Abstract
Termination of transcription is a key process in the regulation of mitochondrial gene expression in animal cells. To investigate transcription termination in sea urchin mitochondria, we cloned the mitochondrial RNA polymerase (mtRNAP) of Paracentrotus lividus and used a recombinant form of the enzyme in a reconstituted transcription system, in the presence of the DNA-binding protein mtDBP. Cloning of mtRNAP was performed by a combination of PCR with degenerate primers and library screening. The enzyme contains 10 phage-like conserved motifs, two pentatricopeptide motifs and a serine-rich stretch. The protein expressed in insect cells supports transcription elongation in a promoter-independent assay. Addition of recombinant mtDBP caused arrest of the transcribing mtRNAP when the enzyme approached the mtDBP-binding site in the direction of transcription of mtDNA l-strand. When the polymerase encountered the protein-binding site in the opposite direction, termination occurred in a protein-independent manner, inside the mtDBP-binding site. Pulse-chase experiments show that mtDBP caused true transcription termination rather than pausing. These data indicate that mtDBP acts as polar termination factor and suggest that transcription termination in sea urchin mitochondria could take place by two alternative modes based on protein-mediated or sequence-dependent mechanisms.
INTRODUCTION
Maintenance and expression of the mitochondrial genome is accomplished by both nuclear and mitochondrial encoded proteins. Transcription of mitochondrial DNA (mtDNA) is carried out by a basic machinery consisting of the mitochondrial RNA polymerase (mtRNAP) and accessory factors, all encoded by nuclear genes (1,2).
Complete sequences have been reported for the mtRNAP of many different eukaryotes including yeasts (3), protozoans (4,5), fungi (6,7), insects (8), vertebrates (9) and higher plants (10,11). Despite the endosymbiotic origin of mitochondria, organelle RNA polymerases do not resemble the bacterial multi-subunit enzyme. Instead, they consist of a single catalytic subunit and display a strong sequence similarity to the single-polypeptide RNA polymerase of bacteriophages T3 and T7. It is likely that the mtRNAP gene was acquired from an ancestor bacteriophage, early in eukaryotic evolution, at the time of the mitochondrial endosymbiosis (12). The similarity between phage and mtRNAPs occurs in 10–12 conserved domains, in the carboxy-terminal portion of the protein, that were initially identified by aligning the mitochondrial S. cerevisiae and bacteriophage RNA polymerases (3,4,13). The catalytic role of those motifs was defined for the T7 enzyme that has been crystallized and extensively studied (14,15). Unlike bacteriophage RNA polymerases, all the mitochondrial enzymes exhibit a highly divergent extension at the amino-terminal region that accounts for the size variability among the mtRNAPs. The N-terminal extension and the phage-type conserved domains are considered the typical hallmarks of the mtRNAPs.
The mitochondrial transcription machinery has been extensively studied in budding yeast and in humans. In S. cerevisiae, the core RNA polymerase (Rpo41p) forms a heterodimer with the specificity factor sc-mt-TFB (Mtf1p), to constitute a competent holoenzyme for accurate promoter recognition (16). The transcription machinery in mammals is more complex as it requires the mtRNAP and additional transcription factors. Transcription initiation is assisted by the high-mobility group-box protein TFAM and either TFB1M or TFB2M, two proteins homologous to sc-mtTFB (1,2,17). Those proteins have been highly purified in recombinant form from insect cells and used to develop a basal in vitro transcription system in the presence of a promoter-containing DNA fragment (18). Termination of transcription in mammals is mediated by the factor mTERF through its interaction with a tridecamer sequence placed downstream of the 3′-end of the ribosomal gene unit (19). The biochemical characteristics of the human termination factor have been investigated in the basal reconstituted transcription system. The relevant conclusion of this study was that the recombinant human mTERF is fully active in a monomeric and non-phosphorylated form to promote bidirectional arrest of transcription in vitro (20). Interestingly, characterization of the recombinant rat mTERF suggested that phosphorylation is required for transcription termination activity (21).
The peculiar organization of mitochondrial genomes in invertebrates prompted many studies addressed to clarify the transcription mechanisms in such systems. In sea urchin it was demonstrated that the mitochondrial genome is transcribed through partially overlapping transcription units, probably initiating at multiple points (22,23). We identified the mitochondrial DNA-binding protein mtDBP, homologous to human mTERF, that binds two sequences, one located in the main non-coding region (NCR), at the 3′-end of the D-loop structure, and the other at the border of the oppositely transcribed genes ND5 and ND6 (24). By in vitro heterologous transcription assays we showed that mtDBP is able to terminate transcription with a biased polarity depending on the direction of the approaching RNA polymerase (25). These findings aroused interest in the mechanism by which mtDBP terminates transcription and prompted us to develop a homologous in vitro transcription assay that is competent for termination.
Here we report the cDNA cloning and functional characterization of the mitochondrial RNA polymerase from sea urchin Paracentrotus lividus. The cloned mtRNAP has a predicted length of 1421 amino acids and displays the shared features of mtRNAPs. The protein, expressed in insect cells, is able to support RNA synthesis on a non-selective template. Transcription experiments performed in the presence of the termination factor mtDBP, also expressed in insect cells, show that the transcribing mtRNAP is arrested in a protein-dependent manner only when the orientation of mtDBP-binding site is opposite to the direction of elongation. When the enzyme travels in the other direction, transcription is not affected by the bound protein but rather stops prematurely inside the protein-binding site.
MATERIALS AND METHODS
Amplification, cloning and sequencing of P. lividus mtRNAP cDNA
The initial 216-bp fragment of mtRNAP cDNA was generated by PCR amplification on a λ Uni-Zap cDNA library from P. lividus embryos at the four blastomere stage. Primers were two inosine-containing oligonucleotides designed on the highly conserved amino-acid sequences CNGLQHYA and FPPNFIH found in boxes V and VIII, respectively (Figures 1 and 2). The reaction was carried out in a 50 μl volume, in the presence of 1 × 107 phages, 500 μM dNTPs, 2 μM each primer and 2.5 U of Expand Long Template PCR System (Roche). After heating at 94°C for 5 min, the reaction was subjected to 30 cycles of 94°C for 1 min, 45°C for 1 min, 50°C for 30 s, 55°C for 10 s, 68°C for 1 min. A 1 μl sample of the PCR mixture was reamplified as before, and a 3 μl sample was then subjected to a third round of PCR, performed in the same conditions, using the same primer For (I.For1) and a nested, inosine-containing primer (I.Rev2) designed on the highly conserved amino-acid sequence KQTVMTVV placed in box VI. The 216-bp product was gel-purified and inserted into pGEM-T easy vector (Promega). To extend the sequence, additional amplification reactions were carried out in the presence of either two primers both degenerate, or a combination of degenerate and specific primers (Figure 1). PCR products were cloned into PCR 2.1 TOPO-TA Cloning vector (Invitrogen). Additional cDNA sequences were obtained by screening about 8 × 105 plaques of the P. lividus cDNA library with the known cDNA portion. For 3′-RACE, egg poly(A)+ RNA was prepared as described (24). One microgram of poly(A)+ RNA was reverse-transcribed with a 36mer oligonucleotide containing (dT)15 (24), using the ThermoScript Reverse Transcriptase (Invitrogen). The specific product was obtained by performing two rounds of PCR using PfuUltra High Fidelity DNA polymerase (Stratagene); primers were the oligo(dT) and the specific primer For (nt 3823–3845 on the coding sequence of P. lividus mtRNAP cDNA) in the first PCR reaction, and the oligo(dT) and the specific nested primer Sp.For5 (nt 4069–4092) in the second reaction. 5′-RACE was carried out on 1 μg of egg poly(A)+ RNA using the SMART RACE cDNA amplification kit (Clontech) and the specific primer Rev (nt 1807–1783). Two rounds of PCR followed: the first reaction with the company supplied primer For UPM (Universal Primer Mix) and the same primer Rev, the nested reaction using the company supplied nested primer NUP (Nested Universal Primer) and the specific primer Sp.Rev1 (nt 1758–1735). The specific 5′-RACE product was identified by Southern blot hybridization. The 5′-end of the cDNA was further extended by screening a P. lividus λ FIX II genomic library using as a probe the cDNA fragment isolated from the cDNA library. Several positive clones were isolated; for sequencing, λ DNA was purified using the High Pure Lambda Isolation Kit (Roche). The very 5′-end of the cDNA plus upstream sequence was obtained by PCR screening of some positive plaques using the Expand Long Template PCR System (Roche) in the presence of the specific primer Sp.Rev2 (nt 333–310) and primer T7. The pBluescript recombinant plasmid isolated from the cDNA library and the cDNA amplification products inserted into TA vectors were sequenced manually using the Thermo Sequenase (USB). The recombinant λ DNA and the full-length mtRNAP cDNA were subjected to automated sequencing on both strands, performed by the CRIBI sequencing facility at the Università degli Studi, Padova, Italy, and by the Sequence Laboratories SEQLAB, Gottingen, Germany.
