Abstract
The Sleeping Beauty (SB) transposon is a promising gene transfer vector that integrates nonspecifically into host cell genomes. Herein, we attempt to direct transposon integration into predetermined DNA sites by coupling a site-specific DNA-binding domain (DBD) to the SB transposase. We engineered fusion proteins comprised of a hyperactive SB transposase (HSB5) joined via a variable-length linker to either end of the polydactyl zinc-finger protein E2C, which binds a unique sequence on human chromosome 17. Although DBD linkage to the C-terminus of SB abolished activity in a human cell transposition assay, the N-terminal addition of the E2C or Gal4 DBD did not. Molecular analyses indicated that these DBD-SB fusion proteins retained DNA-binding specificity for their respective substrate molecules and were capable of mediating bona fide transposition reactions. We also characterized transposon integrations in the presence of the E2C-SB fusion protein to determine its potential to target predefined DNA sites. Our results indicate that fusion protein-mediated tethering can effectively redirect transposon insertion site selection in human cells, but suggest that stable docking of integration complexes may also partially interfere with the cut-and-paste mechanism. These findings illustrate the feasibility of directed transposon integration and highlight potential means for future development.
INTRODUCTION
Many different gene delivery vectors have been developed through the years to achieve lifelong gene expression in target cells via the stable integration of a therapeutic transgene into host cell chromosomes. Unfortunately, the systems currently in use in clinical gene therapy trials rely entirely on random or nonspecific integration. This lack of insertion site specificity can result in undesired targeting events, leading to attenuated gene expression due to position-effect variegation or to deregulated gene expression via insertional mutagenesis. Notably, in a recent gene therapy trial, a randomly integrating retroviral vector caused leukemia in three children following its integration near—and subsequent activation of—the LMO-2 proto-oncogene (1–3). While the vast majority of commentaries regarding these tragic events suggest a long list of complex factors contributing to the development of leukemia, there remains an undeniable uncertainty regarding the true risk of insertional mutagenesis in each particular instance. Accordingly, the advent of new strategies to effectively direct integration events in human cells remains a goal of significant clinical value.
Retroviral vectors can reliably insert a gene into the chromosomes of a target cell (4). Although these integrations normally occur globally in the genome, previous work has shown the potential to manipulate retroviral target site selection using a molecular tethering strategy. In these studies, the viral integrase enzyme that mediates retroviral integration was fused to a heterologous DNA-binding domain (DBD) protein and shown to facilitate biased integration into a defined 20–60-bp region encompassing the DBD-specific target site. This basic concept was initially demonstrated in vitro using a variety of different chimeric proteins, including ones comprised of the avian sarcoma virus (ASV) integrase and the LexA DBD (5), the Moloney murine leukemia virus (MLV) integrase and the Sp1 DBD (6), the human immune-deficiency virus type-I (HIV-1) integrase fused to the DBDs of LexA (7), lambda repressor (8) and Zif268 (9) or most recently, to the custom-designed zinc-finger protein E2C (10), which recognizes a unique site in the human genome (11,12). Although the extension of this in vitro work to human cells was significantly delayed due to the technological challenge of making an infectious virus encoding the hybrid protein, this barrier was recently overcome by packaging the chimeric integrase in trans (13). Using this new method, hybrid E2C-based integrases have also recently been shown to mediate directed integration within the human genome (14). While these studies demonstrate the tremendous potential of such an approach, alternative recombinases may actually be better suited to the pursuit of site-selective integration via hybrid DBD–recombinases. This is because retroviral DNA integration has since been found to be heavily biased towards transcriptionally active genes (15–18) and that the viral integrase protein, by physically interacting with specific chromatin-associated factors (e.g. p75/LEDGF and p300), can contribute actively to this process (19–23). These inherent mechanisms present an unfortunately high mutagenic risk that would likely remain in the presence of any integrase-based enzyme, which could compromise the potential utility of any directed integration strategy.
The stable transfer of foreign DNA into host genomes can also be achieved using a DNA transposon such as the Sleeping Beauty (SB) transposable element (24). This non-viral element has been shown to be a promising vehicle for therapeutic gene delivery and can integrate efficiently into mammalian host cell chromosomes using a well-defined transposition mechanism (25). Certain stages of this transposition process involve functional- and biochemical-related activities with that of retroviral DNA integration and is initiated by the sequence-specific binding of the SB transposase to each of four 32–34 bp direct-repeat (DR) sequences embedded within the transposon terminal inverted repeats (IRs) (26). These associated transposase molecules mediate the stable pairing of the transposon ends via subunit multimerization, resulting in transposon excision from the donor site and subsequent re-insertion into a new target site. Transposition is catalyzed entirely by the SB transposase, which is comprised of two major functional domains: (i) an N-terminal bipartite DNA-binding domain that mediates multimerization, nuclear transport and binding to transposon end sequences and (ii) a retroviral integrase-related C-terminal catalytic core that mediates all the DNA cleavage and strand transfer reactions required for transposition. SB element transposition always occurs into a TA target dinucleotide, which is subsequently duplicated upon insertion by cellular DNA repair pathways (27,28). Although a consensus AT-rich palindrome is highly favored at the site of transposon insertion, SB target site selection appears to be determined by structural constraints rather than at the level of primary DNA sequence (29–32). In addition, while the SB transposase can physically associate with at least three known host cell components (28,33,34), it is unlikely that any of these binding partners actively biases SB target site selection towards a specific sub-region of the genome. Indeed, recent studies have shown that, in contrast to most retroviral-based vectors, SB integrates fairly randomly in mammalian cells, without any discernable preference for actively transcribed genes (32). These properties collectively make SB a suitable candidate for development into a targeted integration system.
Although there are an estimated 200,000,000 potential TA integration sites for an SB transposon in a haploid human genome (30), the covalent linkage of a site-specific DNA-binding component to the transposase enzyme might ultimately help diminish the number of target sites recognized in the cell. Successful targeting into a much more limited subset of potential target sites is, without question, a vastly complex and difficult barrier to overcome and will likely depend heavily on two major achievements: (i) the design of an appropriate DBD-SB fusion protein capable of high-affinity and site-selective DNA binding in the context of human cells and (ii) the development of novel strategies to prevent and/or diminish integration activity at undesirable sites. In this report, we have focused our efforts on tackling the first of these barriers as a necessary first step towards achieving site-selective integration at the genome-level. We describe the engineering of a series of transposase–DNA-binding domain hybrid proteins and have evaluated these chimeras for proper expression, DNA-binding and catalytic activity in human cells. We also assessed the ability of these fusion proteins to direct integration into the vicinity of plasmid-based target DNA molecules. Our proof-of-principle results demonstrate targeted SB transposition in mammalian cells using hybrid transposases and provide valuable new insights into the transposition mechanism.
MATERIALS AND METHODS
Plasmid constructs
The pc-SB10, pc-GFP and pc-N plasmids have been previously described (35). The pc-HSB5 plasmid is derived from pc-HSB3 (36) and encodes an additional alanine-substitution mutation (S270A) in its catalytic core that further improves its transpositional activity in mammalian cells. To generate pc-E2C, we PCR amplified the E2C gene from pMal-c2-E2C (12) (kindly provided by C. Barbas, Salk Institute), treated the resulting product with BamHI-EcoRI restriction endonucleases and ligated it into pc-N. All fusion constructs described in this report were generated using mutagenic primers together with a two-step overlapping PCR-based approach. Resulting full-length fusion products were digested with BamHI-XbaI restriction endonucleases and ligated into the pc-N plasmid. Detailed description of the primer sequences is available upon request. In every plasmid described herein, we confirmed the sequence identity for each modified plasmid region by extensive DNA sequence analysis. For the pc-hE2C-L5-SB variant, we had the E2C, L5 linker and 5′ SB coding sequences from pc-E2C-L5-SB commercially synthesized (DNA 2.0, Menlo Park, CA) using codons for optimized expression in human cells (sequence available upon request) and then subcloned this synthetic fragment back into pc-E2C-L5-SB by BamHI-BsrG1 ligation. Inactivating control mutations into the transposase DNA-binding domain (pc-E2C-L5-SB-G59A) and catalytic core domain (pc-E2C-L5-SB-E279A) were also introduced into the parental pc-E2C-L5-SB vector using mutagenic PCR-based methods. The pc-Gal4-L5-SB construct was made by fusing the Gal4 DNA-binding domain from pBIND with the linker-plus-HSB5 region from pc-hE2C-L5-SB using mutagenic PCR primers. The pc-E2C-L5-SB(N123) and pc-E2C-L5-SB(N123)-G59A plasmids encoding truncated fusion proteins were generated using the 3′N123 primer (5′-GCAGAATTCCTAGAGCAGTGGCTTCTT), digesting the resulting PCR fragments with BamHI-EcoRI restriction endonucleases and ligating them into pc-N.
