Abstract
Presenilin is an essential component of the LIN-12/Notch signaling pathway and also plays a critical role in the genesis of Alzheimer's disease. Previously, a screen for suppressors of the egg-laying defective phenotype caused by partial loss of presenilin activity in Caenorhabditis elegans identified a number of new spr genes that are potentially involved in the regulation of LIN-12/Notch signaling or presenilin activity. Here we report the molecular identity of two spr genes, spr-1 and spr-5. Our genetic analysis indicates that loss of spr-1 elevates lin-12/Notch gene activity in many different cell fate decisions, suggesting that spr-1 is a negative regulator of LIN-12/Notch signaling. Sequence analysis revealed that spr-1 is an ortholog of human CoREST, a known corepressor. SPR-1 is localized to the nucleus and acts in a cell-autonomous manner; furthermore, human CoREST can substitute for SPR-1 in C. elegans. We also show that spr-5 encodes a homolog of p110b, another known member of the CoREST corepressor complex. Our results suggest that the CoREST corepressor complex might be functionally conserved in worms, and we discuss the potential role of SPR-1 and SPR-5 in the repression of transcription of genes involved in, or downstream of, LIN-12/Notch signal transduction.
Keywords: Notch, presenilin, CoREST, lin-12, polyamine oxidase, HDAC
Many cell–cell interactions that specify cell fate during animal development are mediated by receptors of the LIN-12/Notch family. When activated by transmembrane protein ligands of the Delta/Serrate/LAG-2 family produced by neighboring cells, the extracellular domain of LIN-12/Notch is cleaved near the membrane. This event in turn leads to cleavage within the LIN-12/Notch transmembrane domain, releasing the intracellular domain. Presenilins are essential for transmembrane cleavage. After its release, the intracellular domain translocates to the nucleus, where it forms a complex with the transcription factor known as LAG-1 in Caenorhabditis elegans, Suppressor of Hairless in Drosophila, or RBP-J or CBF1 (herein referred to generically as CBF1) in mammals. This complex, which also contains other proteins, can activate the expression of target genes (for review, see Weinmaster 2000).
In the absence of LIN-12/Notch signaling, CBF1 has been shown to function as a transcriptional repressor. Indeed, transcriptional repression of Notch target genes in the absence of Notch signaling is important for correct cell fate patterning in Drosophila (Furriols and Bray 2000; Morel and Schweisguth 2000). Two-hybrid experiments showed that CBF1 can interact with different repressor proteins (Kao et al. 1998; Taniguchi et al. 1998; Hsieh et al. 1999), notably SMRT (silencing mediator of retinoid and thyroid hormone receptors) and histone deacetylase 1 (HDAC-1; Kao et al. 1998). CBF1 contains a repressor domain that becomes masked by association with the active intracellular part of LIN-12/Notch (Hsieh and Hayward 1995; Waltzer et al. 1995; Hsieh et al. 1996) and is needed for SMRT-mediated repression (Kao et al. 1998). Kadesch and colleagues have proposed that the intracellular domain of LIN-12/Notch proteins activates transcription by displacing a corepressor complex, thereby preventing it from interacting with CBF1 (Kao et al. 1998). Further work suggested that it also provides a transcriptional activation domain to recruit coactivators to function with CBF1 (Jarriault et al. 1995; Doyle et al. 2000; Petcherski and Kimble 2000a,b; Wu et al. 2000; Zhou et al. 2000; Kitagawa et al. 2001).
Genetic screens in C. elegans and Drosophila have been effective in identifying conserved components of the LIN-12/Notch pathway and conserved factors that influence the activity of LIN-12/Notch proteins (for review, see Greenwald 1998). In C. elegans, genetic screens, based on suppression or enhancement of lin-12 (sel) mutations, have defined many new sel genes (Sundaram and Greenwald 1993a; Levitan and Greenwald 1995; Tax et al. 1997). Isolation of the worm presenilin sel-12 in such screens provided the first link between LIN-12/Notch signaling and presenilin activity (Levitan and Greenwald 1995); a second presenilin gene in C. elegans, called hop-1, is functionally redundant with sel-12 (Li and Greenwald 1997; Westlund et al. 1999). There is intense interest in presenilin as a potential therapeutic target for the amelioration or prevention of Alzheimer's disease, because presenilin mediates transmembrane cleavage of β-amyloid precursor protein, a critical step in the generation of a peptide that can cause Alzheimer's disease (Kopan and Goate 2000).
Presenilin is also required for the transmembrane cleavage of LIN-12/Notch, an essential step for signal transduction by these receptors (De Strooper et al. 1999; Ray et al. 1999; Struhl and Greenwald 1999, 2001; Zhang et al. 2000). Because presenilin is critical for LIN-12/Notch signaling, the absence of both sel-12 and hop-1 activity causes phenotypes associated with the loss of the two C. elegans LIN-12/Notch genes, lin-12 and glp-1 (Li and Greenwald 1997; Westlund et al. 1999). However, because sel-12 and hop-1 are functionally redundant, mutants that lack only sel-12 presenilin activity [sel-12(−)] have a relatively mild phenotype. sel-12(−) hermaphrodites are unable to lay eggs, a phenotype believed to be due to loss of lin-12 activity in a subset of gonadal cells (Newman et al. 1995; Cinar et al. 2001).
As the sel-12(−) egg-laying defective (Egl) phenotype reflects reduced lin-12 activity, genetic screens for suppressors of the Egl phenotype of sel-12(−) mutants might in principle identify factors that influence lin-12 activity as well as factors that influence presenilin activity per se. These factors are likely to be negative regulators, as the suppressor mutation will have the net effect of elevating LIN-12/Notch or presenilin function. In a previous study, 14 mutations that act as highly penetrant suppressors of the Egl defect of sel-12(ar171) were identified (Wen et al. 2000). These 14 mutations defined at least four genes, called spr, for suppressor of presenilin. The analysis of one of these genes, spr-2, revealed that it encodes the C. elegans ortholog of mammalian SET, a protein that has been implicated in chromatin remodeling, as well as other biochemical processes (Wen et al. 2000).
Here, we report a detailed genetic and molecular characterization of the spr gene spr-1, and the identification of another spr gene, spr-5. SPR-1 and SPR-5 are orthologs of human CoREST and p110b, respectively. CoREST was first identified as a corepressor for the neural-restrictive silencing factor REST (Andres et al. 1999); p110b was identified in specific HDAC-containing complexes with CoREST (Humphrey et al. 2001; You et al. 2001; Hakimi et al. 2002). CoREST and p110b are members of a core corepressor complex, so the observation that their homologs display spr activity suggests that the complex activity has been conserved in C. elegans. We show that spr-1 acts in a cell-autonomous manner in specific lin-12 expressing cells and is a functional ortholog of CoREST. Our genetic analysis indicates that spr-1 activity negatively influences several different LIN-12/Notch signaling events in C. elegans development. Taken together, our results suggest that at least part of the CoREST corepressor complex might be functionally conserved in worms and influence LIN-12/Notch signaling. We discuss the function of SPR-1 and SPR-5 in the context of the important balance of repression and activation in LIN-12/Notch signal transduction.
Results
After a screen of 27,600 mutagenized haploid genomes for suppressors of the sel-12 presenilin Egl phenotype, four spr mutations were mapped to LG V (Wen et al. 2000). At that time, the name spr-1 was assigned to ar200; the remaining three alleles, ar201, ar205, and ar212, were not assigned by complementation testing to the spr-1 gene because under some conditions they display semidominance (detailed further in Materials and Methods). The molecular analysis described below has indicated that all four mutations are indeed alleles of spr-1, and they will be referred to as such throughout the text. Our analysis also indicates that these alleles reduce spr-1 activity. Unless otherwise specified, the allele spr-1(ar200) was used for the genetic analysis. In all cell fate decisions analyzed, spr-1(ar200) displays a wild-type phenotype (see Materials and Methods).
The interaction between spr-1 and sel-12 is not allele-specific
To rule out the possibility that spr-1 acts as an informational suppressor of sel-12(ar171) (W225STOP) we examined its ability to suppress sel-12(ar131) (C60S), another sel-12 loss-of-function allele (Levitan and Greenwald 1995). We found that spr-1(ar200) appears to be an efficient suppressor of sel-12(ar131) (Table 1), indicating that suppression is not sel-12 allele-specific. In addition, we examined the ability of spr-1(ar201) and spr-1(ar205) to suppress the Egl phenotype of sel-12(ar171). The three spr-1 alleles display a similar degree of suppression and appear to suppress sel-12(ar171) at all temperatures (Table 1), suggesting that these spr-1 alleles have a similar effect on spr-1 activity.
Table 1.
spr-1(ar200) suppresses Egl phenotype of differentsel-12 alleles
| Relevant
genotype
|
15°C Egl+/total
(%)
|
20°C Egl+/total
(%)
|
25°C Egl+/total
(%)
|
|---|---|---|---|
| sel-12(ar171)a | 0/39 (0%) | 0/45 (0%) | 2/60 (3.3%) |
| spr-1(ar200); sel-12(ar171)a | 43/44 (97.7%) | 169/179 (94.4%) | 58/64 (90.6%) |
| sel-12(ar131)b | ND | 1/44 (2.3%) | ND |
| spr-1(ar200); sel-12(ar131)b | ND | 60/71 (84.5%) | ND |
| spr-1(ar205); sel-12(ar171)a | 34/40 (85%) | 73/73 (100%) | 91/92 (98.9%) |
| spr-1(ar201); serl-12(ar171)a | 37/38 (97.3%) | 98/100 (98%) | ND |
Egl+, wild-type egg-laying ability (see Materials and Methods). Others are unable to lay eggs (Egl). spr-1 alone appears wild-type (see Materials and Methods). ND, not determined.
Strains also contained unc-1(e538).
