Abstract
Efficient transcription is linked to modification of chromatin. For instance, tri-methylation of lysine 4 on histone H3 (H3K4) strongly correlates with transcriptional activity and is regulated by the Bur1/2 kinase complex. We found that the evolutionarily conserved Ccr4-Not complex is involved in establishing H3K4 tri-methylation in Saccharomyces cerevisiae. We observed synthetic lethal interactions of Ccr4-Not components with BUR1 and BUR2. Further analysis indicated that the genes encoding the Not-proteins are essential for efficient regulation of H3K4me3, but not H3K4me1/2, H3K36me2 or H3K79me2/3 levels. Moreover, regulation of H3K4me3 levels by NOT4 is independent of defects in RNA polymerase II loading. We found NOT4 to be important for ubiquitylation of histone H2B via recruitment of the PAF complex, but not for recruitment or activation of the Bur1/2 complex. These results suggest a mechanism in which the Ccr4-Not complex functions parallel to or downstream of the Bur1/2 kinase to facilitate H3K4me3 via PAF complex recruitment.
INTRODUCTION
Efficient transcription in eukaryotes requires interplay between the RNA polymerase II (pol II) transcription machinery and factors regulating modification of the chromatin (reviewed in: 1). Tri-methylation of lysine 4 on histone H3 (H3K4) occurs co-transcriptionally (2) and is strongly correlated with transcriptional activity (3). H3K4 methylation is mediated by the Set1p histone methyltransferase (HMT) complex in yeast (4–7). In recent years, several factors required for this modification have been identified. It is now apparent that ubiquitylation of histone H2B is necessary for methylation of H3K4 by Set1p (8,9) and of H3K79 by Dot1p (10). H2B ubiquitylation is mediated by the Bre1p-Rad6p E3-E2 pair (11,12).
The Bur1/2 kinase complex is involved in transcription elongation. This CDK–cyclin pair can phosphorylate the C-terminal domain (CTD) of the largest subunit of pol II in vitro (13), but it does not seem to contribute to CTD phosphorylation in vivo (14). This kinase complex is required for H2B ubiquitylation and H3K4 tri-methylation (15), which may involve direct phosphorylation of Rad6p (16). Interestingly, mono- and di-methylation of H3K4 are not affected by BUR1/2 mutations (15). Like the Bur1/2 complex, the PAF complex is implicated in transcription elongation and is essential for efficient H2B ubiquitylation and H3K4 methylation, but not for recruitment of Rad6p (17,18). The PAF complex consists of five subunits (Paf1p, Rtf1p, Ctr9p, Leo1p and Cdc73p) and interacts with pol II (19,20). BUR2 is required for efficient PAF complex recruitment to chromatin, but the mechanism by which this occurs remains unclear (15,16).
The evolutionarily conserved Ccr4-Not complex is composed of nine core subunits and is implicated in various steps of mRNA production and processing (reviewed in: 21,22). The genes encoding the Not-proteins (NOT1-5) were initially identified as negative regulators of transcription initiation. Support for such a role was provided by the observation that several mutations in NOT genes suppress a temperature-sensitive allele of SRB4 (23), which encodes an essential subunit of the mediator co-activator complex (24). However, various reports also indicate a positive role for the Ccr4-Not complex (25–27). For example, this complex is required for transcription of RNR genes following DNA damage or replication stress (27). Genetic and physical interactions of Ccr4-Not complex components with transcription initiation and elongation factors have been described (reviewed in: 21,22). Besides this, Ccr4p and Caf1p represent the major mRNA deadenylases in yeast (28).
To further investigate the role of the Ccr4-Not complex, we performed a genome-wide screen to find non-essential gene deletion mutants that display synthetic genetic interactions with a deletion of NOT4. Here, we describe genetic interactions of components of the Ccr4-Not complex with BUR2 and BUR1. We found that the NOT genes are required to specifically facilitate tri-methylation, but not mono- or di-methylation, of H3K4. Deletion of NOT4 reduced both histone H2B ubiquitylation and PAF complex recruitment, but did not affect Bur1/2 activation or recruitment. Taken together, our results show a novel role for the Ccr4-Not complex in chromatin modification and suggest a mechanism by which it contributes to positive regulation of transcription.
MATERIALS AND METHODS
Yeast genetics, media and plasmids
The yeast strains used in this study and their relevant genotypes are indicated in Table 1. Knock-out, TAP- and mycAVI-tagged strains were constructed by homologous recombination of a PCR product and verified by PCR, phenotypic and/or western blot analysis. Cells were routinely cultured in YPD or SC medium lacking the appropriate amino acids. The GAL1-LacZ fusion and pGR422 plasmids have previously been described (29,30). The pRS306-based NOT4 integrative vector was previously published (27).
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | EUROSCARF |
| KMY57 | Isogenic to BY4741 except not3:KanMX | EUROSCARF |
| KMY58 | Isogenic to BY4741 except not4:KanMX | EUROSCARF |
| KMY59 | Isogenic to BY4741 except not5:KanMX | EUROSCARF |
| KMY60 | Isogenic to BY4741 except caf1:KanMX | EUROSCARF |
| KMY61 | Isogenic to BY4741 except caf40:KanMX | EUROSCARF |
| KMY62 | Isogenic to BY4741 except caf130:KanMX | EUROSCARF |
| KMY107 | Isogenic to BY4741 except ccr4:KanMX | EUROSCARF |
| KMY108 | Isogenic to BY4741 except caf4:KanMX | EUROSCARF |
| KMY109 | Isogenic to BY4741 except caf16:KanMX | EUROSCARF |
| KMY110 | Isogenic to BY4741 except caf120:KanMX | EUROSCARF |
| MY1 | MATa ura3-52, trp1-Δ1, leu2::PET56 gal2 gcn4-Δ1 | (48) |
| KMY102 | Isogenic to MY1 except not1-1 | Gift from M. Collart |
| KMY114 | Isogenic to MY1 except not1-2 | Gift from M. Collart |
| KMY103 | Isogenic to MY1 except not2:KanMX | Gift from M. Collart |
| KMY104 | Isogenic to MY1 except not3:KanMX | Gift from M. Collart |
| KMY97 | Isogenic to MY1 except not4:KanMX | Gift from M. Collart |
| KMY105 | Isogenic to MY1 except not5:KanMX | Gift from M. Collart |
| W303-1B | MATα leu2-3,112 his3-11 trp1-1 can1-100 ade2-1 ura3-1 | (52) |
| KMY2 | Isogenic to W303-1B except not4:KanMX | (53) |
| UCC7164 | MATa ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE2-TEL-VR adh4::URA3-TEL-VIIL | Gift from F. van Leeuwen |
| UCC7183 | Isogenic to UCC7164 except dot1:KanMX | Gift from F. van Leeuwen |
| KMY81 | Isogenic to UCC7164 except not4:KanMX | This work |
| KMY161 | Isogenic to BY4741 except bur2:KanMX | EUROSCARF |
| KMY162 | Isogenic to BY4741 except spp1:KanMX | EUROSCARF |
| YSB787 | MATa bur1:HIS3 ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 (pRS316-BUR1) | (14) |
| KMY140 | Isogenic to YSB787 except not4:KanMX | This work |
| KMY40 | MATα mfa1Δ::MFA1pr-HIS3 his3Δ1 ura3Δ0 lys2Δ0 can1Δ not4:URA3 | This work |
| KMY133 | not4:LEU2 CTR9-HA6:TRP1, seggregant of YJJ1753 X not4:LEU2 (KMY73) | This work |
| KMY136 | Isogenic to KMY133 except not4:LEU2::NOT4:URA3 | This work |
| YZS276 | hta1-htb1Δ::LEU2 hta2-htb2Δ HTA-Flag-HTB1:HIS3 | (9) |
| YZS277 | Isogenic to YZS276 except HTA-Flag-htb1-K123R | (9) |
| YNL019 | Isogenic to YZS276 except not4Δ:KanMX | Gift from B. Strahl |
| YSB770 | MATa BUR1-HA3:TRP1 ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 | (14) |
| YSB813 | MATa BUR2-HA3:TRP1 ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 | (14) |
| KMY199 | Isogenic to YSB770 except not4Δ:KanMX | This work |
| KMY41 | Isogenic to KMY2 except not4L35A:URA3 | (29) |
| KMY200 | Isogenic to YSB813 except not4Δ:KanMX | This work |
| KMY201 | MATα mfa1Δ::MFA1pr-HIS3 bur2Δ:URA3 his3Δ1 ura3Δ0 lys2Δ0 can1Δ | This work |
Phenotypic assays
Here, 10- or 5-fold serial dilutions of the indicated strains were spotted on SC-U −/+ 6-azauracil (6-AU) (100 μg/ml) or SC −/+ 5FOA (0.1%). For the 6-AU sensitivity assay, cells were transformed with pRS316. Growth at 30°C was assessed after 3–4 days.