For our inter-plasmid transposition studies, it was necessary to generate chloramphenicol-based version of each pc-N-based construct by ligating the BglII-PvuII fragment encoding each CMV-driven expression cassette with BamHI-EcoRV-treated pBC-KS(+) vector (Stratagene). The pBind, pACT and p5G-Luc reporter plasmids are all components of Promega's Mammalian Two-Hybrid Kit. To generate SB- (p5SB-Luc), E2C- (p5E-Luc), and mutant (p5mE-Luc) E2C-based reporter/target plasmids, we annealed five pairs of oligonucleotide primers to generate a double-stranded DNA fragment containing five tandem, unidirectional copies of the SB transposon left inner direct repeat (IDR) sequence, an 18-bp e2c recognition sequence from human chromosome 17 (5′-GGGGCCGGAGCCGCAGTG) or a mutated control e2c binding site (5′-AGTTCGAGAGCCGCAGTG) containing five point mutations (underlined) (12). Each assembled fragment was then used in a directional KpnI-NheI ligation to replace the 131-bp region containing the Gal4-binding sites in p5G-Luc. To generate pc-Gal4-AD, pc-hE2C-AD and pc-SB(N123)-AD plasmids, we PCR amplified the VP16 activation domain and NLS regions encoded by pACT and fused this to the carboxy (C)-terminus of the Gal4 DNA-binding domain encoded by pBIND, the human codon optimized E2C protein encoded by pc-hE2C-L5-SB and the HSB5(N123) DNA-binding domain encoded by pc-HSB5. Secondary full-length PCR products were then cloned into the pc-N plasmid (35) by BamHI-XbaI ligation.
To generate the pT/zeo-Cm-R6K plasmid, we first PCR amplified a 453-bp SpeI-XhoI EM2K-bleomycin fragment from pCpG-mcs (InvivoGen) and inserted it into the SpeI-XhoI-treated pT/MCS vector (35). We then digested the resulting plasmid, pT/zeo, with KpnI-XbaI restriction endonucleases and gelisolated a 1355-bp zeocin-marked transposon from an ethidium gel. We also PCR amplified a 406-bp R6K ori fragment from pUni/V5-HisA (Invitrogen) using mutagenic primers and then treated it with KpnI and PacI restriction endonucleases. These two products were then used in a triple-fragment ligation reaction containing a 886-bp, PCR-amplified, XbaI-PacI-treated chloramphenicol-resistant gene generated from pBC-KS(+) (Stratagene). Recombinants were screened following transformation into chemically competent Pir1 cells (Invitrogen).
DNA-binding studies
We performed our electrophorectic mobility shift assays essentially as previously described (36). Briefly, truncated (N123) derivatives of E2C-L5-SB and E2C-L5-SB-G59A fusion proteins were generated using the TNT T7-coupled reticulocyte lysate system (Promega) and then incubated with double-stranded, radiolabelled oligos corresponding to the inner DR from SB's left IR, a canonical e2c site, or a mutated version of e2c (12). Protein–DNA complexes were formed in the presence and absence of cold competitor DNA (500 nM), resolved on a polyacrylamide gel, and analyzed upon gel-drying with a Phosphorimager.
We also analyzed the binding capabilities of full-length fusion proteins in human cells using a luciferase-based reporter system. For these studies, we triple-transfected 8.5 × 104 HeLa cells on 24-well plates with a total of 1 μg plasmid DNA using Superfect (Qiagen). Each transfection contained (i) 50 ng of a reporter vector (p5E-Luc, p5G-Luc or p5SB-Luc), (ii) 15 ng of an activator plasmid (pc-hE2C-AD, pc-Gal4-ADor pc-SB(N123)-AD, respectively) and (iii) 935 ng of a plasmid encoding various specific (experimental) or nonspecific (control) competitor proteins. Two days later, triplicate cell lysates derived from each experimental condition were prepared and analyzed on a luminometer using Promega's Luciferase Assay System to determine the amount of luciferase activity generated relative to pc-GFP transfected cells, which was adjusted to 100%.
Transposition and western blot analyses
We assessed the integration capabilities of each fusion protein using a transposition assay described earlier (35). We analyzed fusion protein expression in human cells by transfecting 8.8 × 105 HeLa cells on 6-cm dishes with 5 μg pc-GFP, pc-SB10, pc-HSB5 or plasmids encoding individual DBD-transposase fusion proteins using Superfect. In some cases, we titrated the amount of pc-HSB5 plasmid delivered in order to equilibrate for diminished steady-state levels of the fusion proteins. For these purposes, cells were transfected instead with 1 μg, 500, 250 or 100 ng pc-HSB5 together with an appropriate amount of pc-N vector to maintain a constant amount of transfected DNA (5 μg) in each instance. In all cases, cells were harvested 40 h later, lysed in the presence of a complete protease inhibitor mix (Boehringer) and analyzed by immunoblotting with a rabbit polyclonal antibody to SB transposase (36).
Excision assays
We monitored transposon excision activity in human cells using a PCR-based approach described earlier (27,37). Transposon excision and DNA repair products were excised from an agarose gel, purified over a Qiagen mini-elute column and subcloned into the pGEM-T easy vector (Promega) prior to transformation into DH10B cells. Recombinant plasmid DNA was isolated from individual ampicillin-resistant bacterial colonies and the inserts sequenced using the T7 primer.
Inter-plasmid targeted transposition studies
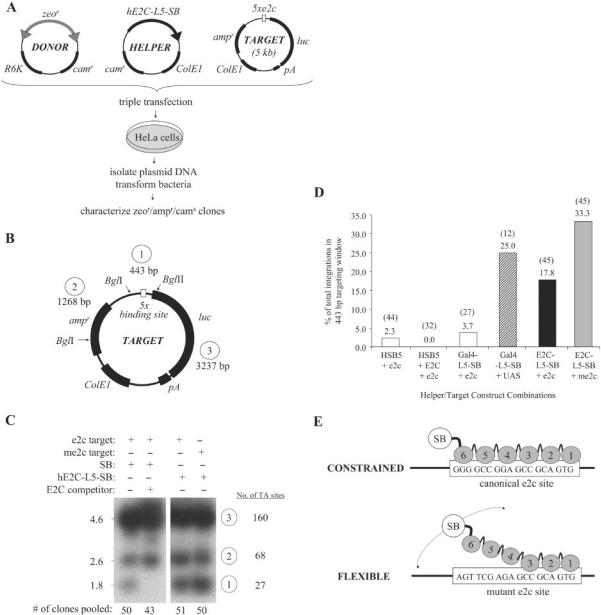
We studied the targeting potential of our fusion proteins by analyzing the transposition of a bleomycin-marked SB element into various predefined target DNA molecules. We triple-transfected 8.8 × 105 HeLa cells on 6-cm dishes with 2.5 μg pT/zeo-cam-R6K donor plasmid together with 2.5 μg each of a target (p5E-Luc or p5mE-Luc as a control) and helper (pBC-hE2C-L5-SB or pBC-Gal4-L5-SB) plasmid using Superfect. As a reference control, we also studied the integration profile in the presence of unfused HSB5 transposase. For these studies, we used 0.3 μg pBC-HSB5 together with 2.2 μg of either pBC-KS(+) or pBC-E2C as the helper plasmid source in our transfection experiments. We then isolated total DNA 2 days later using the Hirt method, transformed it into ElectroMAX ™ DH10B Escherichia coli (Invitrogen), and enriched for inter-plasmid transposition events by plating transformants on LB plates containing Zeocin ™ (50 μg/ml) and ampicillin (100 μg/ml). Individual zeor/ampr colonies were then patched onto LB-Zeo50/amp100 and LB-Chloramphenicol (50 μg/ml) plates and screened for sensitivity to chloramphenicol (cams) to identify integrations specifically into the target plasmid. We then re-patched a similar number of zeor/ampr/cams clones (approximately 50 for each group) onto duplicate master LB-amp100/Zeo50 plates, using one plate to isolate plasmid DNA from individual E. coli colonies and the other to generate pooled integration libraries for each experimental condition. Equivalent amounts (1 μg) of each integration library were double-digested with BglI-BglII restriction endonucleases before resolving each sample by agarose gel electrophoresis. Fragments were then transferred to nitrocellulose by blotting, probed with a 453 bp radiolabelled bleomycin cassette and analyzed using a Phosphorimager. Plasmid DNA isolated from individual clones was sequenced using the transposon-specific primer IR-1 (5′-AGATGTCCTAACTGACTTGCC) to determine the site of integration in the target plasmid.