Strains also contained dpy-11(e224).
spr-1 suppresses the π cell defect caused by sel-12(ar171)
In the gonad of wild-type hermaphrodites during the L3 larval stage, there are twelve descendants of the ventral uterine precursor cells (VUs) that express LIN-12 (Wilkinson and Greenwald 1995). The anchor cell (AC) expresses LAG-2 (Newman et al. 2000), which activates LIN-12 in the six neighboring VU descendants to induce them to become π cells; the six VU descendants that do not have contact with the AC adopt the alternative ρ fate (Newman et al. 1995; Newman and Sternberg 1996). The π cells can be distinguished from the ρ cells by their morphology and position, by their division pattern, and by cell fate markers. The π cells produce 12 daughters. During the L4 stage, eight of these cells fuse with the AC to form the “utse”, a thin membrane at the junction of the vulva and the uterus that will be broken by the first egg layed (Fig. 1A; Newman et al. 1995, 1996). lin-12 and sel-12 loss-of-function mutants lack π cells, whereas more π cells are formed at the expense of the ρ cells in lin-12 gain-of-function mutants (Newman et al. 1995; Cinar et al. 2001). The defect in π cell specification appears to be largely accountable for the Egl phenotype of sel-12 hermaphrodites (Cinar et al. 2001).
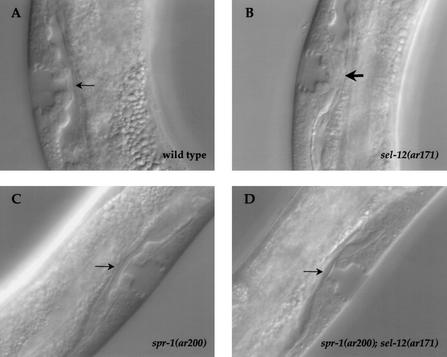
Figure 1.
spr-1; sel-12 hermaphrodites have a wild-type utse. The utse morphology was examined in (A) wild-type N2; (B) sel-12(ar171) (33/35 animals displayed a mutant morphology); (C) spr-1(ar200) hermaphrodites (0/24 animals displayed a mutant morphology); (D) spr-1(ar200); sel-12(ar171) (0/29 animals displayed a mutant morphology). A normal utse is indicated by a thin arrow; an abnormal utse by a thick arrow. Photomicrographs show a lateral view; (A,B) anterior is to the bottom; (C) anterior is to the left; (D) anterior is to the right. Strains containing sel-12(ar171) also contained unc-1(e538).
Using two criteria, we have determined that spr-1(ar200) suppresses the π cell specification defect in sel-12(ar171) mutant. First, we analyzed the expression of a π cell fate marker, lin-11∷lacZ. In wild-type hermaphrodites, lin-11∷lacZ is expressed in the six π cells shortly before they divide, and remains on in their daughters (Newman et al. 1999). The number of cells expressing lin-11∷lacZ is reduced in sel-12(ar171) mutants (Cinar et al. 2001; data not shown), but we found it to be normal in spr-1(ar200) and spr-1(ar200); sel-12(ar171) hermaphrodites (data not shown). Second, we examined the morphology of the vulval–uterine junction in young adults. In sel-12 mutants, a thick tissue is observed at the vulva–uterus junction instead of the utse and the AC remains unfused (Fig. 1, cf. A and B; Cinar et al. 2001). As shown in Figure 1C and D, the morphology of the utse appears wild-type in spr-1(ar200) and spr-1(ar200); sel-12(ar171) mutants.
Suppression of the sel-12 egg-laying defect requires hop-1 activity
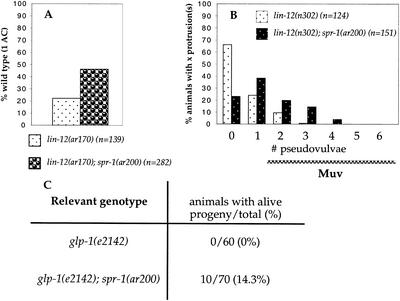
Loss of hop-1 presenilin activity does not cause an overt phenotype, whereas loss of both hop-1 and sel-12 activity causes phenotypes that are similar to those caused by loss of lin-12 and glp-1 activity (Li and Greenwald 1997; Westlund et al. 1999). However, the precise double mutant phenotype observed can be influenced by the presence of maternal presenilin activity (Westlund et al. 1999). For example, the hop-1(0); sel-12(0) hermaphrodites that segregate from a hop-1(0); sel-12(0)/+ mother are viable but Egl, and the progeny they produce die as embryos (called a Mel phenotype, for maternal effect lethal). We found that the phenotype of hop-1(0); spr-1(ar200); sel-12(0) hermaphrodites segregating from a hop-1(0); spr-1(ar200); sel-12(0)/+ mother is Egl and Mel (Table 2). These results indicate that spr-1(ar200) cannot bypass the need for presenilin activity. Such dependence on presenilin activity is expected if spr-1 acts on presenilin activity or in a process that is strictly presenilin-dependent.
Table 2.
spr-1(ar200) does not bypass the need forhop-1 activity
| Relevant
genotype
|
Egl/total (%)
|
Mel/total (%)
|
|---|---|---|
| Wild type | 0% (n = many) | 0% (n = many) |
| spr-1(ar200); serl-12(ar171) | 3/38 (7.9%) | 0/38 (0%) |
| hop-1(ar179); sel-12(ar171)a | 27/27 (100%) | 27/27 (100%) |
| hop-1(ar179); spr-1(ar200); sel-12(ar171)b | 52/52 (100%) | 52/52 (100%) |
Maternal sel 12(+) activity provided by the duplication (see footnotes) enables hop-1(−);sel-12(−) progeny to survive to adulthood and to produce progeny, which arrest as embryos (Mel phenotype). Those hop-1(−); sel-12(−) adults are not able to lay eggs (Egl phenotype). spr-1(ar200) appears wild-type (see Materials and Methods). All strains contained unc-1(e538).
Segregated from a hop(ar179); sel-12(ar171) unc-1(e538); mnDp68 mother.
Segregated from a hop-1(ar179); spr-1(ar200); sel-12(ar171) unc-1(e538); mnDp68 mother.
spr-1 displays genetic interactions with various lin-12 or glp-1 alleles
As the π cell defect of sel-12 mutant hermaphrodites that is suppressed by mutations in spr-1 appears to be due to reduced lin-12 activity (Newman et al. 1995; Cinar et al. 2001), we wondered whether spr-1 could act as a modulator of lin-12/Notch activity in other cells, a role that might be revealed in sensitized backgrounds. If reduced spr-1 activity suppresses the sel-12 Egl phenotype by elevating lin-12 activity, we might observe suppression of other defects caused by lin-12 hypomorphic alleles and enhancement of those caused by lin-12 hypermorphs. Furthermore, if spr-1 functions generally to influence lin-12/Notch activity, we might observe analogous genetic interactions with glp-1 as well. Therefore, we constructed double mutants between spr-1 and lin-12 or glp-1 alleles and examined the consequences for various cell fate decisions. We observed that spr-1 mutation displays genetic interactions with various alleles of lin-12 and glp-1 in various cell fate decisions, suggesting that spr-1 may function generally as a negative regulator of lin-12/Notch activity.
The AC/VU decision
Two gonadal cells, Z1.ppp and Z4.aaa, interact with each other so that one becomes the AC and the other a VU (Kimble 1981). Genetic studies have established that a low level of lin-12 activity is required for the AC fate, whereas high levels of lin-12 activity lead to the VU fate (Seydoux and Greenwald 1989). Therefore, in lin-12 null mutants [lin-12(0)], both Z1.ppp and Z4.aaa become ACs, whereas in lin-12 hypermorphic mutants [lin-12(d)] both become VUs.
lin-12(ar170ts) is a weak lin-12 hypomorphic allele that at 25°C is highly penetrant for the 2AC phenotype associated with loss of lin-12 activity. As shown in Figure 2A, spr-1(ar200) is able to suppress the 2AC defect of lin-12(ar170), suggesting that spr-1 may negatively regulate lin-12 activity during the AC/VU decision.
Figure 2.
spr-1(ar200) displays genetic interactions with lin-12 and glp-1. n, the total number of animals scored. No cell fate defects were observed in spr-1(ar200) hermaphrodites (see Materials and Methods). (A) spr-1(ar200) suppresses the 2 AC defect of lin-12(ar170). lin-12(ar170) and lin-12(ar170); spr-1(ar200) hermaphrodites were scored at 25°C. (B) Percent of animals (y axis) with a given pseudovulvae number (x axis) are shown. Hermaphrodites with two or more pseudovulvae are considered to have a Multivulva (Muv) phenotype (wild-type hermaphrodites have a single vulva and no pseudovulvae). spr-1(ar200) enhances the activity of lin-12(n302), resulting in a Muv phenotype at 14°C. Note that enhancement is very temperature sensitive. The low-penetrance sterility defect of lin-12(n302) is also enhanced (data not shown), but the cellular basis for this defect is not known. (C) spr-1(ar200) suppresses the maternal embryonic lethality (Mel) defect of glp-1(e2142). All strains were scored at 25°C and contained unc-32(e189).
Specification of the fates of the vulval precursor cells
Six initially equivalent cells, consecutively numbered P3.p–P8.p, each have the potential to generate vulval cells and therefore are called vulval precursor cells (VPCs; for review, see Greenwald 1998). Cell–cell interactions specify that only the central three VPCs adopt vulval fates: the AC induces P6.p to adopt a vulval fate termed 1o, whereupon it laterally signals its two neighbors, P5.p and P7.p, to adopt a vulval fate termed 2o. The phenotypes of lin-12 mutants suggest that the receptor for the lateral signal is LIN-12 (Greenwald et al. 1983). When LIN-12 is constitutively active to a high level, as in the strong hypermorph lin-12(n950), most VPCs adopt the 2o fate; the progeny of each 2o VPC organize to form a pseudovulva, so that such hermaphrodites have a distinctive Multivulva phenotype (Muv). However, when LIN-12 is constitutively active to a lesser extent, as in the weaker hypermorph lin-12(n302), the Muv phenotype is not generally evident.
To examine whether spr-1 negatively regulates lin-12 activity during VPC specification, we first asked whether spr-1(ar200) can enhance the activity of lin-12(n302) so as to cause a Muv phenotype. We observed a Muv phenotype in lin-12(n302); spr-1(ar200) hermaphrodites, in contrast to the single mutant lin-12(n302) (Fig. 2B). These results suggest that spr-1 negatively regulates lin-12 activity during VPC specification.
Early embryonic development
Loss of maternal glp-1 activity causes embryonic lethality due to abnormal early blastomere specification (Mel), and the glp-1(e2142) allele appears to preferentially lack this activity (Priess et al. 1987). As shown in Figure 2C, spr-1 mutation is able to suppress partially the Mel phenotype of glp-1(e2142), suggesting that spr-1 is also a negative regulator of glp-1 activity.
