Abstract
When the bacterial ribosome stalls on a truncated mRNA, transfer–messenger RNA (tmRNA) acts initially as a transfer RNA (tRNA) and then as a messenger RNA (mRNA) to rescue the ribosome and add a peptide tag to the nascent polypeptide that targets it for degradation. Ribosomal protein S1 binds tmRNA but its functional role in this process has remained elusive. In this report, we demonstrate that, in vitro, S1 is dispensable for the tRNA-like role of tmRNA but is essential for its mRNA function. Increasing or decreasing the amount of protein S1 in vivo reduces the overall amount of trans-translated proteins. Also, a truncated S1 protein impaired for ribosome binding can still trigger protein tagging, suggesting that S1 interacts with tmRNA outside the ribosome to keep it in an active state. Overall, these results demonstrate that S1 has a role in tmRNA-mediated tagging that is distinct from its role during canonical translation.
INTRODUCTION
Protein synthesis requires quality control systems to ensure accuracy between the genetic information carried by messenger RNAs (mRNAs) and the sequence of the corresponding proteins. In bacteria, when translation proceeds on a truncated mRNA, ribosomes carry on the elongation process until the mRNA 3′-end is reached. There, translation termination is inefficient and ribosomes stall with the nascent polypeptide chain on the peptidyl-tRNA site (P-site), whereas the aminoacyl-tRNA site (A-site) is empty or only partially filled with the incomplete mRNA (Figure 1A). A quality-control mechanism, referred as trans-translation, rescues and recycles the stalled ribosomes and targets the incomplete protein for degradation. This process requires a specific RNA acting as both a transfer and a messenger RNA (tmRNA, ssrA or 10Sa RNA). TmRNA has a tRNA (TLD) and a mRNA (MLD)-like domains. The TLD is aminoacylated with alanine (Ala) by alanyl-tRNA synthetase (AlaRS) and the MLD is a short (27–105 nt-long) internal open reading frame (ORF). During trans-translation, alanyl-tmRNA enters the A-site and, despite the lack of codon–anticodon interaction, the incomplete peptide chain is transferred to its alanine. Translation then switches to the MLD, which encodes a tag that targets the incomplete protein to degradation. During trans-translation, tmRNA acts in concert with specific ligands [for a recent review, see (1)] including 70S ribosomes, AlaRS (2), small protein B (SmpB) (3), elongation factor Tu (EF-Tu) (4) and ribosomal protein S1 (5).
Figure 1.
Ribosomal protein S1 is dispensable for the tRNA-like function of tmRNA. (A) Schematic representation of the first step of trans-translation (trans-peptidation) on polyU mRNAs. The light gray ovals are the ribosomal subunits (the larger for the 50S and the smaller for the 30S). The white ovals are, from right to left, the decoding site (A); the transpeptidyl-site (P) and the exit site (E). tRNAs and tmRNA are depicted as hooks and as a lasso, respectively (for further insights see (1)). (B) Western blots showing the quasi-absence of S1 in purified ‘S1-free’ 70S ribosomes compared to genuine 70S ribosomes. (C) The trans-translation in vitro assay is inactive in the absence of SmpB. (D) Assay of alanine trans-peptidation (asterisk in A), using S1-free 70S ribosomes and increasing amounts of exogenous S1. Trans-peptidation is monitored by the incorporation of 3H-Ala from alanyl-tmRNA into polyPhe and is normalized to the level of translation, measured in parallel by the incorporation of 14C-Phe (section ‘Experimental procedures’). The Y-axis represents the quantities (number of pmoles) of 3H alanine incorporated per assay.
S1, the largest ribosomal protein, binds weakly and reversibly to the head of the 30S ribosomal subunit (6), with an apparent binding constant of 20 nM [(7), and M. Hallier, unpublished Surface Plasmon Resonance data]. S1 consists of six imperfectly repeated motifs, each organized in an oligonucleotide/oligosaccharide binding (OB) fold (8). Whereas the first two N-terminal motifs are required for anchoring S1 to the head of the ribosomal small subunit, the four C-terminal motifs face the solvent side (9).
Protein S1 co-purifies in vivo with an over-expressed tmRNA–SmpB complex (10) and the protein binds tmRNA in vitro with a 10–100 nM apparent binding constant (5,11–13). Its exact role during trans-translation is unclear. Conceivably, S1 could prevent degradation of the tmRNA in the cytoplasm before it is loaded onto the ribosome and/or it could be involved in translation resumption onto tmRNA internal ORF, especially since the latter lacks a Shine Dalgarno element (SD) or an initiation codon (in most species, translation resumes onto an alanine codon). However, S1 stands at the junction of the head, platform and main body of the 30S subunit, at the opposite side of the decoding site (14). This location makes a concomitant binding of S1 to the stalled ribosome and to tmRNA during initial loading very unlikely. Moreover, recent cryo-EM data indicate that tmRNA enters stalled ribosomes in the absence of S1 (15,16), arguing for a role of S1 in trans-translation in solution and not when bound to the ribosomes.