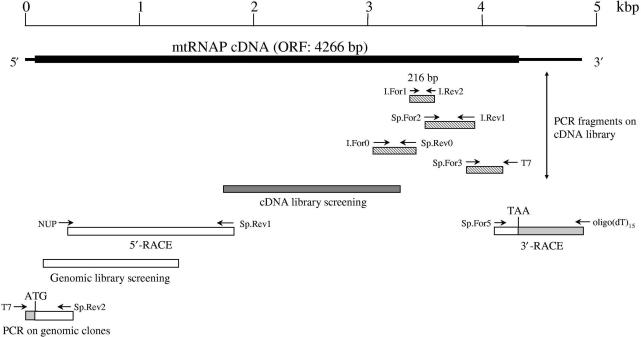
Figure 1.
Cloning strategy of the sea urchin mtRNAP cDNA. The 4266 bp ORF is represented by a thick line, the untranslated sequences by thin lines. Below the complete cDNA are positioned the cDNA fragments isolated by PCR on the cDNA library with degenerate, inosine-containing primers and vector- or internal-specific primers (striped bars); the fragment obtained by hybridization-screening of the cDNA library (heavy-shaded bar); the fragments obtained by RACE, hybridization-screening of the genomic library and PCR on genomic clones (open bars, with the shaded portion indicating the untranslated sequences). The position of the initiating ATG and stop TAA codons is shown. I, inosine-containing primers; Sp, specific primers; NUP (Nested Universal Primer, Clontech), 5′-RACE commercial primer.
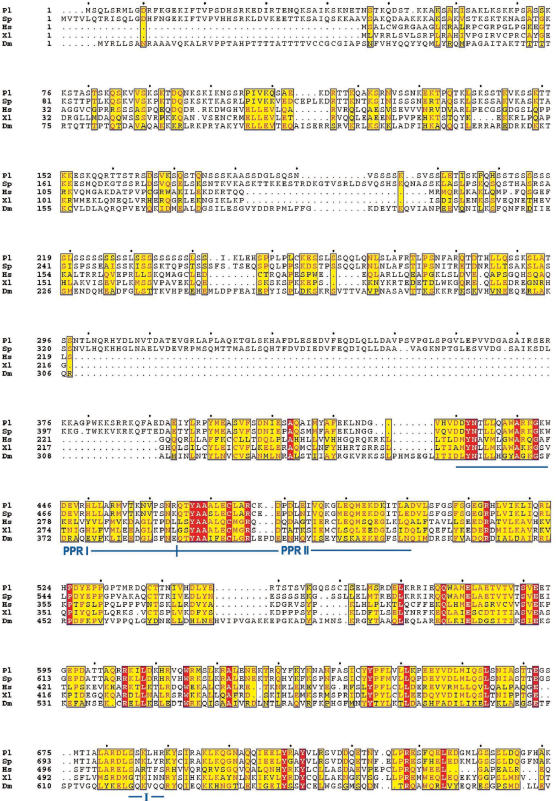
Figure 2.
Multiple alignment of mtRNA polymerases. Amino-acid sequences of mtRNAP from P. lividus (EF068167), Pl; S. purpuratus (29), Sp; H. sapiens (U75370), Hs; X. laevis (AF200705), Xl, and D. melanogaster (AE003587), Dm, were aligned with the ClustalW program. The multiple alignment was performed at NPS@ Web server of the PBIL using GONNET weight matrix and formatted with the Web tool ESPript 2.2. Red boxes include amino acids identical in all sequences; yellow boxes include amino acids similar or identical (red letters) in at least three of the five sequences; black letters, non-conserved residues. The 10 conserved blocks and the two putative PPR motifs are indicated by continuous lines; the DX2GR motif, that is placed inside block III, is indicated by a striped box The amino acids of the P. lividus enzyme implicated in the catalysis, as inferred from T7 mutant studies, are indicated by asterisks.
Construction of the full-length cDNA coding for mtRNAP
The full-length cDNA (GenBank accession no. EF068167) was obtained by reverse-transcription of about 1 μg of egg poly(A)+ RNA with the primer Rev (nt 4266–4246), using the ThermoScript Reverse Transcriptase (Invitrogen). PCR was performed in the presence of the same primer Rev and the primer For (nt 1–24) using PfuUltra High Fidelity DNA polymerase (Stratagene). The PCR product was cloned into PCR 2.1 TOPO-TA Cloning vector (Invitrogen); the insert was sequenced automatically on both strands.
Construction of the baculovirus recombinant expression vectors and protein production in insect cells
pBacPAK9 baculovirus transfer vectors containing either mtRNAP or mtDBP, both lacking the mitochondrial presequence, were prepared by standard DNA manipulation techniques. The cDNA encoding mtRNAP from amino acids 16–1421, and bearing an hepta-histidine tag at its N-terminus, was amplified by PCR on the full-length mtRNAP cDNA, in the presence of the enzyme Expand High FidelityPLUS PCR System (Roche) and proper primers. Primer For contained at its 5′-end an EcoRI site followed by the sequences coding for the added initial methionine and the seven histidines; primer Rev contained an XhoI site at the 5′-end. The cDNA coding for the mature mtDBP was amplified by PCR on the recombinant pBluescript-mtDBP (24) using two specific primers For and Rev bearing at the 5′-end an EcoRI and XhoI site, respectively. MtRNAP and mtDBP amplification products were subjected to restriction digestion, gel-purified and inserted into the EcoRI/XhoI sites of the transfer vector pBacPAK9 (Clontech). Linearized baculovirus DNA and individual recombinant pBacPAK9 plasmids were co-transfected in Spodoptera frugiperda (Sf9) cells according to the manufacturer's protocol (Clontech). Recombinant viruses were plaque purified and evaluated by PCR for the presence of the insert; viral stocks were prepared by three-step growth amplification, as described in the BacPAK manual (Clontech). For protein expression, Sf9 cells (400 ml, about 8 × 108 cells) were grown in suspension at 27°C, in SFM-900 II insect cell culture medium with l-Glutamine (Invitrogen), supplemented with 2% fetal bovine serum (Invitrogen) and 1% Penicillin/Streptomycin antibiotic mixture (Invitrogen). Cells were infected with mtDBP recombinant baculovirus (10 plaque forming unit/cell) or co-infected with mtRNAP and TFB1M recombinant viruses (18) (5 plaque forming unit of each virus/cell). Cells were collected 60–72 h after infection, washed with an equal volume of cold phosphate-buffered saline, frozen in liquid nitrogen and stored at −80°C.