In summary, we have found evidence that spr-1 appears to act as a negative regulator of LIN-12/Notch activity in the AC/VU decision, VPC specification, and early blastomere specification.
Molecular identification of the spr-1 gene
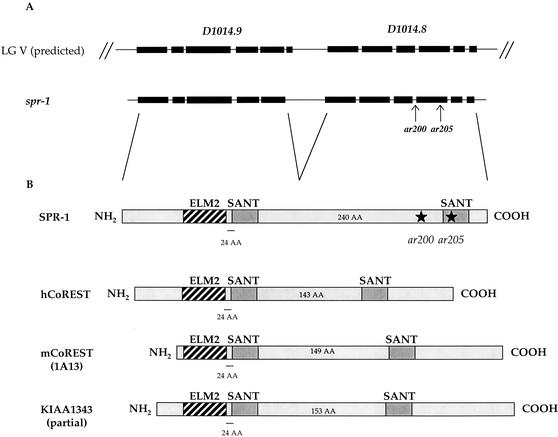
spr-1 had been previously mapped to the dpy-11–rol-3 interval on LG V (Wen et al. 2000). In order to map spr-1 to a smaller interval, we took advantage of single nucleotide polymorphisms (SNPs) in the C. elegans Hawaiian strain CB4856, using SNPs identified by the Genome Project and new ones we identified by sequencing intergenic regions. In this way, we mapped spr-1 between two SNPs, located on Y97E10AL and D1014. This interval encompassed five cosmids, but none of them showed antisuppression when injected into the double mutant spr-1(ar200); sel-12(ar171). However, injection of three overlapping cosmids [DD11, F58G4, and T23D2 (=D1014)] led to larval lethality, so that antisuppression could not be scored. We sequenced the predicted genes in this region in the spr-1(ar200) mutant. One of these genes, D1014.8, contained a G→A mutation (as expected for an EMS-induced mutation), changing the AG splice acceptor site in front of predicted exon 4 into AA. Next, we sequenced the three other spr mutants on LG V and found the same change in ar201 as well as another G→A mutation introducing a stop codon early in predicted exon 4 of gene D1014.8 in ar205 and ar212 (see Fig. 3A).
Figure 3.
spr-1 gene structure and protein product. (A) The relationship between the two previously predicted open reading frames, D1014.8 and D1014.9, to the spr-1 gene structure we determined is shown. spr-1(ar200) affects the splice acceptor at the start of exon 9, resulting in a frameshift that leads to a stop codon. spr-1(ar205) is a G-to-A transition that results in a stop codon in exon 9. (B) SPR-1/CoREST protein family. The domain structure of SPR-1 and of all the mammalian homologs found in the databases is shown. spr-1(ar200) and spr-1(ar205) results in truncation of the SPR-1 protein as shown. The spacing between the ELM2 and the first SANT domain is always 24 amino acids; the spacing between the SANT domains is variable, as indicated. Accession nos.: SPR-1, AAA96286; hCoREST, no. AF155595; mCoREST (murine 1A13), no. X83587; KIAA1343, no. BAA92581. Note that KIAA1343 most likely represents a partial cDNA.
The gene D1014.8 was predicted by GENEFINDER to be in an operon with the 5′ gene D1014.9. When we analyzed the structure of spr-1 by PCR screening of three different cDNA libraries, we found that spr-1 encodes a single mRNA encompassing both D1014.8 and D1014.9 predicted genes. The resulting spr-1 gene structure is shown in Figure 3A.
We confirmed that mutations in the gene D1014.8 correspond to spr-1 by injecting a PCR fragment amplified from genomic DNA comprising the spr-1 coding sequence into spr-1(ar200); sel-12(ar171) worms and found that this product caused antisuppression (see Materials and Methods). In addition, we confirmed that D1014.9 behaved functionally as part of the spr-1 gene by showing that double-stranded RNA from D1014.9 as well as D1014.8 suppressed the Egl phenotype of sel-12(ar171), when delivered by injection (Table 3) or by bacterial feeding (data not shown). These RNA-mediated interference (RNAi) results also suggest that reduced spr-1 activity can suppress the Egl phenotype of sel-12(−) hermaphrodites, implying that spr-1 alleles reduce or eliminate spr-1 activity.
Table 3.
spr-1, but not Y74C9A.4, double stranded RNA suppresses the Egl phenotype ofsel-12(ar171) animals
| Injected
dsRNA
|
Po plates with Egl+ progeny/total
(%)
|
|---|---|
| Mock | 0/20 (0%) |
| spr-1 (D1014.8) | 12/14 (85%) |
| spr-1 (former D1014.9) | 14/17 (82%) |
| Y74C9A.4 | 0/19 (0%) |
sel-12(ar171) unc-1(e538) adult hermaphrodites were injected. Egl+, egg-laying ability was restored in their progeny. D1014.9 and D1014.8 were originally predicted to be two distinct open reading frames, but are both part of the spr-1 gene (see text and Fig. 3).
We have evidence that in ar200, an mRNA is produced that potentially encodes a truncated protein: We cloned a single cDNA corresponding to the mRNA produced in spr-1(ar200) animals and showed that the mutation of the AG splice acceptor site led to aberrant splicing to the immediately adjacent AG dinucleotide, leading to a shift in the reading frame and an early stop in exon 9.
The nature of the spr-1 mutations that were found does not allow us to conclude that ar200 and ar205 are null alleles. It may be that the null phenotype of spr-1 is lethal or sterile, and therefore that null alleles could not be recovered in a genetic screen that requires viability and fertility. However, there is no evidence from RNAi for such phenotypes, and there is no available deficiency for the region to conduct classical tests. It should be noted that RNAi does not always cause complete loss of gene activity, so the lack of a phenotype by RNAi cannot be conclusive. Alternatively, it may be that the null phenotype of spr-1 is wild-type, and a phenotypic effect can only be revealed when spr-1(−) is placed in a sensitized genetic background. A wild-type null phenotype may be evidence that the gene, pathway, or process is redundant (e.g., Greenwald and Horvitz 1980; Thomas 1993).
SPR-1 belongs to a conserved protein family
Analysis of SPR-1 primary sequence using SMART (http://smart.embl-heidelberg.de) suggests that the protein contains an ELM2 (EGL-27 and MTA1 homology domain 2) domain followed by two SANT domains (SWI3, ADA2, NCoR, TFIIIB B" subunit). The function of ELM2 domains is unknown, but they have been observed in several transcriptional repressors (Solari et al. 1999). The SANT domain is also found in transcriptional regulators (Aasland et al. 1996). These observations, plus the homology described further below, suggest that SPR-1 has a role in the modulation of transcription.
BLAST analysis revealed that SPR-1 has been conserved throughout evolution. Proteins with one ELM2 domain followed by two SANT domains (Fig. 3B) appear to represent a small conserved subfamily among proteins with two SANT domains. In particular, the spacing between the ELM2 and the first SANT domain is invariant in the members of this family; the spacing between the two SANT domains appears to vary between 140 and 240 amino acids (see Fig. 3B).
The apparent human ortholog of SPR-1 is known as CoREST, which was isolated on the basis of its ability to bind REST or HDAC-1/-2 (Andres et al. 1999; You et al. 2001). CoREST has been shown to exhibit transcriptional repressor activity when fused to the Gal4 DNA-binding domain and to be necessary for REST-dependent repression of neural-specific gene expression (Andres et al. 1999; You et al. 2001). It copurifies with a limited number of factors including HDAC-1/-2 and p110b in an equimolar ratio (Humphrey et al. 2001; You et al. 2001; Hakimi et al. 2002) and could represent a new corepressor complex. The HDAC-1/-2 interaction domain in CoREST maps to its SANT domains (Ballas et al. 2001; You et al. 2001), which are also found in other transcriptional corepressors and HDAC interacting partners such as N-CoR, SMRT, MTA1, or MTA2. As described below, human CoREST can functionally replace SPR-1, confirming the orthology predicted by sequence analysis.
We note that another gene encoding a protein with two SANT domains and which shows homology to SPR-1 and CoREST exists in worms, Y74C9A.4. However, Y74C9A.4 appears to lack an ELM2 motif. Nevertheless, we assessed whether Y74C9A.4 behaves as a spr gene by analyzing its ability to suppress the sel-12 Egl phenotype when its activity is lowered or lost by injection of Y74C9A.4 double-stranded RNA. We saw no suppression, therefore we provisionally conclude that Y74C9A.4 is not a spr gene (Table 3).
Subcellular localization of SPR-1
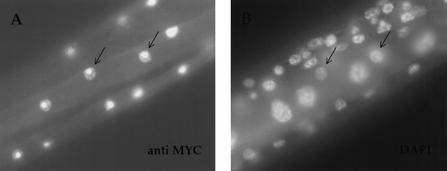
As expression of SPR-1(+) under its own promoter appears to be highly toxic (see Materials and Methods), we expressed a Myc-tagged SPR-1 protein (SPR-1∷MYC) using the cog-2 promoter (see below) in order to analyze its subcellular localization. As shown in Figure 4, nuclear staining is observed when transgenic animals are stained with an anti-MYC antibody. There also appears to be an intensification of staining within the nucleus, in speckles and a large structure that we presume to be the nucleolus (Fig. 4). Nuclear localization has also been observed for CoREST in mammalian cells (Grimes et al. 2000; Humphrey et al. 2001; S. Jarriault and I. Greenwald, unpubl.) and is consistent with a role for SPR-1 in transcriptional repression.
Figure 4.
SPR-1 is localized to the nucleus. Photomicrographs of a spr-1(ar200); sel-12(ar171) unc-1 hermaphrodite expressing a SPR-1∷MYC protein under the cog-2 promoter and stained with an antibody directed against MYC (A) or with DAPI (B) are shown. Arrows point to discrete stained muscle nuclei. Anterior is to the right.
spr-1 functions cell autonomously
To assess whether spr-1 activity is required autonomously for π cell induction, we wanted to analyze the ability of spr-1; sel-12 mutants to lay eggs when SPR-1(+) is delivered specifically in the π cell precursors. In the gonad, the cog-2 promoter has been reported to be active only in the π cell precursors and their daughters, and not in the AC or other gonadal cells (Hanna-Rose and Han 1999). We examined transgenic lines expressing SPR-1∷MYC under the cog-2 promoter in spr-1; sel-12 hermaphrodites and found a high proportion of hermaphrodites unable to lay eggs (Fig. 5). These results suggest that spr-1 functions autonomously in π cell specification, and by extension, spr-1 is likely to function autonomously in other lin-12-mediated cell fate decisions.
