Abstract
To examine whether the transcription factor Sox9 has an essential role during the sequential steps of chondrocyte differentiation, we have used the Cre/loxP recombination system to generate mouse embryos in which either Sox9 is missing from undifferentiated mesenchymal cells of limb buds or the Sox9 gene is inactivated after chondrogenic mesenchymal condensations. Inactivation of Sox9 in limb buds before mesenchymal condensations resulted in a complete absence of both cartilage and bone, but markers for the different axes of limb development showed a normal pattern of expression. Apoptotic domains within the developing limbs were expanded, suggesting that Sox9 suppresses apoptosis. Expression of Sox5 and Sox6, two other Sox genes involved in chondrogenesis, was no longer detected. Moreover, expression of Runx2, a transcription factor needed for osteoblast differentiation, was also abolished. Embryos, in which Sox9 was deleted after mesenchymal condensations, exhibited a severe generalized chondrodysplasia, similar to that in Sox5; Sox6 double-null mutant mice. Most cells were arrested as condensed mesenchymal cells and did not undergo overt differentiation into chondrocytes. Furthermore, chondrocyte proliferation was severely inhibited and joint formation was defective. Although Indian hedgehog, Patched1, parathyroid hormone-related peptide (Pthrp), and Pth/Pthrp receptor were expressed, their expression was down-regulated. Our experiments further suggested that Sox9 is also needed to prevent conversion of proliferating chondrocytes into hypertrophic chondrocytes. We conclude that Sox9 is required during sequential steps of the chondrocyte differentiation pathway.
Keywords: Sox9, Sox5/Sox6, chondrocyte differentiation, osteochondroprogenitors, apoptosis
At the onset of endochondral bone formation, a number of patterning molecules target the mesenchyme by outlining the three-dimensional coordinates that determine the shape, number, and size of the primordia of skeletal elements. In parallel to these patterning effects, the multistep process of endochondral bone formation is initially marked by the acquisition in undifferentiated mesenchymal cells of chondrogenic potency. These committed mesenchymal cells first undergo condensation, by which cells become closely packed, followed by the overt differentiation of cells within these condensations into chondrocytes. These differentiated chondrocytes then sustain a series of sequential changes that include unidirectional proliferation, conversion to hypertrophic chondrocytes, ability to calcify the extracellular matrix, cell death, and replacement by bone. A number of secreted polypeptides are known to cooperatively regulate the rates of proliferation of chondrocytes and their transition to hypertrophy (Karaplis et al. 1994; Lanske et al. 1996; Vortkamp et al. 1996; St-Jacques et al. 1999; Hartmann and Tabin 2000; Vortkamp 2001). However, until recently, much less was known about intracellular events, especially about the transcription factors that control the onset of chondrogenesis in undifferentiated mesenchymal cells as well as the progression of chondrocytic differentiation.
Sox9 is a transcription factor with a high-mobility-group (HMG-box) DNA-binding domain exhibiting a high degree of homology with that of the mammalian testis-determining factor, SRY. During chondrogenesis, Sox9 is expressed in all chondroprogenitors and all differentiated chondrocytes, but not in hypertrophic chondrocytes (Ng et al. 1997; Zhao et al. 1997). The human skeletal dysmorphology syndrome, campomelic dysplasia, which is characterized by hypoplasia of most endochondral bones, is caused by heterozygous mutations in and around Sox9 (Foster et al. 1994; Wagner et al. 1994). Previous studies based on mouse embryo chimeras derived from Sox9−/− embryonic stem (ES) cells showed that the mutant cells were excluded from chondrogenic mesenchymal condensations and could not express chondrocyte-specific markers, such as Col2a1, Col11a2, and Aggrecan (Bi et al. 1999). Other studies showed that Sox9 bound to and activated chondrocyte-specific enhancer elements in the Col2a1, Col11a2, Aggrecan, and CD-RAP genes in vitro, consistent with the notion that these genes were direct targets of Sox9 (Lefebvre et al. 1997; Bridgewater et al. 1998; Xie et al. 1999; Sekiya et al. 2000). Moreover, ectopic expression of Sox9 in transgenic mice activated the endogenous Col2a1 gene in vivo (Bell et al. 1997).
Two other members of the Sox family, Sox5 and Sox6, also play an essential role in chondrocytic differentiation. Sox5 and Sox6 are coexpressed with Sox9 in all chondroprogenitors and all differentiated chondrocytes (Lefebvre et al. 1998). In vitro studies showed that Sox5 and Sox6 cooperate with Sox9 to activate the Col2a1 enhancer and the Col2a1 gene. Whereas Sox5-null and Sox6-null mutant mice have relatively mild skeletal phenotypes, Sox5; Sox6 double-null mutants develop a severe, generalized chondrodysplasia characterized by the virtual absence of cartilage (Smits et al. 2001). In these double mutants, chondrogenic cells are largely arrested at the stage of chondrogenic mesenchymal condensations, and hence, these experiments showed that Sox5 and Sox6 are needed for overt differentiation of chondrocytes.
Although our studies with mouse embryo chimeras derived from Sox9−/− ES cells strongly suggested that Sox9 is needed for chondrogenic mesenchymal condensation (Bi et al. 1999), they did not address a number of critical questions regarding other potential roles of Sox9 during endochondral bone formation. First, given previous speculation about a common progenitor for both the chondrocyte and osteoblast lineages (Grigoriadis et al. 1988; Ahdjoudj et al. 2001) and the expression of Runx2 in chondrogenic mesenchymal condensations (Otto et al. 1997), we asked whether inactivation of Sox9 before mesenchymal condensation affected establishment of the osteoblast lineage in endochondral skeletal elements. Second, whereas the normal levels of Sox9 expression in mesenchymal condensations of Sox5; Sox6 double-null mutants indicated that Sox5 and Sox6 were not required for Sox9 expression (Smits et al. 2001), it was possible that Sox9 might control expression of Sox5 and Sox6. Third, given that specific patterning factors control the axes of limb development, we wanted to examine whether inactivation of Sox9 before mesenchymal condensation would affect expression of these limb-patterning genes. Finally, although in vitro results suggested the possibility that Sox9 was involved during chondrogenesis beyond mesenchymal condensation, there is no reported in vivo evidence that supports this hypothesis.
The perinatal lethality of Sox9 heterozygous mutant mice, which almost perfectly reproduces the skeletal abnormalities of human campomelic dysplasia patients, precludes, however, the generation of Sox9 homozygous null mutants (Bi et al. 2001) and, hence, studies of the functions of Sox9 during embryonic development using conventional genetic methods. To determine the roles of Sox9 during the sequential steps of chondrogenesis and chondrocytic differentiation, we therefore conditionally inactivated the Sox9 gene using the Cre recombinase/loxP (Cre/loxP) recombination system of bacteriophage P1 (Gu et al. 1994) both in undifferentiated mesenchymal cells and after chondrogenic mesenchymal condensation.
Results
Generation of floxed Sox9 mice and Sox9 conditional null mutant mice
The Cre/loxP system was developed in order to restrict inactivation of a target gene in a cell- or tissue-specific manner (Gu et al. 1994). To inactivate Sox9 in chondrogenic cell lineages, we used mice carrying Sox9flox, a Sox9 allele in which the DNA segment that includes exons 2 and 3 was flanked by loxP sites (Fig. 1A). Exons 2 and 3 code for part of the HMG-box DNA-binding domain and the transactivation domain of Sox9. Both heterozygous and homozygous animals were viable and fertile and showed no noticeable phenotypic change. Crossing of the Sox9flox allele with animals expressing the Cre recombinase ubiquitously (deleter-Cre; Schwenk et al. 1995) leads to severe bone defects and perinatal death, a phenotype identical to that found in Sox9 heterozygous mutant mice (Bi et al. 2001; Kist et al. 2002; data not shown), indicating that the recombined allele represents real loss of Sox9 function. Using the mating scheme described in the Materials and Methods section, we produced animals that were heterozygous for Sox9flox carrying either a Prx1–Cre or a Col2a1–Cre transgene. In embryos harboring the Prx1–Cre transgene, Cre is expressed in all mesenchymal cells of limb buds starting around embryonic day 9.5 (E9.5) (Martin and Olson 2000; Logan et al. 2002). In addition, it is also expressed in mesenchymal precursor cells of sternum (data not shown). The Sox9flox heterozygotes that harbor Prx1–Cre are viable and fertile and show the same phenotype as Sox9 heterozygous embryos in their limbs (Bi et al. 2001), that is, hypoplasia of scapula and pelvic bones and bending of radius, ulna, tibia, and fibula, as well as absence of deltoid protuberance of the humerus (H. Akiyama, unpubl.). Ninety-five percent of Sox9flox heterozygotes harboring the Col2a1–Cre transgene died around 10 d after birth, displaying dwarfism and severe kyphosis with compression of cervical and thoracic vertebrae. Only 5% were viable beyond this point and fertile, probably due to mosaicism of Sox9floxdel, the deleted allele of Sox9flox, in chondrocytes.