Protein purification
All operations were performed at 0–4°C. To purify the isolated recombinant mtRNAP from the cells co-expressing mtRNAP and TFB1M, we used Ni+2-NTA agarose affinity chromatography followed by a further purification on Heparin–Sepharose resin (Amersham Biosciences). The frozen pellet deriving from 400 ml cell culture was resuspended in 10 ml of lysis buffer [50 mM sodium phosphate buffer pH 7.8, 10% glycerol, 10 mM β-mercaptoethanol, the following protease inhibitors (Sigma) 1 mM PMSF, 2 mM Pepstatin A, 0.02 mM Leupeptin, 2 mM Benzamidine] and incubated on ice for 20 min. Cells were then lysed by 20 strokes in a Dounce homogenizer using a tight-fitting pestle. NaCl was added to a final concentration of 0.8 M and the homogenate volume was brought to 20 ml with lysis buffer. After incubation at 4°C for 45 min with gentle rotation, the homogenate was centrifuged at 130 000 × g in the Beckman 70.1 Ti rotor for 45 min at 4°C. The supernatant was passed 5× through an 18-gauge needle to shear the DNA; it was then supplemented with imidazole solution (pH adjusted to 7.0) to obtain a final concentration of 10 mM and mixed with 2 ml of Ni+2-NTA agarose beads (QIAGEN) equilibrated in buffer A (50 mM sodium phosphate buffer pH 7.8, 10% glycerol, 0.8 M NaCl, 10 mM β-mercaptoethanol, protease inhibitor mix as before) supplemented with 10 mM imidazole. The suspension was incubated at 4°C for 2 h with gentle rotation. The beads were collected by centrifugation for 10 min at 1500 × g. After removal of the supernatant (flow-through), the beads were washed with buffer A containing 20 mM imidazole for 10 min with gentle rotation. The beads were then collected by centrifugation and the supernatant (wash) removed; the pelleted beads were gently resuspended in 10 ml of buffer A supplemented with 20 mM imidazole and packed into a Poly-Prep chromatography column (Bio-Rad). Proteins bound to the beads were eluted with buffer A containing 250 mM imidazole. After SDS–PAGE and western blot analysis, immunopositive fractions were pooled, diluted to 0.13 M NaCl with buffer B (25 mM Tris–HCl pH 8.0, 0.5 mM EDTA, 10% glycerol, 1 mM DTT, protease inhibitor mix) and subjected to fast-liquid chromatography onto a 1-ml Heparin–Sepharose (Amersham Biosciences) column, previously equilibrated with buffer B containing 0.13 M NaCl. After sample injection, the column was washed with three volumes of buffer B containing 0.13 M NaCl. Bound proteins were eluted with a linear NaCl gradient from 0.13 to 1.2 M in buffer B. Fractions of about 0.4 ml were collected and analysed by SDS–PAGE and immunoblotting. The recombinant mtRNAP eluted at NaCl concentration between 0.75 and 0.9 M, as determined by immunoblotting analysis. To purify the recombinant mtDBP, 200 ml of infected Sf9 cells were lysed as before, the lysate was clarified and passed through an 18-gauge needle, and then loaded onto a 5-ml Heparin–Sepharose (Amersham Biosciences) column, previously equilibrated with buffer C (25 mM Tris–HCl pH 8.0, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM DTT, protease inhibitor mix) supplemented with 0.15 M NaCl. The lysate was diluted to 0.15 M NaCl with buffer C prior to loading; the column was washed with buffer C, and mtDBP was eluted with three column volumes each of a step gradient of 0.3, 0.5 and 0.8 M NaCl in buffer C. The peak fractions of mtDBP were found at 0.5 M NaCl. They were pooled, ultrafiltered with Centricon 100 (Amicon) to eliminate high molecular weight molecules, and the filtrate collected. The purity of mtDBP in the filtrate was about 80% as estimated by SDS–PAGE and Coomassie Brilliant Blue staining.
Western immunoblot analysis
Column fractions were separated on 7.5 or 10% SDS–polyacrylamide mini-gels. Proteins were then electrotransferred to a polyvinylidene fluoride (PVDF) membrane (Immobilon P, Millipore) for 16 h. Membranes were blocked for 1 h with 5% non-fat dry milk in phopsphate-buffered saline containing 0.1% Tween 20; it followed an incubation for 1 h, in phopsphate-buffered saline containing 0.1% Tween 20, with polyclonal affinity-purified antibodies raised against the synthesized peptide CKDPDLEIVQKGLEQMEKDK of mtRNAP (AgriSera, Sweden) or against recombinant human TFB1M (18). The secondary antibody was an anti-rabbit IgG conjugated to horseradish peroxidase (Amersham Biosciences). Signals were generated using the ECL Plus Detection System (Amersham Biosciences). Quantitative analysis was performed with ChemiDoc using Quantity-One software (Bio-Rad Laboratories).
Protein analysis with mass spectrometry
Matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) analysis of in-gel digested proteins was carried out with an Ultraflex TOF/TOF (Bruker). The samples were prepared by in gel-digestion using sequencing grade modified trypsin from Promega. Database searches were done with the MS BioTools software from Bruker by using the Mascot search engine (http://www.matrixscience.com/home.html).
Gel-mobility shift assay
The reactions were set up in a 20 μl volume of 10 mM Tris–HCl, pH 8.0, 300 mM NaCl, 10 mM MgCl2, 0.05 mg/ml BSA, 0.024 mg/ml poly [d(I-C)], 50 fmol of labeled DNA, and the indicated amounts of mtDBP. The DNA probe TermNCR(F) (see later) was labeled at the 5′-termini with polynucleotide kinase (Roche) and [γ−32]PATP. The binding reactions were incubated for 20 min at 23°C and resolved on a 6% polyacrylamide gel in 0.5× Tris–borate–EDTA (TBE); the gel was dried and analyzed by Typhoon 8600 Phosphor Imaging System (Molecular Dynamics).
In vitro transcription reactions
DNA templates were as follows. The 71-bp double-stranded 3′-tailed template, named 71bpDNA, was generated by annealing a 86mer oligonucleotide, bearing a 3′-tail of 15 dC [5′-TTAGAGGGTGAACTCACTGGAACTTGGATGCTTGCATGTGTAATCTT ACTAAGAGCTAATAGAAAGGCTAG(C)15-3′], to a complementary 71mer oligonucleotide. The 98-bp double-stranded 3′-tailed template, named NoTerm, was generated by annealing a 114mer oligonucleotide, bearing a 3′-tail of 16 dC [5′-ATGCTTGTTCCTTTTGATCGTGGTGATTTAGAGGGTGAACTCACTGGAA CTTGGATGCTTGCATGTGTAATCTTACTAAGAGCTAATGAAAGGCTAG(C)16-3′], to a complementary 98mer oligonucleotide. The 98-bp double-stranded 3′-tailed template containing mtDBP-binding site in the NCR of P. lividus mtDNA, named TermNCR(F), was prepared by annealing a 114mer oligonucleotide, bearing a 3′-tail of 16 dC [5′-GTGCTTGTCCTTTCGTACAGAGAGGGGGACATGTTGTGATGGG AAATACAAAAGCCTGAAGGTAGATAGAAACCGACCTGGATTACTCCGGTCTGAAC(C)16-3′; the sequence of P. lividus mtDNA is underlined], to a complementary 98mer oligonucleotide. The template TermNCR(R) was as TermNCR(F) except that the P. lividus mtDNA sequence was reversed. Template TermNCRbis(F) was obtained by annealing a 114mer oligonucleotide, bearing a 3′-tail of 16 dC [5′-TTAGAGGGTGAACTCACTGGAGAGGGGGACATGTTGTGATGGGAAATACAAAAGCCT CTTGCATGTGTAATCTTACTAAGAGCTAATAGAAAGGCTAG(C)16-3′; the sequence of P. lividus mtDNA is underlined], to a complementary 98mer oligonucleotide. Template TermNCRbis(R) was as TermNCRbis(F) except that the P. lividus mtDNA sequence was reversed. Transcription assays were carried out in a 25-μl mixture containing DNA template (2 pmol), 40 mM Tris–HCl, pH 8.0, 25 mM NaCl, 8 mM MgCl2, 2 mM spermidine-(HCl)3, 1 mM DTT, 0.1 mg/ml BSA, 28 U of RNaseOUT (Invitrogen), 1 mM ATP, 0.3 mM CTP, 0.3 mM GTP, 0.0125 mM UTP, 0.5 μl of [α−32]PUTP (800 Ci/mmol) and 5 μl of mtRNAP-containing fraction, corresponding to about 0.15 pmol of mtRNAP. The enzyme concentration in the Heparin–Sepharose fractions was estimated by measuring the intensity of the mtRNAP-containing band in the stained gel and by relating it to the protein content. Incubation was performed at 30°C for 30 min. Transcription termination assays were set-up as before except that, prior to the addition of mtRNAP, the DNA template was incubated for 20 min at 23°C with the indicated amounts of mtDBP or an equivalent volume of mtDBP-containing buffer. Reactions were stopped by addition of 150 μl of stop buffer (10 mM Tris–HCl pH 7.4, 0.5% SDS, 0.2 M NaCl, 10 mM EDTA pH 8.0, 0.14 mg/ml glycogen). Samples were phenol extracted and the nucleic acids were ethanol precipitated. The pellets were dissolved in 150 μl of DNaseI buffer (40 mM Tris–HCl, pH 7.5, 6 mM MgCl2) and incubated with DNaseI (Pharmacia) for 10 min at 37°C. Nucleic acids were ethanol precipitated, the pellets were dissolved in urea gel loading buffer, heated at 80°C for 10 min and separated through a 12% polyacrylamide/7 M urea mini-gel in 1X TBE. Gels were washed twice with water, vacuum-dried and analyzed by phosphorimaging. For pulse-chase experiments, a 100 μl transcription termination reaction was set up in the presence of template TermNCR(F), 70 μCi of [α−32]PUTP (800 Ci/mmol), 48 pmol of mtDBP. After pre-incubation of DNA template with mtDBP for 15 min at 23°C, 20 μl of mtRNAP-containing fraction were added and incubation was continued for 30 min at 30°C. After addition of cold UTP to a final concentration of 2 mM, fractions (20 μl) of the reaction were taken at 0, 30, 100, 200 min, subjected to electrophoresis as before, and analyzed by phosphorimaging.