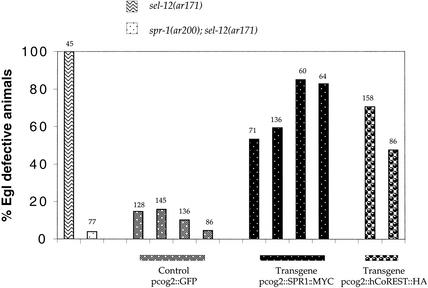
Figure 5.
Expression of SPR-1 or CoREST in π cell precursors complements spr-1(ar200). For transgenic lines, each bar represents an independent array in a spr-1(ar200); sel-12(ar171) background. The total number of animals scored is indicated above each bar. All strains containing sel-12(ar171) also contain unc-1(e538). (pcog-2∷GFP) Control lines, arrays containing plasmid (pWH17). (pcog-2∷SPR1∷ MYC) Expression of SPR-1(+) in π cells complements spr-1(ar200), indicating cell autonomy. (pcog-2∷CoREST∷HA) Expression of human CoREST in π cells also complements spr-1(ar200), indicating that human CoREST can functionally substitute for the C. elegans SPR-1.
Human CoREST can functionally replace SPR-1 in C. elegans
Using the cog-2 promoter to express human CoREST in the π cells, we assayed whether CoREST has SPR-1(+) activity. We found that hCoREST can functionally replace SPR-1, causing spr-1; sel-12 hermaphrodites in which it is expressed to display an increased proportion of Egl hermaphrodites relative to a vector-only control (Fig. 5). These results suggest that SPR-1 and CoREST are true orthologs, and therefore that the information about CoREST function that has been inferred from studies in mammalian cells is likely to be illuminating about SPR-1 function, and vice versa.
One homolog of p110b, another member of the CoREST corepressor complex, displays spr activity in an RNAi assay
The finding that SPR-1 is the ortholog of CoREST suggested that genes encoding other components of the CoREST complex might have spr activity. In addition to the zinc finger protein, REST, a variety of proteins have been recently found to exist in a core complex with CoREST: p110b (a protein with a FAD binding domain and a polyamine oxidase motif, also known as KIAA0601) and histone deacetylases 1 and 2 (HDAC-1/-2; Humphrey et al. 2001; You et al. 2001; Hakimi et al. 2002). Depending on the conditions, additional factors were also found, such as p110a (a zinc finger protein, ZNF217), p40 or BRAF35 (Sox-like putative transcription factors), BCH80 (a CHD3-like molecule), or p80 (a hypothetical, novel protein; Humphrey et al. 2001; You et al. 2001; Hakimi et al. 2002). However, known members of corepressor complexes such as Sin3 (which interacts with REST in HEK-293 cells; Grimes et al. 2000), MTA2, RbAp46 and RbAp48, or MBD2, key components of the Sin3 and NuRD HDAC-1/-2 containing complexes were not coimmunoprecipitated (Humphrey et al. 2001; You et al. 2001).
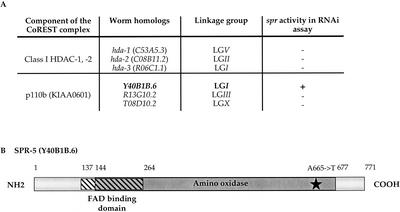
We did BLAST searches to identify putative C. elegans homologs of REST and of the CoREST-interacting partners for which accession numbers were given or readily apparent (ZNF217, p110b, and HDAC-1/-2). Although the zinc finger proteins identified a number of C. elegans zinc finger proteins, none is clearly orthologous to REST or ZNF217 (data not shown). However, we identified three proteins that are highly related to class I HDAC-1/-2, and three proteins that are highly related to p110b (Fig. 6A).
Figure 6.
Identification of Y40B1B.6 as spr-5 by RNAi and sequence analysis. (A) RNAi analysis of C. elegans CoREST complex homologs. Double stranded RNA was delivered to sel-12(ar171) unc-1(e538) hermaphrodites by either feeding (hda-1, hda-2, hda-3) or injection (Y40B1B.6, R13G10.2, T08D10.2). At least 60 individuals fed dsRNA were examined: (−) no Egl+ individuals were seen. The progeny of at least 14 injected hermaphrodites were examined: (−) no Egl+ individuals were seen. For Y40B1B.6, 35/37 injected animals gave many Egl+ progeny. (B) The predicted protein structure of Y40B1B.6 and the position of the mutation found in spr-5(ar197) are shown. The overlapping FAD-binding (hatched box) and polyamine oxidase (gray box) domains are indicated.
We assessed their spr activity by looking for RNAi-induced suppression of sel-12(ar171). As shown in Figure 6A, all three HDAC homologs, and two of the p110b homologs, failed to show spr activity. A negative result by RNAi may not be definitive (see above). Furthermore, it has been shown that both HDAC-1 and HDAC-2 interact with CoREST, raising the possibility of functional redundancy.
In contrast, one of the p110b homologs, Y40B1B.6, displayed strong spr activity. Y40B1B.6 is predicted to have a FAD domain partially overlapping the polyamine oxidase domain, and of the three C. elegans p110b homologs, has the most similar domain structure to human p110b.
Y40B1B.6, the p110b homolog, is spr-5
Y40B1B.6 maps to LG I, where we had previously identified three spr mutations: spr-4(ar208), spr(ar204), and spr(ar197). We sequenced the Y40B1B.6 coding region from all three mutant strains and identified a sequence change associated with spr(ar197). This gene, which has also been identified by Eimer et al. among their collection of suppressors of sel-12(ar171) (S. Eimer, B. Lakowski, and R. Baumeister, pers. comm.) is now designated spr-5. spr-5(ar197) is a G-to-A change at the end of exon 7, resulting in an A665T missense mutation, which affects a conserved amino acid of the polyamine oxidase domain (Fig. 6B). The sequence and RNAi data suggest that spr-5(ar197) results in reduced or absent Y40B1B.6 function.
Although reducing the activity of spr-1 and spr-5 can suppress the sel-12(ar171) mutant phenotype, the nature of our alleles does not resolve whether complete absence of the CoREST/p110b complex causes any visible phenotypes. We note that we did not see any effects on viability, fertility, or egg-laying when we performed the following experiments: spr-1(RNAi) in spr-5(ar197), spr-5(RNAi) in spr-1(ar200), and double RNAi for both spr-1 and spr-5 in both wild-type and spr-1(ar200) backgrounds (data not shown). Therefore, while it remains possible that molecular null alleles of spr-1 and/or spr-5 might cause a visible phenotype, we have not been able to find any evidence that this is so.
Discussion
In this study, we have described a genetic and molecular characterization of the spr-1 gene. Through genetic analysis we showed that lowering spr-1 activity leads to allele-nonspecific suppression of the Egl phenotype of sel-12 hermaphrodites. We also found that suppression is achieved by restoring π cell specification, the cellular defect that underlies the Egl phenotype of sel-12 hermaphrodites (Cinar et al. 2001). When SPR-1(+) is expressed in the π cell precursors of spr-1; sel-12 hermaphrodites, suppression is lost, suggesting that spr-1 function is cell autonomous. Suppression of sel-12 by spr-1 depends on the activity of hop-1 presenilin, suggesting that reduced spr-1 activity does not bypass the need for presenilin activity. Finally, loss of spr-1 increases the activity of lin-12 or glp-1 in all cell fate decisions examined, suggesting that spr-1 functions generally as a negative regulator of LIN-12/Notch activity.
Through molecular analysis, we showed that SPR-1 is orthologous to CoREST, which was first identified as a corepressor for the neural-restrictive silencing factor REST (Andres et al. 1999). A functional, epitope-tagged SPR-1 protein is localized to the nucleus in C. elegans, consistent with a function in transcriptional repression. Moreover, human CoREST is also localized to the nuclei when expressed in worms (see Materials and Methods) and can partially substitute for SPR-1, indicating that the sequence conservation is likely to extend to functional conservation. Finally, using RNAi, we found that another gene, Y40B1B.6,, encoding a homolog of a known member of the CoREST complex, also has spr activity, and that a previously identified spr mutation is in the Y40B1B.6 gene (now known as spr-5). Therefore, at least some of the spr genes are likely to function in the same process, and the functional conservation shown between SPR-1 and CoREST might extend to other members of the CoREST corepressor complex.
How does spr-1 suppress the Egl phenotype of sel-12(−) hermaphrodites?
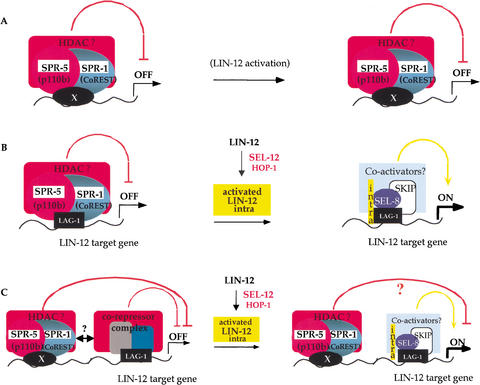
In considering how mutations in spr-1 suppress the Egl phenotype of sel-12 mutants, it must be borne in mind that presenilin is a critical component of the LIN-12/Notch pathway, and that failure of π cell induction is a manifestation of reduced lin-12 activity (Newman et al. 1995; Cinar et al. 2001). Therefore, loss of spr-1 activity might affect presenilin level or activity per se, or affect the level or activity of any limiting component of the LIN-12/Notch pathway. Because SPR-1 is a functional ortholog of CoREST, a protein with demonstrated corepressor activity, one straightforward model postulates that SPR-1 and SPR-5 negatively regulate the transcription of genes encoding members of the LIN-12/Notch pathway or of genes encoding positive regulators of these pathway members (Fig. 7A).
Figure 7.