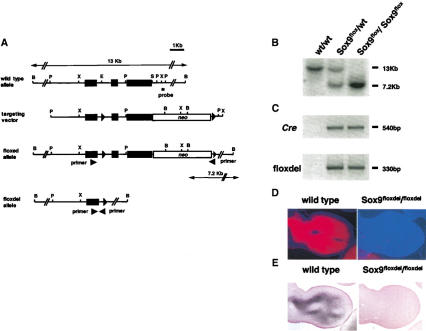
Figure 1.
Targeting strategy for conditional inactivation of the gene for Sox9. (A) Structure of the genomic Sox9 locus, targeting vector, and the homologous recombined allele. Exons are depicted as closed boxes, and intronic sequences are shown as solid lines. The Neo cassette is depicted as an open box. Forward and reverse primers used for PCR genotyping for floxdel alleles are shown as arrowheads. DNA fragments revealed in Southern analysis are indicated as arrows with the restriction enzymes and the probe. B, BamHI; P, PstI; X, XbaI; E, EagI; S, SspI. (B) Southern blot analysis of fetal genomic DNA. Genomic DNA isolated from the skin of wild-type, Sox9flox/wt; Cre, and Sox9flox/flox; Cre was digested with BamHI and then hybridized with the 3′ probe. The wild-type and floxed alleles were detected as 13-kb and 7.2-kb fragments, respectively. (C) PCR genotyping of Cre transgenes and floxdel alleles. (D,E) Expression of Sox9 mRNA (in situ hybridization, D) and Sox9 protein (immunohistochemistry, E) in limb buds of E12.5 wild-type and Sox9flox/flox; Prx1–Cre embryos. Both Sox9 mRNA and Sox9 protein were not detected in mutant limb buds.
In Sox9flox/flox; Prx1–Cre and Sox9flox/flox; Col2a1–Cre embryos, Cre-mediated conversion of Sox9flox to Sox9floxdel occurred and the floxdel allele was detected only in embryos positive for either Prx1–Cre or Col2a1–Cre (Fig. 1B,C), indicating that Cre was capable of recombining the loxP sites within the Sox9 gene in vivo.
Complete absence of cartilage and bone in limbs of Sox9flox/flox; Prx1–Cre mutant embryos
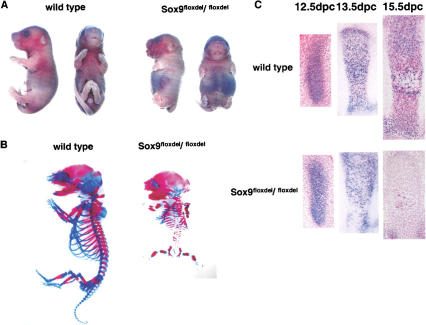
The conditional Sox9 null mutants resulting from expression of the Prx1–Cre transgene, which were recovered with the expected Mendelian frequency, mostly died in the immediate postnatal period from respiratory distress due to the absence of a sternum. Newborns had very short limbs, but they looked otherwise normal (Fig. 2A). Staining of skeletal preparations from newborn mutant mice with alcian blue and alizarin red indicated a virtual absence of all skeletal components in forelimbs and hindlimbs (Fig. 2B). Occasionally, only rudiments of scapula, pubis, and ishium were detected. In situ hybridization and immunohistochemistry indicated a complete absence of Sox9 RNA and protein in mutant limb buds, an indication that the inactivation of the Sox9 genes was complete (Fig. 1D,E).
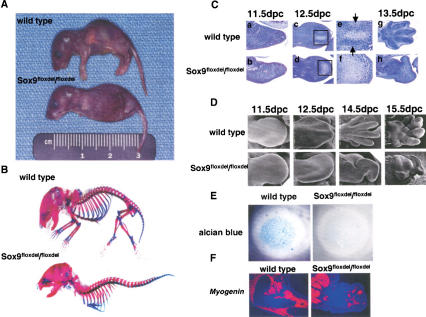
Figure 2.
Analysis of skeletal phenotypes in Sox9flox/flox; Prx1–Cre mice. (A) Gross appearance of newborn mice. Mutant newborns had very short limbs. (B) Skeletons of newborn mice stained by alcian blue followed by alizarin red. Cartilage and bone in the mutant limbs were completely absent. (C) Histological analysis of limb buds stained by hematoxylin and Treosin at 11.5 dpc (a,b), 12.5 dpc (c,d), and 13.5 dpc (g,h). Mutant limb buds had no discernible chondrogenic mesenchymal condensations at 12.5 dpc (c,d). Boxed regions at 12.5 dpc are shown at a high magnification (e,f). The arrows indicate chondrogenic mesenchymal condensations. (D) Morphological analysis of limb buds by scanning electron microscope. In the mutant limb buds, no distinct digit formation was observed, and the outgrowth of the mutant limb buds was arrested. (E) Micromass culture of dissociated mutant mesenchymal cells shows no alcian-blue positive cartilaginous nodules 7 d after plating. (F) Expression of Myogenin RNA in limb buds of E15.5 wild-type and mutant embryos. Myogenin-positive cells were present in the mutant limb buds.
Histological analysis of wild-type limb buds showed condensed mesenchymal cells at 12.5 days postcoitum (dpc), in which the digits differentiate into chondrocytes at 13.5 dpc (Fig. 2C). In contrast, mutant limb buds had no discernible chondrogenic mesenchymal condensations at 12.5 dpc, and, thereafter, no cartilages or bones were detectable. The lectin peanut agglutinin (PNA) bound to chondrogenic mesenchymal condensations of radius and ulna but not yet in the digit condensations in E12.5 wild-type embryos. In E12.5 mutant limb buds, there was very little localized binding of PNA (data not shown). Consistent with the absence of mesenchymal condensations in the mutant limb buds, no alcian blue-stainable cartilage nodules were observed in an in vitro high-density micromass culture using E12.5 mutant limb mesenchymal cells (Fig. 2E), indicating that Sox9−/− cells lost their chondrogenic potency even if conditions of cell–cell interactions were provided in the micromass culture. Histological examination of the mutant limb buds at 15.5 dpc revealed a small number of muscle fibrils and apparently normal skin development (data not shown). Although myogenin-positive cells were present in the mutant limb buds, an indication that myoblasts migrated into Sox9-deficient limb buds, the organization of muscle bundles were very abnormal (Fig. 2F).
Scanning electron microscope analysis of mutant limb buds at 11.5 dpc showed a normal paddle shape with typical apical ectodermal ridges. In E12.5 mutant limb buds, no distinct digit formation was observed, and the outgrowth of the mutant limb buds was arrested by 13.5 dpc. The malformation of the limb buds grew progressively more severe with time, although prominent pads were seen on the palmer surface (Fig. 2D).
To further characterize the Sox9 mutant limb buds, we examined the expression of Runx2, which is required for osteoblast differentiation (Otto et al. 1997). In E12.5 wild-type forelimb buds, Runx2 transcripts were detected in scapula, humerus, radius, and ulna, but not in metacarpals and digits (Fig. 3A). In contrast, little or no expression of Runx2 was detected in the mutant limb buds, strongly suggesting that Sox9 is needed for Runx2 expression at this stage. In wild-type E15.5 limbs, Runx2 was expressed at high levels in osteoblasts of bone collars and bone trabeculae and at lower levels in prehypertrophic and hypertrophic chondrocytes, whereas Bone sialoprotein (Bsp), an early marker of osteoblast differentiation (Chen et al. 1992), was expressed in osteoblasts of bone collars and bone trabeculae. In mutant E15.5 limbs, no expression of Runx2 and Bsp was detectable in limb mesenchymal tissues (Fig. 3B). We conclude that no osteoblasts were present in Sox9 mutant limbs.