RESULTS
cDNA cloning of the P. lividus mitochondrial RNA polymerase
The cDNA for the sea urchin mtRNAP was isolated by using a combination of techniques including PCR, RT-PCR, screening of cDNA and genomic libraries, as depicted in Figure 1. The first portion of the cDNA was obtained by PCR on a cDNA library from P. lividus embryos at four-cell stage, using degenerate, inosine-containing oligonucleotide primers, designed on the most conserved regions of bacteriophage-type organelle RNAPs. These experiments produced a 216-bp cDNA fragment that was sequenced and shown to contain the conserved motifs V and VI of the phage-type mtRNAPs. Then, to further extend the cDNA sequence, we employed primer pair consisting of internal-specific primers and either vector-specific primers or external degenerate primers designed on the well-conserved regions. The obtained fragments were used to screen the cDNA library and isolate a clone of about 1500 bp that, together with the previous PCR sequences, produced a merged sequence of about 2500 bp. The complete 3′-end sequence, containing the stop codon TAA and a 3′-untranslated region of 590 bp, was obtained by 3′-RACE using egg poly(A)+ RNA as template and an oligo(dT) primer. To obtain the 5′-end of the cDNA we employed 5′-RACE, however it produced a sequence lacking the initiating methionine. The missing portion plus a short upstream non-coding region of 108 bp were obtained by screening a genomic library and performing PCR on the positive clones. The continuous full-length cDNA was obtained by RT-PCR on polyadenylated RNA, using primers complementary to the ends of the open reading frame (ORF) sequence.
Analysis of the amino-acid sequence of the P. lividus mtRNAP
The P. lividus mtRNAP ORF is 4266 bp long (accession no. EF068167) and predicts a 1421 amino-acid product, whose sequence is reported in Figure 2. Sequence analysis with MitoProt II program (26) predicts that the N-terminal portion is likely to contain the mitochondrial localization sequence, as expected for a nuclear-encoded protein that has to be imported into the mitochondrion. According to the prediction program, the potential cleavage site is placed between residues 15 and 16; therefore the mature protein should be 1406 amino-acid long, with a calculated molecular weight of 159 700 Da and an isoelectric point of 9.19. The amino-acid sequence alignment of the sea urchin mtRNAP with those of S. purpuratus, H. sapiens, X. laevis and D. melanogaster (Figure 2) revealed a remarkable similarity in correspondence of 10 conserved phage-type motifs, indicated as boxes I–X, placed in the carboxy-terminal region. All the proteins have the stop codon at the same position in box X. Within the conserved domains are placed the specific amino acids that were proven to be functionally critical for the T7 enzyme by mutant analysis (27). On this basis it can be inferred that the most crucial residues in the sea urchin mtRNAP should be Asp1106 and Asp1334 that would be involved in binding divalent metal ions at the active site, Lys1175 and His1333, possibly implicated in phosphodiester bond formation, Tyr1183 and Gly1184, necessary to distinguish rNTPs from dNTPs, and Phe1420, required for ribonucleotide binding. Inside box III we detected the conserved motif DX2GR. This sequence was found in many DNA-dependent RNA polymerases and in the bacteriophage T7 RNAP where it was shown to be involved in stabilizing the RNA:DNA hybrid during early stages of transcription initiation (28). As for the known organelle RNAPs, the sea urchin enzyme also displays a poorly conserved amino-terminal region that accounts for the large size of the protein. The N-terminal region of the P. lividus enzyme harbors a series of 29 serines (position 210–242) interrupted by one threonine and three leucines (Figure 2). A similar sequence was found in corresponding position of the S. purpuratus protein (29). About 200 amino acids downstream of the serine-rich region there are two putative pentatricopeptide repeat (PPR) motifs (30) (Figure 2). These sequences are conserved among the analyzed organisms despite the low conservation of the amino-terminal extensions.
Expression of the P. lividus mtRNAP in baculovirus-infected insect cells and activity assay
By RT-PCR with proper specific primers, we produced a continuous full-length cDNA, which codes for the mature form of the protein bearing at the N-terminus an hepta-histidine tag to allow affinity-purification of the protein. It has been reported that N-terminal tags keep the mtRNAPs functionally competent, whereas C-terminal tags tend to inactivate the catalytic centers, since the functionally critical residues are placed in the conserved carboxy-terminal region (31).
The tagged sea urchin mtRNAP was expressed in insect cells using the baculovirus expression system, as it provides a variety of co-translational and post-translational modifications of the recombinant proteins. Furthermore, this eukaryotic system allows simultaneous expression of multiple cDNAs. Recombinant baculoviruses were constructed as described in the ‘Materials and Methods’ section, and used to infect Sf9 insect cells. Both the soluble and insoluble fractions of the cell lysate were analyzed by immunoblotting using rabbit antibodies raised against a synthetic 20 amino-acid peptide of the sea urchin mtRNAP, chosen in the non-conserved N-terminal region of the sequence to obtain a highly specific immunoreaction. The immunoblot analysis indicated that only 10–20% of the recombinant protein was recovered in the soluble fraction, while the majority of the expressed protein was enclosed in the insoluble portion of the lysate (not shown). Falkenberg and colleagues (18) have reported that the human mtRNAP was insoluble when expressed on its own but, instead, formed a soluble heterodimer complex when co-expressed with TFB1M or TFB2M. Therefore, in order to increase the solubility of the sea urchin enzyme, we co-infected the Sf9 cells with the sea urchin mtRNAP and the human TFB1M recombinant baculoviruses.
Comparison by western blotting analysis of the cleared lysate and the remaining pellet indicated that about 50% of the co-expressed sea urchin mtRNAP was recovered in the soluble fraction (not shown). The lysate was then subjected to Ni2+-NTA agarose affinity chromatography. Western blot analysis of the eluted fractions revealed a main product migrating with an apparent molecular weight of about 150 kDa, a size close to the predicted molecular mass of the recombinant mtRNAP (Figure 3A). The additional bands present in the lysate and in the flow-through, but not in the eluted fractions, are likely to be truncated forms deriving from proteolytic degradation occurring at the N-terminus. This event removes the histidine tag, thus preventing the truncated forms from binding to the resin. None of the bands cross-reacted with preimmune serum, furthermore they were absent in the nickel-affinity fractions from an uninfected cell lysate (not shown). No co-expressed TFB1M protein was detected with specific antibodies (not shown). The immunoreactive fractions were pooled and further purified by Heparin–Sepharose chromatography on FPLC. The proteins eluted by a linear salt gradient from 0.13 to 1.2 M NaCl were separated on a 7.5% SDS–PAGE followed by Coomassie Blue staining. As shown in Figure 3B, the fractions eluting at 0.75–0.85 M NaCl (fractions 13–17), which was the same salt concentration at which the recombinant human mtRNAP eluted from the column (18), revealed few products including a band migrating with an apparent mass around 150 kDa. This product was identified as the sea urchin mtRNAP by means of two independent analyses. Western blotting showed that only the 150 kDa band was able to react with the mtRNAP antiserum (Figure 3C); furthermore, analysis by MALDI-TOF mass spectrometry of the major stained bands, excised from the gel and subjected to trypsin digestion, revealed that only the highest molecular weight product produced three ion peaks corresponding to the sea urchin mtRNAP.