Models for SPR-1 and SPR-5 function. (A) SPR-1 and SPR-5 are part of a corepressor complex affecting the transcription of known members of the LIN-12/Notch pathway. Although no evidence exists, hop-1 transcription could, for example, be affected in a cell-specific way. Alternatively, the complex could repress the transcription of a subset of crucial genes for π cell fate which would not get activated in lin-12(−) or sel-12(−) mutants unless this repression is released. (B) SPR-1/CoREST and SPR-5/p110b are part of the LAG-1-associated corepressor complex. After ligand binding leads to the presenilin-dependent release of the intracellular domain of LIN-12 (yellow, noted “intra”), the corepressor complex is released and a coactivator complex formed, leading to activation of the target genes. (C) SPR-1 and SPR-5 form a distinct corepressor complex, which may interact with the LAG-1 repressor complex. The corepressor complex containing SPR-1 and SPR-5 is targeted to the DNA by a factor X, and is distinct from the LAG-1 complex. This complex, which can comprise more factors, may cooperate with the LAG-1 complex to potentiate repression. Association of LAG-1 and LIN-12(INTRA) might or might not release the CoREST corepressor complex as well.
Compatible with this model, a simple hypothesis to account for hop-1-dependent suppression of sel-12 by reduced spr-1 activity is that hop-1(+) is derepressed in the π cells. We attempted to examine the level of hop-1 transcription in a spr-1 mutant background. As a detectable hop-1 transcriptional reporter gene has been elusive (X. Li, pers. comm.), we were unable to address this question with cellular resolution. Instead, we probed a Northern blot containing polyA(+) RNA prepared from hermaphrodites at the L3 and L4 stages (the time of π cell induction and utse formation) for the level of hop-1(+) mRNA expression. Hermaphrodites of genotype sel-12(ar171), which are egg-laying defective, appeared to have higher levels of hop-1(+) RNA than spr-1(ar200); sel-12(ar171) hermaphrodites, which are able to lay eggs, when normalized to an inf-1 (CeIF-4A) probe (data not shown). These results, although not definitive, suggest that a simple and general derepression of hop-1 transcription is unlikely to account for the phenotypic suppression observed.
Another simple hypothesis to account for suppression of the Egl phenotype of sel-12 is that in spr-1 mutants, lin-12(+) is derepressed in the π cells or more generally, or that lag-2(+) is derepressed in the anchor cell or more generally. However, we did not detect any ectopic expression, nor any increase in the level of expression, of the transcriptional reporters lin-12∷lacZ or lag-2∷lacZ in a spr-1(−) background (S. Jarriault and I. Greenwald, unpubl.; see Materials and Methods for more details). In contrast to our results with spr-1, strong and ectopic derepression of a lag-2 reporter gene has been observed in a mutant that lacks one of the histone deacetylases, with concomitant developmental defects (Dufourcq et al. 2002). Although it remains possible that loss of spr-1 activity leads to derepression of a gene whose activity is limiting for presenilin or LIN-12/Notch signal transducing activity in many different cell fate decisions, there is another class of models for the role of SPR-1 that fits with a large body of work suggesting that the balance between repression and activation of LIN-12/Notch target genes is important. These models will be considered further below.
Transcriptional repression and LIN-12/Notch signaling
Many pathways appear to activate target gene transcription by promoting a switch from a repressed state to an activated state, displacing a corepressor activity upon signaling (for review, see Barolo and Posakony 2002). This switch appears to be the case for the LIN-12/Notch signaling pathway. Prior to ligand binding, a corepressor complex is associated with the DNA binding protein CBF1 (see Introduction), which is bound to regulatory regions of LIN-12/Notch targets genes (Kao et al. 1998). Several proteins with demonstrated repressor activity have been shown to interact with CBF1 and negatively regulate Notch target genes in cultured cells. These proteins include KyoT (Taniguchi et al. 1998), CIR (Hsieh et al. 1999), SMRT, NcoR, and HDAC-1 (Kao et al. 1998). It is not clear, however, what the roles of these different repressors are in vivo, if all of them are recruited simultaneously to Notch target genes, to what extent they could act redundantly, or if spatio-temporal specificities exist.
Ligand binding leads to the presenilin-dependent transmembrane cleavage event that releases the intracellular domain of LIN-12/Notch, which translocates to the nucleus. There, it appears to mask the repressor domain of CBF1 (Waltzer et al. 1995; Hsieh et al. 1996) thereby displacing the CBF1-associated corepressor proteins (Kao et al. 1998). The LIN-12/Notch intracellular domain also appears to recruit coactivator proteins. These coactivators include Mastermind in Drosophila and mammals (Petcherski and Kimble 2000b; Wu et al. 2000; Kitagawa et al. 2001), and its likely counterpart, SEL-8 in C. elegans (Doyle et al. 2000; Petcherski and Kimble 2000a).
A reduction in presenilin activity leads to decreased release of the intracellular domain of LIN-12/Notch, and therefore to a decrease in signal transduction (De Strooper et al. 1999; Struhl and Greenwald 1999). In principle, then, it would be more difficult to convert CBF1 from a repressor to an activator. By removing repressor activity, the balance between activator and repressor might be restored. In sel-12 mutants, reduced presenilin activity results in the Egl phenotype; perhaps in spr mutants reduced repressor activity makes it easier for target genes to be activated by the reduced level of lin-12 signal transduction. In the next section, we consider models for how SPR-1 and SPR-5 might be involved in the repression of LIN-12 target genes, based on the available biochemical information about CoREST in mammalian cells as well as our analysis of spr-1 in C. elegans.
SPR-1/CoREST, SPR-5/p110b, and potential regulation of LIN-12/Notch target genes
One model for how SPR-1 and SPR-5 might contribute to repression of LIN-12 target genes is that the SPR-1/SPR-5 complex acts as a corepressor of LAG-1 in the absence of LIN-12/Notch activation, and is displaced upon LIN-12/Notch activation (Fig. 7B). This model is analogous to what has been proposed for CBF1 and SMRT (Kao et al. 1998). Although we have not been able to detect an interaction between CoREST and CBF1 in mammalian cells (S. Jarriault, unpubl.), it may be that the presence of CBF1 in association with the CoREST complex is evident only under certain conditions.
Another model is that SPR-1/CoREST and SPR-5/p110b are instead part of a corepressor complex that binds to LIN-12/Notch target genes through a sequence that is distinct from the LAG-1/CBF1 binding site. Therefore, there may be a specific DNA binding protein (denoted as X in Fig. 7C), which recruits the SPR-1/CoREST complex to LIN-12/Notch target genes. This model allows for potential cooperativity between the corepressor complex associated with LAG-1/CBF1 and the SPR-1/CoREST complex associated with factor X. CoREST has been shown to associate with at least two different zinc finger DNA-binding proteins, REST and ZNF217, suggesting that CoREST may have multiple DNA binding protein partners. In this context, it is interesting to note that Lakowski et al. have found that spr-3 and spr-4 encode zinc finger proteins (B. Lakowski, S. Eimer, and R. Baumeister, pers. comm.). Although these do not appear to be orthologous to either REST or ZNF217, they might represent the hypothetical DNA binding factor X that targets SPR-1 and SPR-5 to DNA.
Finally, the SPR-1/CoREST–SPR-5/p110b complex may be involved in more global or long-range repression, perhaps involving dynamic deacetylation of chromatin, rather than being specifically associated with LIN-12/Notch target genes. Perhaps a crucial target gene in the π cells is particularly susceptible to this long-range negative regulation, making the Egl phenotype of sel-12(−) hermaphrodites a sensitive background with which to detect subtle relief from a more generalized repression.
We note that the models involving recruitment of the SPR-1/SPR-5 complex to target genes by factor X could be applied either to LIN-12/Notch target genes directly or to intermediate genes in a genetic regulatory cascade. In addition, for all of the models, the nature of the target genes that would be sensitive to a reduction in CoREST/p110b-mediated repression is likely to be influenced by the number of the DNA binding sites allowing recruitment of this complex to regulatory regions and the relative contribution of the CoREST and potential other corepressor complexes to repression.
We also note that these models are not incompatible with the observation that spr-1(ar200) does not have lin-12(d)-associated cell fate transformations. In a sel-12(+) background, activation of a subset of LIN-12/Notch target genes may not be sufficient to cause a phenotype. Furthermore, there is precedent for transcriptional repressors to associate with more than one corepressor complex. Indeed, REST has two domains that recruit corepressor complexes: one recruits CoREST and the other recruits Sin3. When either of these domains is fused to a heterologous DNA-binding domain, or if either one of these domains is deleted, repression by the mutant REST protein is indistinguishable from repression by wild-type REST, suggesting that the corepressor complexes are redundant (Ballas et al. 2001). In addition, loss of the retinoblastoma (Rb) homolog lin-35 does not result in a visible phenotype in C. elegans, apparently because of functional redundancy with a group of novel proteins; roles for lin-35 Rb-mediated repression are only reveled in sensitized genetic backgrounds (Lu and Horvitz 1998).
All SPR proteins may be part of a single transcriptional repression complex
Five spr genes have been identified to date; all encode nuclear proteins (or proteins that are predicted to be nuclear), indicating that they may be physically as well as functionally associated. As we have discussed here, SPR-1 and SPR-5 correspond to CoREST and p110b, respectively, which have been found in a complex and mediate transcriptional repression. Previously, SPR-2 was shown to be orthologous to SET, a protein that has many biochemical activities attributed to it; intriguingly, much of the evidence points to a role in chromatin structure (summarized in Wen et al. 2000). Finally, Lakowski et al. have shown that SPR-3 and SPR-4 contain multiple zinc fingers (B. Lakowski, S. Eimer, and R. Baumeister, pers. comm.). Therefore, it is possible that a core complex consisting of SPR-1/CoREST, SPR-5/p110b, and possibly a histone deacetylase, is recruited to the regulatory region of target genes through its interaction with zinc finger proteins in worms. These target genes may be LIN-12/Notch target genes, as proposed above, or other genes that influence LIN-12/Notch signaling or cell fate specification. Loss of one of the members of such a complex might allow a weak LIN-12/Notch signal to activate the transcription of a subset of target genes that would otherwise not have been activated in the sel-12 background.
