Abstract
L1Tc is the best represented autonomous LINE of the Trypanosoma cruzi genome, throughout which several functional copies may exist. In this study, we show that the first 77 bp of L1Tc (Pr77) (also present in the T. cruzi non-autonomous retrotransposon NARTc, in the Trypanosoma brucei RIME/ingi elements, and in the T. cruzi, T. brucei and Leishmania major degenerate L1Tc/ingi-related elements [DIREs]) behave as a promoter element that activates gene transcription. The transcription rate promoted by Pr77 is 10–14-fold higher than that mediated by sequences located upstream from the T. cruzi tandemly repeated genes KMP11 and the GAPDH. The Pr77 promoter-derived mRNAs initiate at nucleotide +1 of L1Tc, are unspliced and translated. L1Tc transcripts show a moderate half life and are RNA pol II dependent. The presence of an internal promoter at the 5′ end of L1Tc favors the production of full-length L1Tc RNAs and reinforces the hypothesis that this mobile element may be naturally autonomous in its transposition.
INTRODUCTION
Trypanosoma cruzi is the etiological agent of Chagas’ disease, which affects between 16 and 18 million people, primarily in Central and South America (1). Apart from its impact on human health, the T. cruzi parasite has been extensively studied because of the interesting molecular characteristics shown by the members of the Trypanosomatidae. It is known that the transcription of protein-coding genes and mRNA maturation processes in this parasite involve mechanisms that are distinct from those in most higher eukaryotes. In fact, most trypanosome mRNAs are synthesized as polycistronic precursors, and mature mRNAs are generated by trans-splicing and polyadenylation. The trans-splicing process implies joining two separate RNA precursor transcripts (2). Consequently, mature mRNAs possess a common capped (cap4) 39-nt non-coding mini-exon sequence (called the splice leader sequence [SL]) at the 5′ end. Polyadenylation is coupled to downstream trans-splicing (3,4). It has also been reported that at least 50% of the T. cruzi genome consists of repeated sequences, such as retrotransposons and gene families of surface proteins. These repeated sequences have been correlated with the significant genomic polymorphism and the high degree of plasticity this parasite shows (5,6). The retrotransposons (both the LTR-retrotransposons and non-LTR retrotransposons) possessed by this parasite may be involved in the generation and maintenance of tandem gene structures as well as in the regulation of gene expression (7). The non-LTR retrotransposons (SINE and LINE) are interspersed replicating elements found in almost all eukaryotes (8). These retroelements are flanked by a variable length target site duplication sequence, are polyadenylated, and lack the long terminal repeat (LTR) observed in retrovirus and LTR-retrotransposons. LINE elements encode the enzymes involved in their own transposition, whereas the non-coding SINE transcripts are recognized by the LINE retrotransposition machinery and mobilized by the LINE-encoded proteins (8–10).
The mobilization of non-LTR retrotransposon elements occurs by a mechanism termed target-primed reverse transcription (TPRT), in which the RNA encoded by the element is reverse transcribed. The newly synthesized DNA copy is integrated at a new site in the genome (11). Retrovirus and LTR-retrotransposons use long terminal repeats to synthesize complete cDNAs which maintain their promoters (12). However, non-LTR retrotransposons do not possess LTRs. Since the initial step in the LINE self-retrotransposition process requires transcription of the full length of the element to guarantee a transcriptionally competent (and autonomous) new version, it has been proposed that these elements ought to contain an internal promoter. To date, the type of RNA polymerase that participates in the transcription of LINEs has not been clearly identified, although both RNA polymerases II and III have been implicated in the transcription of human LINE-L1, and recent data strongly suggest that L1 transcription is mediated by the former (13).
L1Tc is the best represented LINE in T. cruzi, and it has been found in most, if not all, of the chromosomes of the T. cruzi strains analyzed to date. It is actively transcribed in all three stages of the parasite's life cycle (14). At least 15 theoretically retrocompetent L1Tc elements have been identified in the T. cruzi genome (5). L1Tc has been reported as being associated with a gene coding for a transporter protein belonging to the ABC family (15), as being integrated into the coding sequence of the DNAj gene endowed with chaperone activity (16), as present in the expressed RHS multigene family located in a subtelomeric region, and as associated with SINE-like sequences (17–19). It codes for all the enzyme machinery involved in its retrotransposition, including AP endonuclease (20), 3′ phosphatase, 3′ phosphodiesterase (21), reverse transcriptase (22), RNAse H (23) and a nucleic acid chaperone (24).
The absence of reports on the presence of promoters in genes encoding proteins in the T. cruzi genome, together with the significant role of active LINE elements, raises the question of how the transcription of L1Tc occurs. In the present work we show that the first 77 base pairs of L1Tc, which are identical to the first 77 bp of NARTc (a highly represented non-autonomous retrotransposon of the T. cruzi genome) (25), drive the expression of a downstream gene (CAT), and that transcription initiates at nucleotide +1. Interestingly, the transcripts produced are unspliced, indicating that a full-length RNA is generated. Similar results were observed after analysis of the expression of the endogenous L1Tc transcripts. The results shown are consistent with the existence of an internal promoter in the L1Tc element. Via run-on experiments, the L1Tc promoter was shown to be RNA pol II dependent.
MATERIALS AND METHODS
Oligonucleotides
5′R77 (5′ GCATAGATATCCCTGGCTGAG 3′) [sense, L1Tc 1–11]
3′R77 (5′ GCATTAAGCTTCAGCAGGCGC 3′) [antisense, L1Tc 68–77]
CAT1sense (5′ CCCGCCTGATGAATGCTC 3′) [sense, 191–208 from the CAT start codon]
CAT1 (5′ GAGCATTCATCAGGCGGG 3′) [antisense, 191–208 from the CAT start codon]
CAT2 (5′ CTGAGACGAAAAACAT 3′) [antisense, 424–439 from the CAT start codon]
SLTc (5′ CGCTATTATTGATACAGTTTCTG 3′) [sense, 8-30 from the T. cruzi splice leader sequence]
L1Tc239 (5′ ACATACGGCACGCAAACGG 3′) [antisense, L1Tc 221–239]
L1Tc152 (5′ TGTAAATGGCTCCATCT 3′) [antisense, L1Tc 136–152]
L1Tc22 (5′ GCTCAGCCGGCCACCTC 3′) [sense, L1Tc 6–22]
kmp182 (5′ TTGTCGGTGTGCTCCTGAATC 3′) [antisense, 162–182 from the KMP11 start codon]
kmp21 (5′ TTCCTCAAGAGTGGTGGC 3′) [antisense, 4–21 from the KMP11 start codon]
CAT90 (5′ TTGAGCAACTGACTGAAATG 3′) [antisense, 71–90 from the CAT start codon]
SK (5′ CGCTCTAGAACTAGTGGATC 3′)
CAT-32f (5′ CGACGAGATTTTCAGGAG 3′) [sense, −32 to −15 from the CAT start codon]
CAT43r (5′ GGGATATATCAACGGTGGTA 3′) [antisense, 24–43 from the CAT start codon]
R77 (5′ CCCTGGCTCAGCCGGCCACCTCAACGTGGTGCCAGGGTCTAGTACTCTTT
GCTAGAGAGGAAGCTAAGCGCCTGCTG 3′)
MCS (5′ CGACGGTATCGATAAGC 3′) [antisense, 3–19 downstream Pr77 in the pTEX(−)pR77CAT vector; 3–19 downstream 5′UTRKMP11 region in the pTEX(−)p5′KMP11CAT vector]
L1Tc70 (5′ CGCTTAGCTTCCTCTCTAGC 3′) [antisense, L1Tc 51–70]
The EcoRV and Hind III restriction sites are underlined. The gene name and position to which the primers map in each case are indicated in brackets.
Plasmid constructs
The chloramphenicol acetyltransferase coding gene (CAT) was excised from the pMSGCAT vector (Pharmacia) by SalI digestion and cloned into a SalI-digested pTEX expression vector (26) to produce the pTEXCAT clone. The 5′ upstream region of the gGAPDH I gene (p) was removed from this vector by digestion with SacI and BamHI enzymes, and subjected to Klenow treatment and religation to generate the pTEX(−)pCAT plasmid. The 77 nucleotides located at the L1Tc 5′ end were amplified by PCR using pSPFM55 (accession number X83098 in the GenBank database) (14) as a template, and the 5′R77 and 3′R77 primers. These include the EcoRV and HindIII restriction sites, respectively. The amplified fragment was digested with EcoRV and HindIII and directly cloned into pTEX(−)pCAT digested with the same enzymes to produce pTEX(−)pR77CAT. The 5′ upstream sequence from the T. cruzi KMP11 locus (nucleotides −576 to +37 from the first ATG) was excised from the PS1 clone (accession number AF167435 in the GenBank database) (27) by Klenow-treated-SalI digestion and HindIII digestion, and cloned into an EcoRV and HindIII-digested pTEX(−)pCAT vector to produce the pTEX(−)p5′KMP11CAT transfection vector. Correct cloning was confirmed in all cases by DNA sequencing.