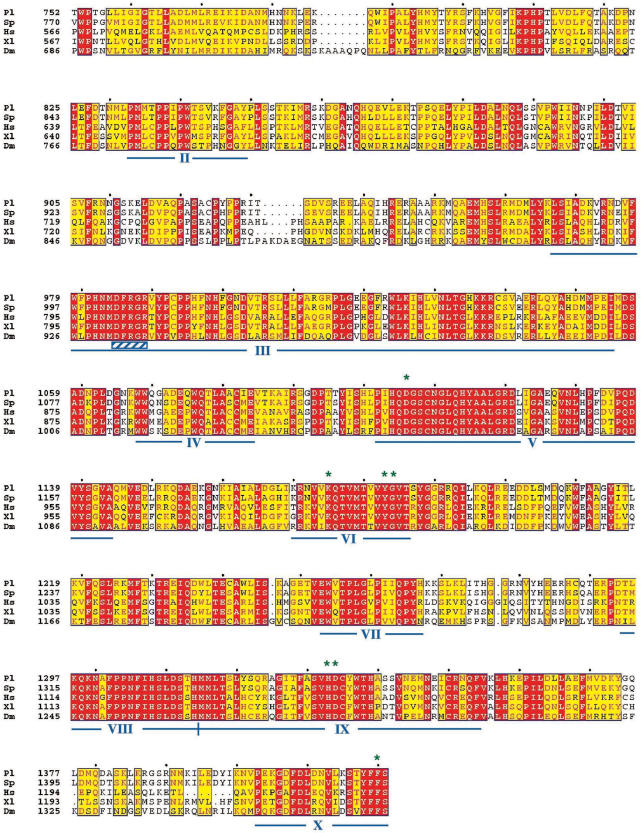
Figure 3.
Purification of the sea urchin mtRNAP from baculovirus-infected insect cells and functional assay. (A) Purification of mtRNAP by metal chelate affinity chromatography. The soluble portion of the insect cell lysate expressing the sea urchin mtRNAP was purified by Ni2+-NTA chromatography; cleared lysate, C.lys, flow-through, FT, wash, W, 3–5, fractions eluted at 250 mM imidazole, were separated on a 10% SDS–PAGE and revealed by immunoblotting as described in ‘Materials and Methods’ section. (B) Purification profile of mtRNAP as obtained by Heparin–Sepharose chromatography. Peak fractions from Ni2+-NTA column were pooled and subjected to Heparin–Sepharose chromatography. Input to the column (I) and fractions eluting between 0.75 and 0.9 M NaCl were analyzed by 7.5% SDS–PAGE and Coomassie Brilliant Blue stained. The molecular weight marker Precision Plus Protein Standards (Bio-Rad) is shown (M). The arrow inside the picture indicates the mtRNAP-containing band, as assessed by MALDI-TOF analysis. (C) Immunoblotting assay of input to the column (I) and Heparin–Sepharose eluted fractions. (D) Transcriptional activity of purified mtRNAP. The indicated Heparin–Sepharose fractions were assayed in the presence of [α-32]PUTP, as described in ‘Materials and Methods’ section. On the top it is shown the diagram of the 71-bp tailed template, named 71bpDNA, with the open bar referring to the duplex DNA portion and the thin line to the 3′-tail. Run-off transcripts are indicated by arrowed line. Radiolabeled transcripts were separated on a 12% polyacrylamide/7M urea mini-gel followed by phosphorimaging analysis. 15 + R, fraction 15 treated with RNase A. RNA markers corresponding to the 10 nt ladder are indicated on the left.
Based on the immunoreactivity and total protein content of cell lysate and chromatographic fractions (Figure 3A and C), an overall 100-fold purification of mtRNAP was estimated (about 12-fold after the Ni2+-NTA column and about 8-fold after the Heparin–Sepharose column).
Next, we tested if the purified mtRNAP could catalyze RNA synthesis on a duplex DNA. We employed the tailed template assay that is based on the observation that the presence of a single-stranded 3′-tail on a duplex DNA allows specific initiation by most of the purified RNA polymerases, independently of accessory factors and promoter recognition. This kind of assay has been particularly useful to study the transcription elongation properties of nuclear and mitochondrial RNA polymerases as well as the arrest of their elongation activities (18,32,33). In the experiment shown in Figure 3D, we employed a tailed template (71bpDNA) comprising a double-stranded DNA portion of 71 bp preceded by a single-stranded 3′ extension of 15 dC. Addition to the assay of the high-salt Heparin–Sepharose eluted fractions produced a prominent RNase sensitive product, corresponding to a run-off transcript, whose size (about 75 nt) was consistent with initiation of transcription at the oligo(dC) terminus. The transcription activity coincided with the peak of the immunoreactive high-molecular weight proteins eluting from the Heparin–Sepharose column (Figure 3, compare panels C and D). Furthermore, transcription activity was absent in the column fractions deriving from an uninfected cell lysate, which had been subjected to the same purification scheme as the recombinant insect cells (not shown), thus ruling out the possible contribution of a contaminant RNA polymerase activity.
Recombinant mtDBP expressed in insect cells promotes transcription termination unidirectionally in a basal transcription system
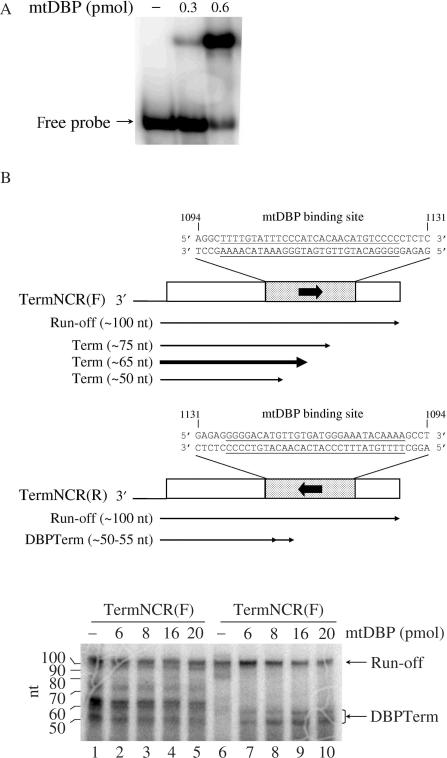
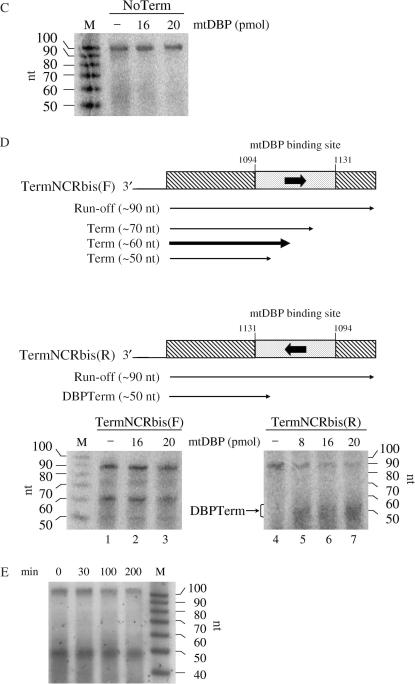
To investigate the transcription termination properties of mtDBP in the presence of the sea urchin mtRNAP, we set up a minimal promoter-independent transcription assay containing the proteins mtRNAP and mtDBP. The transcription factor mtDBP was expressed in insect cells and purified to a high degree by Heparin–Sepharose chromatography. To confirm that the recombinant mtDBP retained its binding activity, we performed EMSA experiments and showed that the protein is able to efficiently bind its target site in the NCR (Figure 4A). The recombinant mtDBP and mtRNAP were then employed in a promoter-independent transcription assay in the presence of 3′-tailed DNA constructs. They were obtained by annealing two partially complementary oligonucleotides to yield a duplex of 98 bp with a 3′ overhang of 16 dC. The duplex region contained the sequence contacted by mtDBP in the NCR and flanking sequences that are unrelated to P. lividus mtDNA (25). The protein-binding site was placed in either orientation downstream of the polymerase entry site to produce the forward template, named TermNCR(F), and the reverse template, named TermNCR(R). As shown in Figure 4B, lane 6, in the absence of mtDBP the recombinant mtRNAP synthesized on the reverse template a transcript of about 100 nt whose size corresponded to that of a full-length run-off transcript initiating a few nucleotides upstream of the tail–duplex junction. When increasing amounts of recombinant mtDBP were added to the assay (lanes 7–10), we observed a decrease in the intensity of the run-off band and the gradual appearance of two bands whose length (about 50–55 nt) corresponded to that of molecules arrested at the protein-binding site. These transcripts could represent terminated molecules produced by mtDBP. A different result was obtained with the template TermNCR(F), which bears the protein-binding site in the opposite orientation. As shown in Figure 4B, lane 1, transcription in the absence of mtDBP produced not only the run-off transcript, but also at least three shorter prematurely terminated molecules ranging in size from about 50 to about 75 nt. Addition of increasing amounts of mtDBP did not substantially alter the transcription profile, nor the ratio of run-off to prematurely terminated transcripts. A control experiment (Figure 4C) with a template of the same length but lacking the mtDBP-binding site (NoTerm) showed no terminated transcripts, even in the presence of the highest amount of mtDBP. This result indicates that transcription termination requires the presence of mtDBP-binding site.