The spr gene screen has not been saturated (Wen et al. 2000), and other genes are likely to be found to have spr activity, either through conventional genetic screens or RNAi screens. The key challenges for the future will be to ascertain whether the proteins defined by these genes are part of a single complex and to find the target genes with which this complex (or complexes) is associated. The only potential LIN-12 target gene known to play a role in π cell specification is lin-11 (Gupta and Sternberg 2002), which appears to be unaffected in a spr-1 mutant background (S. Jarriault and I. Greenwald, unpubl.). As more potential LIN-12 target genes become available for study, this question may be addressed through biochemical as well as genetic approaches. Finally, it will be interesting to assess whether the SPR-1/CoREST, SPR-5/p110b corepressor complex is also involved in repression of the LIN-12/Notch signaling in other organisms as well.
Materials and methods
Genetic materials and methods
Standard methods were used for handling, maintenance, and genetic analysis. Experiments were conducted at 20°C unless otherwise indicated. The wild-type parent for most strains used in this study is C elegans var. Bristol strain N2. The relevant genes and alleles used in this study are:
LG I: hop-1(ar179), spr-4(ar208), spr(ar204), spr-5(ar197) (Wen et al. 2000; this study).
LG III: lin-12(n302, n950) (Greenwald et al. 1983), lin-12(ar170ts) (Hubbard et al. 1996), glp-1(e2142ts) (Priess et al. 1987).
LG V: spr-1(ar200, ar201, ar205, ar212) (Wen et al. 2000; this study).
LG X: sel-12(ar131, ar171) (Levitan and Greenwald 1995).
In addition, the following integrated transgenes have been used: nIs2 (containing lin-11∷lacZ; Freyd 1991), arIs11 (containing lin-12∷lacZ; Wilkinson et al. 1994), arIs13 (containing lag-2∷lacZ; Wilkinson et al. 1994), arIs41 (containing LIN-12∷GFP; Levitan and Greenwald 1998), arIs51 (containing pcdh-3∷gfp; Pettitt et al. 1996; X. Karp and I. Greenwald, unpubl.).
Additional information about these alleles, as well as about markers used for mapping or for facilitating genetic analysis mentioned in the text, can be found through WormBase (http://www.wormbase.org).
Mutant analysis and scoring
Strains were grown and scored at 20°C unless otherwise specified and assessed for phenotypes as follows:
Egg-laying ability: L4 larvae were picked to individual plates and scored for 3 consecutive days. An animal was scored as Egl+ if it showed robust egg-laying and Egl if it laid none or only a few eggs and “bagged” during this period of time.
utse: Late L4 larvae or young adults were examined using Nomarski microscopy.
Anchor cells: L3 larvae were scored using Nomarski microscopy.
Vulval development: The number of pseudovulval protrusions was scored under the dissecting microscope.
Embryonic lethality: L2 larvae grown at 20°C were transferred to individual plates at the nonpermissive temperature for glp-1(e2142), 25°C, and scored for their ability to give live progeny.
Genetic characterization of spr-1 mutants
None of the spr-1 mutants appeared to display obvious phenotypes. Unmarked spr-1(ar200) chromosomes used in this study have been backcrossed two and seven times. In particular, a spr-1(ar200) mutant that had been backcrossed seven times did not display any defect in the following scorings: 20/20 had a wild-type vulva and were Egl+ and fertile (>100 progeny; 15°C), 30/30 were Egl+ (20°C), 20/20 had one AC and did not appear to display any embryonic lethality (25°C). Therefore, spr-1(ar200) and N2 appear indistinguishable for those cell fate decisions.
spr-1 alleles all display semidominance in their ability to suppress the egg-laying defects of sel-12(ar171). The extent of this semidominance depends on the genotype of the mother, that is, whether the mother is heterozygous or homozygous for sel-12(ar171). More specifically, 9/54 (16.7%) of dpy-11(e224) spr-1(ar201)/+; sel-12(ar171) unc-1(e538) hermaphrodites were Egl+ when segregating from a heterozygous sel-12 mother whereas 2/61 (3.3%) of dpy-11(e224) spr-1(ar201)/+; sel-12(ar171) unc-1(e538) hermaphrodites were Egl+ when segregating from a homozygous sel-12 mother at 20°C.
We note that we did not observe suppression of the egg-laying defect caused by the partial loss-of-function allele lin-12(n676n930). Although we previously used this assay (and others) to evaluate spr-2 (Wen et al. 2000), we now discount a negative result in this assay because the egg-laying defect of this allele reflects the sum of several different cellular defects, so that more than one may need to be corrected for egg-laying ability to be restored (Sundaram and Greenwald 1993b).
Detailed genetic mapping of spr-1
SNPs from LG V were used for mapping spr-1(ar200). SNPs were identified in C. elegans var. Hawaii strain CB4856 by the Genome Sequencing Consortium (http://genome.wustl.edu/projects/celegans/chrom5_layout.html; vd50e09.s1 on cosmid T05B11; vr92g12.s1 on C37C3; vd50e05.s1 on F20D6; vm25h11.s1 on K07C11; and vc88c08.s1 on F21F8) or by us (SJ1C13498T, SJ2A13705, SJ3C13798A, SJ4ΔA16984, SJ5A17355C, and SJ6C18005T on cosmid F40A3; SJ7C28575T on F26D11; SJ8ICATTGGCT1363 and SJ9TA3428–29CΔ on Y97E10AL; SJ10T15326C on ZC513; SJ11C7063T, SJ12G19662T, and SJ13T20903 on CO9H5; SJ14Δ19850–19944 on D1014. Numbers refer to position of the SNPs in the cosmid sequences; Δ indicates deletion and the bold I indicates insertion). The mutant strain dpy-11(e224) spr-1(ar200) rol-3(e754); sel-12(ar171) unc-1(e538) was crossed to CB4856 and 109 Dpy non-Rol recombinants were isolated. Analysis of these recombinants allowed us to restrict the spr-1 interval to the region between SJ9TA3428–29CΔ on Y97E10AL and SJ14Δ19850–19944 on D1014.
Antisuppression experiments and sequencing
Cosmid DNA or PCR amplified genomic DNA spanning the determined spr-1 region (see above) was injected at 5 or 10 μg/mL each, alone or in pools, together with pRF4 (150 μg/mL) or pmyo3∷gfp (20 μg/mL) and pBluescript (≤200 μg/mL) into recipient strain spr-1(ar200); sel-12(ar171) unc-1(e538). As a control, each marker was injected with pBluescript (≤200 μg/mL). Transgenic lines were established and their egg-laying ability was checked for antisuppression, indicative of spr-1(+) activity. None of the pools that gave viable transgenic lines resulted in detectable antisuppression (data not shown). However, transgenic lines could not be established for cosmids DD11, F58G4, and T23D2 (=D1014); these cosmids appeared to cause larval lethality, so we sequenced candidate genes in the region. spr-1 was associated with mutations in D1014.8, as described in the Results section. A PCR fragment amplified from C. elegans genomic DNA with oligos 5′-D1014–8505 (CAGATGACCAG AAAGTGGTC) and 3′-D1014–12185 (GAGAGAGAAAAGA CACGGCC) and extending from the stop codon of predicted D1014.5 to the ATG of predicted D1014.3 was then injected at (20 μg/mL) together with pmyo3∷gfp (20 μg/mL) and pBluescript (≤100 μg/mL) into recipient strain spr-1(ar200); sel-12(ar171) unc-1(e538). Two transgenic lines were obtained that displayed antisuppression (line 1, 22/36 F2 and 50/73 F3 were Egl; line 2, 12/16 F2 and 26/46 F3 were Egl. All transgenic F2s were scored and around 13% in both lines arrested as young larvae). We note that when this PCR fragment was cloned into pCR-XL-TOPO (Invitrogen), and the GFP cDNA was introduced at the ATG or before the TAG of spr-1, we where not able to obtain viable lines displaying antisuppression, and no expression of SPR-1∷GFP as visualized in live worms or by anti-GFP antibody staining was observed (data not shown). Therefore, overexpression of SPR-1 under the control of its own promoter appears toxic. Nevertheless, fusion proteins similarly tagged at the C terminus, like SPR-1∷MYC or hCoREST∷HA, are functional when expressed under the control of a heterologous promoter (see text).
cDNA cloning and correction of the GENEFINDER prediction for D1014.8 and D1014.9
In order to determine spr-1 gene structure, and in the absence of available ESTs, three cDNA libraries were screened to clone spr-1 cDNA. Two rounds of nested PCRs were performed using: (1) spr-1 primers 3′-race1 (CCAGAACCATCAGACTCGC), 3′-race12 (GAGTAATTTCCTGATCTGTCC), and 5′-race9 (GAT CAGCGCTCTGTCATCG) and the library-specific primers 5′pPC86 (TATAACGCGTTTGGAATCACT) and 3′pPC86 (GTAAATTTCTGGCAAGGTAGAC) recommended by the manufacturer (ProQuest library from Life Technologies) or T3 and T7 (Stratagene's embryonic and mixed-stage libraries); (2) spr-1 primers 3′-race2 (5′-CTTCCGGTCGCCTGTGCGG-3′), 3′-race13 (5′-CGTTGTTGAGATGGGCGTGG-3′), 5′-race8 (5′-CAACTGTGGAGAAAAAGCTG-3′), and 3′-spr-1 (5′-GTAAA TTGAATCTTCTTCTTCTTCCATATCAGCCAAGCCTTTC TGG-3′) or an oligo(dT)(12–18) primer (GIBCO-BRL) and the library-specific primers 5′pPC86, 3′pPC86, or SK. This allowed the cloning of three overlapping fragments of cDNA that were pieced together by PCR (note that no spr-1 cDNA fragment could be amplified from the Life Technologies library). The resulting sequenced cDNA encompasses previously predicted genes D1014.9 and D1014.8 and represented the only mRNA species we could amplify (accession no. AAA96286). We communicated the spr-1 gene structure to WormBase which was then corrected accordingly. The cDNA corresponding to the mRNA species present in spr-1(ar200) mutants was synthesized using an oligo(dT)(12–18) primer (GIBCO-BRL) from total spr-1(ar200); sel-12(ar171) unc-1(e538) RNA and further amplified with primers 5′D1014.8/T3 (GCGCGCAAT TAACCCTCACTAAAGGCTTCTTCCATATCAGCCAGCC) and 3′D1014.8/T7 (CGCGCGTAATACGACTCACTATAGG GCTAACGCGATGAATCGTCCAG). Control experiments were performed similarly on total RNA fromsel-12(ar171)unc-1(e538) worms. The products were then sequenced and the consequences of the G-to-A mutation for the mutant transcript analyzed.