Protein S1 is essential for cell viability (17) and autoregulates its own synthesis (18), so that its concentration cannot be easily manipulated in vivo (see subsequently). We therefore developed an in vitro method to assess precisely its role in trans-translation. Assays were set up with purified recombinant proteins (19) and ribosomes depleted from protein S1 (20). We took advantage of the fact that polyU genuinely lacks a termination codon, so that its translation in the presence of Phe-tRNAPhe yields stalled ribosomes, wherever translation initiates on the polyU RNAs. We found that S1 is dispensable for the codon-independent transfer of the stalled nascent peptide chain to the alanine from alanylated-tmRNAAla but is required for the translation of tmRNA internal ORF. The N-terminal domain of the protein that is required for ribosome binding is dispensable for trans-translation, indicating that the role of S1 during tmRNA-mediated protein tagging takes place outside from the ribosome, presumably when in complex with soluble tmRNA. Also, tmRNA-mediated protein tagging is reduced in vivo when the expression levels of protein S1 are either reduced or increased, compared to wild-type cells. Altogether, these results indicate that S1 plays an important role during trans-translation that is distinct from its documented role during protein synthesis.
RESULTS AND DISCUSSION
Protein S1 is dispensable for codon-independent trans-peptidation but required for the mRNA function of tmRNA
Mechanistically, trans-translation proceeds in two steps: (i) codon-independent transfer of the stalled polypeptide to the alanyl-tmRNA via peptide bond formation (tRNA role of tmRNA), and then (ii) ORF switching from the original mRNA to tmRNA (mRNA role of tmRNA). These two steps were dissociated in vitro to monitor the influence of the S1 protein on each of them. To monitor the role(s) of protein S1, we started from purified S1-free 70S ribosomes (Figure 1B), and added back variable amounts of pure recombinant S1 to the reaction mixture.
To assay the tRNA role of tmRNA, we monitored the incorporation of labeled alanine from Ala-tmRNA to polyPhe in the absence of Ala-tRNA. Translation then cannot resume on the tmRNA ORF and the assay measures only codon-independent trans-peptidation. To confirm that the signal is only due to trans-translation, a control experiment was carried out in the absence of SmpB, a protein indispensable to the process. No signal is detected in the absence of SmpB (Figure 1C). As shown in Figure 1D, trans-peptidation takes place in the absence of S1 protein, and is slightly stimulated by it (by ∼25% from 0.3 to 1.2 μM of S1 versus 0.3 μM of S1 free-ribosomes or tmRNA), leading to the incorporation from ∼3 to 4 pmoles of 3H alanine.
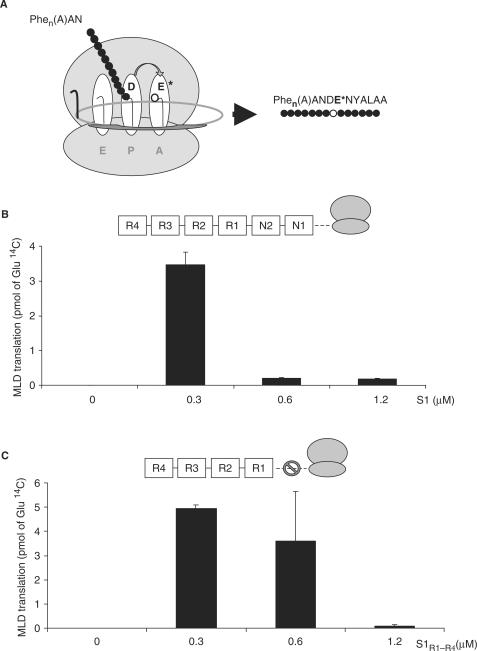
To monitor specifically the effect of S1 on translation of the tmRNA ORF, we added all purified tRNAs and aminoacyl-tRNA synthetases and recorded the incorporation of glutamic acid, the fourth amino acid of this ORF (Figure 2A). This incorporation was undetectable in the absence of S1, and passed through a maximum of ∼3 to 4 pmoles, for a same concentration of S1 versus the ribosomes. Therefore, S1 is essential for the glutamic acid incorporation within the tag-reading frame but becomes detrimental when present in excess (Figure 2B). Increasing the concentration of S1 leads to the binding of more than one protein per tmRNA (11,12), which might be detrimental for the translational activity (MLD function) of tmRNA.
Figure 2.
Ribosomal protein S1 is essential for the mRNA function of tmRNA. (A) Trans-translation of polyU RNA; the position of Glutamic acid, the fourth amino acid of the tmRNA tag, is emphasized by an asterisk. (B) Incorporation of 14C Glu during trans-translation in the presence of S1-free ribosomes and increasing amounts of exogenous S1 is normalized to the level of translation. The Y-axis represents the quantities of pmoles of 14C glutamic acid incorporated per assay. (C) Incorporation of 14C Glu during trans-translation in the presence of S1-free ribosomes and increasing amounts of exogenous S1R1–R4 is normalized to the level of translation. The Y-axis represents the quantities (number of pmoles) of 14C glutamic acid incorporated per assay.