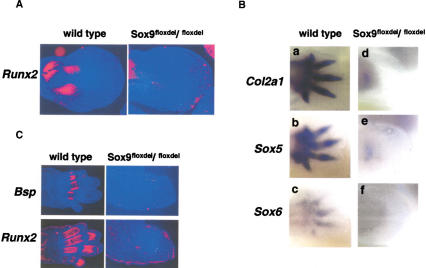
Figure 3.
Expression of early markers of osteoblast differentiation and chondrogenic mesenchymal condensations in limb buds of Sox9flox/flox; Prx1–Cre mice. (A) The limb buds of E12.5 wild-type and mutant embryos were hybridized with the Runx2 probe. Runx2 RNA was not detected in the mutant limb buds. (B) The limb buds of E15.5 wild-type and mutant embryos were hybridized with Bsp or Runx2 probes. Bsp and Runx2 transcripts were not detectable in mutant limb mesenchymal tissues. (C) Whole-mount RNA in situ hybridization showed lack of expression of Col2a1 (a,d), Sox5 (b,e), and Sox6 (c,f) in E12.5 mutant limb buds.
Complete inhibition of Sox5 and Sox6 in limb buds of Sox9flox/flox; Prx1–Cre mutant embryos
To characterize the complete defect in chondrogenic mesenchymal condensations in mutant mice, we analyzed the expression of genes known to be expressed in condensed mesenchymal cells. Sox5 and Sox6 are coexpressed with Sox9 during chondrogenesis starting before mesenchymal condensation, and their gene products are required for overt chondrocyte differentiation (Lefebvre et al. 1998; Smits et al. 2001). Expression of Sox5 and Sox6, as well as of Col2a1, an early marker of chondrogenic cells, was essentially abolished in the mutant limb buds (Fig. 3C). This indicated that Sox9 is needed for the expression of Sox5 and Sox6.
No evidence for patterning defects in limb buds of Sox9flox/flox; Prx1–Cre mutant embryos
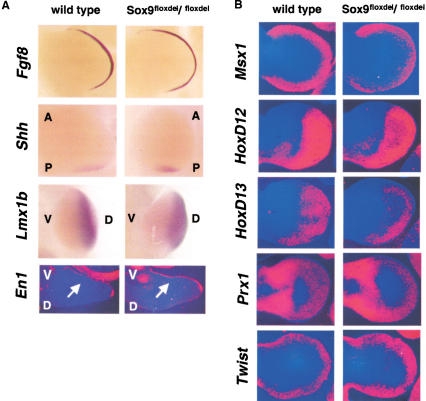
To determine whether patterning defects occurred in Sox9-deficient limb buds, we examined the expression of a series of marker genes characteristic of each of the vertebrate limb axes (Cohn and Tickle 1996; Capdevila and Belmonte 2001). Expression of fibroblast growth factor 8 (Ffg8), a marker of the apical ectodermal ridge, was identical in wild-type and mutant E12.5 limb buds (Fig. 4A). Likewise, expression of Sonic hedgehog (Shh), a posterior marker, was very similar in wild-type and mutant limb buds. Expression of the dorsal marker Lmx1b and that of the ventral marker En1 were also comparable in wild-type and mutant limb buds. In addition, expression of Msx1, HoxD12, HoxD13, Prx1, and Twist were essentially identical in wild-type and mutant limb buds (Fig. 4B). Thus, the absence of Sox9 in E12.5 mutant limb buds did not cause misexpression of a series of limb patterning genes, further supporting the conclusion that Sox9 is a true differentiation, not a patterning factor.
Figure 4.
Expression of markers of limb patterning in Sox9flox/flox; Prx1–Cre mice. The normal expression in limb buds of E11.5 (A) and E12.5 (B) mutant embryos of Fgf8, Shh, Lmx1b, En1, Msx1, HoxD12, HoxD13, Prx1, and Twist revealed no patterning defects in the mutant limb buds. A, anterior; P, posterior; V, ventral; D, dorsal. The arrows indicate the expression domain of En1.
Sox9 is necessary for survival of mesenchymal cells in limb buds of Sox9flox/flox; Prx1–Cre mutant embryos
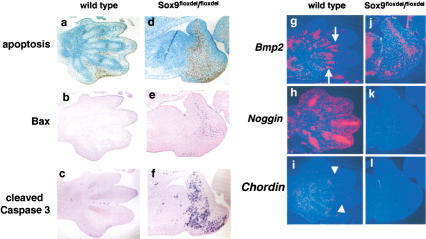
The severe malformation and the arrest of outgrowth of mutant limb buds could have been due to an increase in apoptosis within the population of mesenchymal cells. To test this hypothesis, we performed TUNEL assays in E12.5 and E13.5 limb buds. In the limb buds of wild-type embryos, no apoptotic cells were seen at 12.5 dpc (data not shown), but starting at 13.5 dpc, apoptosis was observed in the interdigital mesenchymal cells (Fig. 5a). In the mutant limb buds, no apoptosis was observed at 12.5 dpc, but extensive apoptosis was detected in mesenchymal cells at 13.5 dpc, in the area in which wild-type embryo chondrogenic mesenchymal condensations differentiate to form cartilage (Fig. 5d). Furthermore, immunohistochemistry showed that the expression of Bax was dramatically up-regulated in mutant limb buds and indicated large accumulation of cleaved caspase 3 in this domain of the mutant limb buds (Fig. 5b,c,e,f). These observations suggested that in the absence of chondrogenesis Sox9−/− mesenchymal cells do not survive in the distal part of limb buds. Accumulating evidence shows that bone morphogenetic protein (BMP) signals induce, and BMP antagonists inhibit cell apoptosis in the interdigital spaces (Ganan et al. 1996; Zou and Niswander 1996; Merino et al. 1999a). In E13.5 wild-type limb buds, Bmp2 was expressed in condensed cells, perichondrium, and mesenchymal cells in the vicinity of interdigital spaces (Fig. 5g). Noggin and Chordin, BMP antagonists, were expressed in chondrocytes and perichondrium of the digits, respectively (Fig. 5h,i). In contrast, in the mutant limb buds, Bmp2 was expressed in a broad band of mesenchymal cells that paralleled the broad band of apoptotic cells (Fig. 5j), but expression of Noggin and Chordin was abolished (Fig. 5k,l). These results indicate that Sox9 controls, either directly or indirectly, expression of Noggin and Chordin, and provide evidence for the hypothesis that Sox9−/− mesenchymal cells are not protected by Noggin and Chordin from apoptotic signals generated by BMPs.
Figure 5.
Analysis of apoptosis in Sox9flox/flox; Prx1–Cre mice. TUNEL (a,d), immunohistochemistry of Bax (b,e) and cleaved caspase 3 (c,f), and in situ hybridization of Bmp2 (g,j), Noggin (h,k), and Chordin (i,l) in limb buds of E13.5 wild-type and mutant embryos, respectively. The arrows indicate the expression of Bmp2 in the vicinity of interdigital spaces. The arrowheads indicate the expression of Chordin.