Epimastigote culture and transfection procedure
Y strain T. cruzi epimastigotes were grown at 28°C in liver infusion tryptone (LIT) medium supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Flow Lab., Irvine, UK). Plasmid DNAs from the pTEX(−)pR77CAT, pTEX(−)p5′KMP11CAT, pTEX(−)pCAT and pTEXCAT vectors were purified using the Wizard Plus Maxipreps kit (Promega). Transfection was performed with 100 μg of each vector as previously described (28). The transfectants were selected in the presence of 100 μg/ml of G418. To increase the plasmid copy number, the G418 concentration was increased up to 250 and 500 μg/ml.
Northern, Southern and DNA dot blotting
The cytoplasmic RNA from T. cruzi transfectants was purified as previously described (29). DNA from transformed and non-transformed parasites was isolated by standard methods (30). Cytoplasmic RNA (5 µg) was size fractionated on 1% agarose/formaldehyde gels. Genomic DNA (2 µg) was HindIII-digested and resolved on 0.8% agarose gels. Both RNA and DNA were transferred to Z-probe membranes (Bio-Rad) using a 10 × SSC solution. Hybridization was performed using the method of Thomas et al. (27). The probes used were a Sal/Sal fragment from the pMSGCAT vector (Pharmacia) corresponding to the CAT coding sequence, a DNA fragment coding for the T. cruzi 18S small RNA subunit (18S) (31), and a 273-nt DNA fragment corresponding to the T. cruzi KMP11 coding sequence (27). The hybridization products were visualized and quantified using a phosphorimager (Storm, Pharmacia). All analyses were performed using the ImageQuant program (Molecular Dynamics). The local background (PSL-BG) was subtracted from the photo-stimulated luminescence (PSL) to obtain PSLcorr. The transcription efficiency was calculated as:
 |
The total RNA and poly(A)+ RNA fractions from 15 × 107 transfectants were isolated and purified using, respectively, the total RNA Mini Kit (Bio-Rad) and the QuickPrep micro mRNA Purification Kit (Amersham Biosciences). Total and polyadenylated RNA (7 and 1 μg respectively) samples were loaded onto 1% agarose/formaldehyde gels and transferred to Z-Probe nylon membranes (Biorad) (three replicates). One membrane was hybridized under the conditions described above using the CAT-coding region sequence as probe. The other two were hybridized for 2 h in 6 × SCC, 0.1% SDS (w/v), 100 mM Tris HCl, pH = 8 buffer and 100 μg/ml herring sperm DNA at 53°C with radiolabeled CAT antisense and CAT sense primers (CAT1 and CAT1sense respectively). These primers have complementary sequences, were used at the same concentration, and showed the same specific activity. Post-hybridization washes were performed in hybridization buffer at 48°C. The CAT probe was radiolabeled using [α-32P]-dCTP and the Random Prime Labelling System (Amersham Biosciences). The antisense CAT1 and sense CAT1 oligonucleotides were 5′end-labeled with [γ-32P]-dATP as described in Heras et al. (24). The hybridization products were visualized using a phosphorimager and quantified using the ImageQuant program as described above.
The ability of each radiolabeled oligonucleotide to detect a similar amount of a complementary DNA was determined by DNA dot blotting. Three-fold dilutions of the pTEXCAT vector (from 196 to 2.4 ng) and of the antisense CAT1 or CAT1sense oligonucleotides (from 196 to 0.8 ng) were denatured by treatment with 0.4 N NaOH and 10 mM EDTA for 10 min at 95°C or 10 min at 65°C respectively, and loaded onto Zeta-Probe Blotting Membranes (Biorad) using a Millipore dot blot manifold. Filters were hybridized with the radiolabeled antisense and sense CAT1 oligonucleotides as described above. The membranes were also visualized using a phosphorimager and quantified using the ImageQuant program as above.
RT-PCR
To analyze the 5′ end of the CAT mRNAs, 0.8 μg of poly(A)+ RNA from pTEXCAT-, pTEX(−)p5′KMP11CAT- and pTEX(−)pR77CAT-transformed parasites were reverse transcribed using the CAT2 primer and the ThermoScript RT-PCR System (Invitrogen, California) according to the manufacturer's instructions. PCR was performed using the Expand High Fidelity PCR System (Roche) and employing 2 μl of the synthesized cDNA as the template DNA along with the SLTc and CAT1 primers. The synthesized cDNA from pTEX(−)pR77CAT transfectants was also PCR amplified using 5′R77 and CAT1 primers as a positive control. As a negative control, the product from a parallel reaction without the reverse transcriptase was used. The reaction conditions were 10 cycles at 94°C for 30 s, 38°C for 1 min and 72°C for 1 min, and 25 cycles at 94°C for 30 s, 56°C for 1 min, and 72°C for 1 min.
To analyze the 5′ end of the L1Tc mRNAs (Figure 7), 0.6 μg of RQ1 RNAase-free DNAse-treated cytoplasmic RNA from epimastigotes were reverse transcribed using the L1Tc239 primer. PCR was performed employing the above-mentioned synthesized cDNA as the DNA template and SLTc and L1Tc152 as primers. PCR was also performed with L1Tc22 and L1Tc152 primers or without reverse transcriptase as RT-PCR positive and negative controls respectively. Trans-spliced KMP11 transcripts were also reverse transcribed using the kmp182 oligonucleotide, and PCR performed using the SLTc and kmp21 primers as a control of RNA integrity and proper sample processing. The PCR reaction conditions were 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min. All the amplified products were cloned into the pGEM T®-easy vector (Promega) and sequenced.
Figure 7.
Analysis of the 5′ end of T. cruzi L1Tc transcripts. (A) Primer extension analysis performed with RNase-free-DNase-treated cytoplasmic RNA from non-transformed T. cruzi epimastigotes (Yc) and radiolabeled L1Tc70 or L1Tc152 antisense primers (which map at L1Tc 5′ end at positions 51–70 and 136–152 respectively). Products were size-fractionated on urea-polyacrylamide sequencing gels. (MW) Molecular weight marker expressed in bp. DNA sequencing reactions (MW marker) were used to determine band sizes throughout. The arrows indicate the size of the extended products. (B) Sequence of 5′ untranslated region of L1Tc. The sequences of oligonucleotides used in primer extension analysis, the L1Tc70 and L1Tc152 primers, are underlined and double underlined respectively. The arrows indicate the last nucleotide of the extended products, 70 nt and 149 nt. (C) Ethidium bromide staining of RT-PCR products. RNase-free- DNase-treated cytoplasmic RNA from non-transformed T. cruzi parasites (Yc) was employed in all reactions as a template. For detection of the L1Tc transcript, 5′end reverse transcription was performed with the antisense L1Tc primer mapping at position 221–239 from L1Tc (L1Tc239) followed by PCR using the L1Tc152 primer (the antisense L1Tc primer which maps closer to the L1Tc 5′ end than L1Tc239 primer) and the SLTc primer (lane Yc-SL/L1Tc152), or alternatively PCR-amplified using the L1Tc152 primer and the L1Tc22 sense primer, which maps at position 6–22 from L1Tc (lane C+). A similar reaction without reverse transcriptase was used as a negative control (lane C−). For detection of the KMP11 transcript 5′ends, reverse transcription was performed with the antisense kmp182 primer mapping at position 162–182 from the KMP11 start codon, followed by PCR performed with Kmp21, which maps at position 4–21 from the KMP11 start codon, and SLTc primers (lane Yc-SL/kmp21). A similar reaction without reverse transcriptase was used as a negative control (lane C−). The size of the amplified bands is indicated at the right of the figure.
T. cruzi soluble protein extraction and western blotting
Soluble proteins were extracted from parasites in the logarithmic phase of growth. The protein concentration was determined by the standard method described by Heras et al. [43]. Briefly, parasites were recovered by centrifugation at 2500 rpm for 30 min, washed in 1 × PBS, and resuspended in lysis buffer (50 mM Tris-HCl, pH 7.4, 50 mM NaCl, 0.05% NP-40 supplemented with protease inhibitors, 1 µg/ml leupeptin, 0.7 µg/ml pepstatin and 1 mM PMSF). Total extracts were sonicated for 45 s. The soluble fraction proteins were recovered by centrifugation at 10 000 rpm for 20 min. Soluble proteins (30 µg) were separated by 12% SDS-PAGE and transferred to PVDF membranes (Millipore) using the Miniprotean system (Bio-Rad). Western blots were performed according to standard techniques, employing anti-chloramphenicol acetyl transferase antibodies (Sigma) at a dilution of 1:1000 and anti-rabbit IgG peroxidase conjugate (Sigma) at a dilution of 1:5000. The blots were subjected to peroxidase and luminol/enhancer solutions using the SuperSignal® West Pico Chemiluminescent Reagent kit (Sigma), and subsequently exposed to Kodak X-Omat autoradiographic film.