A mutant S1 protein inactive in ribosome binding enhances trans-translation in vitro
Of the six contiguous S1 motifs, the two N-terminal ones are required for ribosome binding and the next three (R1–R3) for mRNA binding (6). To determine whether S1 participates to trans-translation via its association to the stalled ribosome, we used a N-terminal truncated S1 variant that only retains motifs R1–R4 (S1R1–R4). This variant fails to bind ribosomes in vivo whereas it binds tmRNA (12,13). S1R1–R4 stimulates glutamic acid incorporation in vitro as the intact protein (Figure 2C), indicating that this stimulation involves binding to tmRNA independently of the ribosome. The maximal stimulation persists at higher amounts of S1R1–R4 compared with S1 (compare Figure 2B and C), probably reflecting the weaker affinity of S1R1–R4 for tmRNA (12).
Overall effect of protein S1 on trans-translation
To record the overall effect of S1 upon trans-translation, we again recorded the incorporation of labeled Ala, but in the presence of all factors required for the translation of the tmRNA ORF. In that case the incorporation of Ala reflects both trans-peptidation and translation of the four Ala codons of the ORF (ANDENYALAA) (Figure 3A). Ala incorporation is slightly detected in the absence of S1, presumably reflecting the sole trans-peptidation step (Figures 1D and 3B). At similar concentrations of S1 and S1 free-ribosomes, Ala incorporation is maximal (Figure 3B), as observed for Glu (Figure 2B). This incorporation of ∼15 pmoles is consistent with the ∼3 pmoles of alanine or glutamic acid obtained at the same concentration of S1 (Figures 1D and 2B, respectively) and accounts for the incorporation of five alanines, one carried by the TLD and the other four from the tag. Overall trans-translation leads to the incorporation of five Ala, to be compared to the single one (carried by tmRNA) incorporated during trans-peptidation or the single Glu incorporated during trans-translation. Increasing further the concentration of S1 decreases Ala incorporation, but not as drastically as for Glu (Figure 2B). The higher rate of Ala incorporated at the highest concentration (1.2 μM) of S1 probably reflects the sum of the incorporation of the first alanine (transpeptidation, Figure 1D) plus the translation of the first alanine codon from tmRNA internal ORF.
Figure 3.
Overall effect of S1 and S1R1–R4 on protein tagging. (A) Same as Figure 2A, except that Ala residues are shown in bold. (B) Incorporation of 14C Ala from alanyl-tmRNA and alanyl-tRNAAla during trans-translation in the presence of S1-free ribosomes and increasing amounts of exogenous S1 is normalized to the level of translation. The Y-axis represents the number of pmoles of 14C alanine incorporated per assay. (C) Similar as in Figure 3B, except that a truncated S1 protein (S1R1–R4) lacking the first 193 N-terminal aminoacids and therefore unable to bind ribosomes, was used (12). The Y-axis represents the quantities of 3H Alanine incorporated per assay.
Mutant S1R1–R4 also stimulates alanine incorporation in vitro as the intact S1 protein does (Figure 3C). As previously observed for glutamic acid incorporation, maximal stimulation requires higher amounts of S1R1–R4 compared with wild type S1. Altogether, these results indicate that the role of protein S1 during trans-translation is distinct from its documented role during protein synthesis. Gram-positive bacteria lack ribosomal protein S1, whereas trans-translation is detected in Bacillus subtilis. In B. subtilis, the ypfD gene expresses a protein containing four repeated RNA-binding motifs (21), each corresponding to four of the six repeated motifs present in S1 from Escherichia coli. The B. subtilis S1 mimic has sequence similarity to the four C-terminal domains of S1 from E.coli, corresponding to mutant S1R1–R4. As shown in this report, mutant S1R1–R4 can sustain trans-translation in vitro. Therefore, the ypfD gene may be required for translation of tmRNA internal ORF during B. subtilis trans-translation.
In vivo modulation of ribosomal protein S1 concentration affects trans-translation
The in vitro results indicate that the amount of ribosomal protein S1 added in the assays can modulate trans-translation. To evaluate the effect of varied concentrations of ribosomal protein S1 on trans-translation in vivo, we used a modified chromosomal ssrA allele (ssrAH7) encoding a histidine stretch within the tmRNA ORF that replaces the terminal aminoacids, which normally constitutes an essential recognition signal for the proteases (22). Resulting his-tagged trans-translated proteins are not degraded and can be isolated on Ni2+ columns and quantified relatively to the amount of total proteins extracted from strains expressing various levels of S1 (see subsequently). In order to avoid the influence of S1 decrease or increase on protein synthesis, the same amounts of total proteins were applied to each of the columns for all the strains used. Trans-translated proteins were separated from bulk proteins onto an imidazole gradient on a Ni-NTA column and immunoblots using anti-His antibodies were performed on each collected fraction (Supplementary Data). The negative control prepared from a strain that does not express tmRNA-His7 (and contains an endogenous level of S1) did not display any significant signal (data not shown). On the other hand, equivalent elution profiles were obtained for the strains expressing tmRNA-His7 and various levels of S1 (Supplementary Data). In each case, fractions 3 were found to contain most of the tagged proteins. These fractions were pooled and applied to a desalting column to remove excess of imidazole. Afterwards, protein amounts were measured by UV absorbance at 280 nm (section ‘Experimental procedures’).