Chromosomal integration site analysis
In order to assess the chromosomal targeting capabilities of the E2C-SB fusion protein, we first transfected 4.4 × 105 HeLa cells with 2 μg pT/neo and 0.75 μg pc-GFP together with 0.25 μg of either pc-HSB5 (control) or pc-hE2C-L-SB using Superfect. We then split these cells 2 days later onto multiple 10-cm dishes containing DMEM and the antibiotic G418 (600 μg/ml). Cells were growth-selected in G418 for 2 weeks, at which point G418-resistant cells were pooled and used to isolate genomic DNA from multiple independent integration libraries. We then characterized the genomic distribution of transposon integrations from three independent samples derived from each experimental condition using a ligation-mediated (LM)-PCR approach described earlier (38). PCR products were ligated to the pGEM-T easy vector (Promega), transformed into DH10B cells (Invitrogen) and analyzed using the IR-1 sequencing primer. Novel flanking genomic sequences were mapped to the human genome using an Ensembl blast homology search.
RESULTS
Site-selectivity of individual DNA-binding domain (DBD) proteins in human cells
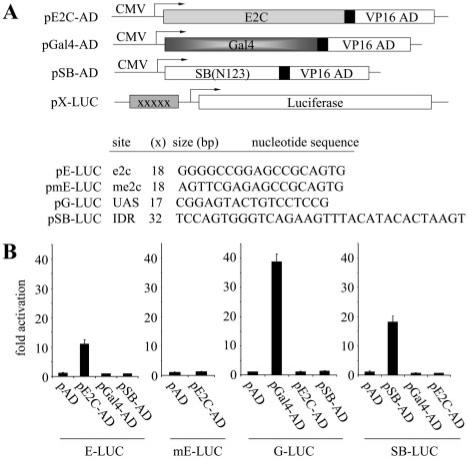
The successful application of hybrid transposases for site-directed integration will rely to a large degree on the affinity and specificity of the DNA-binding domains used to bias integration site-selection in the cell. We therefore focused initial work on identifying DBDs capable of high-affinity, sequence-specific DNA-binding within the context of human cells. For these purposes, we studied two different zinc-finger proteins: (i) the well-characterized Gal4 DBD (amino acids 1–147), which binds with high affinity to upstream activating sequences (UAS) and (ii) the synthetic polydactyl zinc-finger protein E2C, which recognizes a unique 18-bp DNA sequence 5′-GGG GCC GGA GCC GCA GTG-3′ in the human genome, located in the 5′ untranslated region (UTR) of the erbB-2 gene (chr.17q12) (12,39). This E2C DNA-binding protein was constructed from six Sp1C-based, predefined, and modular zinc-finger domains, each recognizing a specific 3-bp subsite (11). As an internal control, we also analyzed the SB transposase DBD (amino acids 1–123), which binds to short 32–34 bp direct repeats (DRs) encoded by the ends of the SB transposon (26). Each of these DBDs was fused to the herpes virus VP16 transcriptional activation domain (AD) (amino acids 411–456) and evaluated for its ability to activate expression of a reporter gene (Figure 1A). These assays were performed in HeLa cells using reporter constructs containing five consecutive binding sites for E2C, Gal4 or SB cloned upstream of a minimal promoter. Although performed with plasmid DNA, this assay permits rapid assessment of the specificity of different DBDs within the background of the human genome. Consistent with site-specific DNA-binding by E2C, expression of E2C-AD resulted in significant luciferase gene activation from the e2c reporter plasmid (p5E-Luc) relative to an empty-AD vector control (Figure 1B, left panel). The specificity of this reaction was further confirmed first by the absence of any appreciable gene activation in the presence of two additional control proteins (Gal4-AD and SB(N123)-AD), and second, by the lack of any significant E2C-AD-mediated reporter activation in the presence of the p5mE-Luc control plasmid, which contains 5 bp substitution mutations within the 5′ end of each of the five encoded e2c target sites (Figure 1B, left two panels). Similar results consistent with site-specific DNA-binding activity were also obtained upon expression of the Gal4-AD and SB(N123)-AD proteins in the presence of their cognate reporter plasmids, p5G-Luc and p5SB-Luc, respectively (Figure 1B, right two panels). Taken together, these results indicate that E2C, Gal4 and SB(N123) are all independently capable of highly specific substrate recognition in the context of human cells, suggesting that each may be suitable for the development of a site-selective transposase–DBD fusion protein.
Figure 1.
Fusion protein strategy to achieve site-directed transposon integration. (A) Reporter assay for monitoring DNA-binding specificity of candidate DNA-binding domains (DBD). Prospective DBDs were fused to the VP16 activation domain (AD) and expressed from the strong CMV promoter. These activator plasmids were co-transfected into HeLa cells together with a reporter plasmid containing a luciferase gene, a minimal promoter element, and five upstream binding sites for the DBDs (XXXXX) such that co-delivery of an appropriate activator and reporter plasmid results in activation of the downstream luciferase reporter gene. E2C, a synthetic polydactyl zinc-finger protein that recognizes the e2c target site; Gal4, the DNA-binding domain from the Gal4 protein that binds to upstream activator sequences (UAS); SB(N123), the SB transposase N-terminal 123 amino acid DNA-binding domain that binds to the SB transposon IDR (inner direct repeat). (B) DNA-binding specificity of independent protein domains within the context of human cells. Each graph displays luciferase activity relative to transfection with empty vector (pAD). Bars represent the average (mean ± st.dev.) obtained from three independent transfection experiments.
Design of DBD–SB fusion proteins
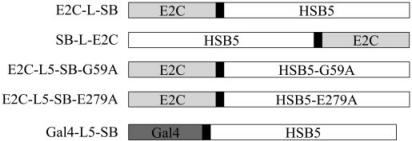
The various transposase–DBD fusion enzymes constructed and used in this study are shown in Figure 2. These fusions make use of a hyperactive transposase variant, called HSB5, in place of the canonical SB10 gene. This mutant HSB5 protein contains alanine substitution mutations within its DNA-binding (K13A, K33A and T83A) and catalytic core (S270A) domains that enhance the stability of the transposase–transposon complex and importantly, mediates ∼10-fold greater transposition activity relative to SB10. A hyperactive mutant was chosen for these studies since preliminary studies suggested that addition of even short epitope tags (i.e. 6 × His) to either end of SB10 severely compromised its enzymatic capabilities (data not shown). In the absence of any a priori information regarding the domain configuration most conducive to supporting optimal protein activity, we constructed fusions in which the E2C DBD was fused to either end of full-length HSB5 (E2C-SB and SB-E2C). These fusions were made with and without an intervening flexible peptide linker comprised of variable numbers of repeating glycine–glycine–serine (-GGS-)n triplets. This was done in the hope of identifying an optimal peptide linker that enabled independent folding of each functional domain contained within the fusion protein. As controls in our studies, we also made E2C-based fusion constructs containing single-amino-acid substitution mutations that ablate its ability to either bind transposon end sequences (E2C-L5-SB-G59A) or to catalyze DNA transposition (E2C-L5-SB-E279A) (36). Finally, a fusion comprised of full-length HSB5 and the Gal4 DBD was also constructed to determine whether the optimized conditions and components identified with the E2C-based constructs might also be applicable to other potential DBD-SB fusion constructs.