In vivo elongation assay
Cells transformed with the GAL1-LacZ plasmid were grown in SC-U medium containing 2% raffinose overnight, collected and subsequently shifted to a medium containing 2% galactose. Samples were taken at the indicated time points. RNA extraction and northern blot analysis were performed as described previously (27). PCR product probes spanning the ORFs were radiolabeled using the Redi-prime II kit (Invitrogen).
Western blotting and antibodies
Samples were taken from cells grown in YPD, and extracts were prepared as described previously (31). Proteins were separated by SDS-PAGE and analyzed by western blot. Antibodies against H3K4me1 (Ab8895), H3K4me2 (Ab7766), H3K4me3 (Ab8580), H3K79me3 (Ab2621) and the C-terminus of H3 (Ab1791) were obtained from Abcam. H3K36me2 (#07–369) and H3K79me2 (#07–366) antibodies were from Upstate Biotechnology. TBP antiserum was a kind gift from P.A. Weil. Antibodies H5 and H14 were used to detect the serine 2, serine 5 phosphorylated forms of the CTD of RNA pol II. The CTD-specific antibody 8WG16 was used to detect RNA pol II. TAP-tagged proteins were detected using IgG-peroxidase conjugates. Antibodies against the HA-tag (12CA5 and 3F10) or the FLAG-tag (M2, sigma) were used to detect Ctr9-HA, Bur1-HA, Bur2-HA and FLAG-H2B, respectively. Immunoblots in Figure 2B were quantified using ImageQuant software and represented as fraction of WT after normalization on total H3 levels.
Figure 2.
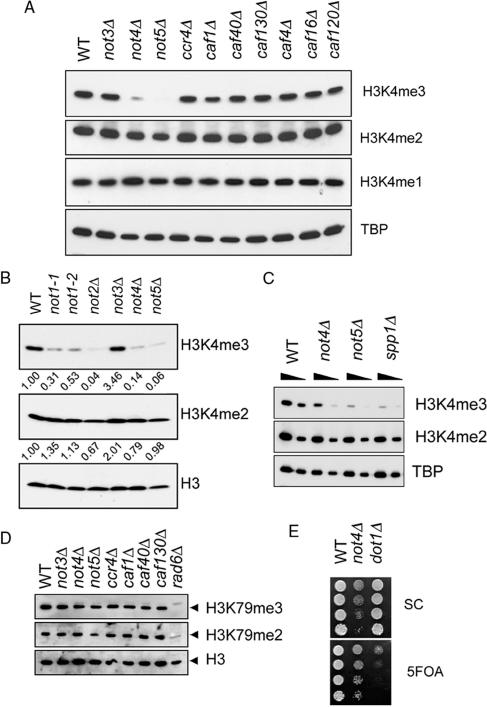
Specific requirement for the NOT genes for global H3K4 tri-methylation. (A) Histone H3K4 mono-, di- and tri-methylation levels were determined in lysates from logarithmically growing BY4741-based CCR4-NOT gene deletion strains. Protein extracts of the indicated strains were separated by SDS-PAGE (15%) and subjected to western blotting using H3K4me1-, H3K4me2- and H3K4me3- specific antibodies. TBP levels were determined as a loading control. (B) MY1-based NOT mutant strains were used to determine H3K4me2, H3K4me3 and total H3 levels as in (A). H3K4me3 and H3K4me2 levels were quantified using ImageQuant software and represented as relative to WT after normalization to total H3 levels. (C) Direct comparison of H3K4me2 and H3K4me3 levels in BY4741, not4Δ, not5Δ and spp1Δ strains. Analysis was performed as in (A). (D) H3K79me2 and H3K79me3 levels in the indicated BY4741 deletion strains. Analysis was performed as in (A). (E) Silencing assay using strains, containing URA3 in the telomeric region of the left arm of chromosome 7, lacking NOT4 or DOT1. Cells were spotted in 5-fold serial dilutions on SC plates or on SC plates containing 5FOA.
Chromatin immunoprecipitation (ChIP)
Cell cultures and extract preparation were essentially performed as described previously with minor modifications (27). Briefly, protein A-coupled agarose beads were incubated with the antibodies indicated above for 30 min at 4°C, washed and subsequently incubated with extracts of cross-linked cells for 2–3 h at 4°C. Beads were washed, DNA eluted and cross-links reversed at 65°C overnight. DNA was isolated using a PCR purification kit (Qiagen) and analyzed by SYBR-green-based quantitative PCR analysis. Not4-mycAVI was purified using streptavidin-coated Dyna-beads (Dynal) and subjected to stringent washing conditions (3% SDS in TE) before reverse cross-linking. Further analysis was performed as above. Data is represented as percentage of input with a region from the HMR locus as a control.
Northern blot analysis and reverse transcriptase qPCR
RNA extraction and Northern blots were performed as described previously (27). Equal amounts of total RNA were used to prepare cDNA using random hexamers and the SuperScript II kit (InVitrogen) according to the manufacturer's protocol. A dilution series of genomic DNA was used to determine the cDNA levels by SYBR-green-based quantitative PCR analysis.
