 10.1074/jbc.270.12.6843. [DOI] [PubMed] [Google Scholar]
- 4.Howe JA, Newport JW. A developmental timer regulates degradation of cyclin E1 at the midblastula transition during Xenopus embryogenesis. Proc. Natl. Acad. Sci. USA. 1996;93:2060–2064. doi: 10.1073/pnas.93.5.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chevalier S, Couturier A, Chartrain I, Le Guellec R, Beckhelling C, Le Guellec K, Philippe M, Ford CC. Xenopus cyclin E, a nuclear phosphoprotein, accumulates when oocytes gain the ability to initiate DNA replication. J. Cell Sci. 1996;109(Pt 6):1173–1184. doi: 10.1242/jcs.109.6.1173. [DOI] [PubMed] [Google Scholar]
- 6.Slevin MK, Lyons-Levy G, Weeks DL, Hartley RS. Antisense knockdown of cyclin E does not affect the midblastula transition in Xenopus laevis embryos. Cell Cycle. 2005;4:1396–1402. doi: 10.4161/cc.4.10.2035. [DOI] [PubMed] [Google Scholar]
- 7.Audic Y, Anderson C, Bhatty R, Hartley RS. Zygotic regulation of maternal cyclin A1 and B2 mRNAs. Mol. Cell Biol. 2001;21:1662–1671. doi: 10.1128/MCB.21.5.1662-1671.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartley RS, Rempel RE, Maller JL. In vivo regulation of the early embryonic cell cycle in Xenopus. Dev. Biol. 1996;173:408–419. doi: 10.1006/dbio.1996.0036. [DOI] [PubMed] [Google Scholar]
- 9.Richter JD. Cytoplasmic polyadenylation in development and beyond. Microbiol. Mol. Biol. Rev. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- 11.Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- 12.Wu L, Good PJ, Richter JD. The 36-kilodalton embryonic-type cytoplasmic polyadenylation element-binding protein in Xenopus laevis is ElrA, a member of the ELAV family of RNA-binding proteins. Mol. Cell Biol. 1997;17:6402–6409. doi: 10.1128/mcb.17.11.6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Good PJ. A conserved family of elav-like genes in vertebrates. Proc. Natl. Acad. Sci. U.S.A. 1995;92:4557–4561. doi: 10.1073/pnas.92.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng B. In: Methods in Cell Biology. Kay B, Peng H, editors. Vol. 36. San Diego, CA: Academic Press; 1991. pp. 657–658. [Google Scholar]
- 16.Nieuwkoop PD, Faber J. Normal Table of Xenopus Laevis (Daudin) Amsterdam: North-Holland Pub. Co.; 1967. [Google Scholar]
- 17.Audic Y, Garbrecht M, Fritz B, Sheets MD, Hartley RS. Zygotic control of maternal cyclin A1 translation and mRNA stability. Dev. Dyn. 2002;225:511–521. doi: 10.1002/dvdy.10191. [DOI] [PubMed] [Google Scholar]
- 18.Westmark CJ, Malter JS. Up-regulation of nucleolin mRNA and protein in peripheral blood mononuclear cells by extracellular-regulated kinase. J. Biol. Chem. 2001;276:1119–1126. doi: 10.1074/jbc.M009435200. [DOI] [PubMed] [Google Scholar]
- 19.Audic Y, Omilli F, Osborne HB. Postfertilization deadenylation of mRNAs in Xenopus laevis embryos is sufficient to cause their degradation at the blastula stage. Mol. Cell Biol. 1997;17:209–218. doi: 10.1128/mcb.17.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 21.Ma WJ, Chung S, Furneaux H. The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res. 1997;25:3564–3569. doi: 10.1093/nar/25.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meisner NC, Hackermuller J, Uhl V, Aszodi A, Jaritz M, Auer M. mRNA openers and closers: modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. Chembiochem. 2004;5:1432–1447. doi: 10.1002/cbic.200400219. [DOI] [PubMed] [Google Scholar]