Figure 4.
Transcription termination assays with recombinant mtRNAP and mtDBP. (A) Gel mobility shift analysis of recombinant mtDBP purified from baculovirus-infected insect cells. The assay was performed as described in ‘Materials and Methods’ section, with the indicated amounts of purified mtDBP and 50 fmol of end-labeled probe TermNCR(F) (see below). (B) Effect of mtDBP on the elongation activity of mtRNAP on templates TermNCR(F/R). (Top) Scheme of the 98-bp 3′-tailed DNA constructs, named TermNCR(F) and TermNCR(R), used in the transcription assays. The filled boxes indicate the 38-bp region in the Non-Coding Region (NCR) of P. lividus mtDNA (the sequence and nucleotide position are shown); the mtDBP-binding site, as from DNase I footprinting analysis (35), is underlined. The arrow enclosed in the boxes marks the orientation of mtDBP target site with respect to transcription direction. The open boxes represent the flanking sequences (41 and 19 bp, respectively), which are unrelated to sea urchin mtDNA. Run-off and terminated transcripts are indicated by arrowed lines. The thickness of the lines referring to the terminated molecules (Term), obtained with TermNCR(F) template, represents the relative abundance of the transcripts. (Bottom) Transcription termination assay. Transcription reactions were performed as described in ‘Materials and Methods’ section in the presence of 2 pmol of the indicated template, about 0.15 pmol of recombinant mtRNAP and the indicated amounts of mtDBP. DBPTerm refers to the terminated products generated by mtDBP, indicated by the line with two consecutive arrows in the top scheme. RNA markers corresponding to the 32P-5′-end labeled RNA ladder, Decade Markers, Ambion, are shown on the left. (C) Transcription assay performed on template NoTerm. The construct is a 98-bp 3′-tailed DNA lacking mtDBP-binding site. Reactions were performed as in (B). (D) Transcription assay on templates TermNCRbis(F/R). (Top) Schematic representation of the 98-bp 3′-tailed constructs, named TermNCRbis(F) and TermNCRbis(R). The regions flanking the mtDBP-binding site, indicated with striped boxes, are different in sequence from those of TermNCR(F/R). The sequence and nucleotide position of the region containing the mtDBP-binding site are as in (B). The thickness of the lines that represent the terminated molecules (Term), obtained with the forward template, indicates the relative abundance of the transcripts. (Bottom) Transcription termination assay. Reactions were performed as reported in (B). DBPTerm, RNA molecules terminated by mtDBP. (E) Pulse-chase of transcription elongation by mtRNAP in the presence of mtDBP. Assays were carried out as described in ‘Materials and Methods’. After a pulse-label of 15 min with [α−32]PUTP, the reaction was chased with a 2300-fold excess of unlabeled UTP. Samples (20 μl) were taken at the time points indicated and analyzed on a 12% polyacrylamide/7M urea, followed by phosphorimaging analysis.
Next, we wished to investigate whether the protein-independent termination occurring at the mtDBP-binding site in the forward orientation was somewhat affected by the sequences flanking the target site. To test this, we performed transcription experiments with a new template, named TermNCRbis(F), in which the mtDBP-binding site had been embedded in a different sequence context (25). As shown in Figure 4D, the transcription profile obtained with this construct was similar to that obtained with the TermNCR(F) DNA and was not affected by the presence of mtDBP (Figure 4, compare panel D, lanes 1–3 with panel B, lanes 1, 4, 5). The slight size discrepancy between these transcripts and those obtained with TermNCR(F) template is due to the variability of the transcription initiation site on the tailed template DNAs (34). The obtained result rules out that the sequences adjacent to the mtDBP-binding site could play a major role in promoting arrest of the mtRNAP, and rather suggests that the NCR-binding sequence in the forward orientation could influence by itself transcription termination. Transcription on the reverse template TermNCRbis(R) in the absence of mtDBP generated molecules that were progressively converted into terminated molecules in the presence of increasing amounts of mtDBP (Figure 4D, lanes 4–7). Finally, we examined whether the terminated transcript observed on the TermNCR(R) template were produced by a protein-mediated transcription termination or by RNA polymerase pausing. To test this hypothesis, we performed a pulse-chase transcription experiment on the reverse template. After a 15-min pulse of radiolabeled UTP, a chase of 200 min with a 2300-fold excess of cold UTP followed. The results of the experiment, reported in Figure 4E, showed that the ratio of terminated to run-off transcript did not change appreciably, with the shorter transcript persisting for up to 200 min of incubation, a time longer than the half-life of mtDBP–DNA complex (150 min) (35). These results indicate that the shorter transcripts are likely formed by transcription termination rather than by pausing of the transcribing enzyme.
DISCUSSION
Transcription termination is a key process in the regulation of mitochondrial gene expression in animal cells; moreover, termination factors play a role that often goes beyond the simple interruption of RNA synthesis. Mitochondrial termination factors belong to a large conserved protein family from animals and plants, the so-called mTERF family (36,37). It has been recently reported that the human termination factor mTERF binds simultaneously to the 3′-end of the ribosomal gene unit and to a sequence located in the mtDNA control region; this binding allows the formation of a loop structure that is thought to be responsible for the recycling of the mtRNAP and, ultimately, for the high rRNA/mRNA ratio (38). In Drosophila, studies on the mTERF homologue, DmTTF, showed that the depletion of the protein remarkably affects mitochondrial transcription: it increases the level of the transcripts located downstream of the protein-binding sites and, interestingly, decreases that of the transcripts located upstream of the two protein target sites (39). In sea urchin the termination factor mtDBP displays also a contra-helicase activity, functioning as negative regulator of mtDNA replication (40). To provide further insights into the mechanism of transcription termination in sea urchin, we cloned the cDNA of the sea urchin mtRNAP and employed the recombinant enzyme and the transcription factor mtDBP, both expressed in insect cells, to set up a promoter-independent in vitro transcription system.
cDNA cloning of the mtRNAP revealed that the protein, with its predicted 1421 amino-acid sequence, falls within the largest organelle RNAPs which comprise the S. purpuratus mtRNAP (1439 amino acids) (29) and the P. falciparum enzyme (1503 amino acids) (4). The sequence displays the typical features of organelle RNA polymerases such as 10 phage-type conserved sequence blocks, placed in the C-terminal part of the protein which harbors the catalytic domains. The order of these motifs and their relative spacing are highly conserved; furthermore they contain several amino acids that are critical for the polymerase activity, as inferred from mutant studies on the T7 bacteriophage enzyme. Box III displays the conserved DX2GR motif that is common to nuclear polymerases and to the mitochondria related T7 RNAP. The sea urchin mtRNAP exhibits an extension at the N-terminus which is highly variable in both length and sequence among the known organelle RNAPs. This region may be devoted to establish specific protein–protein interactions; the variability of this domain probably parallels changes that have occurred in the corresponding interacting factors. Rodheffer and Shadel demonstrated that the amino-terminal domain of yeast mtRNAP interacts with the matrix protein Nam1p and with the inner membrane protein Sls1p, to provide a functional link between transcription and translation (41). The amino-terminal region of sea urchin mtRNAP contains two tandemly repeated 35 amino acid motifs (PPR). These sequences, which have been detected also in other animal mtRNAPs, may be involved in coupling RNA processing or translation activities to transcription, through protein–protein or RNA–protein interactions. The leucine-rich PPR-motif containing protein (LRPPRC) and the heterogeneous nuclear ribonucleoprotein K were proposed as possible candidates for interacting with the N-terminus of human mtRNAP (30). An additional feature of the P. lividus enzyme is the presence of a stretch of serines at the N-terminus. Polyserine segments have been found in a broad variety of proteins serving disparate functions. In the sea urchin mtRNAP the serine-rich region may function as a flexible linker between the N- and C-termini; alternatively, its evolutionary selection might suggest unique, though still unknown, functions. Similarly, a polyglutamine stretch has been found in the N-terminal portion of the Neurospora crassa mtRNAP, but its function is still to be defined (6).