Figure 2.
Schematic overview of the fusion proteins described in this report. Each construct contains an enzymatic transposase domain fused to a site-specific DNA-binding domain protein. For the synthetic zinc-finger protein E2C, both possible configurations (N- and C-terminal fusions) were tested. HSB5, a hyperactive variant of the SB10 transposase; G59A, an alanine substitution mutation in the DNA-binding domain of SB that disrupts its ability to bind transposon DNA; E279A, an alanine substitution mutation in the catalytic core of SB that disrupts its excision and integration activity. Black boxes denote flexible inter-domain linkers of variable lengths (0–21 amino acids).
Expression and catalytic activity of chimeric transposases in human cells
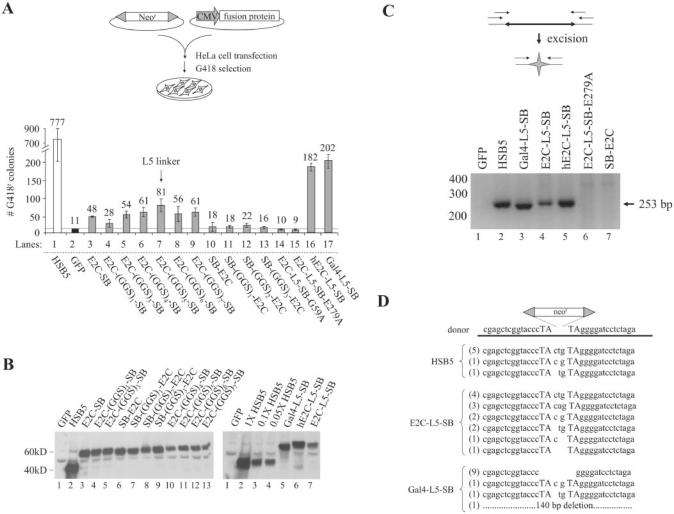
Fusion constructs were each cloned under the control of a strong cytomegalovirus (CMV) promoter and used in a standard transposition assay to evaluate the relative level of integration activity for each protein (Figure 3A, upper panel). To do this, we transfected HeLa cells with a plasmid encoding a neomycin-marked SB transposon (pT/neo), together with plasmids encoding the green fluorescent protein (GFP) as a control, HSB5 or one of many different fusion proteins. We then grew these co-transfected cells in the presence of the antibiotic G418 (600 μg/ml) for 2 weeks, at which point individual G418-resistant (G418r) colonies were subsequently fixed, stained and counted to determine the number of integration events achieved under each experimental condition. Results are shown in Figure 3A (lower panel) and indicate that fusions containing the E2C and Gal4 DBD linked to the N-terminus of SB retain some level of integration activity relative to the negative (GFP) control. Depending on the length of the intervening linker, however, integration efficiencies with the E2C-linker-SB constructs varied from 3- to 8-fold over background, whereas all of the C-terminal SB-linker-E2C fusion constructs were completely inactive. Importantly, western blot analyses showed that all of these fusion proteins were equally expressed in transfected HeLa cells (Figure 3B), suggesting that the observed variations in activity were not due to expression profile differences. Among the active fusions we studied, maximal integration activity reached ∼10% of HSB5 activity and was achieved using the E2C-L5-SB fusion protein, which contains an ‘L5 linker’ comprised of 5 consecutive (-GGS-) triplets. These data suggest that the N-terminus of SB is a much better acceptor site for protein linkage studies, and indicate that the addition of a flexible inter-domain linker can improve transposase activity under certain circumstances.
Figure 3.
Optimizing expression and activity of chimeric transposase proteins in human cells. (A) Upper panel, schematic overview of SB transposition assay. Lower panel, fusion protein activity in human cells. Fusion proteins containing variable numbers (n) of a flexible inter-domain (GGS)n peptide linker were tested for transpositional activity in HeLa cells. The (Gly–Gly–Ser)5 linker supporting the highest level of integration activity was designated L5 for the sake of simplicity. The average number of integration events obtained from three independent transfections is shown (mean ± st.dev.). (B) Western blot analysis of fusion protein expression. Transfected HeLa cells were harvested two days post-transfection, lysed and subjected to immunoblot analysis using a polyclonal antibody against the SB transposase. The right panel shows an attempt to normalize HSB5 and fusion protein expression in the cell by transfecting diminishing amounts of the HSB5 plasmid (1×, 0.1× or 0.05×) relative to fusion protein constructs. (C) Excision activity of chimeric transposases. HeLa cells were transfected with a neomycin-marked (neor) transposon plasmid together with plasmids encoding GFP, HSB5 transposase, or selected fusion proteins. Hirt DNA samples were prepared 2 days later and used as template in a series of nested PCR reactions. Transposon excision and subsequent DNA repair by the host enables the amplification of a diagnostic 253 bp PCR excision-and-repair product. (D) Sequence analysis of transposon excision and repair products generated from HSB5- and DBD-SB-mediated transposition. Target TA dinucleotides are capitalized. Number of events of each type are shown in parentheses.
Although there was no significant variation in expression among the different fusion proteins, the steady-state levels for the E2C-based fusion proteins appeared to be ∼10- to 20-fold lower relative to that of unfused HSB5 transposase (Figure 3B, right panel, compare lanes 2–4 with lane 7). Since this lower expression could theoretically contribute to the reduced integration potential observed with the fusion proteins, we attempted to increase the expression of our best E2C-L5-SB fusion protein in human cells to see if this might result in higher overall activity. To do this, we re-synthesized the E2C gene and its flexible linker using codons that optimize for high-level protein expression in human cells. This was done because E2C was originally isolated in bacterial-based screens and because previous work has suggested that codon optimization can, in some instances, dramatically improve heterologous protein expression in human cells (40,41). Interestingly, when we studied the resulting humanized hE2C-L5-SB fusion protein, we observed a 2-to-3-fold increase in both its expression and integration activity in transfected HeLa cells (see Figure 3A, lower panel, compare bars 7 and 16; and Figure 3B, right panel, lanes 6 and 7). These data support the notion that fusion protein expression and/or stability may be a limiting factor in E2C-SB-mediated integration strategies.
We also tested whether the low-level integration activity observed with our fusion protein constructs was indeed indicative of DNA transposition rather than some alternative integration mechanism (i.e. illegitimate recombination). To do this, we studied the integration capabilities of two additional fusion constructs that differed from E2C-L5-SB only by the presence of inactivating single amino-acid substitution mutations within either the transposase DNA-binding (E2C-L5-SB-G59A) or catalytic core (E2C-L5-SB-E279A) domain (36). Despite proper protein expression, both of these mutant fusion proteins were unable to support any detectable integration activity relative to the GFP control (data not shown; and Figure 3A, lower panel, bars 14 and 15), which was consistent with fusion protein integration activity being transposase-mediated. As a secondary assessment, we also investigated whether fusion protein expression was associated with transposon excision from the co-transfected donor plasmid. This was done using a nested PCR-based assay to detect specific products of transposon excision and double-strand break repair in transfected cells. As shown in Figure 3C, in contrast to the GFP control, we could readily detect the expected 253 bp excision and repair product in the presence of either HSB5 or those fusion proteins that were previously found to be active in our transposition experiments. In sharp contrast, neither the catalytic mutant E2C-L5-SB-E279A nor the C-terminal fusion SB-E2C showed any evidence for excision activity in this assay, which is consistent with our integration data. We cloned and sequenced these excision and repair products and detected a 1–3 bp TA-flanked footprint at the vast majority of HSB5- and E2C-L5-SB-specific excision sites (Figure 3D), which is further evidence of a transposase-mediated excision process (27). Interestingly, however, 83% (10/12) of the excision and repair products generated in the presence of the Gal4-L5-SB fusion protein completely lacked any TA-flanked footprint (Figure 3D). This is consistent with the reproducibly smaller PCR product generated in our excision assays (Figure 3C, compare lane 3 with 2, 4 and 5) and suggests that the cleavage characteristics for this particular fusion protein may be somewhat atypical. Collectively, these data indicate that fusion proteins consisting of novel DBDs and the SB transposase retain the ability to catalyze authentic DNA transposition in human cells, albeit at reduced frequencies.