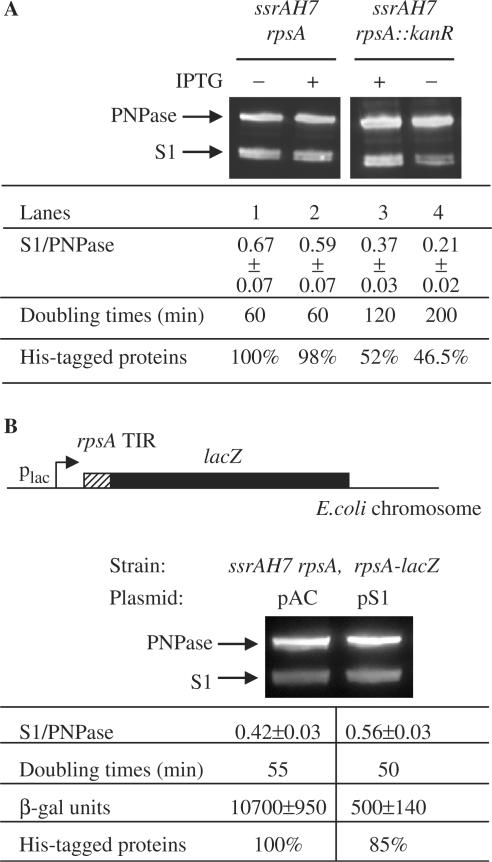
Since ribosomal protein S1 is indispensable for cell survival, it was not possible to reproduce in vivo the zero S1 condition obtained in vitro. Various alterations in the gene coding for S1 have been assayed in vivo, showing that both the NH2-terminal domain and R3 are essential for function (23). Nevertheless, we first varied the concentration of S1 by constructing a strain from which the essential ribosomal protein can be depleted using the same approach as previously described (24). For this purpose, the chromosomal rpsA gene (encoding S1) was deleted from the ssrAH7 strain that carries the pJFR32 plasmid where rpsA is controlled by the IPTG inducible lac promoter (section ‘Experimental procedures’) (24). As expected, removal of the inducer strongly increased the doubling time of the ssrAH7 rpsA::kanR strain (Figure 4A, lane 4). After 4 h of depletion, the ribosomal protein S1 was found to be 3-fold less abundant than in the wild-type ssrAH7 rpsA strain as judged by Western blot analyses (Figure 4A; compare lanes 1 and 4). Importantly, the deleted but complemented strain ssrAH7 rpsA::kanR (+IPTG) also displayed less amount of S1 (1.8-fold) indicating that expression of the rpsA gene from the pJFR32 plasmid (24) is not sufficient to restore a wild-type level of S1 as well as a wild-type doubling time (Figure 4A; compare lanes 3–1).
Figure 4.
Quantification of the trans-translated proteins in cells expressing varied S1 concentration. (A) Strains depleted for S1 contain less his-tagged proteins. Lanes 1 and 2: control strain ssrAH7 rpsA grown in the absence (−) or presence (+) of IPTG, respectively. Lanes 3 and 4: rpsA deleted strain (ssrAH7 rpsA::kanR) grown in the presence (+; complementation condition) or absence of IPTG (−; depletion condition), respectively. IPTG induces expression of the extra plasmid-borne rpsA copy present in the strains (section ‘Experimental procedures’). Western blot analysis was performed for samples of the cultures used to purify his-tagged protein. Blot was revealed with S1 antibodies mixed with PNPase antibodies to normalize quantification. The S1/PNPase ratio was determined by quantification of the S1 and PNPase signals on a chemi-smart 5000 (Vilbert-Lourmat). Doubling time of each strain is indicated. Amounts of purified his-tagged protein were measured by absorbance (section ‘Experimental procedures’). Percentage of his-tagged proteins was determined after normalization to the amount of total proteins extracted from each strain and strain ssrAH7 rpsA (lane 1) was used as reference. (B) Excess of free S1 affects the accumulation of trans-translated proteins. Up: schematic representation of the rpsA-lacZ chromosomal fusion used as translational reporter. Western blot analysis was performed as described earlier. β-Galactosidase activities from rpsA-lacZ translational fusion are given in β-galactosidase units corresponding to nanomoles of ONPG hydrolyzed per min and per mg of total protein. The values shown are averages of three independent assays. Presence of the pS1 plasmid strongly decreases β-galactosidase synthesis (20-fold) due to S1 autogenous control. Doubling time is given for both strains. Percentage of his-tagged protein was determined as in B, and strain ssrAH7 rpsA, rpsA-lacZ containing the control plasmid pAC, was used as reference.
In these conditions of lower abundance of ribosomal protein S1, we observed a ∼2-fold decrease of trans-translation as judged by quantification of the in vivo his-tagged proteins that were purified from the corresponding strains (Figure 4A, lanes 3 and 4 compared to lane 1). Moreover, the lower level of trans-translation (46.5%) was obtained for the strain containing the lower detectable amount of ribosomal protein S1 (Figure 4A, lane 4). To exclude that the lower levels of his-tagged proteins result from the influence of S1 on protein synthesis and consequently on trans-translation, all the measures were performed at identical growth rates for each strain.