RNA ligase-mediated amplification of the cDNA ends
To characterize the 5′ end of the T. cruzi L1Tc transcripts, cytoplasmic RNAs from the T. cruzi pTEX(−)pR77CAT and pTEX(−)p5′KMP11CAT transfectants were treated with several enzymes to guarantee their monophosphate state and consequently their capacity to bind to a RNA linker. A method similar to that described by Bruderer (32) was employed. Briefly, 15 μg of cytoplasmic RNA from each transfectant were treated with 20U of RQ1 RNAase-free DNAse (Promega) in a final volume of 200 μl. The RNA was subsequently decapped, dephosphorylated and then phosphorylated by treatment with tobacco acid pyrophosphatase (TAP) (Epicentre), alkaline phosphatase (AP) (Roche) and T4 polynucleotide kinase (PNK) (Biolabs) respectively. An RNA linker derived from the EcoRI-digested pBluescripts KS+ plasmid (Stratagene) was synthesized in vitro as described by Heras et al. (24) and ligated to the previously generated monophosphate transcripts using T4 RNA ligase (Roche) following the method described by Bruderer (32). cDNA was synthesized using the CAT1 primer and employed as a template in PCR using SK and CAT90 as primers. Crude PCR products were directly cloned into the pGEM-T®easy vector (Promega). Selection was performed after IPTG-X-gal induction by blue/white screening. White colonies were numbered and grown in triplicate on LB-agar plates and transferred to nitrocellulose membranes. One membrane was hybridized in situ with the CAT coding fragment obtained by PCR using CAT-32f and CAT43r as primers and the pMSGCAT vector as a template. It was then labeled using a random prime kit (Amersham). Two other membranes were in situ hybridized with the SLTc and R77 primers labeled at the 5′ end. Probe labeling and hybridization were performed as described above.
Primer extension analysis
About 10 µg of RNase-free DNase I cytoplasmic RNA or 1 µg of poly(A)+ RNA from transformed and non-transfected T. cruzi parasites were extended using an MCS primer (complementary to the multi-cloning region located between the Pr77 or 5′UTRKMP11 region and the CAT sequence in the pTEX(−)pR77CAT and pTEX(−)p5′KMP11CAT vectors respectively), the L1Tc70 or L1Tc152 primers (complementary primers to L1Tc mRNA), and the RT from the avian myeloblastosis virus, as described by Teran et al. (33). Primers were 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. Reactions were resolved in urea–6.5% polyacrylamide sequencing gels.
Treatment of parasites with actinomycin D
RNA synthesis was inhibited by treating the parasites with actinomycin D as previously described (27). Cytoplasmic RNA was determined by northern blotting. The region between nucleotides 1844 and 3737 of L1Tc (accession code X83098) was employed as a probe. The abundance of the L1Tc mRNAs was standardized relative to the abundance of the T. cruzi KMP11 and ribosomal S6 subunits. All probes were radiolabeled using [α-32P]-dCTP, employing the Random Prime Labelling System (Amersham Biosciences).
Nuclear run-on transcription assay
Nuclei were isolated and purified from T. cruzi epimastigotes in the logarithmic growth phase and incubated with 83 µM of tagetitoxin (Tagetin™, Epicenter Biotechnologies), 25 µg/ml of α-amanitin (Sigma) or 0.2% N-lauryl sarcosine (sarkosyl; Sigma) for 30 min at 4°C. Nuclear run-on transcription reactions were performed using isolated nuclei, as described by Marañón et al. (31). The labeled nascent RNA was hybridized to 3 µg of linearized DNAs (see Figure 9 for details) adsorbed onto Z-probe membranes (Biorad). The hybridization conditions and post-hybridization wash conditions were those as described earlier (29).
Figure 9.
Nuclear run-on assay to identify the RNA polymerase responsible for transcription of the T. cruzi L1Tc retroelement (A). Detection of nascent RNAs sensitive and resistant to RNA polymerase inhibitors by dot blotting. Nascent transcripts from epimastigotes in the logarithmic phase of growth (non-treated) were hybridized to the following membrane-bound cloned DNAs: T. cruzi 18S rDNA (18S) (31), the T. cruzi α-tubulin gene (pTcα3) (58), the T. cruzi tRNA gene (tRNA), the sequences coding for full-length L1Tc (L1Tc) or L1Tc endonuclease (NL1Tc), L1Tc reverse transcriptase (RT), and the L1Tc nucleic acid chaperone protein (C2-L1Tc). Run-on was also performed with nuclei incubated in the presence of 1mg/ml of α-amanitin, 0.6% of sarkosyl and 8 μM of tagetitoxin. (B) Quantification of the L1Tc nascent RNAs from untreated nuclei and nuclei treated with different RNA polymerase inhibitors. The percentage (%) of the nascent transcripts sensitive and resistant to different RNA polymerase inhibitors was calculated relative to the total amount of the nascent transcripts from untreated nuclei detected in each case. Scanning of the autoradiograms was performed with an Image Quant program (Molecular Dynamics).
RESULTS
The 77 nucleotides at the 5′end of L1Tc activate transcription of a reporter gene
While the first 77 bp of the T. cruzi L1Tc (autonomous LINE) and NARTc (non-autonomous retrotransposon) elements are 100% identical, the rest of the L1Tc and NARTc sequences share only 54% similarity (25). To test the ability of Pr77 to induce gene transcription, the pTEX(−)pR77CAT vector was generated (Figure 1). Several constructs were also produced to allow comparative analyses of CAT transcription. Vectors containing the CAT gene cloned at the pTEX expression site (pTEXCAT vector), and into the pTEX vector lacking the 5′UTR of the gGADPH I gene (p) (pTEX(−)pCAT vector), were used as controls. The sequence located at the 5′ end of the T. cruzi KMP11 locus was also tested and cloned upstream of the CAT gene to generate the pTEX(−)p5′KMP11CAT transfection vector. T. cruzi (Y strain) epimastigotes were electroporated with either pTEXCAT, pTEX(−)pCAT, pTEX(−)pR77CAT or pTEX(−)p5′KMP11CAT vectors. Figure 2A shows the CAT cytoplasmic RNA level of each transfectant as determined by northern-blot analysis using the 32P-labeled CAT coding-region as a probe. A hybridization band showing the expected size of the CAT mRNA, roughly 1200 nt, was detected in pTEXCAT-, pTEX(−)p5′KMP11CAT- and pTEX(−)pR77CAT-transformed parasites. However, no CAT hybridization band was detected in pTEX(−)pCAT transfectants (Figure 2A). As shown in Figure 2A, the CAT expression level was significantly higher in pTEX(−)pR77CAT-transformed parasites than in pTEXCAT or pTEX(−)p5′KMP11CAT transfectants.
Figure 1.
Diagrammatic illustration of the pTEXCAT, pTEX(−)pCAT, pTEX(−)pR77CAT and pTEX(−)p5′KMP11CAT transfection vectors. (a) The chloramphenicol acetyltransferase coding gene (CAT) was cloned into the pTEX expression vector (26) to produce the pTEXCAT clone. (b) The pTEX(−)pCAT vector was generated by deleting the promoter region (the 5′ untranslated region of the GAPDH gene) from the pTEXCAT plasmid. (c) Pr77 was amplified from L1Tc and cloned at the 5′ end of the CAT gene in the pTEX(−)pCAT vector to produce the pTEX(−)pR77CAT clone. (d) The 5′ UTR region from the KMP11 gene tandem repeat was also inserted at the 5′ end of the CAT gene generating the pTEX(−)p5′KMP11CAT transfection vector. The central part of the figure shows the T. cruzi L1Tc (ACC: AF208537) and NARTc (ACC: AF215898) retroelements. The L1Tc represented is formed by a single long ORF (gray box) and is flanked by untranslated regions (striped boxes). The first 77 bp of L1Tc (Pr77), which are 100% identical to the first 77 bp of NARTc retrotransposons, are indicated by a white box. The regions from NARTc that, respectively, share 54% similarity and 85% homology to L1Tc, are also indicated. The four tandemly repeated copies of KMP11 gene are represented as light gray boxes and the 5′UTR region cloned as a white box.
Figure 2.
Abundance of CAT transcripts in T. cruzi transfectants. (A) northern blot analysis of CAT transcripts from T. cruzi transfectants. 5µg of cytoplasmic RNA from parasites transfected with pTEX(−)p5′KMP11CAT, pTEX(−)pR77CAT, pTEX(−)pCAT and pTEXCAT were resolved on 1% agarose-formaldehyde gels and immobilized on a quaternary amine-derived nylon membrane. The filter was hybridized with the radiolabeled CAT coding region as a probe. After phosphorimage analysis, the filter was rehybridized with T. cruzi 18S rDNA (18S) (31). The size of the hybridization bands is indicated in kb (MW). (B) Southern blot analysis of HindIII-digested genomic DNA from non-transformed parasites (Yc) and parasites transfected with pTEX(−)p5′KMP11CAT, pTEX(−)pR77CAT, pTEX(−)pCAT and pTEXCAT vectors. Radiolabeled DNA corresponding to the CAT coding region was used as a probe. After phosphorimage analysis, the filter was rehybridized with the radiolabeled coding sequence of KMP11 as a probe. The 0.6-kb hybridization band corresponding to the KMP11 gene is shown at the bottom of the figure. MW, molecular weight markers in bp. (C) Quantification of CAT RNA in T. cruzi transfectants. Photo-stimulated luminescence was determined using phosphorimaging employing ImageQuant software (Molecular Dynamics).