The recombinant P. lividus mtRNAP was shown to be fully able to catalyze the synthesis of RNA in a promoter-independent transcription assay. Such system was chosen since the promoter sequences of the sea urchin mtDNA as well as the factors required for specific transcription initiation are not known. Addition of increasing amounts of mtDBP to the assay caused arrest of the elongating enzyme in correspondence of the protein–DNA complex, in a dose-dependent manner and without the aid of additional factors. Such termination by mtDBP occurs only when the orientation of the NCR-binding site is opposite to the direction of RNA elongation (reverse template). Instead, on the forward template, transcription termination occurs in a protein-independent manner, inside the sequence contacted by mtDBP. These results were confirmed by the observation that the transcription termination profiles remained substantially unvaried when the sequences flanking the mtDBP-binding site were changed. Furthermore no terminated transcripts were obtained on a template lacking the protein-binding site, even in the presence of mtDBP. This rules out the possibility that the free mtDBP could bind the polymerase and promote premature arrest of transcription.
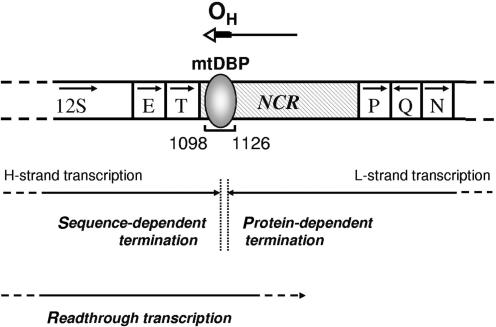
Figure 5 illustrates the two modes of transcription termination in the non-coding region of sea urchin mtDNA. The mtDBP-dependent transcription termination occurs when the enzyme is transcribing the L-strand and could serve to produce the H-strand replication primer. The sequence-dependent transcription termination occurs when the RNA polymerase is transcribing the H-strand and could form a precursor of the 12S rRNA. Early work on the mapping of mature and precursor sea urchin mitochondrial RNAs allowed the identification of an RNA species whose 3′-end mapped in proximity of the mtDBP-binding site in the NCR (22). Sequence-specific termination signals have been described for the T7 bacteriophage RNAP. Similarly to what observed for the sea urchin mtRNAP, the terminator element functions in a context-independent manner and is utilized only in one orientation (42). A protein-independent transcription termination has been recently reported by Pham et al.; they found that, in a reconstituted human mitochondrial transcription system, L-strand transcription prematurely terminates in proximity of the G-rich Conserved sequence Box II (43).
Figure 5.
Schematic diagram showing the mitochondrial transcription termination in sea urchin NCR. The region of P. lividus mtDNA containing the NCR, with the bound mtDBP, and some adjacent genes is shown. Numbers mark the position of the mtDBP-binding site. Arrows inside the open boxes indicate the transcription direction of the genes. Arrow on the top indicates the direction of H-strand DNA synthesis; the possible DNA portion of the newly synthesized strand is indicated as a bold line, the remaining RNA portion (primer) as a thin line (44). OH, DNA replication origin. Arrows below the scheme represent sequence-dependent and protein-dependent termination events, with points of transcription arrest indicated by dashed vertical lines. H-strand readthrough transcription can cause dislodging of mtDBP and resumption of H-DNA synthesis (40).
We have recently reported that mtDBP displays also a contrahelicase activity functioning as negative regulator of the D-loop extension. We demonstrated that the passage of the RNA polymerase, which is transcribing the mtDNA H-strand, through the mtDBP–DNA complex, causes the dislodging of the bound protein; this event would allow the resumption of the mtDNA synthesis (40). The results presented in this study show that run-off and termination transcripts are formed at a roughly comparable amount on the forward template (Figure 4B) and that their ratio is not affected by the presence of the highest amount of bound mtDBP. Therefore it is likely that the constant mtRNAP readthrough may be functional for mtDBP dislodging and then for the resumption of mtDNA replication (Figure 5). Finally, the pulse-chase transcription experiment (Figure 4E) indicates that mtDBP promotes authentic termination of transcription rather than pausing since in the presence of an excess of unlabeled precursor the ratio of terminated to run-off transcripts does not change appreciably with time. A similar behavior was described for the human transcription termination factor mTERF (20).
Altogether, the data reported in this study suggest that, in the main regulatory region of sea urchin mtDNA, two alternative modes of transcription termination take place, depending on the direction of transcription: in one case termination seems to be directed by the DNA-bound mtDBP, in the other, by sequence-specific elements present in the mtDBP contacted region. Based on the results deriving from this and previous studies of ours (25,40), it is tempting to speculate that the proposed mechanism of transcription termination may be functional to coordinate DNA replication and transcription in sea urchin mitochondria.
ACKNOWLEDGEMENTS
The authors would like to thank N.G. Larsson and C.M. Gustafsson (Department of Laboratory Medicine, Division of Metabolic Diseases, Karolinska Institutet) who generously allowed P.L.P. to spend a few months in their laboratory. P.L.P. is very grateful to the people of the above laboratory who offered great support, in particular to Martina Gaspari for helping with mtRNAP purification, Olga Khorosjutina for MALDI-TOF analysis and C.M. Gustafsson for constructive criticism to the experiments. The authors wish to thank F. Bruni for helping in bioinformatic analysis and F. Fracasso for technical assistance. We thank M. Branno (Stazione Zoologica Anton Dohrn, Napoli, Italy) for kindly providing the P. lividus λ FIX II genomic library and the P. lividus embryo λ Uni-Zap cDNA library. This work was supported by grants from: M.I.U.R.-COFIN PRIN 2005; M.I.U.R.-Contributo straordinario di ricerca/Aree obiettivo 1 (to I.B.B.E.); ‘Università di Bari’: Progetto di Ricerca di Ateneo. The visit of P.L.P. to the Karolinska Institutet was supported by a Short-Term Fellowship from Federation of European Biochemical Societies (FEBS).
Funding to pay the Open Access publication charges for this article was provided by M.I.U.R.-COFIN PRIN 2005.
Conflict of interest statement. None declared.