It is known that excess of S1 can repress its own translation (18). In other words, the rpsA gene is autoregulated and consequently a S1 overexpression is hardly achievable in vivo. This is exemplified in the first attempt we made where the ssrAH7 rpsA strain containing the pJFR32 plasmid was grown in the presence of IPTG to induce expression of the extra plasmid-borne rpsA copy (Figure 4A, lane 2). Indeed, this condition did not allow overexpression of S1 and, on the contrary, we observed a slight decrease of the S1 signal (Figure 4A, compare lanes 1 and 2).
In a second attempt to increase endogenous expression of the S1 protein we exploited the fact that the presence of an excess of free S1 in the cell can be followed using a chromosomal fusion between the rpsA translation initiation region (TIR) and the lacZ gene as a translational reporter system (25; Figure 4B). This rpsA-lacZ chromosomal fusion leads to a β-galactosidase synthesis that is driven at the transcription level by the lac promoter, and most importantly, at the translational level by the rpsA TIR. As a result, when excess of free S1 is present in the cell, the β-galactosidase activity is decreased due to S1 autogenous control (25). The rpsA-lacZ fusion was thus introduced in the ssrAH7 rpsA strain (section ‘Experimental procedures’) and the resulting strain (ssrAH7 rpsA, rpsA-lacZ) was transformed by the control plasmid pACYC184 (pAC) or its pS1 derivative that expresses S1 under the control of its own promoter (26). β-Galactosidase activity was measured for both strains. As expected, presence of the pS1 plasmid strongly decreases β-galactosidase activity indicating that the pS1 containing strain does harbor an excess of free S1 protein, as also independently demonstrated by western blot analyses (Figure 4B). The cells doubling-time is not influenced by the overexpression of S1 (50 min versus 55 min, Figure 4B). His-tagged proteins were isolated from the same strains except that in this case the rpsA-lacZ expression was not IPTG induced to avoid S1 titration by the rpsA-lacZ transcripts. A decrease of the trans-translated proteins (from 100 to 85%) was observed in the pS1-containing strain (Figure 4B) that reflects trans-translation inhibition by excessive amounts of S1, as observed in vitro (Figures 2B and 3B), probably by preventing efficient decoding of the tag-reading frame. These results suggest that the concentration of S1 in vivo is optimally set up for efficient trans-translation and that minor up- or down- regulations of S1 are deleterious for ribosome rescue. However, in vitro, the highest inhibitory effect was obtained with a 200% excess of S1 (Figure 3B). A same extent of trans-translation inhibition was impossible to reach in vivo, since excess amounts of S1 could hardly exceed 130% (Figure 4B, compare the two lanes). In accordance with the present study, the use of an inducible plasmid overexpressing the S1 protein, or protein fragments, leads to a same extent of variation of the in vivo tagging activity of tmRNA, from 25 to 35% (12). Altogether, these data suggest that the S1 protein, by playing its role outside the ribosome, may bind tmRNA early during trans-translation (5,11).
EXPERIMENTAL PROCEDURES
Chemicals and enzymes
All the common reagents and enzymes were purchased from Sigma, except for tRNAs, Creatin phosphate and Creatine kinase, obtained from Roche (Meylan, France), Trichloroacetic acid (TCA) from Acros Organics (Noisy le Grand, France), Urea from Eurobio (Les Ulis, France) and radiolabeled amino acids from Perkin-Elmer (Courtabœuf, France).
Purification of the ‘PURE System’ factors
The translation factors necessary to the cell free in vitro protein synthesis were purified according to Shimizu and co-workers (19). Briefly, His-tagged proteins were overexpressed in E.coli cells transformed with their respective plasmids. After lysis the extracts were centrifuged and the supernatants applied on Ni2+-Sepharose chelating columns (HisTrap HP, Amersham) by using the AKTA Basic system (Amersham). After elution by a 10–300 mM linear gradient of imidazole, the fractions containing the proteins were pooled and concentrated on centrifugal filter devices (Amicon® Ultra-15, Millipore) before being dialyzed against a stock buffer containing 30% of glycerol. All the proteins were frozen in small aliquots at −80°C.
Cloning, expression and purification of trans-translation specific components
Escherichia coli tmRNA was overproduced in vivo in strain JM109(DE3) transformed with a derivative of plasmid pGEMEX-2 carrying the ssrA gene under the control of the T7 RNA polymerase promoter. Purification was performed according to Ushida et al. (27). SmpB was purified from the E.coli strain BL21(DE3)/pet-21a-SmpB, as described before (28). His-tagged ribosomal protein S1 was purified under denaturing conditions (5) and slowly refolded directly on the HisTrap column by using a linear gradient of Urea, from 6 to 0 M (60 ml for 10 h). The protein was then eluted and dialyzed against the stock buffer as described earlier.