Sequence-specific DNA-binding of E2C-L5-SB and Gal4-L5-SB fusion proteins
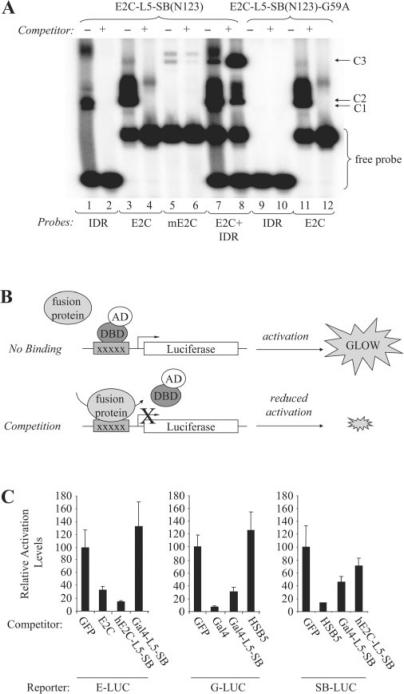
The earlier work demonstrating transposon excision and re-integration implies that these hybrid transposase proteins are still able to bind transposon end sequences despite the presence of a rather bulky DBD at their N-terminus. Here, we experimentally tested this hypothesis by characterizing the DNA-binding capabilities of our active fusion proteins. To do this, we first produced a truncated E2C-L5-SB(N123) derivative of the fusion protein by in vitro transcription and translation and then used it in a standard electrophoretic mobility shift assay (EMSA) to investigate its capacity to bind E2C and SB target recognition sequences. Use of the truncated N123-based polypeptides was necessary in these studies since full-length SB will aggregate in gel shift analyses. Consistent with our integration data, we observed a protein–DNA complex in the presence of an SB inner direct repeat (IDR) target site and the E2C-L5-SB(N123) protein (Figure 4A, lane 1, C1 complex). The specificity of this complex was verified both by competition with an excess of unlabelled IDR oligonucleotide and by the absence of any similar complex in the presence of the mutant E2C-L5-SB(N123)-G59A protein (Figure 4A, lanes 2 and 9, respectively). Importantly, both of these fusion proteins were capable of binding the 18-bp target e2c site in a sequence-specific manner (Figure 4A, lanes 2, 4, 11 and 12, C2 complex). This was further demonstrated by the absence of any detectable protein–DNA complex in the presence of a control e2c target site (me2c) containing five base pair differences (Figure 4A, lanes 5 and 6). Finally, when the E2C-L5-SB(N123) protein was mixed with both potential target sites (e2c and IDR), we observed a third protein–DNA complex with greatly reduced mobility compared to either of the two other complexes (Figure 4A, lane 7, C3 complex). Moreover, the intensity of this third band greatly increased in the presence of excess unlabelled IDR oligonucleotide (Figure 4A, lane 8, C3 complex). A reciprocal study in which excess unlabelled e2c DNA was used instead of IDR resulted in complete competition of the C2 and C3 complexes, suggesting that the affinity of the hybrid protein for e2c may be significantly greater than for the SB binding site (data not shown). Nevertheless, these results are consistent with the expected behavior of a protein with dual-specificity DNA-binding capabilities, and suggest that this fusion protein is capable of binding simultaneously to both e2c and IDR target sites. Such findings are encouraging since active tethering of integration complexes will theoretically require coordinated binding of the hybrid transposase to both transposon and e2c target sites if the protein is to have any discernable effect on insertion site selection.
Figure 4.
DNA-binding activity of the E2C-L5-SB fusion protein. (A) Analysis of E2C-L5-SB fusion protein DNA-binding activity by electro-mobility shift assay. N123-based peptides corresponding to truncated forms of E2C-L5-SB and E2C-L5-SB-G59A (control) fusion proteins were produced by in vitro transcription and translation, and then incubated with 32P-radiolabelled double-stranded DNA probes (IDR, e2c or mutant e2c). Protein–DNA complexes were formed in the presence and absence of excess unlabelled competitor DNA, resolved by electrophoresis through a gel, and visualized by autoradiography. The competitor in lane 8 is unlabelled IDR DNA. C1, fusion peptide–IDR complex; C2, fusion peptide–e2c complex; C3, fusion peptide/IDR/e2c trimeric complex. (B) Schematic of competition assay to monitor the DNA-binding activity of full-length hE2C-L5-SB and Gal4-L5-SB fusion proteins within human cells. HeLa cells were transfected with luciferase reporter plasmids together with limiting amounts of an activator plasmid encoding their respective trans-activator protein (hE2C-AD, Gal4-AD or SB(N123)-AD). Cells also received an excess of plasmids encoding various experimental and control proteins to test whether any could compete for protein binding at the target sites, thereby reducing the level of luciferase trans-activation in the cell. (C) Reporter assay. Each graph displays luciferase activation levels relative to transfection with a control vector (pc-GFP). Bars represent the average (mean ± st.dev.) obtained from three independent transfection experiments.
One limitation of the earlier in vitro work is that it might not be reflective of how the full-length fusion protein behaves within the complex milieu of a human cell. Therefore, we developed an alternative cell-based strategy to evaluate the DNA-binding activities of our active hybrid transposases. To do this, we employed a mammalian one-hybrid-based approach in which various proteins could be individually tested for their ability to reduce the level of luciferase reporter gene activation via competitive DNA-binding (Figure 4B). Using this general strategy, we detected a 70–80% reduction in reporter gene activation levels upon expression of the hE2C-L5-SB and Gal4-L5-SB fusion proteins in the presence of their cognate reporter plasmids (Figure 4C, left and middle panels). In sharp contrast, we did not observe any significant change in reporter gene activation levels from the e2c- and Gal4-based reporters upon expression of Gal4-L5-SB and HSB5, respectively (Figure 4C, fourth bar in left and middle panels). These latter results indicate that the competition observed in this assay does indeed originate through site-specific DNA-binding activity. Importantly, we also observed competition from the IDR-based reporter in the presence of full-length Gal4-L5-SB and hE2C-L5-SB fusion proteins, although the level of competition was only ∼35–50% as compared to the 90% reduction observed upon HSB5 expression (Figure 4C, right panel). The estimated 2- to 3-fold diminished capacity of these fusion proteins to bind their transposon-specific substrate relative to that of either of the DBD-specific targets could indicate that the adjoining DBD interferes somewhat with the transposase DNA-binding domain. This notion is supported by our in vitro competition studies and could explain the reduced integration capacity of these chimeras since such an effect would diminish the formation and/or stability of active transposase–transposon complexes. Nevertheless, our data clearly indicate that the hE2C-L5-SB and Gal4-L5-SB fusion proteins are both competent for dual-specificity DNA-binding, suggesting that DBD–SB fusion proteins might prove useful in redirecting transposon insertion site selection in the cell.