The relative plasmid copy number present in each transfectant was determined in the same parasites used for RNA isolation and northern blot analysis to avoid differences in the quantity of transfection vectors, which could have influenced the CAT transcription level. For this purpose, total DNA from the same amount of each transfectant was purified and digested with HindIII. The DNA was analyzed by Southern blotting using radiolabeled CAT and KMP11 coding sequences as probes (Figure 2B). Quantification of the hybridization bands showed the differences in the level of the CAT transcripts not to be a consequence of differences in the transfectant plasmid load (Figure 2A and B). Figure 2C shows that level of CAT mRNAs to be between 10-and 14-fold higher in pTEX(−)pR77CAT transfectants than in the pTEXCAT- and pTEX(−)p5′KMP11CAT-transfected parasites respectively. This suggests that the Pr77 sequence actively induces the transcription of a reporter gene, producing a larger quantity of gene transcripts than that induced by other sequences that also activate transcription.
To confirm that the detected CAT RNAs in the pTEX(−)pR77CAT-transfected parasites were driven by the Pr77 region and not by the gGAPDH intergenic region downstream of the CAT gene and upstream of the NEO gene in the pTEX vector, the presence of CAT transcripts from coding and non-coding strands was determined by northern blotting. Total RNA from pTEX(−)pR77CAT transfectants was separately hybridized with a pair of 5′ end-labeled complementary oligonucleotides, the antisense CAT1 and CAT1sense probes (Figure 3B and C). The same amount of RNA was hybridized with radiolabeled dsDNA corresponding to the CAT gene coding sequence (Figure 3A). In the membranes hybridized using dsDNA CAT and the antisense CAT1 primer as probes, a single hybridization band of a size compatible with that expected for CAT mRNA, and with approximately the same intensity, was observed (Figure 3A and B). However, no band was detected in the membrane hybridized with the CAT1sense probe (Figure 3C). A similar result was obtained when the RNA from pTEX(−)p5′KMP11CAT- and pTEXCAT -transfected parasites was hybridized with the dsDNA CAT probe and with the CAT1 antisense and sense probes (Figure 3A–C). The ability of both radiolabeled oligonucleotides to detect similar amounts of the complementary DNA strand under the experimental conditions employed was corroborated by dot-blotting. Thus, serial dilutions of the pTEXCAT vector DNA and of the antisense CAT1 or CAT1sense oligos were made, and all these DNAs fixed onto the membrane. As shown in Figure 3D and E, both probes showed the same capacity to hybridize with CAT DNA; hybridization dots of the same intensity were obtained with each. Similar results were obtained when poly(A)+ RNA from the transfected parasites was used in northern blot analyses (data non shown). Thus, the results suggest that all the detected CAT transcripts corresponded solely to the transcription of the sense CAT strand.
Figure 3.
Analysis of activation of sense and antisense transcription of CAT in T. cruzi transfectants. (A–C) Detection of sense and antisense CAT RNA by northern blot analysis. 7 µg of total RNA from parasites transfected with pTEX(−)pCAT, pTEX(−)p5′KMP11CAT, pTEXCAT and pTEX(−)pR77CAT vectors were loaded in triplicate and size-fractioned on 1% agarose/formaldehyde gels and subsequently immobilized on nylon membranes. Each membrane was hybridized with the radiolabeled CAT coding sequence (dsDNA CAT probe) (A), a 5′ end labeled antisense CAT oligo mapping at position 191–208 from the CAT start codon (antisense CAT1 probe) (B), and a 5′ end radiolabeled sense CAT oligo (CAT1sense probe) mapping at position 191–208 from the CAT start codon (C) as probes. The size of the hybridization bands is indicated in kb (MW). (D and E) Hybridization of sense and antisense CAT1 primers to the CAT sequence. Three-fold dilutions, from 196 ng to 2.4 ng for the pTEXCAT vector, and from 196 ng to 0.8 ng for the CAT1sense (D) or antisense CAT1 (E) oligos, were denatured and fixed on Zeta-Probe Blotting Membranes. The DNA filters were hybridized with 5′ end labeled antisense CAT1 oligo (D) or 5′ end labeled CAT1sense oligo (E) as probes.
The unspliced Pr77-derived transcripts are translated
We next examined whether the CAT transcripts derived from the Pr77 promoter in the pTEX(−)pR77CAT transfectants lacked the splice leader sequence, and also whether they contained the Pr77 sequence. RT-PCR using the purified poly(A)+ RNA fraction from the pTEX(−)pR77CAT, pTEXCAT and pTEX(−)p5′KMP11CAT transfectants was performed as described in the Materials and Methods section (see diagram in Figure 4A). As shown in Figure 4B, an amplification band of the expected size was observed when the pTEXCAT and pTEX(−)p5′KMP11CAT transfectants were used (Figure 4B). Cloning and sequencing of the amplified bands showed that CAT transcripts contained, respectively, the SL sequence at the KMP11 and the GADPH splicing acceptor sites as described earlier (27,34). However, when poly(A)+ RNA from pTEX(−)pR77CAT transfectants was employed, no band was amplified with the SLTc and CAT1 primers despite the fact that the amount of CAT mRNA was sufficient for reverse transcription and subsequent PCR-amplification using primer mapping at the 5′end of the Pr77 sequence (5′R77) and the CAT1 oligo (Figure 4B (C+)).
Figure 4.
Detection of the splice leader sequence at the 5′ end of CAT transcripts in T. cruzi transfectants. (A) Diagram of reverse transcriptase-PCR assay. Poly(A)+ RNAs purified from pTEX(−)pR77CAT, pTEXCAT and pTEX(−)p5′KMP11CAT transfectants were employed as templates for reverse transcription of CAT mRNAs using a CAT antisense primer that maps at position 424–439 from the CAT start codon (CAT2 oligo). PCR was subsequently performed using the SLTc primer and the CAT1 oligo which maps at position 191–208 from the CAT start codon. The amplified products were sequenced. (B) Ethidium bromide staining of amplicons derived from CAT transcripts. Amplification products were resolved in 2% agarose gel. The poly(A)+ RNA purified fractions from pTEX(−)pR77CAT, pTEXCAT and pTEX(−)p5′KMP11CAT transformants employed as templates for reverse transcription are indicated. As a positive control, the cDNA synthesized employing the CAT2 primer and polyadenylated RNA from pTEX(−)pR77CAT transfectants was PCR-amplified using 5′R77 as a forward primer mapping at position 1–11 from L1Tc, and the CAT1 oligo (C+). The sample with no reverse transcriptase was used as a negative control (C−). (MW): molecular weight marker in kb. (C) Analysis of CAT expression in T. cruzi transfectants. Expression of the CAT gene was detected by western blot analysis employing 30 µg of total soluble proteins from T. cruzi transfectants (pTEX(−)pCAT, pTEX(−)pR77CAT, grown in 250 and 500 µ/ml of G418, and pTEXCAT, grown in 500 µg/ml G418) and non-transfected parasites. Anti-chloramphenicol acetyl transferase antibodies were used (Sigma) in all cases. The molecular weight marker (kDa) is shown at the right side of the figure. The arrow indicates the 26 kDa CAT protein.
The next question to be answered was whether the unspliced Pr77-derived CAT transcripts were able to be translated. To answer this, total protein extracts were purified from the transfectants and analyzed by western blotting employing an anti-CAT antibody. As shown in Figure 4C, this antibody recognized a 26 kDa protein in the pTEX(−)pR77CAT and pTEXCAT parasites. However, no CAT protein was detected either in the pTEX(−)pCAT transfectants or in the wild type parasites (Figure 4C). The CAT expression level was significantly higher in the parasites that contained a spliced messenger (pTEXCAT transfectants) than in those that had unspliced CAT transcripts (the pTEX(−)pR77CAT transfectants).
To further corroborate that the Pr77-derived CAT transcripts from pTEX(−)pR77CAT transfectants were unspliced, the approach outlined in Figure 5A was followed. Accordingly, an RNA linker was ligated to the cytoplasmic RNA of the pTEX(−)pR77CAT transfectants. The RNA 5′ ends were reverse transcribed, PCR amplified, and then cloned into the pGEM vector. RNA isolated from the pTEX(−)p5′KMP11CAT transfectants was employed as a positive control. Screening was then performed by hybridizing to the CAT, R77 and SLTc probes. Interestingly, no hybridization with the SLTc probe was seen for any clone derived from the pTEX(−)pR77CAT transfectants. Ten colonies showing positive hybridization with either the R77 probe (the pTEX(−)pR77CAT transfectants) or the SL probe (the pTEX(−)p5′KMP11CAT transfectants) were analyzed after sequencing. The results shown in Figure 5B indicate that the SL sequence had been added to the CAT transcripts derived from the KMP11 5′ UTR region at the KMP11 splicing acceptor site. However, the CAT transcripts derived from Pr77 lacked the SL sequence at their 5′ end and appeared to be initiated at the Pr77 sequence, which is part of the CAT mRNA 5′ end. In all cases the CAT transcripts were partially truncated at their 5′ end, with truncation beginning at position +5, +8, +10, +14 or +21 of the SL sequence (KMP11-derived CAT transcripts), or at nucleotide +48, +53, +67 or +68 of Pr77 (Pr77-derived CAT transcripts). The greater degree of truncation observed in the CAT transcripts derived from the Pr77 sequence could be a consequence of the absence of a splice leader sequence in the Pr77 derived transcripts; this could influence transcript stability.