In vitro trans-translation assays
The translation assays were prepared essentially using the method described by Shimizu et al. (19). Slight modifications were nevertheless introduced to the method in order to detect optimal yields of trans-translated proteins. Prior to triggering trans-translation, polyUridines RNAs were first translated for 30 min in standard conditions. Some free S1 is likely to bind the polyU RNAs. However, S1 binds the 30S subunits or tmRNA one order of magnitude tighter than to polyU RNAs (6). SmpB, tmRNA and various amounts of S1 were then added in that order for 30 additional minutes in a final volume of 50 μl. The trans-translation factor mix was the same as previously described (29) except that it contains 0.3 μM ribosomes, 0.3 μM tmRNA, 0.6 μM SmpB and 1.2 μM elongation factor EF-Tu. The bulk of tRNAs (Roche) was adjusted to 4 μM and release factors (RFs) were removed in order to standardize the results. No putrescin was used during the reaction. Different radiolabeled amino acids were incorporated to the reaction mixture in order to monitor the various steps of the reaction: (i) 14C phenylalanine (18.5 kBq corresponding to 1 nmol) was used for measuring the rate of translation of the polyU RNA (ii) 14C alanine (9.25 kBq corresponding to 1.5 nmole) for measuring the first step of trans-peptidation of tmRNA (in that case the bulk of tRNAs was replaced by pure tRNAPhe, 313 nM) (iii) 14C glutamic acid (0.9 kBq corresponding to 30.75 nmole) for measuring the rate of translation of the internal ORF of tmRNA (iv) 3H alanine (92.5 MBq corresponding to 29.25 nmole) for measuring the global rate of trans-translation, including the first step of trans-peptidation of tmRNA as well as the four alanines encoded by the internal ORF of tmRNA. In each case the corresponding cold amino acid was removed from the reaction mixture. At the end of the respective reactions, the rates of incorporation of the radiolabeled aminoacid were measured by filter binding assays.
In parallel to trans-translation, the corresponding rate of translation was measured for each assay by using 14C Phenylalanine instead of radiolabeled alanine or glutamic acid. The amount of phenylalanine incorporated was then taken into account to normalize the data and eliminate the influence of S1 on polyU RNA translation. Normalization factors were calculated by dividing the level of translation for each assay by the level of translation for the minus S1 assay. The value of each of the labeled amino acids (Ala, Glu) incorporated at each concentration of protein S1 was divided by its own normalization factor.
Filter binding assays
Reactions were terminated by the addition of 1 ml 5% ice-cold TCA, heated for 20 min at 90°C and incubated for 30 min on ice (30). Incorporated radioactive aminoacids were measured by filtering under vacuum suction on nitrocellulose filters (Schleicher and Schuell), followed by rapid washing with 3 × 2 ml 5% ice-cold TCA and 1 × 1 ml 96% ethanol. Membranes were dried and the holding radioactivity measured by scintillation counting (WALLAC 1409). Background values for each point were measured by using the corresponding samples without any polyU RNAs.
S1 detection by western blotting
In order to detect remaining S1 protein from ribosomes, 0.3 pmol 70S purified samples were electrophoresed on 10% SDS PAGE gels and transferred to PVDF membranes (Amersham). Western blot analysis was performed using specific anti-S1 antibodies as previously described (31).
Bacterial strains and measurement of β-galactosidase activity
The PhB2677 strain used in this work contains a chromosomal ssrAH7 allele encoding the peptide tag ANDENYHHHHHHH (22). To make a strain from which the essential ribosomal protein S1 can be depleted, we introduced the chromosomal rpsA::kan deleted allele into PhB2677 along with the pJFR32 (plac rpsA) plasmid that contains an IPTG inducible wild-type rpsA gene (24). First, strain PhB2677 has been transformed with plasmid pJFR32 and resulting ampicilin resistant strain (AmpR) was transduced with a P1 lysate from strain JFR175 [rpsA::kanR, pJFR32 (24)]. Kanamycin resistant (kanR) transductants were selected in the presence of IPTG (1 mM). The resulting ssrAH7, rpsA::kan, pJFR32 strain (hereafter ssrAH7 rpsA::kan) was shown to be dependent of the presence of IPTG for growth and presence of the rpsA::kan deleted allele was checked by PCR analysis. To deplete S1, the IPTG inducer was removed from an exponential growing culture (OD600 = 0.23) by filtration and cells were rapidly resuspended in an IPTG-free medium. Growth was continued for 4 h and cells harvested (OD600 = 0.61). For control, the same strain was continuously grown in the presence of IPTG (1 mM) but a filtration step has been equivalently done and growth continued before harvesting cells. Depletion of S1 has been checked by western blot analyses.
To make strains that expressed increased level of S1, we either grown strain PhB2677 containing the pJFR32 plasmid (hereafter ssrAH7 rpsA) in the presence of IPTG (1 mM) or transformed the PhB2677 strain containing the chromosomal rpsA-lacZ fusion (see below) with the pS1 plasmid, a derivative of pACYC184 (pAC) that expresses S1 under the control of its own promoter (26). Increased level of S1 was either checked by Western blotting, or we used β-galactosidase activity measurements to assess the presence of excess free S1 protein: due to autoregulation, higher concentration of free S1 leads to lower activity of the TIR of the chromosomal rpsA-lacZ fusion (25). The resulting chloramphenicol resistant strain (hereafter ssrAH7 rpsA, rpsA-lacZ pS1) was used for β-galactosidase activity measurements. As control, we used the PhB2677 rpsA-lacZ strain transformed by pAC (hereafter ssrAH7 rpsA, rpsA-lacZ pAC) that expressed endogenous level of ribosomal protein S1. β-Galactosidase activity was assayed as described previously (25). Each measurement was done at least in triplicate. Strains were grown at 37°C in LB medium supplemented with chloramphenicol (20 μg/ml) and IPTG (500 μM) and collected at OD600 of 0.8.