Site-directed transposition in human cells
We next tested whether stable DNA tethering mediated by our fusion proteins could alter transposase target site selection in human cells using predefined target DNA molecules. This plasmid-based system involved the use of three different DNA molecules: (i) a chloramphenicol-resistant (camr) donor plasmid encoding a bleomycin-marked transposon, (ii) a camr helper plasmid encoding fused or unfused transposase and (iii) an ampicillin-resistant (ampr) target plasmid containing multiple tandem DBD recognition sites (Figure 5A). When all three of these plasmids are co-delivered to mammalian cells, the transposon is efficiently excised by the transposase protein and, in rare instances, hops somewhere into the target plasmid. Target-specific inter-plasmid transposition events can then be isolated using a bacterial-based genetic selection (i.e. by screening for ampr/zeor/cams bacterial colonies) and subsequently analyzed at the molecular level. Using this general approach, we induced transposition into e2c- and mutant e2c-containing target plasmids by expressing the HSB5 or individual DBD-fusion proteins and then isolated target-specific integration events for each experimental group. As an initial assessment of the targeting potential, we first generated representative integration libraries for each group by pooling similar numbers of ampr/zeor/cams colonies, and then cutting 1 μg of each library DNA sample with BglI and BglII restriction endonucleases (Figure 5B). All samples were then resolved on an agarose gel, subjected to Southern blot hybridization using a transposon-specific probe, and visualized upon Phosphorimager analysis. Under these experimental conditions, the intensity of the smallest (1.8 kb) transposon-specific band observed in each group serves as a relative indicator of the frequency of transposon integration into a 0.4 kb targeting window. Results of a portion of these studies are shown in Figure 5C, with the first lane representing the integration distribution obtained with HSB5 and thus in the absence of any active DNA tethering mechanism. Interestingly, we were able to completely block HSB5-mediated integration into the e2c-containing fragment by co-expressing the E2C DBD protein in trans (Figure 5C, lanes 1 and 2, lower bands). These findings are consistent with E2C binding in the cell and suggest that DNA-binding proteins may be able to functionally interfere with and/or modify transposon insertion site selection in mammalian cells. More importantly, we found that the hE2C-L5-SB fusion protein catalyzed substantially increased transposition into the experimental targeting window relative to the unfused transposase (Figure 5C, compare lanes 1 and 3, lower bands). Surprisingly, however, we observed a similar, if not better, level of targeting with this fusion protein into an otherwise equivalent mutant e2c-containing plasmid (Figure 5C, lanes 3 and 4, lower bands). Since this mutant target site is still expected to support base pair contacts with the first three fingers of E2C, these data could indicate that the E2C DBD is still capable of interacting with the partially mutated e2c control site in the context of a transposase hybrid. While these protein–DNA interactions would undoubtedly be of significantly diminished affinity relative to E2C and its cognate site, the more transient nature of these interactions could actually facilitate regional integrations following physical association with the target site. To further validate these findings, however, we mapped insertion sites from individual members from our integration libraries to quantitatively assess targeting in each experimental group. This 443-bp region contains a total of 27 potential target sites, with 23/27 located 5′ of the target sites. Accordingly, most of the integration events we studied ultimately mapped 5′ to the target sites. Compared to unfused HSB5 transposase, the Gal4-L5-SB and hE2C-L5-SB fusion proteins directed 11- and 8-fold more integration events into the 443 bp targeting window, respectively (Figure 5D, bars 1, 4 and 5). Moreover, consistent with our earlier findings, we also observed even better hE2C-L5-SB-mediated targeting in the presence of a mutant e2c-containing target plasmid, with the overall integration bias approaching ∼15-fold that of HSB5 (Figure 5D, bars 1 and 6). Among all the targeted events, we did not observe any obvious clustering or apparent preference for one particular TA site over another. Collectively, these data demonstrate that fusion protein-mediated tethering can adequately redirect transposon integration in human cells.
Figure 5.
Site-directed transposition in human cells. (A) Schematic overview of plasmid-based assay for investigating site-directed transposition. Transposition was initiated by transfecting human HeLa cells with a plasmid encoding chimeric or unfused HSB5 transposase, together with a donor plasmid encoding a bleomycin-marked (zeor) transposon and a counter-selectable chloramphenicol-resistance (camr) gene. These plasmids were co-delivered with an ampicillin-resistant (ampr) target plasmid containing five tandem DBD recognition sequences and allowed to undergo transposition. Low-molecular weight plasmid DNA fractions were isolated 2 days later and transformed into DH10B E. coli. Replication of the R6K origin-containing donor plasmid is strictly dependent on expression of the pir1 gene product, which is absent in this bacterial strain. Ampr/zeor bacterial colonies were patched onto LB-camr plates to screen for inter-plasmid transposition events specific for the target plasmid (i.e. cams). Both pooled and clonal ampr/zeor/cams populations of bacteria were amplified, plasmid DNA isolated and the locations of transposon insertions relative to the target sites determined by restriction site analyses and DNA sequence analyses, respectively. (B) Target plasmid features. Positions of BglI and BglII restriction endonuclease recognition sites are shown, as are the sizes for each resulting DNA fragment. (C) Southern blot analysis of targeted integration. For each experimental condition, 500 ng of plasmid DNA isolated from pooled ampr/zeor/cams bacterial colonies (n = 43–51) was treated with BglI-BglII restriction enzymes. Samples were resolved on an agarose gel, transferred to nitrocellulose, hybridized to a 32P-radiolabelled probe corresponding to the left SB transposon inverted repeat, and resulting bands visualized upon autoradiography. In one instance, excess E2C DNA-binding domain was co-expressed with transposase protein to determine whether associated proteins could inhibit SB target site DNA capture. The lower band intensity under each experimental condition represents a qualitative assessment of the relative frequency of transposition of the 1.35 kb zeor-marked element into the 443 bp targeting window. (D) Targeted transposition frequencies. Recombinant target DNA molecules were isolated from individual ampr/zeor/cams colonies and sequenced using an internal transposon-specific primer. Bars denote the percentage of total integrations occurring within the 443-bp targeting window, whereas the numbers in parentheses denote the actual number of integrations analyzed in each group. (E) Flexing model for targeted transposition. Multiple changes in both DNA- and protein conformation are necessary to complete a fullcycle of transposition. A protein that remains too tightly bound to DNA, such as hE2C-L5-SB to the canonical e2c site, cannot efficiently catalyze these reactions. In the case of the mutant e2c site, however, only fingers 1 through 3 of hE2C-L5-SB retain the capacity for DNA-binding. This would essentially improve the flexibility of the transposase domain, which might enhance the acquisition and/or manipulation of neighboring target sites.
We attempted to ascertain why the hE2C-L5-SB fusion protein would mediate enhanced targeting in the presence of a partially mutated target site. Based on the physical makeup of this mutant substrate, we hypothesized that stable docking of the transposase to DNA might be interfering, either directly or indirectly, with some stage of the cut-and-paste transposition mechanism. This notion is based on the knowledge that transposase proteins generally require many coordinated conformational changes in order to effectively catalyze a complete transposition cycle. Therefore, to further explore this phenomenon, we measured the absolute frequency of inter-plasmid transposition by our DBD–SB fusion proteins using both cognate and non-cognate target plasmids for each. Interestingly, with either of two DBD-SB fusion proteins (hE2C-L5-SB and Gal4-L5-SB), we observed approximately 3- to 4-fold less total target plasmid integrations whenever an appropriate DBD-binding site was present on the plasmid (Table 1). These data suggest that stable docking to DNA may partially impair normal transposase function and indicate that a less-tightly constrained transposase molecule might support better transpositional activity (and site-directed integration) than a more strongly-associated transposase molecule (Figure 5E). Considering that the hE2C-L5-SB protein is probably much more transiently associated with the partially mutated e2c site than with the cognate e2c site, our data suggest that flexible transposase tethers may further improve the performance of site-directed transposition modalities.
Table 1.
Inter-plasmid transposition frequencies in the presence and absence of stable transposase docking
| Transposase | Target DNA | Stable tpase docking? | Transposition frequencya |
|---|---|---|---|
| HSB5 | p5E-Luc | – | 400 × 10−8 |
| E2C-L5-SB | p5mE-Luc | −/+ | 27 × 10−8 |
| E2C-L5-SB | p5E-Luc | + | 8 × 10−8 |
| Gal4-L5-SB | p5E-Luc | – | 18 × 10−8 |
| Gal4-L5-SB | p5G-Luc | + | 4 × 10−8 |
aWe divided the number of ampr/zeor/cams colonies obtained from three independent transformations by the total number of ampr colonies.
hE2C-L5-SB-mediated integrations in the human genome
We studied whether our E2C-based fusion protein could redirect transposition into regions adjacent to the endogenous e2c site on human chromosome 17. To do this, we generated pools of stably transfected HeLa cells following co-transfection with a neomycin-marked transposon (pT/neo) and plasmids expressing either the hE2C-L5-SB fusion protein or HSB5 as a control, and then isolated genomic DNA from pooled G418r colonies. We then used a high-throughput ligation-mediated (LM-) PCR approach to recover host chromosomal DNA sequences immediately flanking the 5′ ends of integrated transposons present within each integration library (38). PCR-amplified junction fragments were cloned, sequenced and then mapped to a unique location in the human genome using the Ensembl-based server. Of the 67 total hE2C-L5-SB-mediated integrations we analyzed, only 6% (4/67) mapped to human chromosome 17, as compared to 4% (2/55) in the HSB5 control group, with the remaining 63 hE2C-L5-SB-mediated integrations being distributed randomly throughout the genome (data not shown). Moreover, none of the four hE2C-L5-SB-mediated integrations into chromosome 17 mapped within a 1 kbp-window surrounding the endogenous e2c site. These results suggest that, at least under the experimental conditions employed here, the hE2C-L5-SB fusion protein cannot adequately redirect integrations into the vicinity of e2c.