RNAi
Each RNA strand was synthesized according to the manufacturer (Stratagene) using as templates PCR fragments amplified from N2 genomic DNA with the following primers: spr-1 (corresponding to previously predicted D1014.8), 5′D1014.8/T3 (GCGCGCAATTAACCCTCACTAAAGGCTTCTTCCATAT CAGCCAGCC) and 3′D1014.8/T7 (CGCGCGTAATACGAC TCACTATAGGGCTAACGCGATGAATCGTCCAG); spr-1 (corresponding to previously predicted D1014.9), 5′D1014.9/T3 (GCGCGCAATTAACCCTCACTAAAGGGAAATTGCCTCT ACTGCTTTGG) and 3′D1014.9/T7 (CGCGCGTAATAC GACTCACTATAGGGCGATGGAGAATCTGCTCAATCCG); Y74C9A.4, 5′Y74C9A.4/T3 (GCGCGCAATTAACCCTCAC TAAAGGCAATATCCCGACGGAACCC) and 3′Y74C9A.4/T7 (CGCGCGTAATACGACTCACTATAGGGCATTCCCAA TTTCTTCCAGC); Y40B1B.6, 5′Y40B1B.6E4/T3 (GCGCGCAATTAACCCTCACTAAAGGGATATCGAAAACGAAAA AGTG) and 3′Y40B1B.6E4/T7 (CGCGCGTAATACGACT CACTATAGGGCCCATTTTCCTTAAACTTTC); R13G10.2, 5′R13G10.2E5/T3 (GCGCGCAATTAACCCTCACTAAAGGCCAGTTACCAATTTCCGGCAC) and 3′R13G10.2E5/T7 (CGCGCGTAATACGACTCACTATAGGGTCTAACATACC TTTCCAG); T08D10.2, 5′T08D10.2E4/T3 (GCGCGCAAT TAACCCTCACTAAAGGCTCTCGCAGAGGTTGCCCG) and 3′T08D10.2E4/T7 (CGCGCGTAATACGACTCACTATAGG GCTCGAGCAACTTGGCTTTTCAG). Each RNA strand was purified using QIAGEN RNeasy columns and annealed in 0.5× injection buffer (Mello and Fire 1995). RNAi was performed as in Fire et al. (1998). dsRNA or annealing buffer was microinjected into the pseudocoelomic space of young adults. Injected hermaphrodites were cultured individually and their progeny were scored for their ability to lay eggs. Alternatively, DNA was cloned into the vector pPD129.36 (see below) and transformed into bacterial strain HT115 (Timmons and Fire 1998; Timmons et al. 2001). Eggs or bagging mothers were placed on a lawn of such bacteria, and their ability to lay eggs was scored as adults.
Plasmid constructions
All constructs were sequenced.
RNAi constructs
The spr-1 cDNA fragment amplified from Stratagene's library with primers 3′-race2 and SK was subcloned into pCR-XL-TOPO (Invitrogen), excised with EcoRI and cloned into EcoRI digested vector pPD129.36 (Timmons et al. 2001). Exon E3 of C53A5.3 (hda-1), exon 2 of C08B11.2 (hda-2), and exon 2 of R06C1.1 (hda-3) were amplified from genomic DNA using primers 5′hda1–25/RI (CGGAATTCCGCACGGTAA GCGCCGTGTCG) and 3′hda1–1340/RI (CGGAATTCCGCT GCTCTCTTCGCATCGC), 5′-C08B11.2E2/RI (CGGAATTCC TTCATCCAATGAAACCTCAAC) and 3′-C08B11.2E2/RI (CG GAATTCCGACAAAGCAAATTGACC), or 5′-R06C1.1-E2/RI (CCGGAATTCCCTTTAAATCTCCAGTTC) and 3′-R06C1.1-E2/RI (CCGGAATTCCAGCGTCCCGCCCGTGCTTC), respectively, and cloned into EcoRI digested pPD129.36.
Expression constructs
pcog-2∷SPR1∷MYC was obtained by amplifying spr-1 cDNA with oligos 5′-Xma/spr-1 (5′-TC CCCCCGGGGGGTGTAGAAAATGGATTTGTATGACGAT GATGG-3′) and 3′-spr-1MYC/NcoI (5′-CCCATGCCATGGC TACAGATCCTCTTCTGAGATGAGTTTTTGTTCAATTGA ATCTTCTTCTTCTTCC-3′) and cloned into XmaI–NcoI digested pWH17 (pcog-2∷GFP; Hanna-Rose and Han 1999). pcog-2∷CoRest∷HA was obtained by amplifying human CoREST cDNA (You et al. 2001), previously subcloned in pCDNA3.1 with an HA tag added in frame before the stop codon, with oligos 5′-CoREST/Xma (5′-TCCCCCCGGGGGGAAAATG GTGGAGAAGGGCCCCGAGGTC-3′) and 3′-HA/NcoI (5′-CCCATGCCATGGTCAAGCATAATCAGGAACATC-3′) and cloned into XmaI–NcoI digested pWH17.
Expression in π cells and antibody staining
pcog-2∷SPR1∷MYC or pcog-2∷CoREST∷HA (40 μg/mL) were injected together with pWH17 (40 μg/mL) and pRF4 (150 μg/mL) into the germ line of spr-1(ar200); sel-12(ar171) unc-1(e538) (Mello and Fire 1995). As a control, pWH17 (80 μg/mL) and pRF4 (150 μg/mL) were injected alone. Four, two, and six transgenic lines were obtained, respectively, and their ability to lay eggs was analyzed by scoring all the transgenic animals of the broods segregating from at least two mothers or alternatively all the transgenic animals layed during the two first days of egg-laying. We note that one of the six control lines exhibited comparatively a higher level of Egl animals. Expression of the cog-2∷gfp reporter was observed in live transgenic animals. Synchronized transgenic populations were stained according to Bettinger et al. (1996) using antibodies against the MYC tag (146.7 μg/mL, monoclonal 9E10, SIGMA) or the HA tag (2 μg/mL, monoclonal 12CA5, Roche) and secondary antibodies (Jackson ImmunoResearch Laboratories, nos. 115-165-146 and 115-095-146) together with DAPI, and both SPR1∷MYC and CoRest∷HA proteins were found to be expressed in the nuclei of the expected cells. To analyze lin-11 expression in π cells, synchronized strains containing nIs2 (see text) were stained according to Bettinger et al. (1996) using antibodies against LacZ protein (Promega, no. Z3781) and secondary antibodies (Jackson ImmunoResearch Laboratories, no. 115-165-146) together with DAPI. As a staining and staging marker, MH27 antibodies against JAM-1 (Francis and Waterston 1991) were also used on a fraction of the fixed animals. Worms were mounted in a drop of antifade (Molecular Probes) and analyzed with a fluorescence microscope (Zeiss Axioplan 2).
LacZ activity staining
It has been reported previously that LacZ staining of the lin-12∷lacZ transcriptional reporter arIs11 appears faster in lin-12(d) compared to lin-12(+) worms (Wilkinson et al. 1994). To assess whether spr-1(ar200) affected lin-12 or lag-2 expression, we analyzed the speed and intensity of LacZ staining of the transcriptional reporters arIs11 and arIs13, respectively (Wilkinson et al. 1994). Synchronized populations were fixed and stained for various periods of time (2.5 h, 4.5 h, 6.5 h, and overnight) according to A. Fire (1992). Staining was analyzed for the following cell fate decisions: AC/VU decision (lag-2), VPC specification (lin-12), and π cell specification. No difference in staining speed or intensity was observed between spr-1(ar200) and control worms.
Acknowledgments
We thank Angie You and Stuart Schreiber for the human CoREST-FLAG plasmid, and Marty Chalfie, Alan Coulson, Wendy Hanna-Rose, Andy Fire, Barth Grant, and Theresa Stiernagle for providing useful plasmids, primers, or nematode strains. We are grateful to Carlos Bais, Oliver Hobert, Xantha Karp, and Daniel Shaye for a critical reading of the manuscript; Anna Newman for communicating helpful information prior to publication; Bernard Lakowski and Stefan Eimer for discussion and permission to cite unpublished information; and Veronique Hermitte and Richie Ruiz for valuable technical assistance. Some of the strains used in this study were provided by the Caenorhabditis Genetics Center. This work was supported by a postdoctoral long-term fellowship from the Human Frontier Science Program awarded to S.J., NIH grant NS35556 awarded to I.G., and by the Howard Hughes Medical Institute. S.J. is a postdoctoral associate and I.G. is an investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL greenwald@cancercenter.columbia.edu; FAX (212) 305-1721.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1022402.