Severe chondrodysplasia in Sox9flox/flox; Col2a1–Cre mutant embryos
Inactivation of Sox9 by Prx1–Cre-mediated recombination, which took place in undifferentiated limb bud mesenchymal cells before chondrogenic mesenchymal condensation occurred, revealed Sox9's essential roles in formation of chondrogenic mesenchymal condensations and in cell survival. However, the question remains whether Sox9 is necessary for overt chondrocytic differentiation after formation of chondrogenic mesenchymal condensations. To address this question, we inactivated the Sox9 gene in condensed mesenchymal cells and differentiated chondrocytes using Col2a1–Cre transgenic mice. Only a few heterozygous Sox9flox/wt; Col2a1–Cre mice survived beyond 10 d after birth and were able to mate. Very few newborn homozygous Sox9flox/flox; Col2a1–Cre mice were found, indicating that embryos died before birth and were resorbed. However, until 16.5 dpc, Sox9flox/flox; Col2a1–Cre mutant embryos were recovered at the expected Mendelian ratio. E12.5 mutants were undistinguishable from wild-type embryos (data not shown). However, from 14.5 dpc on, the mutants were easily distinguished, as they featured a round head with a short snout, short trunk, tail, and limbs, and a prominent abdomen, a phenotype that was very similar to that of the Sox5; Sox6 double-null mutant embryos (Fig. 6A) (Smits et al. 2001). Sox9flox/flox; Col2a1–Cre mutant embryos were characterized by a very severe and generalized chondrodysplasia (Fig. 6B). Alcian blue and alizarin red staining of skeletal preparations of E17.5 mutant embryos indicated that all of the skeletal elements formed by endochondral bone formation, including ribs, limbs, and vertebrae, were extremely small. Alcian blue staining was severely reduced, indicating that cartilage formation was markedly defective. In contrast, skeletal elements formed by intramembranous ossification lacked significant abnormality (Fig. 6B).
Figure 6.
Severe, generalized chondrodysplasia in Sox9flox/flox; Col2a1–Cre mice. (A) Gross appearance of E18.5 embryos. (B) Skeletons of E17.5 mice stained by alcian blue followed by alizarin red. Mutant embryos were characterized by a very severe and generalized chondrodysplasia. (C) Expression of Sox9 protein in metacarpals of E12.5, E13.5, and E15.5 wild-type and mutant embryos, respectively. The inactivation of Sox9 occurred over a short window of time starting in condensed mesenchymal cells.
To assess the onset and efficiency of Cre-mediated gene deletion during limb bud development in Sox9flox/flox; Col2a1–Cre embryos, we examined Sox9 expression in the limb buds by immunohistochemistry using anti-Sox9 antibody. In wild-type embryos, Sox9 is expressed in condensed mesenchymal cells and then in the different chondrocytic cellular layers of the cartilage growth plate, except in hypertrophic chondrocytes, in which its expression is shut off. As shown in Figure 6C, in E12.5 mutant metacarpals, Sox9 was expressed in condensed mesenchymal cells. From 13.5 dpc, the number of cells expressing Sox9 was significantly less than in wild-type metacarpals, and at 15.5 dpc, only a few cells expressed lower levels of Sox9 in the mutant, a result that was consistent with results of RNA in situ hybridization (data not shown). Thus, the inactivation of Sox9 occurred over a short window of time starting in condensed mesenchymal cells.
Histological analysis of Sox9flox/flox; Col2a1–Cre mutant embryos
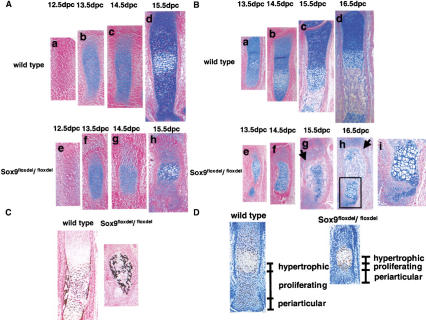
Figure 7 shows the progression of chondrocyte differentiation and bone formation for two representative skeletal elements, metacarpals and radius. In E12.5, Sox9flox/flox; Col2a1–Cre metacarpals, chondrogenic mesenchymal condensations occurred normally (Fig. 7A, a,e). At 13.5 and14.5 dpc, when deletion of the Sox9 genes was proceeding, Sox9−/− mutant cells remained mainly as condensed cells (Fig. 7A, b,c,f,g). Until 14.5 dpc, most chondrogenic cells accumulated little alcian blue-stainable extracellular matrix. At 15.5 dpc, hypertrophic chondrocytes appeared in the mid portion of metacarpals, but no orderly columns of proliferating chondrocytes were observed (Fig. 7A, d,h). The size of the metacarpals was also much reduced compared with wild-type metacarpals. In E13.5 and E14.5 mutant radius, cells with the aspect of hypertrophic chondrocytes surrounded by alcian blue-stainable extracellular matrix were also detected, although many cells remained in a condensed mesenchyme, indicating that some Sox9-expressing cells had entered the chondrocytic differentiation pathway. Again, in contrast to wild-type skeletal elements, the orderly parallel columnar structures of proliferating chondrocytes were not observed (Fig. 7B). Occasionally, small clusters of cells resembling proliferating chondrocytes were detected (Fig. 7B, g,h,i). In addition, bone formation took place, although it produced very deformed skeletal elements, a consequence of the deformity of cartilage primordia. Formation of bone trabeculae indicated that osteoblast differentiation occurred. In addition, mineralization took place in the malformed skeletal elements (Fig. 7C). We also noted that in the mutants, a thin layer of periosteum-like cells formed around the zone containing bone trabeculae and hypertrophic chondrocytes, separating this zone from the area of mesenchymal condensations (Fig. 7B, g,h).
Figure 7.
Histological analysis in Sox9flox/flox; Col2a1–Cre mice. (A,B) Alcian blue and nuclear fast-red staining of metacarpals (A) and radius (B) during chondrocytic differentiation showed abnormal endochondral bone formation. The arrows indicate a thin layer of periosteum-like cells surrounding bone trabeculae and hypertrophic chondrocytes (B, g,h). A boxed region in B, h is shown at a high magnification as B, i. (C) Staining by von Kossa's method visualized mineral deposition in radius of E15.5 wild-type and mutant mice. (D) PCNA staining showed only a few PCNA-positive cells (brown nuclei) in metacarpals of E15.5 mutant embryos.
Proliferative cell nuclear antigen (PCNA), a good marker for proliferating chondrocytes, was abundantly positive at 16.5 dpc in periarticular and columnar chondrocytes of wild-type growth plates. In contrast, in mutant growth plates, periarticular cells were PCNA negative, and only a few cells adjacent to hypertrophic chondrocytes were PCNA positive, indicating that the size of proliferating zones was dramatically reduced (Fig. 7D).
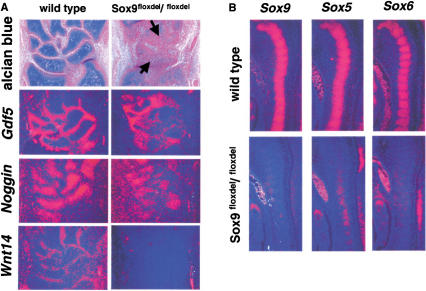
Formation of almost all joints was markedly delayed in the mutants; this was very striking in the carpal area, in which most carpal skeletal elements were still fused together at 15.5 dpc (Fig. 8A). In the mutant limbs, the expression of Noggin, which is required for the formation of articular joints (Brunet et al. 1998), was dramatically reduced, and that of Gdf5 and Wnt14, markers of joint formation (Storm and Kingsley 1996; Hartmann and Tabin 2001), was also down-regulated, suggesting that Sox9 is involved, directly or indirectly, in joint formation.
Figure 8.
In situ hybridization analyses of markers of joint formation and of downstream genes of Sox9 in chondrocytes. (A) Alcian blue and nuclear fast red staining and RNA in situ hybridization of Gdf5, Noggin, and Wnt14 in carpal cartilage primordia of E15.5 wild-type and mutant mice. The arrows indicate fusion of carpal bones. (B) Sox5 and Sox6 are downstream genes of Sox9 in chondrocytes. RNA in situ hybridization showed absence of expression of Sox5 and Sox6 in chondrocytes of E12.5 Sox9flox/flox; Col2a1–Cre mice.
Severe down-regulation of Sox5 and Sox6 expression in Sox9flox/flox; Col2a1–Cre mutant embryos
During chondrogenesis, Sox5 and Sox6 play essential roles during overt chondrocytic differentiation after mesenchymal condensation (Smits et al. 2001). Inactivation of the Sox9 gene in limb buds before chondrogenic mesenchymal condensation abolished Sox5 and Sox6 expression (Fig. 3C). Here, we examined whether Sox5 and Sox6 expression was also down-regulated when the Sox9 alleles were inactivated after mesenchymal condensations had been established. In E12.5 wild-type embryos, cells in sclerotomes overtly differentiated into chondrocytes and expressed high levels of Sox9, Sox5, and Sox6 (Fig. 8B). In contrast, cells in sclerotomes of mutant mice showed a dramatic decrease in transcript levels, demonstrating that Sox5 and Sox6 are genetically downstream of Sox9 in chondrocytes.