The chromosomal rpsA-lacZ fusion has been introduced in the PhB2677 strain by P1 mediated transduction in two steps. First, the Zah281::Tn10 transposon was introduced near the rpsA-lacZ fusion of strain HfrG6 rpsA(-91)::lacZ (25) by selecting for tetracycline resistance (Tetr). The rpsA-lacZ fusion was then P1 transduced into PhB2677 strain by selecting again for Tetr. At each step, strains were confirmed by PCR analyses. Strain ssrAH7 rpsA, rpsA-lacZ was then transformed with the indicated plasmids.
In vivo tagging activity
The E.coli strains expressing the ribosomal protein S1 at different levels were grown at 37°C in LB medium. The cells were harvested at OD600 0.4–0.8 and washed once in 10 mM Tris-HCl, 100 mM Na-phosphate, pH 7.4 before freezing. After lysis, equal amounts of total proteins from each bacterial strain were applied to a Ni2+-NTA chromatography column under denaturing conditions. His-tagged trans-translated proteins were eluted by a linear gradient of imidazole and recovered in 1 ml fractions (Supplementary Data) (22). A small aliquot of each fraction was electrophoresed in SDS-PAGE gels and submitted to western blot using anti-His antibodies (Ref 34660, Qiagen) in order to detect the presence of tagged proteins. The positive fractions were then pooled and 900 μl applied to a 5 ml HiTrap desalting column (Amersham) in order to discard the imidazole that could interfere with the measurement of protein concentration using absorbance at 280 nm. The following buffer was used to elute the tagged proteins: 100 mM NaH2PO4, 10 mM Tris-HCl (pH 7). The amounts of trans-translated proteins were then measured by peak integration of the desalting column absorbance curve at 280 nm, after subtracting the control curve (Supplementary Data).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
MS and PS are supported by Ph.D. fellowships from INSERM and the ‘Région Bretagne’, and from the Fondation pour la Recherche Médicale (FRM), respectively. This work was supported by grants from Région Bretagne (PRIR Grant no 691 and CRB 2004-1483), ACI BCMS 136 and ANR program MIME 2006. Authors are grateful to Dr M. Dreyfus for helpful discussions and comments on the manuscript. We thank Dr T. Ueda (Nagoya University, Japan) for providing the clones to produce the components of the PURE system, Drs V. Ramakrishnan (MRC, Cambridge) for providing S1-free 70S ribosomes, RT. Sauer (MIT, USA) for the clones expressing S1 variants, P. Bouloc (Orsay, France) for the PhB2677 (ssrAH7) strain and M. Hallier for the SPR data. Funding to pay the Open Access publication charges for this article was provided by INSERM.
Conflict of interest statement. None declared.
REFERENCES
- 1.Saguy M, Gillet R, Metzinger L, Felden B. tmRNA and associated ligands: a puzzling relationship. Biochimie. 2005;87: 897–903. doi: 10.1016/j.biochi.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 2.Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. USA. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karzai AW, Susskind MM, Sauer T. SmpB, a unique RNA-binding protein essential or the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudinger-Thirion J, Giegé R, Felden B. Aminoacylated tmRNA from Escherichia coli interacts with procaryotic elongation factor Tu. RNA. 1999;5:989–992. doi: 10.1017/s135583829999101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wower J, Zwieb C, Guven SA, Wower I. Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome. EMBO J. 2000;19:6612–6621. doi: 10.1093/emboj/19.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subramanian AR. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid. Res. Mol. Biol. 1983;28:101–142. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- 7.Draper DE, Von Hippel PH. Interaction of Escherichia coli ribosomal protein S1 with ribosomes. Proc. Natl. Acad. Sci. USA. 1979;76:1040–1044. doi: 10.1073/pnas.76.3.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bycroft M, Hubbard T.JP, Proctor M, Freund S.VM, Murzin AG. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 9.Giorginis S, Subramanian AR. The major ribosome binding site of Escherichia coli ribosomal protein S1 is located in its N-terminal segment. J. Mol. Biol. 1980;141:393–408. doi: 10.1016/0022-2836(80)90253-3. [DOI] [PubMed] [Google Scholar]
- 10.Karzai AW, Sauer RT. Protein factors associated with the SsrA-SmpB tagging and ribosome rescue complex. Proc. Natl. Acad. Sci. USA. 2001;98:3040–3044. doi: 10.1073/pnas.051628298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bordeau V, Felden B. Ribosomal protein S1 induces a conformational change of tmRNA; more than one protein S1 per molecule of tmRNA. Biochimie. 2002;84:723–729. doi: 10.1016/s0300-9084(02)01442-6. [DOI] [PubMed] [Google Scholar]