Tandem affinity purification
TAP-tag-mediated protein purifications were performed essentially as described (32). Briefly, 20 l of YPD culture was grown to OD600 ∼ 2–3, washed and lysed in E-buffer (20 mM HEPES-KOH pH 8, 350 mM NaCl, 10% glycerol, 0.1% Tween-20). Lysates were cleared by centrifugation in a Beckman 50.2Ti rotor (45 000 r.p.m., 45 min, 4°C). An aliquot of lysate was used for purification over a 200 μl IgG sepharose column (IgG-sepharose fast flow, Pharmacia). Proteins were bound by rotating at 4°C for 2 h and subsequently washed with 35 ml E-buffer and 10 ml TEV protease cleavage-buffer (10 mM Tris-HCl pH 8, 150 mM NaCl, 0.1% Tween-20, 0.5 mM EDTA and 1 mM DTT). TEV protease (100 U) cleavage was performed in 1 ml at 18°C for 2 h. The TEV eluate was bound to 100 μl calmodulin affinity resin (Stratagene) in binding buffer (10 mM Tris pH 8, 150 mM NaCl, 1 mM MgAc, 1 mM imidazole, 2 mM CaCl2, 0.1% Tween-20, 10% glycerol and 10 mM β-mercaptoethanol) rotating at 4°C for 1 h. The column was washed with 25-ml binding buffer and bound proteins were recovered in elution buffer (10 mM Tris-HCl pH 8, 150 mM NaCl, 1 mM MgAc, 1 mM imidazole, 2 mM EGTA, 0.1% Tween-20, 10% glycerol and 10 mM β-mercaptoethanol). A fraction of the purified proteins was precipitated as described (33), separated on a 4–12% SDS-PAGE gradient gel (NuPage, Invitrogen), stained with Biosafe (BioRad) and processed for mass spectrometric analysis.
Tandem mass spectrometry
In-gel proteolytic digestion of Coomassie-stained bands was performed essentially as described (34), using trypsin (Roche). Samples were subjected to nanoflow liquid (LC) chromatography (Agilent 1100 series) and concentrated on a C18 precolumn (100 μm ID, 2 cm). Peptides were separated on an analytical column (75 μM ID, 20 cm) at a flow rate of 200 nl/min with a 60 min linear acetonitrile gradient from 0 to 80%. The LC system was directly coupled to a QTOF Micro tandem mass spectrometer (Micromass Waters, UK). A survey scan was performed from 400 to 1200 a.m.u./s and precursor ions were sequenced in MS/MS mode at a threshold of 150 counts. Data were processed and subjected to database searches using Proteinlynx Global Server version 2.1 (Micromass, UK) or MASCOT software (Matrixscience) against SWISSPROT and the NCBI non-redundant database, with a 0.25-Da mass tolerance for both precursor ion and fragment ion. The identified peptides were confirmed by manual interpretation of the spectra.
In vitro methylation assay
Set1p complexes (10–20 μl TEV eluate) from WT or not4Δ cells were incubated with 10 μg purified histones (Sigma) or 2.5 μg H3K4 (methylated) tail peptides (Abcam, Ab7228, Ab1340, Ab7768 and Ab1342, respectively), 1.5 μCi 3H-S-adenosyl methionine in buffer (50 mM Tris-HCl pH 8, 10 mM MgCl2 and 10 mM β-mercaptoethanol) at 30°C for 30 min. After separation by SDS-PAGE, the gels were dried and exposed to X-ray films.
RESULTS
The Ccr4-Not and Bur1/2 complexes interact genetically
To obtain more insight into the function of the Ccr4-Not complex, we performed a genome-wide survey to find synthetic genetic interactions with a null allele of NOT4. We identified several interactions with genes implicated in transcription elongation by RNA polymerase II (K.W.M., A.I. and H.T.M.T., submitted for publication). Most striking was the interaction between NOT4 and BUR2. Subsequent tetrad analysis of double deletions of BUR2 and Ccr4-Not components showed additional synthetic lethal interactions with NOT2 and CCR4, but not with NOT3, CAF40 and CAF130 (Figure 1A). Notably, deletion of the gene encoding Bur1p, the cyclin-dependent kinase partner of Bur2p, results in a strong growth defect and this strain was not represented in the library used for the genetic screen. To test a genetic interaction between NOT4 and BUR1 in a direct manner, we introduced a plasmid-based WT or temperature-sensitive mutant of BUR1 (bur1-23) into either NOT4 or not4Δ strains in which the sole copy of BUR1 was expressed from a URA3 plasmid (pRS316). Plasmid-shuffle analysis demonstrated synthetic lethality between a deletion of NOT4 and bur1-23 (Figure 1B).
Figure 1.
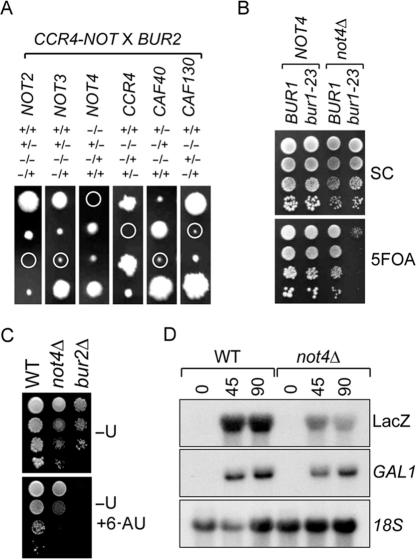
Ccr4-Not complex components genetically interact with BUR1 and BUR2 and play a role in transcription elongation. (A) Tetrad analysis of bur2Δ (KMY201) and BY4741 Ccr4-Not deletion or not4Δ (KMY40) BY4741bur2Δ diploids. Circles indicate double knock-out strains. Relevant genotypes of the haploid strains obtained are indicated (B) Plasmid-shuffle analysis of bur1-23 and not4Δ double-mutant strains containing pRS316-BUR1. Here, 10-fold serial dilutions of the indicated strains were spotted on SC and SC containing 0.1% 5FOA. (C) 6-Azauracil sensitivity assay. Here, 10-fold serial dilutions of the indicated strains were spotted on SC-U and SC-U containing 6-AU (100 μg/ml). (D) In vivo transcription elongation analysis of a long GC-rich reporter. BY4741 and not4Δ (KMY58) strains transformed with a GAL1pr-LacZ plasmid (pGR422) were grown in a medium containing raffinose and shifted to a medium containing galactose for 0, 45 and 90 min. Northern blot analysis on LacZ and endogenous GAL1 mRNA was performed using 18S rRNA as a control.
Considering that the Bur1/2 complex was found to play a role in transcription elongation (13,14), not4Δ and bur2Δ strains were first tested for sensitivity to 6-AU. In agreement with observations linking the Ccr4-Not complex to transcription elongation (25), cells lacking NOT4 or BUR2 were 6-AU sensitive (Figure 1C). Strikingly, a strict correlation was observed between the 6-AU sensitivity of Ccr4-Not gene deletions and synthetic lethality with BUR2 (25). This suggests that this synthetic lethality may be due to defects in transcription elongation. To extend these observations, we investigated requirement of NOT4 for transcription of a long and GC-rich reporter gene. This experimental setup has previously been used to study transcription elongation defects (29,35). Figure 1D shows that in cells lacking NOT4, the GAL1-promoter-driven LacZ reporter gene was inefficiently transcribed, whereas the endogenous GAL1 gene was expressed to WT levels. Taken together, these experiments show that Ccr4-Not complex components genetically interact with both BUR2 and BUR1 and confirm a role for this complex in transcription elongation (25).
Efficient H3K4 tri-methylation is dependent on the Ccr4-Not complex
The Bur1/2 complex is implicated in the regulation of tri-methylation of H3K4 (15,16). The genetic interactions between NOT4, BUR2 and BUR1 prompted us to investigate the involvement of the Ccr4-Not complex in this process by analyzing global levels of H3K4me1, H3K4me2 and H3K4me3 using specific antibodies. Strains deleted for NOT4 or NOT5 displayed a strong reduction of H3K4 tri-methylation, but not of mono- or di-methylation (Figure 2A). However, deletion of NOT3, CCR4 or any of the Ccr4-associated factors (CAFs) did not give rise to this effect (Figure 2A). It is worth noting that several CCR4-NOT gene deletion phenotypes are not evident in not3Δ cells (26,27; Figure 1A and data not shown).