Figure 5.
Analysis of the 5′end of CAT mRNAs derived from the 5′UTRKPM11 region and the Pr77 promoter. (A) Outline of the method used. Cytoplasmic RNA from T. cruzi transfected with pTEX(−)pR77CAT and pTEX(−)p5′KMP11CAT (used as a control) was decapped with tobacco acid pyrophosphatase (TAP), dephosphorylated with alkaline phosphatase enzyme (AP) and phosphorylated by polynucleotide kinase (PNK) treatment. The changes produced in each case at the 5′ end of the treated RNAs are indicated. A linker RNA was ligated at the 5′ end of the transcripts using T4 RNA ligase. cDNA was synthesized by reverse transcription (RT) using a CAT gene specific primer (CAT1) that maps at position 191–208 from the CAT start codon, followed by PCR-amplification using a second CAT gene antisense primer (CAT90) mapping at position 71–90 from the CAT start codon, and finally an oligo corresponding to the sequence of the linker RNA. PCR products were cloned into the pGEM-T®-easy vector. Plasmid DNAs from the transfected bacteria selected by positive hybridization to CAT (ds DNA) and SLTc or R77 probes were sequenced. (B) Sequence at the 5′ end of processed CAT transcripts in pTEX(−)p5′KMP11CAT (a) and pTEX(−)pR77CAT (b) transfectants. The sequences shown are those downstream of the RNA linker sequence. Arrows indicate the first nucleotide of the 5′end of the CAT transcripts derived from the 5′UTRKMP11 and Pr77 sequences. The position of the first nucleotide of each transcript, with respect to the 5′end of SL and Pr77 sequences, is shown in italics. The number below the arrows corresponds to the quantity of clones starting at this position. Sequences corresponding to the splice leader sequence (SL), 5′UTRKMP11 (5′UTRKMP11CAT), L1TcPr77 (Pr77) and multi-cloning site – CAT region (MCS-CAT) are indicated.
The Pr77 region carries the transcription initiation site
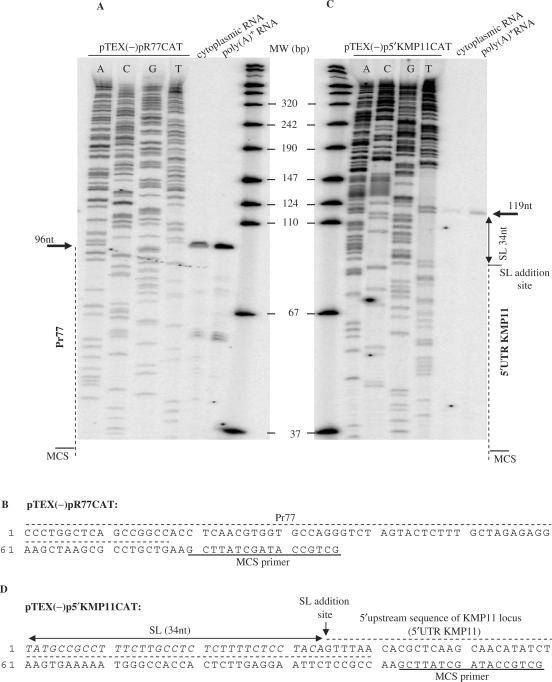
To further characterize the 5′ end of the CAT transcripts and the initiation site of the Pr77-derived CAT mRNA transcripts in the pTEX(−)pR77CAT transfected parasites, primer extension analysis was performed. Both the cytoplasmic RNA and the purified poly(A)+ RNA fraction from the pTEX(−)pR77CAT transfectants were extended using an oligonucleotide complementary to the polylinker sequence located between the Pr77 and CAT sequences in the transfection vector (Figure 6A). An extension product of 96 nt was generated after the extension reaction when either T. cruzi cytoplasmic RNA or the polyadenylated RNA fraction was used as a template. The length of this fragment corresponds to the expected size for the initiation of Pr77-derived CAT transcription at the first nucleotide of Pr77 (Figure 6B). This indicates that transcription mediated by the Pr77 sequence is initiated at nucleotide +1 and corroborates the evidence that the Pr77-derived transcripts lack the splice leader sequence. A primer extension assay performed with a cytoplasmic RNA and a poly(A)+ RNA purified fraction from pTEX(−)pKMP11CAT transfectants, used as a control, generated an extended fragment of approximately 119 nt. This is the expected size of the fragment when taking into account the addition of a spliced-leader sequence to the KMP11 trans-splicing acceptor site (Figure 6C and D). These results are consistent with the RT-PCR data shown in Figures 4 and 5. Thus, the Pr77 sequence actively promotes gene transcription, at least in the extrachromosomal context of this assay, and it behaves as an internal promoter.
Figure 6.
Analysis of the transcription initiation site of the CAT gene in pTEX(−)pR77CAT and pTEX(−)p5′KMP11CAT transfectants. (A and C) Primer extension analyses were performed employing cytoplasmic RNA or the purified polyadenylated fraction of RNA from pTEX(−)pR77CAT (A) and pTEX(−)p5′KMP11CAT (C) transfectants, along with a radiolabeled MCS primer that maps 3–19 nucleotides downstream of the Pr77 and 5′UTRKMP11 sequences in the pTEX(−)pR77CAT and pTEX(−)p5′KMP11CAT vectors respectively. Reactions were subsequently resolved in urea-polyacrylamide gels. The sequencing of pTEX(−)pR77CAT (A) and pTEX(−)p5′KMP11CAT (C) plasmids was performed following the Sanger method using an MCS antisense primer; samples were loaded onto the left side of the gels as size standards. The sequences corresponding to L1TcPr77 (Pr77), to the splice leader (SL) and to KMP11 5′UTR are indicated at both sides of the gels. Arrows indicate the position of the extended products. The radiolabeled molecular weight (MW) marker is expressed in bp. (B and D) Deduced sequence of the extended products in the primer extension assays(B) The Pr77 sequence (Pr77) of the pTEX(−)pR77CAT transfectants (D) Sequence of the splice leader (SL) and the 5′UTR of KMP11 in pTEX(−)p5′KMP11CAT transfectants. The splicing acceptor site is indicated by an arrow. The oligonucleotide used for primer extension analysis and employed for DNA sequencing is underlined. The SL sequence is indicated in italics.
L1Tc endogenous transcripts are unspliced and their transcription is initiated at, or close to, nucleotide +1 of L1Tc
To further explore the role of the Pr77 region in transcription in its native context and as part of the L1Tc mRNA, and to know at which location T. cruzi the L1Tc transcription is initiated, primer extension analysis was performed using cytoplasmic RNA from epimastigotes as a template along with two oligonucleotides complementary to L1Tc (L1Tc70 and L1Tc152). Figure 7A shows the L1Tc70 primer produced a 70-nt-long extension. The size of this fragment is consistent with the initiation of transcription at nucleotide +1 of L1Tc. A similar result was obtained when the L1Tc152 primer was used since a fragment of approximately 150 nt in length was produced (Figures 7A and B). However, in this case it could not be excluded that transcription had begun at nucleotide +2 to +4.
To determine the composition of the extended products derived from L1Tc endogenous transcripts, and to corroborate the absence of the splice leader sequence in the L1Tc mRNA, reverse transcription of the latter was performed, and PCR amplification of the transcript 5′ end undertaken as shown in Figure 4A. This was performed using cytoplasmic RNA from epimastigotes (Yc) as a template. As expected, the SLTc primer was unable to amplify the 5′end of the L1Tc mRNAs but it was able to amplify the N-terminal end of the spliced KMP11 transcripts (Figure 7C). In contrast, the 5′ end of the L1Tc mRNAs, after being reverse transcribed, were amplifiable when a primer mapping at the 5′ end element was used in PCR. This generated a fragment of the expected size as deduced from the previously performed experiments (Figure 7C). These results were confirmed by sequencing the amplified products.
L1Tc transcript stability
The stability of the unspliced L1Tc transcripts was then tested. Epimastigote cultures were treated with actinomycin D and the inhibition of RNA synthesis measured by the fall in [3H]-uridine incorporation into RNA (Figure 8) and by northern blotting using 32-P labeled L1Tc-RT as a probe. In contrast with the highly stable mRNAs of KMP11 (27) and the ribosomal S6 gene, the result shown in Figure 8 indicates that the L1Tc mRNAs have a moderate half-life of about 1 h.
Figure 8.
Kinetics of L1Tc mRNA stability. Cytoplasmic RNA from T. cruzi epimastigotes in the logarithmic phase of growth treated with actinomycin D (10 µg/ml) was isolated at the time intervals indicated (in hours).%U = percentage of [3H] uridine incorporation. The filter was hybridized with the region encoding reverse transcriptase activity of L1Tc (RT), and subsequently rehybridized with the T. cruzi KMP11-coding region (KMP11) and 6S rDNA (Ribosomal S6).