In Sox5; Sox6 double-null mutant embryos, genes for cartilage extracellular matrix components, including Col2a1, Aggrecan, and Cartilage oligometric protein (Comp), were markedly down-regulated. In metacarpals of Sox9flox/flox; Col2a1–Cre mutants, these genes were also severely down-regulated (Fig. 9A). Some cells adjacent to and in the zone of hypertrophic chondrocytes expressed these marker genes in mutant embryos, probably as a consequence of (1) the delayed inactivation of the Sox9 alleles in some chondrogenic cells, and (2) the relatively long half-life of the mRNAs for these genes. In contrast, in E15.5 mutants, the levels of Col10a1 expression were comparable with those in wild-type embryos, consistent with the presence of cells that have the aspect of hypertrophic chondrocytes in the mutant embryos.
Figure 9.
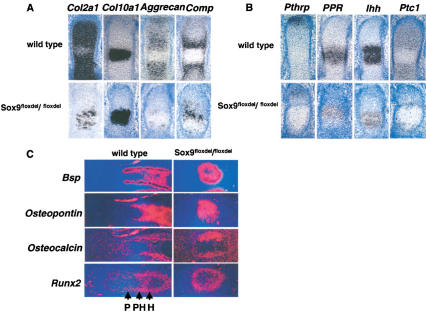
Expression of markers of chondrocytes, Ihh/Pthrp regulatory signaling molecules, and markers of osteoblast differentiation in Sox9flox/flox; Col2a1–Cre mice. (A) Expression of Col2a1, Aggrecan, and Comp, but not Col10a1 was dramatically down-regulated in E15.5 mutant metacarpals. (B) RNA in situ hybridization showed down-regulation of Pthrp, PPR, Ihh, and Ptc1 in E15.5 mutant metacarpals. (C) Expression in radius of E15.5 mutant mice of Bsp, Osteopontin, Osteocalcin, and Runx2 revealed normal osteoblast differentiation. P, proliferating; PH, prehypertrophic; H, hypertrophic.
Reduced signaling by the Indian hedgehog (Ihh)-Parathyroid hormone-related peptide (Pthrp) loop in Sox9flox/flox; Col2a1–Cre mutant embryos
Ihh and Pthrp control the rate of chondrocyte proliferation and the transition of these cells to hypertrophy (Vortkamp et al. 1996); moreover, Ihh coordinates these processes with the onset of osteogenesis in bone collars (St-Jacques et al. 1999). Figure 9B shows that in E15.5 mutant metacarpal skeletal elements, Pthrp receptor (PPR) and Ihh, which are expressed in prehypertrophic chondrocytes (Vortkamp et al. 1996), were down-regulated. Patched 1 (Ptc1), a transcriptional target of Ihh signaling expressed in proliferating chondrocytes and osteoblasts of bone collars (Vortkamp et al. 1996), was expressed only in a small number of cells in the mutant growth plates. Expression of Pthrp, another target of Ihh signaling, which is expressed in periarticular cells (Vortkamp et al. 1996), was also clearly down-regulated. Thus, transcripts for Ihh, PPR, Ptc1, and Pthrp were present at lower levels in the mutants than in the wild-type embryos. Lower levels of Ihh expression probably contributed to the severe decrease in cell proliferation, whereas the reduced levels of Pthrp may have contributed to an accelerated transition to hypertrophy.
No inhibition of osteoblast differentiation in Sox9flox/flox; Col2a1–Cre mutant embryos
Figure 9C shows that the levels of expression of Runx2 in E15.5 mutant radius were comparable with those in wild-type embryos, as was the case for Bsp and Osteopontin. Moreover, Osteocalcin expression, which is just initiated at this stage in wild-type embryos, was also present in the mutants. We conclude that although the skeletal elements were extremely deformed, the process of osteoblast differentiation occurred normally in the mutants.
Discussion
Sox9 is essential for cartilage and bone formation in limb buds
After the onset of limb bud development, Sox9 expression is induced in a subpopulation of undifferentiated pluripotent mesenchymal cells. These Sox9-expressing cells then undergo a process of cell aggregation, followed by overt differentiation into chondrocytes. Inactivation of Sox9 in early limb buds, before condensations appeared, produced very short deformed limbs and resulted in a complete lack of chondrogenic mesenchymal condensations, subsequent cartilage formation, and bone development. The absence of Sox9 in limb buds did not, however, produce patterning defects. The patterns of expression of markers that characterize the limb bud axes were essentially identical to those of wild-type embryos. In addition, the patterns of expression of Msx1, Twist, Hoxd12, Hoxd13, and Prx1 in E12.5 mutant limb buds were all similar to those in wild-type embryos. Moreover, characteristic footpads that are normally present on the ventral side of limbs were observed in E15.5 mutants.
Inactivation of Sox9 in limb buds both before and after mesenchymal condensations abolished expression of Sox5 and Sox6, indicating that Sox9 was required for expression of these two related Sox genes. In addition, inactivation of Sox9 before chondrogenic mesenchymal condensations produced no transcripts for Runx2, which is ordinarily expressed at late stages of condensations and is required for osteoblast differentiation (Otto et al. 1997). This indicates that Sox9 is needed for Runx2 expression in early limb buds. However, if Sox9 was inactivated after mesenchymal condensations, Runx2 expression and osteoblast differentiation took place. Thus, in the absence of Sox9 in early limb buds, there are no mesenchymal condensations, no chondrocyte differentiation, no establishing of the osteoblast lineage, and no osteoblast differentiation. These results are consistent with the hypothesis that in endochondral skeletal elements, Sox9 is needed to establish osteochondroprogenitor cells that will produce both chondrocytes and osteoblasts. We speculate that in endochondral skeletal elements, chondrocyte and osteoblast progenitors begin to segregate from each other at the stage of mesenchymal condensation. Once the mesenchymal condensations were established, inactivation of Sox9 did not prevent osteoblast differentiation. We have no evidence to indicate that either Sox5 and Sox6 or Runx2 are direct targets of Sox9.
Sox9 regulates cell survival of limb bud mesenchyme
In wild-type E13.5 embryos, mesenchymal cells in interdigital spaces undergo apoptosis. Inactivation of Sox9 in Sox9flox/flox; Prx1–Cre limb mesenchyme resulted in markedly increased apoptosis, an increased expression of Bax, and an increase in cleaved caspase 3 production. This cell death presumably contributed to the severe deformity of the mutant limbs. Cells within chondrogenic mesenchymal condensations and differentiating cartilages are unaffected by the apoptotic signals, which cause formation of interdigital spaces in wild-type limb buds. Our hypothesis is that the broad uninterrupted domain of apoptosis in Sox9 mutant limb buds is due to normal proapoptotic signals, which, in wild-type limb buds, generate the interdigital spaces. Previous studies strongly suggest that BMPs have a proapoptotic activity in interdigital mesenchymal cells. BMP-beads implanted in chick embryonic limbs accelerate interdigital apoptosis (Ganan et al. 1996), whereas expression of a dominant-negative type I BMP receptor or exogenous administration of recombinant Gremlin, a BMP antagonist, reduces interdigital apoptosis (Zou and Niswander 1996; Merino et al. 1999b). The broad band of Bmp2 expression, which parallels the broad band of apoptosis in the mutant limb buds, strongly supports the view that BMPs have a proapoptotic activity. In addition, the absence of Noggin and Chordin expression also provides indirect evidence that in wild-type limb buds, BMP antagonists expressed in digit chondrocytes protect the cells in and around the cartilages from proapoptotic BMP signals. Apoptosis in Sox9 null mutant limb buds is unlikely to be an inherent property of Sox9−/− cells, which would result from an inability to differentiate. In limbs of chimeric embryos derived from Sox9−/− ES cells, the mutant cells survive (Bi et al. 1999).