DISCUSSION
In this work, we have fused site-selective DNA-binding domains (DBD) to the Sleeping Beauty transposase as a necessary first step towards achieving targeted integration into predefined sites in the human genome. These hybrid proteins showed new DNA-binding specificity and were capable of mediating bona fide transposition reactions in human cells. These chimeric proteins could direct SB transposition into the proximity of a DBD-specific target site, which to our knowledge, represents the first demonstration of targeted DNA transposition in human cells.
Fusion proteins comprised of the SB transposase and two different zinc-finger proteins (SB-Sp1 and SB-ZNF202) were recently reported by Wilson et al. (42). These proteins, while functional, were never evaluated for target site selection and were found to recognize sequence motifs that are particularly widespread in mammalian genomes (42). In contrast to these earlier fusion proteins, our use of E2C as the target-specifying component offers important advantages in terms of both specificity and versatility, especially since its modular organization should enable the mixing and matching of its respective zinc fingers to generate proteins with new DNA-binding specificities (12,43,44). Compared to unfused transposase, our new fusion proteins were capable of directing ≥8-fold more transposition events into a narrowly defined window encompassing the DBD-binding sites. While this current level of targeting is certainly not expected to be sufficient to achieve genome-level specificity, it represents a critical first step towards achieving such a goal. Although site-directed transposition is believed to occur in cis via a regionally bound hybrid protein, alternative mechanisms can be envisioned, such as the promotion of adjoining site usage via localized DNA distortions. Notably, we have shown that co-expression of the E2C protein in trans inhibited, not stimulated, targeted DNA capture by unfused transposase, which seems inconsistent with an indirect mechanism.
Attempts to achieve targeted integration using a hybrid DBD–transposase (IS30) enzyme has also been recently described by Szabo et al. (45). Interestingly, while these authors also observed directed transposition into target DNA molecules in bacteria, these hybrid proteins were generally inactive in the context of zebrafish embryos, in which integration now occurred almost exclusively by illegitimate recombination (45). This seminal work suggested that controlled modification of DNA transposition might not be a straight-forward process and raised general concerns regarding whether regional tethering of transposes might promote instability of neighboring DNA regions. Although we were able to readily detect integration by DNA transposition with our hybrid transposase molecules, we also detected atypical footprints in the presence of our Gal4-based chimera and observed significantly diminished integration activity upon stable transposase docking. Moreover, although we did not see a similar tendency towards increased illegitimate recombination in human cells, the co-stimulation of such alternative pathways in human cells cannot be presently excluded. If occurring, however, these processes must require stable tethering and enzymatic activity since our catalytically inactive fusion protein (E2C-L5-SB-E279A) supported random integration frequencies equivalent to our GFP control.
Our long-term objective is to achieve targeted integration in the human genome and yet, despite the promising results we obtained in our plasmid-based system, we failed to find any evidence indicative of targeted integration within the endogenous e2c locus. This result was not unexpected and could have been influenced by many different factors, the first being that a single e2c binding site may not be sufficient to permit site-selective tethering of integration complexes at the genome-level. This notion seems at odds, however, with the recent demonstration of E2C-dependent transcriptional repression at this site (12,39) and the reported targeting of retroviral DNA integration at this locus via an E2C-IN fusion protein (14). Alternatively, it is possible that the E2C domain mediates such tight binding at the target site that it prohibits the transposase domain from ever accessing an adjoining TA dinucleotide. This notion remains within reason in light of the high GC-content of the e2c locus and the rather low-abundance and suboptimal spacing of potential TA acceptor sites in the immediate adjoining regions (within a 400-bp region surrounding e2c, the GC-content is 75% and there are only four TA sites, the closest of which is 78 bp away). Furthermore, we have observed reduced transposition frequencies into plasmids whenever stable transposase docking was possible, suggesting that the targeting capability of the fusion protein can be significantly influenced by the physical constraints imposed during target site attachment. A third possibility is that the E2C DNA-binding domain itself exhibits relaxed site-specificity in the context of the full-length E2C-SB hybrid protein, a notion consistent with the high-frequency targeting we observed with E2C-SB and the mutated e2c target plasmid. If true, then the reduced stringency would undoubtedly permit the fusion protein to bind ‘pseudo’ e2c sites located throughout the genome, thereby greatly masking any site-directed events. Finally, it is entirely possible, if not likely, that the transposase portion of the hybrid protein binds DNA randomly via a sequence-independent DNA-binding domain, and in doing so, quickly commits to a target site rather than dissociating prior to integration (46). This activity would invariably obstruct the function of the fused sequence-specific DNA-binding domain, thereby enabling vast numbers of integrations outside of the predicted targeting window. Clearly, reducing this nonspecific DNA-binding activity remains a formidable challenge in future work, especially in light of the present uncertainty regarding what role, if any, transposase nonspecific DNA-binding activity plays in SB target site acquisition. Although it might be possible to significantly reduce this activity through site-directed mutagenesis without comprising the transposition process, this will undoubtedly require more basic research into the nonspecific DNA-binding component of the transposase. Insight gained from such studies would greatly clarify mechanisms of SB target site selection and would significantly aid the development of a second generation of hybrid proteins, ones with substantially increased site-specificity.
In addition to these important considerations, there remains an infinite number of potential ways to achieve further improvements in the activity and/or targeting efficiency of such hybrid proteins. For instance, the design and testing of other hybrid transposases containing different custom-built zinc-finger proteins (44) and alternative transposase enzymes (47–49) might enable higher-affinity DNA binding and/or better transposase folding in the context of a chimeric enzyme. Additionally, new variations in the linker length and/or content could potentially enhance the flexibility and transpositional capabilities of the anchored complex. Earlier studies with integrase-based fusion proteins suggest that nonspecific integration events increase considerably at high protein concentrations (7,9), suggesting that limited expression and/or activity of the hybrid protein, such as through the delivery of mRNA instead of DNA (50), might further reduce the frequency of integration at undesirable sites. Moreover, mechanisms to efficiently pre-dock the fusion protein to the target site prior to transposon substrate delivery may drastically improve the frequency of targeted integration in the cell (46).
In summary, despite a growing number of site-specific integration strategies currently in early stages of development (51–57), the achievement of true site-selectivity at the genome-level remains an unmet challenge. Our results demonstrating directed integration in human cells represents an important step towards this end. With continued development, controlled DNA transposition may someday provide a feasible and versatile approach to achieve targeted genome modification, which could prove invaluable for many basic research and gene therapy applications.