L1Tc is transcribed by RNA polymerase II
To identify the type of RNA polymerase(s) involved in L1Tc transcription, run-on transcription assays were performed with nuclei isolated from epimastigotes treated with different (and specific) RNA polymerase inhibitors. Nuclei were treated with a α-amanitin (1 mg/ml) (to which RNA polymerase I is resistant but RNA polymerase II is sensitive), 0.6% sarkosyl detergent (which disrupts RNA pol II complexes) (35) and tagetitoxin (which partially inhibits RNA pol III-dependent expression) (36). The nascent-labeled transcripts were hybridized to the full length L1Tc sequence and to those encoding the NL1Tc, RTL1Tc and C2-L1Tc proteins which had been previously fixed to a nylon membrane. Plasmid DNAs containing either 18S rRNA, α-tubulin or tRNA-coding genes, the transcription of which is mediated by RNA polymerase I, II and III respectively, were also fixed to the membrane and used as controls. Figure 9A and B show how the transcription of all the analyzed genes, with the exception of the 18S ribosomal gene, was strongly inhibited by α-amanitin treatment, and completely abolished by sarkosyl. However, the transcription of the 18S ribosomal gene was slightly reduced when nuclei were treated with α-amanitin and sarkosyl. tRNA transcripts were the only transcripts inhibited (44%) by treatment with 83 μM tagetitoxin (Figure 9A and B). Interestingly, tagetitoxin treatment of the nuclei seemed to upregulate transcription of the 18S, α-tubulin and L1Tc genes. Together, the results suggest that L1Tc is transcribed by RNA polymerase II. NL1Tc, RTL1Tc and C2-L1Tc, encoded by L1Tc, seem to be transcribed in equimolecular amounts (Figure 9A, see lane for untreated nuclei) and by RNA polymerase II since they were all inhibited by α-amanitin and sarkosyl, but were not negatively affected by tagetitoxin (Figure 9A and B).
DISCUSSION
Mobile retrotransposons make up a large percentage of most of the known eukaryotic genomes. These elements replicate through a round of transcription and reverse transcription, generating a copy at a new genomic location. The most abundant retrotransposons in the T. cruzi genome are the autonomous L1Tc and non-autonomous NARTc elements, which together make up some 10% of the T. cruzi genome. We previously reported the first 77 bp of L1Tc and NARTc elements to be 100% identical, while the remainder of NARTc shares an identity of only 54% with L1Tc. The 13 nucleotides sequence located at the 3′ end of both elements are 85% conserved (25). Pr77 is also conserved in T. brucei ingi/RIME; 76.6% similarity is maintained despite the low level of conservation between L1Tc and ingi at the nucleotide level (25). Leishmania major also contains a 72 bp sequence that is 65% identical to the first 77 bp of L1Tc and NARTc, and 76.6% identical to the homologous region of ingi/RIME (25). This conserved sequence always appears to be present at the 5′ end of all full-length degenerate L1Tc/ingi-related elements (DIREs) in L. major, and to be the only conserved sequence of the T. cruzi, T. brucei and L. major full-length DIREs (37). These elements have accumulated a high degree of degeneration that, theoretically, ought to render them unable to encode proteins for their own transposition. However, it cannot be excluded that, after their transcription, DIREs might be mobilized in trans by the retrotransposition machinery encoded by active elements such as ingi in T. brucei and L1Tc in T. cruzi (37).
Since an internal promoter is expected to mediate the transcription of retrotransposons, and despite the fact that the consensus promoter sequences remain undescribed in trypanosomatids, we were interested in determining whether the Pr77 sequence could drive the transcription of a reporter gene. Pr77 was found to drive the expression of the CAT gene 10–14-fold times more strongly than that mediated by sequences naturally located upstream of the KMP11 and GADPH loci. These sequences are expected to promote transcription of tandemly repeated genes in T. cruzi since GADPH has been extensively shown to promote transcription of foreign genes in T. cruzi transformants (26). Pr77-derived CAT mRNA overproduction was not a consequence of the transcription mediated by the pTEX plasmid since the detected CAT RNA was sense orientated and not detected in pTEX(−)pCAT transfectants. The Pr77-derived CAT mRNA overproduction was not a consequence of a higher concentration of the transfection vector in T. cruzi. In fact, in all transfectants, there was a high concentration of plasmid DNA. Also, a sequence including the Pr77 fragment has previously been shown to mediate overexpression of NL1Tc in T. cruzi stable transfectants (38). A similar approach was used to detect promoter activity associated with the 5′ end of T. brucei ingi/RIME elements. However, neither the first 500 bp of ingi, nor the 250 bp corresponding to the entire Rime A sequence, nor the first 55 bp of ingi/RIME were able to mediate CAT expression. This correlated with the absence of detectable CAT transcription (39). Thus, it is suggested that the previously described abundant and diverse, small, antisense RNAs specific to the ingi/RIME elements are involved in the downregulation of the expression of these elements. However, and despite the fact that RNAi elements are operational in T. brucei, neither T. cruzi nor L. major appear to have the full RNAi machinery. This has led to the suggestion that T. cruzi, and perhaps other trypanosomatids, use an alternative strategy for silencing retroelements (5).
The sequence located at the 5′ end of human L1 has also been shown to promote transcription and to drive the expression of beta-galactosidase in a variety of transiently transfected cell types in tissue culture (40). The most critical sequences are located within the first 100 bp of L1Hs, although sequences within the first 668 bp also contribute significantly to the overall level of expression (40). The present data clearly show that Pr77 actively mediates transcription of the CAT gene, implying that Pr77 may well mediate L1Tc transcription. It cannot be excluded, however, that read-through transcription from neighboring genes may also produce L1Tc transcripts. It is noteworthy that LINE elements might be translated from external promoters, even in the presence of a functional LINE promoter (41). Interestingly, Pr77 contains the conserved CGT(G/T) motif found close to the transcription initiation sites of some non-LTR retrotransposons in insects (42,43). This sequence acts as an efficient binding site for a specific protein (42) and is involved in transcription of the silkworm Bombyx mori TRAS1 non-LTR retrotransposon (43). Further, the location of this conserved core in L1Tc Pr77 is identical to that seen in ingi and RIME. In addition, an upstream sequence almost complementary to the conserved core is also present in all three. These two sequences have been shown to be involved in the binding of an unknown 50 kDa nuclear protein present in both bloodstream and procyclic forms of T. brucei (39). Remarkably, this conserved core is also present in a region located 30–40 bp downstream of the transcriptional start site of several Drosophila and mammalian non-TATA genes which are expressed in a regulated fashion and are known to have transcriptionally important downstream elements (42).
Several experiments have shown that the Pr77-derived transcripts lack a splice leader sequence, while transcripts such as those derived from the 5′UTRKmp11 gene are efficiently spliced and bear the SL at the expected splicing acceptor signal of the gene. The present data show that, in the natural environment of T. cruzi, the L1Tc transcripts also appear to be unspliced. Thus, the moderate L1Tc transcript stability—deduced from transcription inhibition with actinomycin D and the results of northern blot analysis—might be related to its unspliced nature. As far as is known, most genes in trypanosomes are transcribed polycistronically and then cleaved into functional mRNAs by trans-splicing of a capped 39-nucleotide leader RNA derived from a short transcript. However, trans-splicing within a specific transcript produces loss of the sequence upstream of the splicing acceptor site. Sequence analysis of the 5′ end of 19 cDNA clones derived from T. brucei RIME and ingi elements has shown that none contain the mini-exon sequence found at the 5′ end of all known functional trypanosome RNAs (37). Thus, the unspliced nature of the non-LTR retroelement transcripts seems to be a general characteristic of these LINEs in trypanosomatids, which evolved to conserve the full length of their transcripts and thus guarantee the autonomous character of newly inserted copies.
The present data indicate that the Pr77-derived CAT transcripts are initiated at or close to nucleotide +1 of Pr77, as deduced from primer extension analyses performed with cytoplasmic and polyadenylated RNA fractions from Pr77CAT T. cruzi transfectants (Figures 5B and 6). L1Tc endogenous transcripts are also predominantly initiated at nucleotide +1 of L1Tc, as observed in the primer extension assays employing L1Tc-specific antisense primers and cytoplasmic RNA from wild-type T. cruzi epimastigotes. This explains the presence of a highly abundant 5 kb L1Tc messenger the length of the complete genomic element in epimastigotes of T. cruzi (14). Thus, the existence of unspliced L1Tc transcripts starting at nucleotide +1 seems to ensure that full-length retrotransposed L1Tc RNAs retain an intact internal promoter. This result is consistent with that deduced from the analysis of L1Tc/NARTc distributions over 10 kb-long contigs generated from the T. cruzi genome database, suggesting that most elements are generated by reverse transcription of full-length L1Tc/NARTc transcripts. In fact, approximately 47.25% of L1Tc elements and 85.7% of NARTc are full-length elements. The remaining elements are 5′ end truncated (49.7% of L1Tc and 7.15% of NARTc) or 3′end truncated (1.68% of L1Tc and 7.14% of NARTc). This truncation may be a consequence of unfinished reverse transcription (44).