To extend these observations, two mutant alleles of the essential NOT1 gene and deletions of the other NOT genes were analyzed in a different genetic background. Under permissive conditions, both not1 alleles displayed a clear decrease in overall H3K4me3 levels (50–70% reduction as shown by quantification of the results), whereas growth of these cells was not affected. Moreover, deletion of NOT2, NOT4 or NOT5 reduced H3K4me3 levels (86–94% reduction; Figure 2B). In contrast, only a slight decrease in H3K4me2 levels was observed in these strains (Figure 2B). It has been reported that Set1p complexes lacking Spp1p are unable to tri-methylate H3K4 (36,37). We found that deletion of NOT4 or NOT5 resulted in H3K4me3 levels comparable to deletion of SPP1 (Figure 2C). In addition, H3K79me2 and H3K79me3 levels in extracts of CCR4-NOT deletion strains were essentially not affected (Figure 2D), although a minor decrease in H3K79me2 in cells lacking NOT5 was observed. As expected, deletion of RAD6 led to a severe decrease in H3K79me2 and H3K79me3 levels (10).
These results suggest a specific requirement for the NOT genes for efficient H3K4 tri-methylation. Similarly, BUR1 and BUR2 are required for tri-methylation, but not for mono- or di-methylation of H3K4 (15). Dot1p, the HMT specific for methylation of H3K79 plays an important role in silencing of genes near telomeres (38). Deletion of SET1 results in a similar defect in this process (4,39). Notably, integrity of telomeric silencing was not affected by deletion of NOT4 (Figure 2E). This is consistent with the notion that H3K4me3 is not required for telomeric silencing (37) and our observation that NOT4 is specifically required for tri-methylation of H3K4.
Taken together, these results show that the NOT-module of the Ccr4-Not complex is essential for efficient H3K4 tri-methylation but not for H3K79 methylation or telomeric silencing.
H3K4 tri-methylation on the PYK1 locus is affected by deletion of NOT4
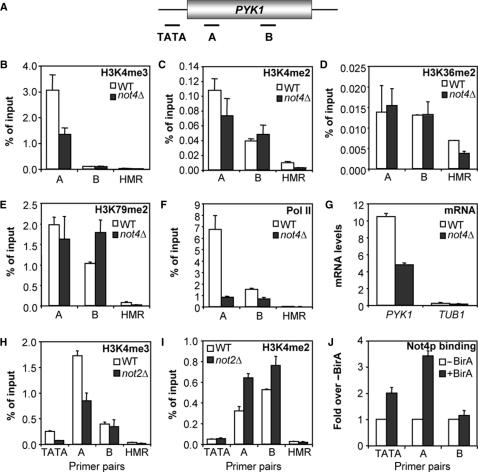
To investigate the role of NOT4 in the regulation of histone H3 methylation on active genes, ChIP experiments on the constitutively active and highly expressed PYK1 gene were performed using extracts of WT and not4Δ cells. Using specific antibodies H3K4me3, H3K4me2, H3K36me2 and H3K79me2 levels at the 5′ and the middle region of the PYK1 ORF were examined (Figure 3A–E). Not4Δ cells displayed a clear decrease in the level of H3K4me3 at the 5′ end of the PYK1 ORF compared to WT (Figure 3B), whereas H3K4me2 levels were much less affected by deletion of NOT4 (Figure 3C). Further analysis showed that H3K36me2 and H3K79me2 levels were not decreased in not4Δ cells (Figure 3D and E). In addition, pol II occupancy was determined using antibodies against the CTD of Rpb1p as a measure for transcriptional activity. In parallel, PYK1 and TUB1 mRNA levels were measured by quantitative RT-PCR in WT and not4Δ cells. Both pol II association to the PYK1 ORF and PYK1 transcript levels were diminished in the absence of NOT4 (Figure 3F and G). Extending on these results, we determined the H3K4 methylation status of cells deleted for NOT2. Indeed, examination of not2Δ cells confirmed a requirement for NOT2 in the regulation of H3K4me3 specifically (Figure 3H and I). Notably, essentially identical results were obtained when analyzing the PGK1 locus (data not shown), in line with a broader role for NOT4 in regulating H3K4me3 levels.
Figure 3.
NOT4 is required for H3K4 tri-methylation but not di-methylation of the PYK1 ORF. (A) Schematic representation of the PYK1 locus and the amplicons used in (B–H). (B) ChIP analysis of the PYK1 ORF in BY4741 and not4Δ strains. Exponentially growing cells were subjected to ChIP analysis using H3K4me3 antibodies. (C) As in (A), using H3K4me2 antibodies. (D) As in (A), using H3K36me2 antibodies. (E) As in (A), using H3K79me2 antibodies. (F) As in (A), using CTD antibodies (8WG16). (G) Quantitative reverse-transcriptase PCR analysis of PYK1 and TUB1 mRNA levels in BY4741 WT and not4Δ strains. (H) ChIP analysis of H3K4me3 levels in MY1 WT and not2Δ cells. (I) ChIP analysis of H3K4me2 levels in MY1 WT and not2Δ cells. (J) Not4p is recruited to the 5′ region of the PYK1 ORF. Strains expressing mycAVI-tagged Not4p were transformed with a BirA expression plasmid and subjected to ChIP analysis. Signals were normalized to an empty plasmid control.
To assess the direct involvement of Not4p in the regulation of PYK1 expression, ChIP analysis using a chromosomally expressed tagged form of NOT4 was performed. The encoded protein (Not4p-mycAVI) was biotinylated in vivo by co-expression of the E. coli-derived BirA biotin ligase (40). Phenotypic analysis indicated that the Not4p-mycAVI fusion protein was fully functional (data not shown). Streptavidin-coated beads were used to capture the biotinylated Not4 proteins from chromatin extracts of cells containing either a plasmid expressing BirA or an empty vector. Interestingly, Not4p was detected on the 5′ end of the PYK1 ORF and the region spanning the TATA box (Figure 3J), coinciding with the region targeted for tri-methylation by the Set1p complex (see Figure 3B and H). Taken together, these ChIP experiments confirm the specificity of the H3K4me3 defect and indicate that Not4p is physically present on the 5′ region of the PYK1 ORF. In addition, deletion of NOT4 resulted in decreased pol II occupancy and transcript levels, suggesting direct regulation of PYK1 expression by Not4p through tri-methylation of H3K4.