Absence of mesenchymal condensations in Sox9 null mutants
The molecular mechanisms underlying the major cellular event of chondrogenic mesenchymal condensation remain largely unknown. A number of patterning factors must act at this step, as chondrogenic mesenchymal condensations prefigure the shape of future skeletal elements. In addition, in vitro experiments have suggested that several other molecules, including cell-adhesion molecules, are involved in this process (Hall and Miyake 2000). We have found that expression levels of N-cadherin and NCAM in E12.5 mutant limb buds are comparable with those in wild-type limb buds (H. Akiyama, unpubl.). These results indicate that Sox9 is not needed for expression of N-cadherin and NCAM, which have been proposed previously to mediate cell adhesion of chondrogenic mesenchymal cells (Widelitz et al. 1993; Oberlander and Tuan 1994). Although Sox9 was reported recently to specifically bind an N-cadherin promoter region and to increase its activity in vitro (Panda et al. 2001), other transcription factors must control the levels of N-cadherin in vivo. The absence of alcian blue-stainable cartilage nodules in an in vitro high-density micromass culture using E12.5 mutant limb mesenchymal cells (Fig. 2E) indicates that even if conditions of cell–cell interactions exist in the micromass culture, Sox9−/− cells are unable to differentiate. We hypothesize that Sox9 controls expression of the cell–cell interaction machinery by a mechanism that does not involve changes in expression of N-cadherin and NCAM. Perhaps Sox9 controls expression of other genes needed for mesenchymal condensations.
Sox9 is needed for chondrocyte differentiation after mesenchymal condensation
During endochondral bone formation in wild-type embryos, condensed mesenchymal cells differentiate into chondrocytes, and then these cells proliferate unidirectionally, forming parallel columns characteristic of cartilage growth plates. In E12.5 homozygous Sox9flox/flox embryos harboring the Col2a1–Cre transgene, chondrogenic mesenchymal condensations were normal and the levels of Sox9 in the cells of these condensations were comparable with those in wild-type digits. However, in E13.5 mutants, the number of Sox9-positive cells was markedly reduced, and by 15.5 dpc, almost all cells had become negative for Sox9. Thus, the Sox9flox alleles were inactivated over a window of time beginning at or immediately after the stage of chondrogenic mesenchymal condensations. The very severe chondrodysplasia and dwarfism of the mutants, which were the result of this inactivation, were characterized by an almost complete absence of cartilage in the endochondral skeleton. This was in contrast with a completely normal membranous skeleton. In the mutant endochondral skeletal elements, the majority of cells remained in mesenchymal condensations, and no parallel columns of proliferating chondrocytes were present. The severe decrease in cell proliferation was further demonstrated by the marked reduction in PCNA staining. Sox9 might either control genes needed for cell proliferation or, alternatively, the decrease in cell proliferation could be a consequence of the lack of cartilage extracellular matrix components. However, hypertrophic chondrocytes, which expressed high levels of Col10a1, were often present in the center of skeletal elements. These results are consistent with the following interpretation: if inactivation of Sox9 occurred in mesenchymal condensations, cells remained at this stage of differentiation, failing to produce a characteristic cartilage extracellular matrix. Those cells in which the Sox9 genes were not inactivated in the condensations, proceeded into the chondrocyte differentiation program. However, as the Sox9 genes were promptly deleted, cells rapidly became hypertrophic. On the basis of this interpretation, our results indicate that Sox9 is required for overt chondrocyte differentiation and for subsequent chondrocyte proliferation. The severe down-regulation of chondrocyte marker genes including Col2a1, Aggrecan, and Comp are in agreement with this conclusion. We propose that Sox9 also prevents conversion of proliferating chondrocytes into hypertrophic chondrocytes. This hypothesis is supported by several lines of evidence. In the wild-type growth plate, expression of Sox9 is abolished in hypertrophic chondrocytes; in addition, the levels of Sox9 are higher in the prehypertrophic zone than in the proliferative zone. In Sox9 heterozygous mice, the zone of hypertrophic chondrocytes was wider than in wild-type embryos, consistent with the notion that the levels of Sox9 needed to inhibit hypertrophic chondrocyte maturation are not attained in the prehypertrophic cells of Sox9 heterozygous mutants (Bi et al. 2001). The presence of abundant hypertrophic chondrocytes in the Sox9flox/flox; Col2a1–Cre mutants is consistent with our hypothesis that Sox9 prevents transition into hypertrophic chondrocytes. Recent preliminary experiments that generated embryos in which Sox9 was overexpressed in chondrocytes indicated that hypertrophic chondrocyte differentiation was delayed in these embryos (H. Akiyama, unpubl.).
The rate of chondrogenic differentiation in the cartilage growth plate as well as the timing and location of osteogenesis initiation in the perichondrium/periosteum are controlled by the Ihh/Pthrp regulatory loop (Vortkamp et al. 1996). Ihh, which is expressed mainly in prehypertrophic chondrocytes and in the immediately adjacent zone of hypertrophic chondrocytes, stimulates chondrocyte proliferation in the growth plate and osteogenesis in the periosteum (St-Jacques et al. 1999); in addition, it induces Pthrp expression in periarticular chondrocytes. Pthrp and its receptor PPR, which is expressed at high levels in prehypertrophic chondrocytes (Karaplis et al. 1994; Lanske et al. 1996), regulate the switch from a proliferative to a postproliferative state by delaying transition of chondrocytes into hypertrophy. In Sox9flox/flox; Col2a1–Cre mice, the levels of expression of Ihh and its two downstream target genes Pthrp and Ptc1, as well as PPR, were down-regulated. Our results are consistent with the notion that in the mutant mice, the zone of prehypertrophic chondrocytes is markedly reduced. We believe that the resulting down-regulation of Ihh and PPR and of the Ihh/Pthrp signaling loop is an indirect consequence of the absence of Sox9. The expression of Ptc1, even in a narrow range, indicated that Ihh signaling was functioning in the mutant mice. In addition, although bone collar formation was delayed and disorganized, cartilage replacement by bone occurred normally. Perhaps the lower levels of Ihh in the Sox9flox/flox; Col2a1–Cre are sufficient to induce osteogenesis but insufficient to stimulate normal chondrocyte proliferation.
Joint formation is initiated by the arrest of chondrogenic differentiation in the prospective joint interzone. Wnt14 induces the expression of Gdf5 and Chordin in this distinct zone, and regulates the subsequent processes of joint formation (Hartmann and Tabin 2001). Noggin is expressed in differentiated chondrocytes. Noggin-deficient mice do not express Gdf5 and fail to initiate joint formation (Brunet et al. 1998), indicating that Noggin is needed for joint formation after chondrogenic mesenchymal condensation. In Sox9flox/flox; Col2a1–Cre mice, joint formation does not take place properly, and mesenchymal condensations remain partly fused to each other. Furthermore, expression of Noggin and Gdf5 is down-regulated and that of Wnt14 is abolished. One possible explanation is that the severe chondrodysplasia of Sox9flox/flox; Col2a1–Cre embryos results in a low expression of Noggin, which, in turn, would inhibit expression of Gdf5 and Wnt14. The virtual lack of Wnt14 expression raises the possibility that Sox9 may regulate the expression of other signaling molecules controlling Wnt14 expression.
Sox9flox/flox; Col2a-1Cre and Sox5; Sox6 double-null mutants have similar phenotypes
The severe generalized chondrodysplasia of Sox9flox/flox; Col2a1–Cre mice is very similar to the phenotype of Sox5; Sox6 double-null mutant mice (Smits et al. 2001). Whereas chondrocytes in Sox5; Sox6 double-null mutant mice expressed Sox9 at levels comparable with those in wild-type chondrocytes, in Sox9flox/flox; Col2a-1Cre mutants, the levels of Sox5 and Sox6 expression were virtually abolished. Hence, the chondrodysplasia phenotype of Sox9 mutants can, in large part, be accounted for by the severe down-regulation of Sox5 and Sox6. Previous in vitro results have shown that Sox9, Sox5, and Sox6 cooperate with each other in activating the Col2a1 gene (Lefebvre et al. 1998). Hence, we believe that in addition to being required for expression of Sox5 and Sox6, Sox9 has itself an important role in the activation of cartilage matrix genes. The low expression levels of cartilage matrix genes, and the resumption of proliferation after a long delay in Sox5; Sox6 double-null mutant chondrocytes, are likely due to the presence of normal levels of Sox9.