ACKNOWLEDGEMENTS
The authors wish to thank Jacob Mikkelsen, Julie Park, Anton McCaffrey, Hiroyuki Nakai and Brian Garrison for their many thoughtful discussions through the years. We wish to thank our reviewers for their insightful comments and gratefully acknowledge Adam Dupuy for his technical help with the LM-PCR method. We also thank Perry Hackett and Carlos Barbas for kindly providing the polyclonal SB antibody and pMal-c2-E2C plasmid, respectively. This work was made possible through the generous support of a Walter V. Berry research fellowship (S.R.Y) and National Institutes of Health grant DK49022 (M.A.K). Funding to pay the Open Access publication charge was provided by National Institutes of Health grant DK49022.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, De Villartay JP, Thrasher AJ, Wulffraat N, Sorensen R, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N. Engl. J. Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 2.Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, Radford I, Villeval JL, Fraser CC, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 3.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 4.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 5.Katz RA, Merkel G, Skalka AM. Targeting of retroviral integrase by fusion to a heterologous DNA binding domain: in vitro activities and incorporation of a fusion protein into viral particles. Virology. 1996;217:178–190. doi: 10.1006/viro.1996.0105. [DOI] [PubMed] [Google Scholar]
- 6.Peng WJ, Chang CM, Lin TH. Target integration by a chimeric Sp1 zinc finger domain-Moloney murine leukemia virus integrase in vivo. J. Biomed. Sci. 2002;9:171–184. doi: 10.1007/BF02256029. [DOI] [PubMed] [Google Scholar]
- 7.Goulaouic H, Chow SA. Directed integration of viral DNA mediated by fusion proteins consisting of human immunodeficiency virus type 1 integrase and Escherichia coli LexA protein. J. Virol. 1996;70:37–46. doi: 10.1128/jvi.70.1.37-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushman FD. Tethering human immunodeficiency virus 1 integrase to a DNA site directs integration to nearby sequences. Proc. Natl. Acad. Sci. USA. 1994;91:9233–9237. doi: 10.1073/pnas.91.20.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushman FD, Miller MD. Tethering human immunodeficiency virus type 1 preintegration complexes to target DNA promotes integration at nearby sites. J. Virol. 1997;71:458–464. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan W, Zhu K, Segal DJ, Barbas CF, 3rd, Chow SA. Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites. J. Virol. 2004;78:1301–1313. doi: 10.1128/JVI.78.3.1301-1313.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target Sequences. Proc. Natl. Acad. Sci. USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes-Son ML, Chow SA. Integrase-lexA fusion proteins incorporated into human immunodeficiency virus type 1 that contains a catalytically inactive integrase gene are functional to mediate integration. J. Virol. 2000;74:11548–11556. doi: 10.1128/jvi.74.24.11548-11556.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan W, Dong Z, Wilkinson TA, Barbas CF, 3rd, Chow SA. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J. Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 18.Hematti P, Hong BK, Ferguson C, Adler R, Hanawa H, Sellers S, Holt IE, Eckfeldt CE, Sharma Y, et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 20.Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 21.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 22.Shibagaki Y, Chow SA. Central core domain of retroviral integrase is responsible for target site selection. J. Biol. Chem. 1997;272:8361–8369. doi: 10.1074/jbc.272.13.8361. [DOI] [PubMed] [Google Scholar]
- 23.Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, Lusic M, Marcello A, Giacca M. Acetylation of HIV-1 integrase by p300 regulates viral integration. EMBO J. 2005;24:3070–3081. doi: 10.1038/sj.emboj.7600770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 25.Izsvak Z, Ivics Z. Sleeping beauty transposition: biology and applications for molecular therapy. Mol. Ther. 2004;9:147–156. doi: 10.1016/j.ymthe.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Izsvak Z, Khare D, Behlke J, Heinemann U, Plasterk RH, Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J. Biol. Chem. 2002;277:34581–34588. doi: 10.1074/jbc.M204001200. [DOI] [PubMed] [Google Scholar]
- 27.Yant SR, Kay MA. Nonhomologous-end-joining factors regulate DNA repair fidelity during Sleeping Beauty element transposition in mammalian cells. Mol. Cell. Biol. 2003;23:8505–8518. doi: 10.1128/MCB.23.23.8505-8518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izsvak Z, Stuwe EE, Fiedler D, Katzer A, Jeggo PA, Ivics Z. Healing the wounds inflicted by sleeping beauty transposition by double-strand break repair in mammalian somatic cells. Mol. Cell. 2004;13:279–290. doi: 10.1016/s1097-2765(03)00524-0. [DOI] [PubMed] [Google Scholar]
- 29.Vigdal TJ, Kaufman CD, Izsvak Z, Voytas DF, Ivics Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 30.Geurts AM, Hackett CS, Bell JB, Bergemann TL, Collier LS, Carlson CM, Largaespada DA, Hackett PB. Structure-based prediction of insertion-site preferences of transposons into chromosomes. Nucleic Acids Res. 2006;34:2803–2811. doi: 10.1093/nar/gkl301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu G, Geurts AM, Yae K, Srinivasan AR, Fahrenkrug SC, Largaespada DA, Takeda J, Horie K, Olson WK, et al. Target-site preferences of Sleeping Beauty transposons. J. Mol. Biol. 2005;346:161–173. doi: 10.1016/j.jmb.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 32.Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walisko O, Izsvak Z, Szabo K, Kaufman CD, Herold S, Ivics Z. Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1. Proc. Natl. Acad. Sci. USA. 2006;103:4062–4067. doi: 10.1073/pnas.0507683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zayed H, Izsvak Z, Khare D, Heinemann U, Ivics Z. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 2003;31:2313–2322. doi: 10.1093/nar/gkg341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay MA. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat. Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 36.Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational analysis of the N-terminal DNA-binding domain of sleeping beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol. Cell Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu G, Aronovich EL, Cui Z, Whitley CB, Hackett PB. Excision of Sleeping Beauty transposons: parameters and applications to gene therapy. J. Gene. Med. 2004;6:574–583. doi: 10.1002/jgm.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 39.Beerli RR, Dreier B, Barbas CF., III Positive and negative regulation of endogenous genes by designed transcription factors. Proc. Natl. Acad. Sci. USA. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 41.Cherepanov P, Pluymers W, Claeys A, Proost P, De Clercq E, Debyser Z. High-level expression of active HIV-1 integrase from a synthetic gene in human cells. FASEB J. 2000;14:1389–1399. doi: 10.1096/fj.14.10.1389. [DOI] [PubMed] [Google Scholar]
- 42.Wilson MH, Kaminski JM, George AL., Jr Functional zinc finger/sleeping beauty transposase chimeras exhibit attenuated overproduction inhibition. FEBS Lett. 2005;579:6205–6209. doi: 10.1016/j.febslet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 43.Hurt JA, Thibodeau SA, Hirsh AS, Pabo CO, Joung JK. Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc. Natl. Acad. Sci. USA. 2003;100:12271–12276. doi: 10.1073/pnas.2135381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Ann. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 45.Szabo M, Muller F, Kiss J, Balduf C, Strahle U, Olasz F. Transposition and targeting of the prokaryotic mobile element IS30 in zebrafish. FEBS Lett. 2003;550:46–50. doi: 10.1016/s0014-5793(03)00814-7. [DOI] [PubMed] [Google Scholar]
- 46.Miller MD, Bor YC, Bushman F. Target DNA capture by HIV-1 integration complexes. Curr. Biol. 1995;5:1047–1056. doi: 10.1016/s0960-9822(95)00209-0. [DOI] [PubMed] [Google Scholar]
- 47.Kapetanaki MG, Loukeris TG, Livadaras I, Savakis C. High frequencies of Minos transposon mobilization are obtained in insects by using in vitro synthesized mRNA as a source of transposase. Nucleic Acids Res. 2002;30:3333–3340. doi: 10.1093/nar/gkf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miskey C, Izsvak Z, Plasterk RH, Ivics Z. The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 2003;31:6873–6881. doi: 10.1093/nar/gkg910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 50.Wilber A, Frandsen JL, Geurts JL, Largaespada DA, Hackett PB, McIvor RS. RNA as a source of transposase for sleeping beauty-mediated gene insertion and expression in somatic cells and tissues. Mol. Ther. 2006;13:625–630. doi: 10.1016/j.ymthe.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 51.Thyagarajan B, Guimaraes MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 52.Groth AC, Calos MP. Phage integrases: biology and applications. J. Mol. Biol. 2004;335:667–678. doi: 10.1016/j.jmb.2003.09.082. [DOI] [PubMed] [Google Scholar]
- 53.Chen L, Woo SL. Complete and persistent phenotypic correction of phenylketonuria in mice by site-specific genome integration of murine phenylalanine hydroxylase cDNA. Proc. Natl. Acad. Sci. USA. 2005;102:15581–15586. doi: 10.1073/pnas.0503877102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Chalberg TW, Vankov A, Molnar FE, Butterwick AF, Huie P, Calos MP, Palanker DV. Gene transfer to rabbit retina with electron avalanche transfection. Invest. Ophthalmol. Visual Sci. 2006;47:4083–4090. doi: 10.1167/iovs.06-0092. [DOI] [PubMed] [Google Scholar]
- 55.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 56.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 57.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]