Decreased H3K4me3 levels in cells deleted for NOT4 does not strictly correlate with a decrease in RNA polymerase II association
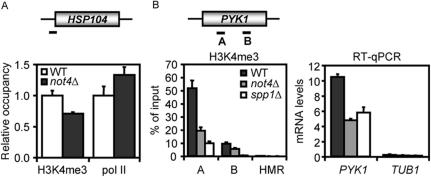
The exact mechanism of recruitment of the Set1 complex to transcribing polymerase is not fully understood. However, it has been suggested that phosphorylation of serine 2 and 5 of the hepta-peptide repeat of the CTD of RNA pol II is important for this (2,7). In this light, the global decrease of H3K4me3 in cells lacking NOT4 could be a consequence of a global loss of RNA pol II CTD phosphorylation. Immunoblot analysis using antibodies against phosphorylated serine 2, serine 5 or total RNA pol II (H5, H14 and 8WG16, respectively) did not reveal a decrease of phosphorylation of these residues in extracts from WT and not4Δ cells (data not shown). This indicates that the role of NOT4 in regulating H3K4me3 levels is independent of RNA pol II phosphorylation. Arguably, the loss of H3K4 tri-methylation could be a consequence, rather than a cause, of decreased RNA pol II recruitment. To address this issue we investigated the HSP104 gene, which allowed determination of the level of H3K4me3 on a gene that is not decreased in transcription in not4Δ cells (data not shown). We observed a decrease of H3K4 tri-methylation of the promoter region of HSP104, albeit less pronounced than what was observed for PYK1 (Figure 4A). In addition, we found a small increase in RNA pol II occupancy, which is in line with a slightly increased transcription rate (Figure 4A and data not shown). Indeed, previous observations suggest that H3K4 methylation is not a critical determinant of HSP gene expression (41). In conclusion, this result shows that a decrease in H3K4me3 in not4Δ cells is not a consequence of decreased RNA pol II association per se.
Figure 4.
Decreased H3K4me3 in not4Δ cells is not necessarily due to decreased RNA polymerase II loading (A) ChIP analysis of H3K4me3 and RNA pol II levels on the HSP104 locus. Data are represented as relative to WT. ChIP efficiencies were 2–3% and 0.15–0.2% for H3K4me3 and 8WG16 antibodies, respectively. (B) Deletion of NOT4 or SPP1 lead to a similar decrease in H3K4me3 and transcript levels on the PYK1 gene. Chromatin extracts of BY4741, not4Δ and spp1Δ strains were subjected to ChIP analysis using H3K4me3-specific antibodies. PYK1 mRNA transcript levels are affected by deletion of NOT4 or SPP1. PYK1 mRNA in total RNA of BY4147, not4Δ and spp1Δ strains was analyzed by quantitative reverse-transcriptase PCR.
To further investigate the role of H3K4me3 in transcription, we compared the levels of H3K4me3 on the PYK1 locus in WT, not4Δ and spp1Δ cells. As shown in Figure 4B, this modification is decreased by deletion of NOT4 or SPP1 to comparable levels. In addition, deletion of SPP1 gave rise to a decrease in mRNA levels reminiscent of deletion of NOT4 (Figure 4B). Similar reductions of mRNA levels were observed for the PGK1 gene (data not shown). This suggests that the diminished PYK1 and PGK1 mRNA levels are a consequence of decreased H3K4 tri-methylation. Taken together, these experiments show that the reduction of H3K4me3 levels in not4Δ cells is not a direct result of reduced RNA pol II association.
NOT4 is not required for HMT activity of the Set1p complex
The observed effect on H3K4me3 levels in not4Δ cells could be the result of NOT4 being required for transcription of genes encoding factors essential for Set1p complex integrity or activity. For instance, deletion of SPP1 results in the formation of a Set1p complex that is unable to tri-methylate H3K4 (16). We investigated this by purifying the Set1p complex from WT and not4Δ strains via a TAP-tagged version of the Bre2p subunit. In addition, we tested mRNA expression levels of Set1p complex subunits by northern blot analysis in strains containing or lacking NOT4. This indicated that neither transcript levels of its subunits nor composition of the Set1p complex are significantly affected by deletion of NOT4 (Figure 5A and B). Notably, the additional bands present in the purification from not4Δ cells were found to be non-specific lysosomal proteins (data not shown), which probably relates to differences in protein concentration between the extracts from WT or not4Δ cells. Using soluble histones as substrate in an in vitro HMT assay, no intrinsic defect in the methylation activity of Set1p complexes purified from not4Δ cells could be detected (Figure 5C). In addition, (pre-methylated) synthetic peptides were used to determine whether Set1 complexes purified from not4Δ cells were active in tri-methylation of a di-methylated substrate. Notably, Set1 complexes from WT and not4Δ cells were equally efficient in modifying this synthetic peptide (Figure 5D).
Figure 5.
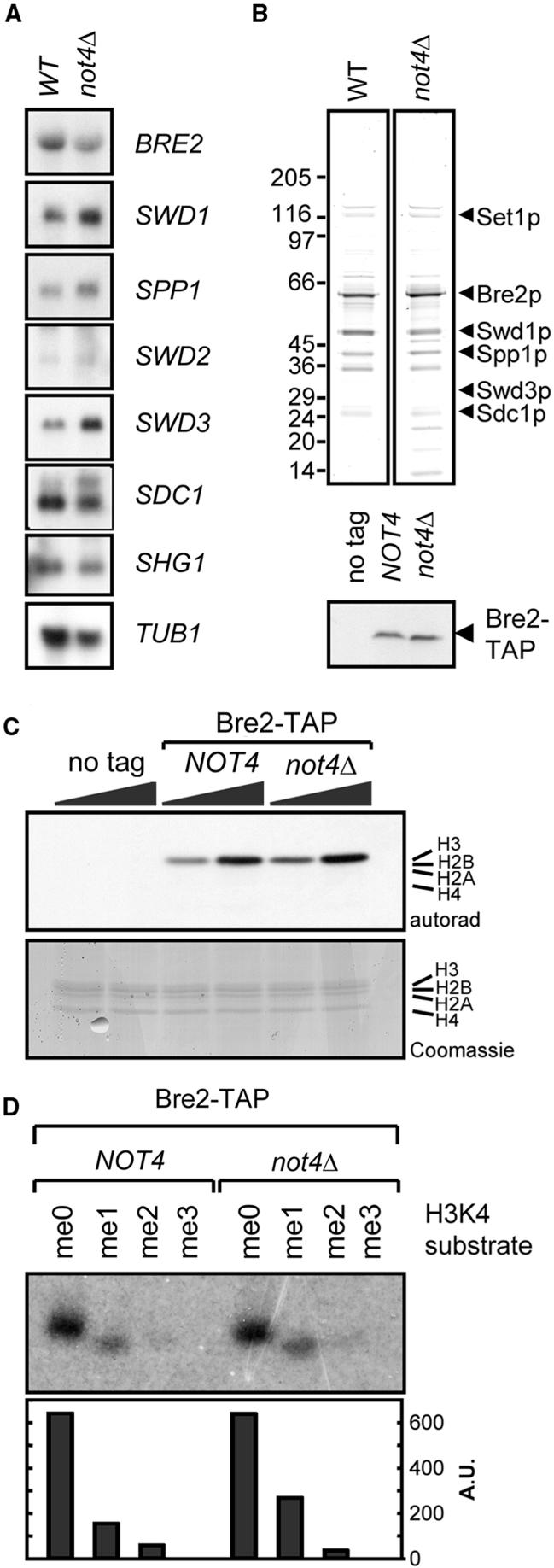
The H3K4me3 defect in not4Δ cells is independent of direct regulation of the Set1p complex. (A) mRNA levels of Set1p complex components. RNA was extracted from exponentially growing BY4741 and not4Δ and subjected to northern blot analysis using the indicated probes. (B) Subunit composition of purified Set1p complexes. Strains containing or lacking NOT4 and expressing a TAP-tagged Bre2p were used to purify Set1p complexes (upper panel). Equal expression of Bre2-TAP proteins was checked by western blot analysis (lower panel). The indicated proteins were identified using LC-MS/MS. Several additional proteins were present in the not4Δ purification when compared to the WT. Mass spectrometry showed that these bands contained non-related lysosomal proteins. We consider this an artifact of the purification, likely caused by differences in the concentration of the lysates of the WT and not4Δ cells. (C) Histone methyl transferase assay using purified Set1p complexes. Increasing amounts of Set1p complex or a mock-purification control were use to methylate soluble histones (from calf thymus) in vitro. Samples were separated on a 15% SDS-PAA gel, stained with Coomassie, dried and exposed to an X-ray film. (D) In vitro methylation of synthetic pre-methylated histone H3-tail peptides. Pre-methylated peptides were used as substrates for in vitro methylation using Set1p complexes purified from WT or not4Δ cells. Samples were separated on a 20% SDS-PAA gel, dried and exposed to an X-ray film. Signals were quantified using ImageQuant software and represented as arbitrary units after background correction.