Conclusion
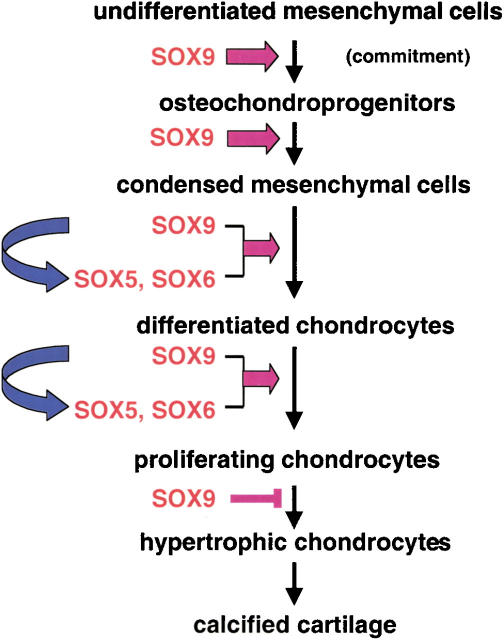
Sox9 is required in several successive steps of the chondrocyte differentiation pathway during endochondral bone formation in vivo and for the proper progression of cells through the sequential steps of this process. Figure 10 presents a model that summarizes several of these different functions. We have presented evidence that Sox9 is needed for the commitment of undifferentiated mesenchymal cells to a cell type that is both a chondroprogenitor and an osteoprogenitor. Sox9 is further required for the establishment of chondrogenic mesenchymal condensations. At this stage, Sox9 also controls antiapoptotic molecules such as Noggin and Chordin that inhibit signals responsible for formation of interdigital spaces. Later, Sox9 is needed for overt differentiation of chondrocytes, for the proliferation of these cells, for the establishment of parallel columns of proliferating chondrocytes in the growth plates of endochondral skeletal elements, and for proper joint formation. Finally, Sox9 also inhibits the transition of chondrocyte into hypertrophic chondrocytes and, hence, controls subsequent endochondral ossification.
Figure 10.
Functions of Sox9 in the successive steps of the chondrocyte differentiation pathway during endochondral bone formation. Sox9 commits undifferentiated mesenchymal cells in the lateral plate mesoderm to osteochondroprogenitors. Sox9 is also needed for chondrogenic mesenchymal condensation, subsequent overt chondrocyte differentiation, and normal chondrocyte proliferation. Overt chondrocyte differentiation and normal chondrocyte proliferation are at least in part mediated by Sox5 and Sox6, the expression of which requires Sox9. Finally, Sox9 inhibits transition of proliferating chondrocytes to hypertrophy. It is also possible that Sox5 and Sox6 participate in inhibiting this last step.
Materials and methods
Introduction of loxP sites into the Sox9 gene and generation of Sox9flox/wt and Sox9flox/flox; Cre mice
A Sox9 clone was isolated from a mouse 129/Sv genomic DNA library. To construct the targeting vector, a 5.5-kb XbaI fragment covering most of the Sox9 gene was subcloned, a loxP sequence (5′-ATAACTTCGTATAGCATACATTATACGAA GTTATGGCCCG-3′) inserted into the EagI site in intron 1, and the fragment extended by adding 2.7 kb of upstream sequences (PstI–XbaI fragment). For the short vector arm, a 1.2-kb fragment starting from the SspI site after the Sox9 polyadenylation signal was subcloned after a second loxP site, and the resulting construct fused with the 5′ vector arm. Finally, an Oct3/4neo cassette was inserted upstream of the second loxP site. Because the Oct3/4 promoter is only expressed in undifferentiated ES cells, applying neomycin selection should result in an enrichment of undifferentiated ES cell clones. 129/SvEv ES clones harboring a Sox9 floxed heterozygous mutation were screened by PCR and confirmed by Southern blot analysis with a Sox9 probe located outside of the homology regions used for gene recombination. Mouse chimeras were generated by C57BL/6 host blastocyst injection of mutant ES cell clones, and chimeras obtained were bred with C57BL/6 mice to Sox9-floxed heterozygous mice. Sox9-floxed heterozygous mice were bred to homozygosity.
In a first cross, Prx1–Cre or Col2a1–Cre transgenic mice (Martin and Olson 2000; Ovchinnikov et al. 2000) were mated with mice heterozygous for the Sox9-floxed allele. The offspring inheriting either Prx1–Cre or Col2a1–Cre and a floxed allele were then mated with Sox9-floxed heterozygous mice to obtain embryos harboring either the Prx1–Cre or Col2a1–Cre transgene together with two Sox9-floxed alleles. Routine mouse genotyping was performed by PCR. The following primer pairs were used for Cre and floxdel alleles: 5′-TCCAATTTACTGACCG TACACCAA-3′ and 5′-CCTGATCCTGGCAATTTCGGCTA3′; 5′-GTCAAGCGACCCATGAACGC-3′ and 5′-TGGTAAT GAGTCATACACAGTAC-3′, respectively.
Histological analysis
Alcian blue and alizarin red staining of skeleton and scanning electron microscope analysis were performed as described previously (Hogan et al. 1994; Chen and Behringer 1995). For the histological analysis, embryos were fixed with 4% paraformaldehyde and embedded in paraffin. Sections of 7 μm were stained with hematoxylin and Treosin, with alcian blue and nuclear fast red, or with the von Kossa reaction and nuclear fast red. Immunohistochemical staining was performed by using peroxidase chromogens (Zymed)/TrueBlue substrate (KPL). The following antibodies were used: rabbit polyclonal anti-Sox9 (1:150), rabbit polyclonal anti-Bax (Santa Cruz;1:200), and rabbit polyclonal anti-cleaved caspase-3 (Cell Signaling Technology; 1:100). Cell proliferation analysis and TUNEL analysis were performed on paraffin-embedded sections using PCNA Staining Kit (Zymed) and ApopTag Plus peroxidase in situ apoptosis detection kit (Intergen) following the manufacturers' protocols. RNA in situ hybridization analysis was carried out as described previously (Conlon and Rossant 1992; Albrecht et al. 1997). Pictures of hybridization signals were taken with a red filter and superimposed with blue fluorescence images of cell nuclei stained with Hoechst 33258 dye.
Micromass culture
For micromass culture, limb buds from E12.5 were isolated and were digested in 0.1% collagenase and 0.1% trypsin for 30 min at 37°C. Cell suspension was placed in DMEM, 10% FCS in 20-μL drops at 2 × 107 cells/mL in 12-well plates. The cells were allowed to attach for 1 h, and then overlaid with 2 mL of DMEM, 10%FCS. After a 7-d culture, micromass cultures were stained with alcian blue for chondrogenic nodules as described previously (Akiyama et al. 1996).
Acknowledgments
We thank Kenneth Dunner Jr. for scanning electron microscopy; Dmitry Ovchinnikov and Richard R. Behringer for Col2a1–Cre transgenic mice; Xiaohong Yang, Andreas Friese, and Danilo Landrock for technical support; and Veronique Lefebvre, Andrew P. McMahon, Randy L. Johnson, Yasuhide Furuta, Tamayuki Shinomura, Jacqueline T. Hecht, and Henry M. Kronenberg for plasmids and probes. We also thank Janie Finch for editorial assistance. This work was funded by NIH grants PO1 AR42919 to B.d.C.; The G. Harold & Leila Y. Mathers Charitable Foundation to B.d.C.; NIDCR grants R29 DE12324 and R01DE013509 to J.F.M.; Grant No. 5-FY00-135 from the March of Dimes to J.F.M.; and the Volkswagenstiftung (grant I/75662) to A.S. We also acknowledge NIH grant CA16672 for DNA sequence analysis and scanning electron microscope analysis.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Corresponding authors.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1017802.