The amount of mRNA derived from a retrotransposon is directly proportional to its transposition rate. However, transcription of almost all trypanosome genes is constitutive. Most of the regulation of gene expression in these organisms seems to occur post-transcriptionally either by controlling the stability of the processed mRNAs or by translational controls (45). Interestingly, the Pr77-derived unspliced transcripts can be translated to generate the corresponding protein. However, as deduced from the amount of CAT protein, the level of protein expression mediated by Pr77 in pTEX(−)pR77CAT transfectants is much lower than that detected in pTEXCAT parasites. The low translation level could be a consequence of the unspliced character of the Pr77-derived transcripts. However, it cannot be excluded that other L1Tc sequences located downstream of Pr77 are required for ribosome stabilization. It has been described that the potentially active L1Tc elements contain an in vivo functional viral-like 2A self-cleaving sequence (L1Tc2A) located downstream of Pr77 and in frame with the proteins that L1Tc encodes (46). The present data suggest that the mechanism of polypeptide cleavage by the L1Tc2A peptide is co-translational and could influence the composition and probably the relative abundance of the proteins that compose the L1Tc mobilization machinery (46).
The transcription of a full-length LINE containing an ‘internal promoter structure’ would lead to the generation of a complete active copy when it is inserted at a new location. Thus, the ‘internal promoter structure’ of human L1 implies that the generation of an active copy occurs when it is inserted at a new location (47). This resembles the promoter of eukaryotic tRNA genes which are transcribed by RNA pol III (48). However the protein coding capacity of the L1 RNA, poly (A) tail, and the in vivo α-amanitin inhibition seen suggest that the non-LTR retrotransposons are transcribed by RNA polymerase II (13). Further, other LINE elements such as Jockey, I factor, F and Doc in Drosophila have been shown to contain an internal pol-II-dependent promoter (49–52). Although the three classical RNA polymerases (pol I, II and III) have been detected in trypanosomatids (53) and it is known that protein-coding genes are transcribed by RNA polymerase II, no RNA polymerase II promoters had been clearly identified to date in these organisms. All previous attempts to identify pol II promoters have failed to provide a full characterization, with the exception of the SL RNA promoter (54). The run-on experiments described in the present work show that the L1Tc transcripts are RNA pol II-dependent since transcription is sensitive to 1 mg/ml α-amanitin and 0.6% sarkosyl treatments, and resistant to 83 µM of tagetitoxin.
The strand-switch regions in trypanosomatids appear to be correlated with non-LTR retrotransposon sites (55). Interestingly, nuclear run-on analyses of L. major chr1 showed that specific RNA-polymerase-II-mediated transcription leading to the production of stable transcripts initiates within the strand switch region and proceeds bidirectionally toward the telomeres, and that non-specific transcription takes place over the entire chr1 but at an approximately 10-fold lower level than that occurring when transcription initiates in the strand-switch area (56). It has been shown that the Pr77 sequence does not associate with retroelements at 24 different genomic positions in T. cruzi (44) and that it is conserved in the L. major genome despite the lack of active mobile elements (37). Further, L1Tc/NARTc elements in T. cruzi are frequently associated with DGF-1, expressed RHS multigene family and with genes coding for surface proteins such as transialidase, mucins, mucin-associated surface proteins and gp63 peptidase (17,57). Thus, the relatively high degree of dispersion that L1Tc and non-coding but transcriptionally active NARTc elements show, reinforces the idea that these non-LTR retrotransposons are responsible for the transcription of adjacent genes and polycistrons.
ACKNOWLEDGEMENTS
We are grateful to Prof. C. Alonso for critical reading of the manuscript, M. Caro for excellent technical assistance, M. García-Cañadas for assistance in western blot assays and Dr W. Teran for assistance in the primer extension assays. This work was supported by BMC2003–00834 from Plan Nacional I + D + I (MEC, Spain) and PAI ref. P05-CVI-01227 (Junta de Andalucía, Spain). S.R.H. was supported by a MEC Predoctoral Fellowship, Spain. Funding to pay the Open Access publication charge was provided by PAI CVI-155 (Junta de Andalucia, Spain).
Conflict of interest statement: None declared.
REFERENCES
- 1.WHO. The World Health Report. Geneva: The World Health Organization; 2002. [Google Scholar]
- 2.Sutton RE, Boothroyd JC. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hug M, Hotz HR, Hartmann C, Clayton C. Hierarchies of RNA-processing signals in a trypanosome surface antigen mRNA precursor. Mol. Cell. Biol. 1994;14:7428–7435. doi: 10.1128/mcb.14.11.7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LeBowitz JH, Smith HQ, Rusche L, Beverley SM. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes. Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 5.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 6.Donelson JE. Genome research and evolution in trypanosomes. Curr. Opin. Genet. Dev. 1996;6:699–703. doi: 10.1016/s0959-437x(96)80023-2. [DOI] [PubMed] [Google Scholar]
- 7.Requena JM, Lopez MC, Alonso C. Genomic repetitive DNA elements of Trypanosoma cruzi. Parasitol. Today. 1996;12:279–283. doi: 10.1016/0169-4758(96)10024-7. [DOI] [PubMed] [Google Scholar]
- 8.Weiner AM. SINEs and LINEs: the art of biting the hand that feeds you. Curr. Opin. Cell. Biol. 2002;14:343–350. doi: 10.1016/s0955-0674(02)00338-1. [DOI] [PubMed] [Google Scholar]
- 9.Kajikawa M, Okada N. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell. 2002;111:433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- 10.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 11.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 12.Boeke JD, Corces VG. Transcription and reverse transcription of retrotransposons. Annu. Rev. Microbiol. 1989;43:403–434. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- 13.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 14.Martin F, Maranon C, Olivares M, Alonso C, Lopez MC. Characterization of a non-long terminal repeat retrotransposon cDNA (L1Tc) from Trypanosoma cruzi: homology of the first ORF with the ape family of DNA repair enzymes. J. Mol. Biol. 1995;247:49–59. doi: 10.1006/jmbi.1994.0121. [DOI] [PubMed] [Google Scholar]
- 15.Torres C, Barreiro L, Dallagiovanna B, Gamarro F, Castanys S. Characterization of a new ATP-binding cassette transporter in Trypanosoma cruzi associated to a L1Tc retrotransposon. Biochim. Biophys. Acta. 1999;1489:428–432. doi: 10.1016/s0167-4781(99)00195-5. [DOI] [PubMed] [Google Scholar]
- 16.Bringaud F, Vedrenne C, Cuvillier A, Parzy D, Baltz D, Tetaud E, Pays E, Venegas J, Merlin G, et al. Conserved organization of genes in trypanosomatids. Mol. Biochem. Parasitol. 1998;94:249–264. doi: 10.1016/s0166-6851(98)00080-2. [DOI] [PubMed] [Google Scholar]
- 17.Olivares M, Thomas MC, Lopez-Barajas A, Requena JM, Garcia-Perez JL, Angel S, Alonso C, Lopez MC. Genomic clustering of the Trypanosoma cruzi non-long terminal L1Tc retrotransposon with defined interspersed repeated DNA elements. Electrophoresis. 2000;21:2973–2982. doi: 10.1002/1522-2683(20000801)21:14<2973::AID-ELPS2973>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 18.Bringaud F, Biteau N, Melville SE, Hez S, El-Sayed NM, Leech V, Berriman M, Hall N, Donelson JE, et al. A new, expressed multigene family containing a hot spot for insertion of retroelements is associated with polymorphic subtelomeric regions of Trypanosoma brucei. Eukaryot. Cell. 2002;1:137–151. doi: 10.1128/EC.1.1.137-151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D, Chiurillo MA, El-Sayed N, Jones K, Santos MR, Porcile PE, Andersson B, Myler P, da Silveira JF, et al. Telomere and subtelomere of Trypanosoma cruzi chromosomes are enriched in (pseudo)genes of retrotransposon hot spot and trans-sialidase-like gene families: the origins of T. cruzi telomeres. Gene. 2005;346:153–161. doi: 10.1016/j.gene.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Olivares M, Alonso C, Lopez MC. The open reading frame 1 of the L1Tc retrotransposon of Trypanosoma cruzi codes for a protein with apurinic-apyrimidinic nuclease activity. J. Biol. Chem. 1997;272:25224–25228. doi: 10.1074/jbc.272.40.25224. [DOI] [PubMed] [Google Scholar]