Taken together, these experiments show that the mechanism underlying the H3K4me3 defect in not4Δ cells is distinct from deregulation of the Set1p complex at the level of transcription, composition or intrinsic activity and specificity.
Deletion of NOT4 reduces ubiquitylation of histone H2B and recruitment of the PAF complex
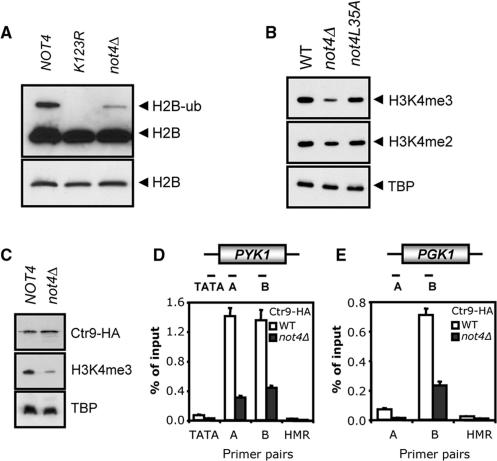
H2B ubiquitylation is required for efficient H3K4 methylation (16). To investigate the role of Not4p in ubiquitylation of H2B, plasmid-based FLAG-tagged H2B was expressed in not4Δ cells. Ubiquitylated H2B could be detected in logarithmically growing WT cells, but H2B-ubiquitin levels were significantly decreased in cells lacking NOT4. As expected, ubiquitylated H2B could not be detected in cells expressing FLAG-tagged H2B-K123R (Figure 6A, 39). Interestingly, Not4p contains a RING-finger motif and displays ubiquitylation activity in vitro (K.W.M., A.I. and H.T.M.T., submitted for publication). Therefore, the H3K4me3 status was determined in cells expressing an inactive (L35A) mutant of Not4p as a measure for H2B ubiquitylation. The H3K4me3 defect of not4Δ cells was fully complemented by the L35A mutant of Not4p, suggesting that its RING-finger is not required for ubiquitylation of H2B (Figure 6B). In addition, in vitro ubiquitylation assays indicated that H2B does not serve as a substrate for Not4p (data not shown).
Figure 6.
Deletion of NOT4 diminishes H2B ubiquitylation and PAF complex recruitment. (A) NOT4 is required for efficient H2B ubiquitylation. Extracts of the indicated strains carrying a FLAG-HTB1 plasmid were separated on a 12.5% SDS-PAGE gel and subjected to western blot analysis using FLAG antibodies. The lower panel shows a shorter exposure of the same blot, indicating equal loading. (B) The ubiquitin-ligase activity of Not4p is not involved in the regulation of H3K4me3 levels. The H3K4 methylation status in extracts from cells expressing NOT4 or not4L35A alleles from the endogenous NOT4 locus and cells lacking NOT4 were subjected to western blot analysis. (C) Ctr9-HA is equally expressed in NOT4 and not4Δ cells. Strains expressing HA-tagged Ctr9p, containing or lacking NOT4, were used to determine Ctr9-HA levels by western blotting. H3K4me3 and TBP levels were used as controls. (D and E) NOT4 is required for PAF complex recruitment to the PYK1 and PGK1 ORFs. Strains from (C) were subjected to ChIP analysis of the PYK1 locus (amplicons are schematically depicted in upper panel). Exponentially growing cells were cross-linked and ChIPs were performed using anti-HA (12CA5) antibodies.
In vivo, H2B ubiquitylation is dependent on recruitment of the PAF complex to chromatin (18). To assess the role of Not4p in the recruitment of the PAF complex, strains expressing HA-tagged Ctr9p and containing or lacking NOT4 were constructed. Ctr9-HA was expressed to equal levels in the two strains, whereas H3K4me3 levels were reduced in the not4Δ cells as expected (Figure 6C). Extracts were prepared from exponentially growing cells and subjected to ChIP analysis. Whereas Ctr9-HA was readily detected in the ORFs of PYK1 and PGK1 in WT cells, binding was severely reduced in cells lacking NOT4 (Figure 6D and E). Taken together, these results show a requirement for NOT4 in H2B ubiquitylation and a role in the regulation of PAF complex recruitment to the PYK1 and PGK1 genes to facilitate tri-methylation of H3K4.
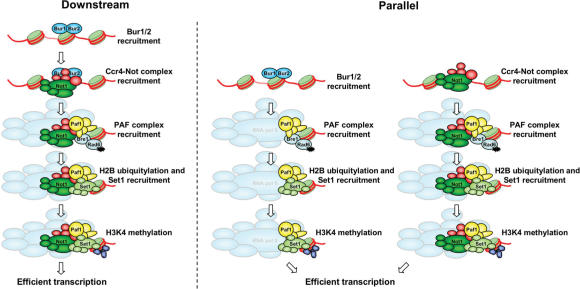
The Ccr4-Not complex functions parallel to or downstream of the Bur1/2 kinase
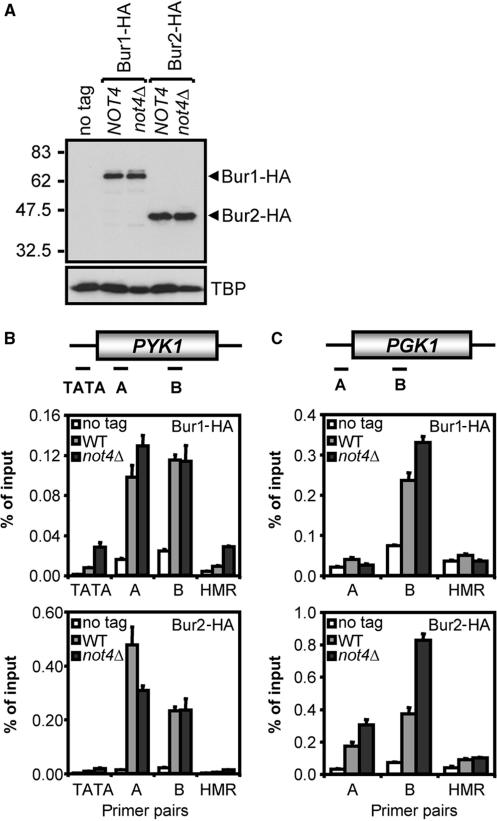
Chromatin association of the PAF complex depends on the Bur1/2 complex (15,16). The requirement for NOT4 for efficient PAF complex recruitment places the Ccr4-Not complex in the pathway containing the Bur1/2 kinase. To investigate this further, strains expressing HA-tagged Bur1p or Bur2p, containing or lacking NOT4, were constructed to determine occupancy of Bur1p and Bur2p. Expression of the HA-tagged versions of Bur1p and Bur2p was equal between WT and not4Δ cells (Figure 7A). ChIP analysis with these strains showed that both Bur1p and Bur2p were found to associate with the PYK1 and PGK1 loci with similar efficiencies (Figure 7B and C). In addition, NOT4 is not required for the activation of the Bur1p kinase since the active phosphorylated form of Bur1p is readily detected in cells lacking NOT4 (Figure 7A). These results place the Ccr4-Not complex parallel to, or downstream of the Bur1/2 kinase and upstream of the PAF complex in a pathway leading to H3K4 tri-methylation.