- 25.Verrotti AC, Thompson SR, Wreden C, Strickland S, Wickens M. Evolutionary conservation of sequence elements controlling cytoplasmic polyadenylation. Proc. Natl. Acad. Sci. USA. 1996;93:9027–9032. doi: 10.1073/pnas.93.17.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon R, Richter JD. Further analysis of cytoplasmic polyadenylation in Xenopus embryos and identification of embryonic cytoplasmic polyadenylation element-binding proteins. Mol. Cell Biol. 1994;14:7867–7875. doi: 10.1128/mcb.14.12.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campos AR, Grossman D, White K. Mutant alleles at the locus elav in Drosophila melanogaster lead to nervous system defects. A developmental-genetic analysis. J. Neurogenet. 1985;2:197–218. doi: 10.3109/01677068509100150. [DOI] [PubMed] [Google Scholar]
- 28.Campos AR, Rosen DR, Robinow SN, White K. Molecular analysis of the locus elav in Drosophila melanogaster: a gene whose embryonic expression is neural specific. EMBO J. 1987;6:425–431. doi: 10.1002/j.1460-2075.1987.tb04772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Good PJ. The role of elav-like genes, a conserved family encoding RNA-binding proteins, in growth and development. Semin. Cell Dev. Biol. 1997;8:577–584. doi: 10.1006/scdb.1997.0183. [DOI] [PubMed] [Google Scholar]
- 30.Myer VE, Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan XC, Steitz JA. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koushika SP, Lisbin MJ, White K. ELAV, a drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr. Biol. 1996;6:1634–1641. doi: 10.1016/s0960-9822(02)70787-2. [DOI] [PubMed] [Google Scholar]
- 35.de Moor CH, Richter JD. Translational control in vertebrate development. Int. Rev. Cytol. 2001;203:567–608. doi: 10.1016/s0074-7696(01)03017-0. [DOI] [PubMed] [Google Scholar]
- 36.Stebbins-Boaz B, Hake LE, Richter JD. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J. 1996;15:2582–2592. [PMC free article] [PubMed] [Google Scholar]
- 37.Paillard L, Maniey D, Lachaume P, Legagneux V, Osborne HB. Identification of a C-rich element as a novel cytoplasmic polyadenylation element in xenopus embryos [In Process Citation] Mech. Dev. 2000;93:117–125. doi: 10.1016/s0925-4773(00)00279-3. [DOI] [PubMed] [Google Scholar]
- 38.Aoki K, Matsumoto K, Tsujimoto M. Xenopus cold-inducible RNA-binding protein 2 interacts with ElrA, the Xenopus homolog of HuR, and inhibits deadenylation of specific mRNAs. J. Biol. Chem. 2003;278:48491–48497. doi: 10.1074/jbc.M308328200. [DOI] [PubMed] [Google Scholar]
- 39.Guo X, Hartley RS. HuR contributes to cyclin E1 deregulation in MCF-7 breast cancer cells. Cancer Res. 2006;66:7948–7956. doi: 10.1158/0008-5472.CAN-05-4362. [DOI] [PubMed] [Google Scholar]
- 40.Richard-Parpaillon L, Cosgrove RA, Devine C, Vernon AE, Philpott A. G1/S phase cyclin-dependent kinase overexpression perturbs early development and delays tissue-specific differentiation in Xenopus. Development. 2004;131:2577–2586. doi: 10.1242/dev.01121. [DOI] [PubMed] [Google Scholar]
- 41.Meng Z, King PH, Nabors LB, Jackson NL, Chen CY, Emanuel PD, Blume SW. The ELAV RNA-stability factor HuR binds the 5′-untranslated region of the human IGF-IR transcript and differentially represses cap-dependent and IRES-mediated translation. Nucleic Acids Res. 2005;33:2962–2979. doi: 10.1093/nar/gki603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colegrove-Otero LJ, Devaux A, Standart N. The Xenopus ELAV protein ElrB represses Vg1 mRNA translation during oogenesis. Mol. Cell Biol. 2005;25:9028–9039. doi: 10.1128/MCB.25.20.9028-9039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.