References
- Aasland A, Stewart AF, Gibson T. The SANT domain: A putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcritional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: A functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Barolo S, Posakony JW. Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes & Dev. 2002;16:1167–1181. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- Bettinger JC, Lee K, Rougvie AE. Stage-specific accumulation of the terminal differentiation factor LIN-29 during Caenorhabditis elegansdevelopment. Development. 1996;122:2517–2527. doi: 10.1242/dev.122.8.2517. [DOI] [PubMed] [Google Scholar]
- Cinar HN, Sweet KL, Hosemann KE, Earley K, Newman AP. The SEL-12 presenilin mediates induction of the Caenorhabditis elegansuterine π cell fate. Dev Biol. 2001;237:173–182. doi: 10.1006/dbio.2001.0374. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- Doyle TG, Wen C, Greenwald I. SEL-8, a nuclear protein required for LIN-12 and GLP-1 signaling in Caenorhabditis elegans. Proc Natl Acad Sci. 2000;97:7877–7881. doi: 10.1073/pnas.97.14.7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourcq P, Victor M, Gay F, Calvo D, Hodgkin J, Shi Y. Functional requirement for histone deacetylase 1 in Caenorhabditis elegansgonadogenesis. Mol Cell Biol. 2002;22:3024–3034. doi: 10.1128/MCB.22.9.3024-3034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A. Histochemical techniques for locating Escherichia coliβ-galactosidase activity in transgenic orgamisms. Genet Anal Tech Appl. 1992;9:151–158. doi: 10.1016/1050-3862(92)90042-4. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Francis R, Waterston RH. Muscle cell attachment in Caenorhabditis elegans. J Cell Biol. 1991;114:465–479. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyd G. “Molecular analysis of the Caenorhabditis elegans cell lineage gene lin-11.” Ph.D. thesis. Cambridge, MA: Massachusetts Institute of Technology; 1991. [Google Scholar]
- Furriols M, Bray S. Dissecting the mechanisms of suppressor of hairless function. Dev Biol. 2000;227:520–532. doi: 10.1006/dbio.2000.9923. [DOI] [PubMed] [Google Scholar]
- Greenwald I. LIN-12/Notch signaling: Lessons from worms and flies. Genes & Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- Greenwald IS, Horvitz HR. unc-93(e1500): A behavioral mutant of Caenorhabditis elegansthat defines a gene with a wild-type null phenotype. Genetics. 1980;96:147–164. doi: 10.1093/genetics/96.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I, Rubin GM. Making a difference: The role of cell–cell interactions in establishing separate identities for equivalent cells. Cell. 1992;68:271–281. doi: 10.1016/0092-8674(92)90470-w. [DOI] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, Horvitz HR. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. 1983;34:435–444. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- Grimes J, Nielsen SJ, Battaglioli E, Miska EA, Speh JC, Berry DL, Atouf F, Holdener BC, Mandel G, Kouzarides T. The co-repressor mSin3A is a functional component of the REST–CoREST repressor complex. J Biol Chem. 2000;275:9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- Gupta BP, Sternberg PW. Tissue-specific regulation of the LIM homeobox gene lin-11 during development of the Caenorhabditis elegansegg-laying system. Dev Biol. 2002;247:102–115. doi: 10.1006/dbio.2002.0688. [DOI] [PubMed] [Google Scholar]
- Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. A core–BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna-Rose W, Han M. COG-2, a Sox domain protein necessary for establishing a functional vulval–uterine connection in Caenorhabditis elegans. Development. 1999;126:169–179. doi: 10.1242/dev.126.1.169. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Hayward SD. Masking of the CBF1/RBPJκ transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD. Truncated mammalian Notch1 activates CBF1/RBPJκ-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard EJ, Dong Q, Greenwald I. Evidence for physical and functional association between EMB-5 and LIN-12 in Caenorhabditis elegans. Science. 1996;273:112–115. doi: 10.1126/science.273.5271.112. [DOI] [PubMed] [Google Scholar]
- Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, Howard BH. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes & Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev Biol. 1981;87:286–300. doi: 10.1016/0012-1606(81)90152-4. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Oyama T, Kawashima T, Yedvobnick B, Kumar A, Matsuno K, Harigaya K. A human protein with sequence similarity to Drosophilamastermind coordinates the nuclear form of notch and a CSL protein to build a transcriptional activator complex on target promoters. Mol Cell Biol. 2001;21:4337–4346. doi: 10.1128/MCB.21.13.4337-4346.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Goate A. A common enzyme connects notch signaling and Alzheimer's disease. Genes & Dev. 2000;14:2799–2806. doi: 10.1101/gad.836900. [DOI] [PubMed] [Google Scholar]
- Levitan D, Greenwald I. Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegansS182 Alzheimer's disease gene. Nature. 1995;377:351–354. doi: 10.1038/377351a0. [DOI] [PubMed] [Google Scholar]
- ————— LIN-12 protein expression and localization during vulval development in C. elegans. Development. 1998;125:3101–3109. doi: 10.1242/dev.125.16.3101. [DOI] [PubMed] [Google Scholar]
- Li X, Greenwald I. HOP-1, a Caenorhabditis eleganspresenilin, appears to be functionally redundant with SEL-12 presenilin and to facilitate LIN-12 and GLP-1 signaling. Proc Natl Acad Sci. 1997;94:12204–12209. doi: 10.1073/pnas.94.22.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Horvitz HR. lin-35 and lin-53, two genes that antagonize a C. elegansRas pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95:981–991. doi: 10.1016/s0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Morel V, Schweisguth F. Repression by suppressor of hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophilaembryo. Genes & Dev. 2000;14:377–388. [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Sternberg PW. Coordinated morphogenesis of epithelia during development of the Caenorhabditis elegansuterine-vulval connection. Proc Natl Acad Sci. 1996;93:9329–9333. doi: 10.1073/pnas.93.18.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, White JG, Sternberg PW. The Caenorhabditis elegans lin-12gene mediates induction of ventral uterine specialization by the anchor cell. Development. 1995;121:263–271. doi: 10.1242/dev.121.2.263. [DOI] [PubMed] [Google Scholar]
- ————— Morphogenesis of the C. eleganshermaphrodite uterus. Development. 1996;122:3617–3626. doi: 10.1242/dev.122.11.3617. [DOI] [PubMed] [Google Scholar]
- Newman AP, Acton GZ, Hartwieg E, Horvitz HR, Sternberg PW. The lin-11 LIM domain transcription factor is necessary for morphogenesis of C. elegansuterine cells. Development. 1999;126:5319–5326. doi: 10.1242/dev.126.23.5319. [DOI] [PubMed] [Google Scholar]
- Newman AP, Inoue T, Wang M, Sternberg PW. The Caenorhabditis elegans heterochronic gene lin-29coordinates the vulval-uterine-epidermal connections. Curr Biol. 2000;10:1479–1488. doi: 10.1016/s0960-9822(00)00827-7. [DOI] [PubMed] [Google Scholar]
- Petcherski AG, Kimble J. LAG-3 is a putative transcriptional activator in the C. elegansNotch pathway. Nature. 2000a;405:364–368. doi: 10.1038/35012645. [DOI] [PubMed] [Google Scholar]
- ————— Mastermind is a putative activator for Notch. Curr Biol. 2000b;10:R471–R473. doi: 10.1016/s0960-9822(00)00577-7. [DOI] [PubMed] [Google Scholar]
- Pettitt J, Wood WB, Plasterk RH. cdh-3, a gene encoding a member of the cadherin superfamily, functions in epithelial cell morphogenesis in Caenorhabditis elegans. Development. 1996;122:4149–4157. doi: 10.1242/dev.122.12.4149. [DOI] [PubMed] [Google Scholar]
- Priess JR, Schnabel H, Schnabel R. The glp-1 locus and cellular interactions in early C. elegansembryos. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Ray WJ, Yao M, Mumm J, Schroeter EH, Saftig P, Wolfe M, Selkoe DJ, Kopan R, Goate AM. Cell surface presenilin-1 participates in the γ-secretase-like proteolysis of Notch. J Biol Chem. 1999;274:36801–36807. doi: 10.1074/jbc.274.51.36801. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Greenwald I. Cell autonomy of lin-12 function in a cell fate decision in C. elegans. Cell. 1989;57:1237–1245. doi: 10.1016/0092-8674(89)90060-3. [DOI] [PubMed] [Google Scholar]
- Solari F, Bateman A, Arhinger J. The Caenorhabditis elegans genes egl-27 and egr-1are similar to MTA-1, a member of a chromatin regulatory complex, and are redundantly required for embryonic patterning. Development. 1999;126:2483–2494. doi: 10.1242/dev.126.11.2483. [DOI] [PubMed] [Google Scholar]
- Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- ————— Presenilin-mediated transmembrane cleavage is required for Notch signal transduction in Drosophila. Proc Natl Acad Sci. 2001;98:229–234. doi: 10.1073/pnas.011530298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M, Greenwald I. Suppressors of a lin-12 hypomorph define genes that interact with both lin-12 and glp-1 in Caenorhabditis elegans. Genetics. 1993a;135:765–783. doi: 10.1093/genetics/135.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Genetic and phenotypic studies of hypomorphic lin-12 mutants in Caenorhabditis elegans. Genetics. 1993b;135:755–763. doi: 10.1093/genetics/135.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Furakawa T, Tun T, Han H, Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol Cell Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tax FE, Thomas JH, Ferguson EL, Horvitz HR. Identification and characterization of genes that interact with lin-12 in Caenorhabditis elegans. Genetics. 1997;147:1675–1695. doi: 10.1093/genetics/147.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH. Thinking about genetic redundancy. Trends Genet. 1993;9:395–399. doi: 10.1016/0168-9525(93)90140-d. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Waltzer L, Bourillot PY, Sergeant A, Manet E. RBP-Jκ repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA2. Nucleic Acids Res. 1995;23:4939–4945. doi: 10.1093/nar/23.24.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmaster G. Notch signal transduction: A real rip and more. Curr Opin Genet Dev. 2000;10:363–369. doi: 10.1016/s0959-437x(00)00097-6. [DOI] [PubMed] [Google Scholar]
- Wen C, Levitan D, Li X, Greenwald I. spr-2, a suppressor of the egg-laying defect caused by loss of sel-12 presenilin in Caenorhabditis elegans, is a member of the SET protein subfamily. Proc Natl Acad Sci. 2000;97:14524–14529. doi: 10.1073/pnas.011446498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlund B, Parry D, Clover R, Basson M, Johnson CD. Reverse genetic analysis of Caenorhabditis elegans presenilins reveals redundant but unequal roles for sel-12 and hop-1in Notch-pathway signaling. Proc Natl Acad Sci. 1999;96:2497–2502. doi: 10.1073/pnas.96.5.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson HA, Greenwald I. Spatial and temporal patterns of lin-12 expression during C. eleganshermaphrodite development. Genetics. 1995;141:513–526. doi: 10.1093/genetics/141.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson HA, Fitzgerald K, Greenwald I. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. eleganscell fate decision. Cell. 1994;79:1187–1198. doi: 10.1016/0092-8674(94)90010-8. [DOI] [PubMed] [Google Scholar]
- Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of Drosophilamastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST–human histone deacetylase complex. Proc Natl Acad Sci. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Nadeau P, Song W, Donoviel D, Yuan M, Bernstein A, Yankner BA. Presenilins are required for γ-secretase cleavage of β-APP and transmembrane cleavage of Notch-1. Nat Cell Biol. 2000;2:463–465. doi: 10.1038/35017108. [DOI] [PubMed] [Google Scholar]
- Zhou S, Fujimuro M, Hsieh JJ, Chen L, Miyamoto A, Weinmaster G, Hayward SD. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol Cell Biol. 2000;20:2400–2410. doi: 10.1128/mcb.20.7.2400-2410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]