Figure 7.
The Ccr4-Not complex functions downstream of or parallel to the Bur1/2 kinase. (A) Bur1-HA and Bur2-HA are equally expressed in WT and not4Δ cells. Extracts of cells expressing HA-tagged Bur1p or Bur2p containing or lacking NOT4, or a non-tagged control, were subjected to western blot analysis using antibodies against the HA-tag, H3K4me3 and TBP. (B) Recruitment of the Bur1/2 complex to the PYK1 locus does not depend on NOT4. WT and not4Δ cells expressing HA-tagged Bur1p or Bur2p, and a no-tag control, were subjected to ChIP analysis using anti-HA (12CA5) antibodies (amplicons are schematically depicted in the upper panel). (C) As in (B), except that the analysis was performed for the PGK1 locus.
DISCUSSION
The Ccr4-Not complex is involved in mRNA biogenesis at different levels (reviewed in 21,22). Here, we show that the NOT genes, in contrast to CCR4 and the CAF genes, are required for global and gene-specific tri-methylation of H3K4 (Figures 2, 3 and 4). In addition, we found that the Ccr4-Not complex is functionally linked to the Bur1/2 kinase complex (Figure 1), which is also specifically required for H3K4 tri-methylation (20). The mechanism by which the Ccr4-Not complex is involved in this histone modification is independent of direct regulation of the Set1p histone methyltransferase complex (Figure 5). Instead, the Ccr4-Not complex functions parallel to or downstream of Bur1/2p at the level of PAF complex recruitment, resulting in decreased ubiquitylation of H2B, in cells lacking NOT4 (Figure 8). Taken together, our results provide a detailed study of the role of the Ccr4-Not complex in the regulation of global H3K4 tri-methylation. Further, they confirm a functional distinction between the cytoplasmic deadenylase activity of the Ccr4p-Caf1p module and the nuclear function of the Not-proteins of the Ccr4-Not complex (42).
Figure 8.
A model indicating how the Ccr4-Not complex may function in establishing H3K4 tri-methylation through regulation of PAF complex recruitment. The Ccr4-Not complex could be controlled by the Bur1/2 kinase to facilitate PAF complex recruitment (downstream). Alternatively, the Bur1/2 and Ccr4-Not complexes function in distinct pathways sharing several components and independently regulate PAF complex recruitment (parallel).
Cooperation between the Bur1/2 and Ccr4-Not complexes in establishing H3K4 tri-methylation
The Bur1/2 complex was shown to be required for H3K4 tri-methylation (15) and this involves phosphorylation of Rad6p (16). The genetic interactions between genes encoding Ccr4-Not complex subunits (NOT2, NOT4 and CCR4) and BUR1 and BUR2, suggest a role for this complex in transcription elongation (Figure 1). Interestingly, deletions of several Ccr4-Not components were shown to exhibit 6-AU sensitivity, suggesting such a role (25). Notably, a strict correlation between the published 6-AU sensitivity for Ccr4-Not components and synthetic lethality with BUR2 was observed. Since the Bur1/2 complex was shown to be involved in the regulation of H3K4me3 levels (15), we determined the levels of various histone H3 lysine methylation marks in cells lacking genes encoding Ccr4-Not components. In contrast to the NOT-gene deletion strains, decreased H3K4me3 levels were not observed in ccr4Δ cells (Figure 2), suggesting that Ccr4p and the Not-module contribute to transcription elongation by distinct mechanisms. An alternative explanation could be that deletion of Ccr4-Not components and BUR2 results in partial loss of subsequent activities within the same pathway, ultimately resulting in the observed synthetic lethality.
Insight into the mechanism by which the Not-proteins function in the pathway leading to H3K4 tri-methylation comes from the observations that PAF complex recruitment and subsequent efficient ubiquitylation of H2B, but not Bur1/2 complex recruitment, are dependent on the presence of NOT4. However, other modifications such as H3K4 mono- and di-methylation or H3K79 di- and tri-methylation were not affected by deletion of NOT4. These results suggest a model in which the pathways leading to H3K4 and H3K79 methylation bifurcate upstream of the Ccr4-Not complex and are in agreement with a recent report showing that BUR2 is only required for H3K4 tri-methylation (15,16). Alternatively, the residual degree of ubiquitylated H2B that is observed in cells lacking NOT4 may be sufficient to sustain H3K79me2/3 and H3K4me2, but not H3K4me3 levels. Support for this hypothesis comes from the observation that cells deleted for PAF1 show significant levels of H3K4 di-methylation but not tri-methylation (43, data not shown).
Regulation of H3K4 tri-methylation by the Ccr4-Not complex and involvement of the PAF complex
The Set1p complex (or COMPASS) represents the sole HMT for H3K4 in yeast (4–6). The observation that transcription, subunit composition, activity and specificity of the Set1p complex are not affected by Not4p suggests that the Ccr4-Not complex functions further upstream in a pathway regulating H3K4 tri-methylation. The finding that Not4p is physically present at the 5′ region of the PYK1 ORF, coinciding with the H3K4 tri-methylation mark and the PAF complex, suggests a direct role for the Ccr4-Not complex in the regulation of this modification. Possibly, recruitment of the Ccr4-Not complex to this region aids localization of the H3K4me3 mark. Another important insight into the mechanism by which the Ccr4-Not complex controls H3K4me3 levels comes from the observation that PAF complex recruitment is severely decreased in the absence of NOT4, whereas Bur1p and Bur2p recruitment and activation are not affected (Figures 6 and 7). The Bur1/2 kinase is known to act upstream of PAF complex recruitment (15). Thus, the Ccr4-Not complex could either act downstream of the Bur1/2 kinase, or in a parallel pathway to regulate PAF complex recruitment (Figure 8). It is interesting to note that both Not3p and Not5p are phospho-proteins (44), but the connection with Bur1/2 complex remains unexplored. In the parallel pathway, the Bur1/2 and Ccr4-Not complexes would act independently of each other in the recruitment of the PAF complex to chromatin. Strikingly, deletion of BUR2 or NOT4 results in only a partial loss of H2B ubiquitylation, PAF complex recruitment and subsequent H3K4 tri-methylation (15, Figures 2 and 6).
Previous experiments have physically linked the PAF complex and Ccr4p (45). However, we and others have failed to detect PAF complex members or Bur1p/2p in Ccr4-Not purifications (46, data not shown). In addition, deletion of PAF1 gives rise to an HU-sensitive phenotype and deregulation of RNR gene transcription (47). Intriguingly, this was also observed for deletions of CCR4-NOT genes (27) and deletion of BUR2 (data not shown), suggesting a functional interplay between these complexes. Alternatively, alteration of other components of the histone code might also contribute to the regulation of H3K4me3 levels by the Ccr4-Not complex. Irrespective of this, it is clear that further experiments are required to elucidate the connections between the Ccr4-Not and PAF complexes.