- 12.McGinness KE, Sauer RT. Ribosomal protein S1 binds mRNA and tmRNA similarly but plays distinct roles in translation of these molecules. Proc. Natl. Acad. Sci. USA. 2004;101:13454–13459. doi: 10.1073/pnas.0405521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada T, Wower IK, Wower J, Zwieb CW, Kimura M. Contribution of the second OB fold of ribosomal protein S1 from Escherichia coli to the recognition of tmRNA. Biosci. Biotechnol. Biochem. 2004;68:2319–2325. doi: 10.1271/bbb.68.2319. [DOI] [PubMed] [Google Scholar]
- 14.Sengupta J, Agrawal RK, Frank J. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl. Acad. Sci. USA. 2001;98:11991–11996. doi: 10.1073/pnas.211266898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valle M, Gillet R, Kaur S, Henne A, Ramakrishnan V, Frank J. Visualizing tmRNA entry into a stalled ribosome. Science. 2003;300:127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- 16.Gillet R, Kaur S, Li W, Hallier M, Felden B, Frank J. Scaffolding as an organizing principle in trans-translation: the roles of proteins SmpB and S1. J. Biol. Chem. 2006;282:6536–6563. doi: 10.1074/jbc.M609658200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitakawa M, Isono K. An amber mutation in the gene rpsA for ribosomal protein S1 in Escherichia coli. Mol. Gen. Genet. 1982;185:445–447. doi: 10.1007/BF00334137. [DOI] [PubMed] [Google Scholar]
- 18.Skouv J, Schnier J, Rasmussen MD, Subramanian AR, Pedersen S. Ribosomal protein S1 of Escherichia coli is the effector for the regulation of its own synthesis. J. Biol. Chem. 1990;265:17044–17049. [PubMed] [Google Scholar]
- 19.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 20.Clemons WM, Jr, Brodersen DE, McCutcheon JP, May JL, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: purification, crystallization and structure determination. J. Mol. Biol. 2001;310:827–843. doi: 10.1006/jmbi.2001.4778. [DOI] [PubMed] [Google Scholar]
- 21.Sorokin A, Serror P, Pujic P, Azevedo V, Ehrlich SD. The Bacillus subtilis chromosome region encoding homologues of the Escherichia coli mssA and rpsA gene products. Microbiology. 1995;141:311–319. doi: 10.1099/13500872-141-2-311. [DOI] [PubMed] [Google Scholar]
- 22.Collier J, Binet E, Bouloc P. Competition between SsrA tagging and translational termination at weak stop codons in Escherichia coli. Mol. Microbiol. 2002;45:745–754. doi: 10.1046/j.1365-2958.2002.03045.x. [DOI] [PubMed] [Google Scholar]
- 23.Schnier J, Stöffler G, Nishi K. Deletion and insertion mutants in the structural gene for ribosomal protein S1 from Escherichia coli. J. Biol. Chem. 1986;261:11866–11871. [PubMed] [Google Scholar]
- 24.Sørensen MA, Fricke J, Pedersen S. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J. Mol. Biol. 1998;280:561–569. doi: 10.1006/jmbi.1998.1909. [DOI] [PubMed] [Google Scholar]
- 25.Boni IV, Artamonova VS, Dreyfus M. The last RNA-binding repeat of the Escherichia coli ribosomal protein S1 is specifically involved in autogenous control. J. Bact. 2000;182:5872–5879. doi: 10.1128/jb.182.20.5872-5879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedersen S, Skouv J, Kajitani M, Ishihama A. Transcriptional organization of the rpsA operon of Escherichia coli. Mol. Gen. Genet. 1984;196:135–140. doi: 10.1007/BF00334105. [DOI] [PubMed] [Google Scholar]
- 27.Ushida C, Himeno H, Watanabe T, Muto A. tRNA-like structures in 10Sa RNAs of Mycoplasma capricolum and Bacillus subtilis. Nucleic. Acids. Res. 1994;22:3392–3396. doi: 10.1093/nar/22.16.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallier M, Ivanova N, Rametti A, Pavlov M, Ehrenberg M, Felden B. Pre-binding of small protein B to a stalled ribosome triggers trans-translation. J. Biol. Chem. 2004;279:25978–25985. doi: 10.1074/jbc.M314086200. [DOI] [PubMed] [Google Scholar]
- 29.Shimizu Y, Ueda T. The role of SmpB protein in trans-translation. FEBS Lett. 2002;514:74–77. doi: 10.1016/s0014-5793(02)02333-5. [DOI] [PubMed] [Google Scholar]
- 30.Jacob Y, Sharkady SM, Bhardwaj K, Sanda A, Williams KP. Function of the SmpB tail in transfer-messenger RNA translation revealed by a nucleus-encoded form. J. Biol. Chem. 2005;280:5503–5509. doi: 10.1074/jbc.M409277200. [DOI] [PubMed] [Google Scholar]
- 31.Boni IV, Isaeva DM, Musychenko ML, Tzareva NV. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic. Acids. Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.