REFERENCES
- 1.Montoya J, Lopez-Perez MJ, Ruiz-Pesini E. Mitochondrial DNA transcription and diseases: past, present and future. Biochim. Biophys. Acta. 2006;1757:1179–1189. doi: 10.1016/j.bbabio.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 2.Gaspari M, Larsson NG, Gustafsson CM. The transcription machinery in mammalian mitochondria. Biochim. Biophys. Acta. 2004;1659:148–152. doi: 10.1016/j.bbabio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Masters BS, Sthol LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Maga JA, Cermakian N, Cedergren R, Feagin JE. Identification and characterization of a Plasmodium falciparum RNA polymerase gene with similarity to mitochondrial RNA polymerases. Mol. Biochem. Parasitol. 2001;113:261–269. doi: 10.1016/s0166-6851(01)00223-7. [DOI] [PubMed] [Google Scholar]
- 5.Grams J, Morris JC, Drew ME, Wang Z, Englund PT, Hajduk SL. A trypanosome mitochondrial RNA polymerase is required for transcription and replication. J. Biol. Chem. 2002;277:16952–16959. doi: 10.1074/jbc.M200662200. [DOI] [PubMed] [Google Scholar]
- 6.Chen B, Kubelik AR, Mohr S, Breitenberger CA. Cloning and characterization of the Neurospora crassa cyt-5 gene. A nuclear-coded mitochondrial RNA polymerase with a polyglutamine repeat. J. Biol. Chem. 1996;271:6537–6544. [PubMed] [Google Scholar]
- 7.Miller ML, Travis JA, Qian F, Miller DL. Identification of a putative mitochondrial RNA polymerase from Physarum polycephalum: characterization, expression, purification, and transcription in vitro. Curr. Genet. 2005;4:259–271. doi: 10.1007/s00294-005-0053-y. [DOI] [PubMed] [Google Scholar]
- 8.Adams MD, Celnicker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 9.Tiranti V, Savoia A, Forti F, D'Apolito MF, Centra M, Rocchi M, Zeviani M. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the expressed sequence Tag database. Hum. Mol. Genet. 1997;6:615–625. doi: 10.1093/hmg/6.4.615. [DOI] [PubMed] [Google Scholar]
- 10.Hess WR, Borner T. Organellar RNA polymerases of higher plants. Int. Rev. Cytol. 1999;109:1–59. doi: 10.1016/s0074-7696(08)62145-2. [DOI] [PubMed] [Google Scholar]
- 11.Hedtke B, Borner T, Weihe A. One RNA polymerase serving two genomes. EMBO Rep. 2000;1:435–440. doi: 10.1093/embo-reports/kvd086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shutt TE, Gray MW. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006;22:90–95. doi: 10.1016/j.tig.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Cermakian N, Ikeda TM, Miramontes P, Lang BF, Gray MW. On the evolution of the single-subunit RNA polymerase. J. Mol. Evol. 1997;45:671–681. doi: 10.1007/pl00006271. [DOI] [PubMed] [Google Scholar]
- 14.Sousa R, Chung YJ, Rose JP, Wang BC. Crystal structure of the bacteriophage T7 RNA polymerase at 3.3 angstrom resolution. Nature. 1993;364:593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- 15.Cheetham GMT, Steitz TA. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305–2309. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- 16.Schinkel AH, Koerkamp MJ, Touw EP, Tabak HF. Specificity factor of yeast mitochondrial RNA polymerase. Purification and interaction with core RNA polymerase. J. Biol. Chem. 1987;262:12785–12791. [PubMed] [Google Scholar]
- 17.Cotney J, Shadel GS. Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J. Mol. Evol. 2006;63:707–717. doi: 10.1007/s00239-006-0075-1. [DOI] [PubMed] [Google Scholar]
- 18.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 19.Kruse B, Narasimhan N, Attardi G. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989;58:391–397. doi: 10.1016/0092-8674(89)90853-2. [DOI] [PubMed] [Google Scholar]
- 20.Asin-Cayuela J, Schwend T, Farge G, Gustafsson CM. The human mitochondrial transcription termination factor (mTERF) is fully active in vitro in the non-phosphorylated form. J. Biol. Chem. 2005;27:25499–25505. doi: 10.1074/jbc.M501145200. [DOI] [PubMed] [Google Scholar]
- 21.Prieto-Martin A, Montoya J, Martinez-Azorin F. Phosphorylation of rat mitochondrial transcription termination factor (mTERF) is required for transcription termination but not for binding to DNA. Nucleic Acids Res. 2004;27:1890–1899. doi: 10.1093/nar/gkh528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cantatore P, Roberti M, Loguercio Polosa P, Mustich A, Gadaleta MN. Mapping and characterization of Paracentrotus lividus mitochondrial transcripts: multiple and overlapping transcription units. Curr. Genet. 1990;32:2059–2068. doi: 10.1007/BF00312615. [DOI] [PubMed] [Google Scholar]
- 23.Elliott DJ, Jacobs HT. Mutually exclusive synthetic pathways for sea urchin mitochondrial rRNA and mRNA. Mol. Cell. Biol. 1989;9:1069–1082. doi: 10.1128/mcb.9.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loguercio Polosa P, Roberti M, Musicco C, Gadaleta MN, Quagliariello E, Cantatore P. Cloning and characterisation of mtDBP, a DNA-binding protein which binds two distinct regions of sea urchin mitochondrial DNA. Nucleic Acids Res. 1999;27:1890–1899. doi: 10.1093/nar/27.8.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Silva P, Loguercio Polosa P, Roberti M, Di Ponzio B, Gadaleta MN, Montoya J, Cantatore P. Sea urchin mtDBP is a two-faced transcription termination factor with a biased polarity depending on the RNA polymerase. Nucleic Acids Res. 2001;29:4736–4743. doi: 10.1093/nar/29.22.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 27.Osumi-Davis PA, de Aguilera MC, Woody RW, Woody A-YM. Asp537, Asp812 are essential and Lys631, His811 are catalytically significant in bacteriophage T7 RNA polymerase activity. J. Mol. Biol. 2000;226:37–45. doi: 10.1016/0022-2836(92)90122-z. [DOI] [PubMed] [Google Scholar]
- 28.Imburgio D, Anikin M, McAllister WT. Effects of substitutions in a conserved DX(2)GR sequence motif, found in many DNA-dependent nucleotide polymerases, on transcription by T7 RNA polymerase. J. Mol. Biol. 2002;319:37–51. doi: 10.1016/S0022-2836(02)00261-9. [DOI] [PubMed] [Google Scholar]
- 29.Sea Urchin Genome Sequencing Consortium. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shadel GS. Coupling the mitochondrial transcription machinery to human disease. Trends Genet. 2004;20:513–519. doi: 10.1016/j.tig.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Matsunaga M, Jang S-H, Jaehning JA. Expression and purification of wilde type and mutant forms of the yeast mitochondrial core RNA polymerase, Rpo41. Protein Expr. Purif. 2004;35:126–130. doi: 10.1016/j.pep.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 32.Kadesch TR, Chamberlin MJ. Studies of in vitro transcription by calf thymus RNA polymerase II using a novel duplex DNA template. J. Biol. Chem. 1982;257:5286–5295. [PubMed] [Google Scholar]
- 33.Kuhn A, Bartsch I, Grummt I. Specific interactions of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature. 1990;344:559–562. doi: 10.1038/344559a0. [DOI] [PubMed] [Google Scholar]
- 34.Dedrick RL, Chamberlin MJ. Studies on transcription of 3′-extended templates by mammalian RNA polymerase II. Parameters that affect the initiation and elongation reactions. Biochemistry. 1985;24:2245–2253. doi: 10.1021/bi00330a019. [DOI] [PubMed] [Google Scholar]
- 35.Roberti M, Mustich A, Gadaleta MN, Cantatore P. Identification of two homologous mitochondrial DNA sequences, which bind strongly and specifically to a mitochondrial protein of Paracentrotus lividus. Nucleic Acids Res. 1991;19:6294–6254. doi: 10.1093/nar/19.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linder T, Park CB, Asin-Cayuela J, Pellegrini M, Larsson NG, Falkenberg M, Samuelsson T, Gustafsson CM. A family of putative transcription termination factors shared amongst metazoans and plants. Curr. Genet. 2005;48:265–269. doi: 10.1007/s00294-005-0022-5. [DOI] [PubMed] [Google Scholar]
- 37.Roberti M, Bruni F, Loguercio Polosa P, Manzari C, Gadaleta MN, Cantatore P. MTERF3, the most conserved member of the mTERF-family, is a modular factor involved in mitochondrial protein synthesis. Biochim. Biophys. Acta. 2006;1757:1199–1206. doi: 10.1016/j.bbabio.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 38.Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–1240. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 39.Roberti M, Bruni F, Loguercio Polosa P, Gadaleta MN, Cantatore P. The Drosophila termination factor DmTTF regulates in vivo mitochondrial transcription. Nucleic Acids Res. 2006;34:2109–2116. doi: 10.1093/nar/gkl181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loguercio Polosa P, Deceglie S, Roberti M, Gadaleta MN, Cantatore P. Contrahelicase activity of the mitochondrial transcription termination factor mtDBP. Nucleic Acids Res. 2005;33:3812–3820. doi: 10.1093/nar/gki693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodeheffer MS, Shadel GS. Multiple interactions involving the amino-terminal domain of yeast mtRNA polymerase determine the efficiency of mitochondrial protein synthesis. J. Biol. Chem. 2003;278:18695–18701. doi: 10.1074/jbc.M301399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He B, Kubarin A, Temiakov D, Chin-Bow ST, Lyakhov DL, Rong M, Durbin RK, McAllister WT. Characterization of an unusual, sequence-specific termination signal for T7 RNA polymerase. J. Biol. Chem. 1998;273:18802–18811. doi: 10.1074/jbc.273.30.18802. [DOI] [PubMed] [Google Scholar]
- 43.Pham XH, Farge G, Shi Y, Gaspari M, Gustafsson CM, Falkenberg M. Conserved sequence box II directs transcription termination and primer formation in mitochondria. J. Biol. Chem. 2006;281:24647–24652. doi: 10.1074/jbc.M602429200. [DOI] [PubMed] [Google Scholar]
- 44.Jacobs HT, Herbert ER, Rankine J. Sea urchin egg mitochondrial DNA contains a short displacement loop (D-loop) in the replication origin region. Nucleic Acids Res. 1989;17:8949–8965. doi: 10.1093/nar/17.22.8949. [DOI] [PMC free article] [PubMed] [Google Scholar]