Positive and negative regulation of transcription by the Ccr4-Not complex
Our results support a global role for the Ccr4-Not complex in the positive regulation of transcription. In addition, mutation of components of this complex results in derepression of a variety of genes (26,48). These seemingly contradicting observations may be reconciled by recent findings of recognition of H3K4-methylated histones by PHD finger containing proteins (49–51). These domains are believed to recruit chromatin-modifying complexes, like NURF and the mSin3a-HDAC1 complex to H3K4-methylated chromatin (49,51). Yeast contains 15 PHD fingers residing in a variety of chromatin-modifying complexes and these motifs display different relative affinities for H3K4me2 and H3K4me3 (50). Specific loss of H3K4 tri-methylation could therefore shift the balance of chromatin regulatory complexes at particular genes. It is unlikely that the sole function of the Ccr4-Not complex is regulation of H3K4 tri-methylation via recruitment of the PAF complex. Besides the well-documented role of the Ccr4p and Caf1p subunits in mRNA deadenylation, multiple observations indicate that the Not-proteins can directly affect TFIID function (reviewed in 21,22). This suggests that the Ccr4-Not complex integrates several steps of mRNA expression. The functional consequences of Ccr4-Not gene deletions are likely to be gene specific as suggested, for example, by our results on the PYK1 and HSP104 genes. Taken together, these findings provide an explanation for the observation that the Ccr4-Not complex can have both negative and positive effects on gene transcription.
In summary, we have identified a role for the Ccr4-Not complex in the regulation of H3K4 tri-methylation levels by acting parallel to or downstream of the Bur1/2 kinase to facilitate PAF complex recruitment and subsequent H2B ubiquitylation. These results provide a connection between the role of the Ccr4-Not complex in the regulation of transcription and histone methylation marks.
ACKNOWLEDGEMENTS
We thank Drs M. Collart, F. van Leeuwen, J. Jaehning, W.W. Pijnappel, S. Buratowski, P.A. Weil, G.P. Prelich, A. Aguilera and B. Strahl for generously sharing reagents, Drs G.S. Winkler and F.C. Holstege for critical reading of the manuscript and members of the Timmers laboratory for helpful discussions, technical advice and assistance. Drs N.J. Krogan and B. Strahl are gratefully acknowledged for sharing results prior to publication. This work was supported by grants from The Netherlands Organization for Scientific Research (NWO-MW Pionier #900-98-142 and NWO-CW #700-50-034), European Union (Improving Human Potential RTN2-2001-00026 and STREP LSHG-CT-2004-502950) and EMBO (ASTF 184.00-02). Funding to pay the Open Access publication charge was provided by the University Medical Center Utrecht.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berger SL. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 2.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 3.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 4.Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 5.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl Acad. Sci. USA. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dehe PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, Rodriguez-Gil A, Mkandawire M, Landsberg K, et al. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J. Biol. Chem. 2006;281:35404–35412. doi: 10.1074/jbc.M603099200. [DOI] [PubMed] [Google Scholar]
- 8.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 9.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 10.Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 11.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang YF, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 12.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 13.Murray S, Udupa R, Yao S, Hartzog G, Prelich G. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol. Cell. Biol. 2001;21:4089–4096. doi: 10.1128/MCB.21.13.4089-4096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keogh MC, Podolny V, Buratowski S. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:7005–7018. doi: 10.1128/MCB.23.19.7005-7018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr. Biol. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 16.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol. Cell. 2005;20:589–599. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 18.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 19.Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764–1774. doi: 10.1093/emboj/21.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collart MA, Timmers HT. The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 2004;77:289–322. doi: 10.1016/S0079-6603(04)77008-7. [DOI] [PubMed] [Google Scholar]
- 22.Denis CL, Chen J. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2003;73:221–250. doi: 10.1016/s0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 23.Lee TI, Wyrick JJ, Koh SS, Jennings EG, Gadbois EL, Young RA. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol. Cell. Biol. 1998;18:4455–4462. doi: 10.1128/mcb.18.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 25.Denis CL, Chiang YC, Cui Y, Chen J. Genetic evidence supports a role for the yeast CCR4-NOT complex in transcriptional elongation. Genetics. 2001;158:627–634. doi: 10.1093/genetics/158.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu HY, Badarinarayana V, Audino DC, Rappsilber J, Mann M, Denis CL. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulder KW, Winkler GS, Timmers HT. DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4-Not complex. Nucleic Acids Res. 2005;33:6384–6392. doi: 10.1093/nar/gki938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 29.Rondon AG, Jimeno S, Garcia-Rubio M, Aguilera A. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 2003;278:39037–39043. doi: 10.1074/jbc.M305718200. [DOI] [PubMed] [Google Scholar]
- 30.Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol. Cell. Biol. 2005;25:6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 32.Logie C, Peterson CL. Purification and biochemical properties of yeast SWI/SNF complex. Methods Enzymol. 1999;304:726–741. doi: 10.1016/s0076-6879(99)04044-6. [DOI] [PubMed] [Google Scholar]
- 33.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 34.Kinter M, Sherman NE. Protein Sequencing Identification Using Tandem Mass Spectrometry. New York: Wiley and Sons; 2000. [Google Scholar]
- 35.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, et al. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 38.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 39.Corda Y, Schramke V, Longhese MP, Smokvina T, Paciotti V, Brevet V, Gilson E, Geli V. Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat. Genet. 1999;21:204–208. doi: 10.1038/5991. [DOI] [PubMed] [Google Scholar]
- 40.van Werven FJ, Timmers HT. The use of biotin tagging in Saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation. Nucleic Acids Res. 2006;34:e33. doi: 10.1093/nar/gkl003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao J, Herrera-Diaz J, Gross DS. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 2005;25:8985–8999. doi: 10.1128/MCB.25.20.8985-8999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell. Biol. 1999;19:6642–6651. doi: 10.1128/mcb.19.10.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlichter A, Cairns BR. Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. EMBO J. 2005;24:1222–1231. doi: 10.1038/sj.emboj.7600607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lenssen E, Oberholzer U, Labarre J, De Virgilio C, Collart MA. Saccharomyces cerevisiae Ccr4-not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol. Microbiol. 2002;43:1023–1037. doi: 10.1046/j.1365-2958.2002.02799.x. [DOI] [PubMed] [Google Scholar]
- 45.Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 47.Betz JL, Chang M, Washburn TM, Porter SE, Mueller CL, Jaehning JA. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics. 2002;268:272–285. doi: 10.1007/s00438-002-0752-8. [DOI] [PubMed] [Google Scholar]
- 48.Collart MA, Struhl K. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- 49.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 Methylated at either lysine 4 or lysine 36. J. Biol. Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 52.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 53.Deluen C, James N, Maillet L, Molinete M, Theiler G, Lemaire M, Paquet N, Collart MA. The Ccr4-not complex and yTAF1 (yTaf(II)130p/yTaf(II)145p) show physical and functional interactions. Mol. Cell. Biol. 2002;22:6735–6749. doi: 10.1128/MCB.22.19.6735-6749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]