- 21.Olivares M, Thomas MC, Alonso C, Lopez MC. The L1Tc, long interspersed nucleotide element from Trypanosoma cruzi, encodes a protein with 3′-phosphatase and 3′-phosphodiesterase enzymatic activities. J. Biol. Chem. 1999;274:23883–23886. doi: 10.1074/jbc.274.34.23883. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Perez JL, Gonzalez CI, Thomas MC, Olivares M, Lopez MC. Characterization of reverse transcriptase activity of the L1Tc retroelement from Trypanosoma cruzi. Cell. Mol. Life Sci. 2003;60:2692–2701. doi: 10.1007/s00018-003-3342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olivares M, Garcia-Perez JL, Thomas MC, Heras SR, Lopez MC. The non-LTR (long terminal repeat) retrotransposon L1Tc from Trypanosoma cruzi codes for a protein with RNase H activity. J. Biol. Chem. 2002;277:28025–28030. doi: 10.1074/jbc.M202896200. [DOI] [PubMed] [Google Scholar]
- 24.Heras SR, Lopez MC, Garcia-Perez JL, Martin SL, Thomas MC. The L1Tc C-terminal domain from Trypanosoma cruzi non-long terminal repeat retrotransposon codes for a protein that bears two C2H2 zinc finger motifs and is endowed with nucleic acid chaperone activity. Mol. Cell. Biol. 2005;25:9209–9220. doi: 10.1128/MCB.25.21.9209-9220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bringaud F, Garcia-Perez JL, Heras SR, Ghedin E, El-Sayed NM, Andersson B, Baltz T, Lopez MC. Identification of non-autonomous non-LTR retrotransposons in the genome of Trypanosoma cruzi. Mol. Biochem. Parasitol. 2002;124:73–78. doi: 10.1016/s0166-6851(02)00167-6. [DOI] [PubMed] [Google Scholar]
- 26.Kelly JM, Ward HM, Miles MA, Kendall G. A shuttle vector which facilitates the expression of transfected genes in Trypanosoma cruzi and Leishmania. Nucleic Acids Res. 1992;20:3963–3969. doi: 10.1093/nar/20.15.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas MC, Garcia-Perez JL, Alonso C, Lopez MC. Molecular characterization of KMP11 from Trypanosoma cruzi: a cytoskeleton-associated protein regulated at the translational level. DNA Cell. Biol. 2000;19:47–57. doi: 10.1089/104454900314708. [DOI] [PubMed] [Google Scholar]
- 28.Cooper R, de Jesus AR, Cross GA. Deletion of an immunodominant Trypanosoma cruzi surface glycoprotein disrupts flagellum-cell adhesion. J. Cell. Biol. 1993;122:149–156. doi: 10.1083/jcb.122.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maranon C, Thomas MC, Puerta C, Alonso C, Lopez MC. The stability and maturation of the H2A histone mRNAs from Trypanosoma cruzi are implicated in their post-transcriptional regulation. Biochim. Biophys. Acta. 2000;1490:1–10. doi: 10.1016/s0167-4781(99)00228-6. [DOI] [PubMed] [Google Scholar]
- 30.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Maranon C, Puerta C, Alonso C, Lopez MC. Control mechanisms of the H2A genes expression in Trypanosoma cruzi. Mol. Biochem. Parasitol. 1998;92:313–324. doi: 10.1016/s0166-6851(98)00003-6. [DOI] [PubMed] [Google Scholar]
- 32.Bruderer T, Tu LC, Lee MG. The 5′ end structure of transcripts derived from the rRNA gene and the RNA polymerase I transcribed protein coding genes in Trypanosoma brucei. Mol. Biochem. Parasitol. 2003;129:69–77. doi: 10.1016/s0166-6851(03)00095-1. [DOI] [PubMed] [Google Scholar]
- 33.Teran W, Felipe A, Segura A, Rojas A, Ramos JL, Gallegos MT. Antibiotic-dependent induction of Pseudomonas putida DOT-T1E TtgABC efflux pump is mediated by the drug binding repressor TtgR. Antimicrob. Agents Chemother. 2003;47:3067–3072. doi: 10.1128/AAC.47.10.3067-3072.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kendall G, Wilderspin AF, Ashall F, Miles MA, Kelly JM. Trypanosoma cruzi glycosomal glyceraldehyde-3-phosphate dehydrogenase does not conform to the ‘hotspot’ topogenic signal model. EMBO. J. 1990;9:2751–2758. doi: 10.1002/j.1460-2075.1990.tb07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Archambault J, Friesen JD. Genetics of eukaryotic RNA polymerases I, II, and III. Microbiol. Rev. 1993;57:703–724. doi: 10.1128/mr.57.3.703-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinberg TH, Mathews DE, Durbin RD, Burgess RR. Tagetitoxin: a new inhibitor of eukaryotic transcription by RNA polymerase III. J. Biol. Chem. 1990;265:499–505. [PubMed] [Google Scholar]
- 37.Bringaud F, Ghedin E, Blandin G, Bartholomeu DC, Caler E, Levin MJ, Baltz T, El-Sayed NM. Evolution of non-LTR retrotransposons in the trypanosomatid genomes: Leishmania major has lost the active elements. Mol. Biochem. Parasitol. 2006;145:158–170. doi: 10.1016/j.molbiopara.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 38.Olivares M, Lopez MC, Garcia-Perez JL, Briones P, Pulgar M, Thomas MC. The endonuclease NL1Tc encoded by the LINE L1Tc from Trypanosoma cruzi protects parasites from daunorubicin DNA damage. Biochim. Biophys. Acta. 2003;1626:25–32. doi: 10.1016/s0167-4781(03)00022-8. [DOI] [PubMed] [Google Scholar]
- 39.García-Salcedo JA, Gijón P, Amiguet-Vercher A, Pays E. Searching for promoter activity in RIME/Ingi retrotransposons from Trypanosoma brucei: binding of a protein to their 5′ extremity. Exp. Parasitol. 2003;104:140–148. doi: 10.1016/j.exppara.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic. Acids Res. 2004;32:3846–3855. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arkhipova IR, Ilyin YV. Properties of promoter regions of mdg1 Drosophila retrotransposon indicate that it belongs to a specific class of promoters. EMBO. J. 1991;10:1169–1177. doi: 10.1002/j.1460-2075.1991.tb08057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi H, Fijiwara H. Transcription analysis of the telomeric repeat-specific retrotransposons TRAS1 and SART1 of the silkworm Bombyx mori. Nucleic Acids Res. 1999;27:1021–2015. doi: 10.1093/nar/27.9.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bringaud F, Bartholomeu DC, Blandin G, Delcher A, Baltz T, El-Sayed NM, Ghedin E. The Trypanosoma cruzi L1Tc and NARTc non-LTR retrotransposons show relative site specificity for insertion. Mol. Biol. Evol. 2006;23:411–420. doi: 10.1093/molbev/msj046. [DOI] [PubMed] [Google Scholar]
- 45.Clayton CE. Life without transcriptional control? From fly to man and back again. EMBO. J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heras SR, Thomas MC, Garcia-Cañadas M, Felipe PD, Garcia-Perez JL, Ryan MD, Lopez MC. L1Tc non-LTR retrotransposons from Trypanosoma cruzi contain a functional viral-like self-cleaving 2A sequence in frame with the active proteins they encode. Cell. Mol. Life Sci. 2006;63:1449–1460. doi: 10.1007/s00018-006-6038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum. Mol. Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 48.Paule MR, White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizrokhi LJ, Georgieva SG, Ilyin YV. jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell. 1988;54:685–691. doi: 10.1016/s0092-8674(88)80013-8. [DOI] [PubMed] [Google Scholar]
- 50.McLean C, Bucheton A, Finnegan DJ. The 5′ untranslated region of the I factor, a long interspersed nuclear element-like retrotransposon of Drosophila melanogaster, contains an internal promoter and sequences that regulate expression. Mol. Cell. Biol. 1993;13:1042–1050. doi: 10.1128/mcb.13.2.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minchiotti G, Contursi C, Di Nocera PP. Multiple downstream promoter modules regulate the transcription of the Drosophila melanogaster I, Doc and F elements. J. Mol. Biol. 1997;267:37–46. doi: 10.1006/jmbi.1996.0860. [DOI] [PubMed] [Google Scholar]
- 52.Contursi C, Minchiotti G, Di Nocera PP. Identification of sequences which regulate the expression of Drosophila melanogaster Doc elements. J. Biol. Chem. 1995;270:26570–26576. doi: 10.1074/jbc.270.44.26570. [DOI] [PubMed] [Google Scholar]
- 53.Kelly S, Wickstead B, Gull K. An in silico analysis of trypanosomatid RNA polymerases: insights into their unusual transcription. Biochem. Soc. Trans. 2005;33:1435–1437. doi: 10.1042/BST0331435. [DOI] [PubMed] [Google Scholar]
- 54.Gilinger G, Bellofatto V. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 2001;29:1556–1564. doi: 10.1093/nar/29.7.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghedin E, Bringaud F, Peterson J, Myler P, Berriman M, Ivens A, Andersson B, Bontempi E, Eisen J, et al. Gene synteny and evolution of genome architecture in trypanosomatids. Mol. Biochem. Parasitol. 2004;134:183–191. doi: 10.1016/j.molbiopara.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 56.Martinez-Calvillo S, Yan S, Nguyen D, Fox M, Stuart K, Myler PJ. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell. 2003;11:1291–1299. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 57.El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, Aggarwal G, Caler E, Renauld H, Worthey EA, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 58.Soares CM, de Carvalho EF, Urmenyi TP, Carvalho JF, de Castro FT, Rondinelli E. Alpha- and beta-tubulin mRNAs of Trypanosoma cruzi originate from a single multicistronic transcript. FEBS Lett. 1989;250:497–502. doi: 10.1016/0014-5793(89)80784-7. [DOI] [PubMed] [Google Scholar]