References
- Ahdjoudj S, Lasmoles F, Oyajobi BO, Lomri A, Delannoy P, Marie PJ. Reciprocal control of osteoblast/chondroblast and osteoblast/adipocyte differentiation of multipotential clonal human marrow stromal F/STRO-1(+) cells. J Cell Biochem. 2001;81:23–38. doi: 10.1002/1097-4644(20010401)81:1<23::aid-jcb1021>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Akiyama H, Hiraki Y, Shigeno C, Kohno H, Shukunami C, Tsuboyama T, Kasai R, Suzuki F, Konishi J, Nakamura T. 1 α,25-dihydroxyvitamin D3 inhibits cell growth and chondrogenesis of a clonal mouse EC cell line, ATDC5. J Bone Miner Res. 1996;11:22–28. doi: 10.1002/jbmr.5650110105. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Eichele G, Helms JA, Lu HC. Visualization of gene expression patterns by in situ hybridization. In: Daston GP, editor. Molecular and cellular methods in developmental toxicology. Boca Raton, FL: CRC Press; 1997. pp. 23–48. [Google Scholar]
- Bell DM, Leung KK, Wheatley SC, Ng LJ, Zhou S, Ling KW, Sham MH, Koopman P, Tam PP, Cheah KS. SOX9 directly regulates the type-II collagen gene. Nat Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgewater LC, Lefebvre V, de Crombrugghe B. Chondrocyte-specific enhancer elements in the Col11a2 gene resemble the Col2a1 tissue-specific enhancer. J Biol Chem. 1998;273:14998–15006. doi: 10.1074/jbc.273.24.14998. [DOI] [PubMed] [Google Scholar]
- Brunet LJ, McMahon JA, McMahon AP, Harland RM. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 1998;280:1455–1457. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- Capdevila J, Belmonte JCI. Patterning mechanisms controlling vertebrate limb development. Annu Rev Cell Dev Biol. 2001;17:87–132. doi: 10.1146/annurev.cellbio.17.1.87. [DOI] [PubMed] [Google Scholar]
- Chen J, Shapiro HS, Sodek J. Development expression of bone sialoprotein mRNA in rat mineralized connective tissues. J Bone Miner Res. 1992;7:987–997. doi: 10.1002/jbmr.5650070816. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes & Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- Cohn MJ, Tickle C. Limbs: A model for pattern formation within the vertebrate body plan. Trends Genet. 1996;12:253–257. doi: 10.1016/0168-9525(96)10030-5. [DOI] [PubMed] [Google Scholar]
- Conlon RA, Rossant J. Exogenous retinoic acid rapidly induces anterior ectopic expression of murine Hox-2 genes in vivo. Development. 1992;116:357–368. doi: 10.1242/dev.116.2.357. [DOI] [PubMed] [Google Scholar]
- Foster JW, Dominguez-Steglich MA, Guioli S, Kowk G, Weller PA, Stevanovic M, Weissenbach J, Mansour S, Young ID, Goodfellow PN, et al. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- Ganan Y, Macias D, Duterque-Coquillaud M, Ros MA, Hurle JM. Role of TGF βs and BMPs as signals controlling the position of the digits and the areas of interdigital cell death in the developing chick limb autopod. Development. 1996;122:2349–2357. doi: 10.1242/dev.122.8.2349. [DOI] [PubMed] [Google Scholar]
- Grigoriadis AE, Heersche JN, Aubin JE. Differentiation of muscle, fat, cartilage, and bone from progenitor cells present in a bone-derived clonal cell population: Effect of dexamethasone. J Cell Biol. 1988;106:2139–2151. doi: 10.1083/jcb.106.6.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase β gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- Hall BK, Miyake T. All for one and one for all: Condensations and the initiation of skeletal development. BioEssays. 2000;22:138–147. doi: 10.1002/(SICI)1521-1878(200002)22:2<138::AID-BIES5>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Hartmann C, Tabin CJ. Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development. 2000;127:3141–3159. doi: 10.1242/dev.127.14.3141. [DOI] [PubMed] [Google Scholar]
- ————— Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell. 2001;104:341–351. doi: 10.1016/s0092-8674(01)00222-7. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lucy E. Manipulating the mouse embryo. 2nd Edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Karaplis AC, Luz A, Glowacki J, Bronson RT, Tybulewicz VL, Kronenberg HM, Mulligan RC. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes & Dev. 1994;8:277–289. doi: 10.1101/gad.8.3.277. [DOI] [PubMed] [Google Scholar]
- Kist R, Schrewe H, Balling R, Scherer G. Conditional inactivation of Sox9: A mouse model for campomelic dysplasia. Genesis. 2002;32:121–123. doi: 10.1002/gene.10050. [DOI] [PubMed] [Google Scholar]
- Lanske B, Karaplis AC, Lee K, Luz A, Vortkamp A, Pirro A, Karperien M, Defize LH, Ho C, Mulligan RC, et al. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science. 1996;273:663–666. doi: 10.1126/science.273.5275.663. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro α1; (II) collagen gene. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M, Martin JM, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre recombinase in the developing mouse limb bud driven by a Prx1 enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Martin JF, Olson EN. Identification of a prx1 limb enhancer. Genesis. 2000;26:225–229. [PubMed] [Google Scholar]
- Merino R, Ganan Y, Macias D, Rodriguez-Leon J, Hurle JM. Bone morphogenetic proteins regulate interdigital cell death in the avian embryo. Ann NY Acad Sci. 1999a;887:120–132. doi: 10.1111/j.1749-6632.1999.tb07927.x. [DOI] [PubMed] [Google Scholar]
- Merino R, Rodriguez-Leon J, Macias D, Ganan Y, Economides AN, Hurle JM. The BMP antagonist Gremlin regulates outgrowth, chondrogenesis and programmed cell death in the developing limb. Development. 1999b;126:5515–5522. doi: 10.1242/dev.126.23.5515. [DOI] [PubMed] [Google Scholar]
- Ng LJ, Wheatley S, Muscat GE, Conway-Campbell J, Bowles JE, Bell DM, Tam PP, Cheah KS, Koopman P. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- Oberlender SA, Tuan RS. Expression and functional involvement of N-cadherin in embryonic limb chondrogenesis. Development. 1994;120:177–187. doi: 10.1242/dev.120.1.177. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–146. [PubMed] [Google Scholar]
- Panda DK, Miao D, Lefebvre V, Hendy GN, Goltzman D. The transcription factor SOX9 regulates cell cycle and differentiation genes in chondrocytic CFK2 cells. J Biol Chem. 2001;276:41229–41236. doi: 10.1074/jbc.M104231200. [DOI] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic AcidsRes. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya I, Tsuji K, Koopman P, Watanabe H, Yamada Y, Shinomiya K, Nifuji A, Noda M. SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J Biol Chem. 2000;275:10738–10744. doi: 10.1074/jbc.275.15.10738. [DOI] [PubMed] [Google Scholar]
- Smits P, Li P, Mandel J, Zhang Z, Deng JM, Behringer RR, de Croumbrugghe B, Lefebvre V. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1:277–290. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes & Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Kingsley DM. Joint patterning defects caused by single and double mutations in members of the bone morphogenetic protein (BMP) family. Development. 1996;122:3969–3979. doi: 10.1242/dev.122.12.3969. [DOI] [PubMed] [Google Scholar]
- Vortkamp A. Interaction of growth factors regulating chondrocyte differentiation in the developing embryo. Osteoarthritis Cartilage. 2001;9:S109–S117. [PubMed] [Google Scholar]
- Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- Widelitz RB, Jiang TX, Murray BA, Chuong CM. Adhesion molecules in skeletogenesis: II. Neural cell adhesion molecules mediate precartilaginous mesenchymal condensations and enhance chondrogenesis. J Cell Physiol. 1993;156:399–411. doi: 10.1002/jcp.1041560224. [DOI] [PubMed] [Google Scholar]
- Xie WF, Zhang X, Sakano S, Lefebvre V, Sandell LJ. Trans-activation of the mouse cartilage-derived retinoic acid-sensitive protein gene by Sox9. J Bone Miner Res. 1999;14:757–763. doi: 10.1359/jbmr.1999.14.5.757. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Eberspaecher H, Lefebvre V, de Crombrugghe B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997;209:377–386. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Zou H, Niswander L. Requirement for BMP signaling in interdigital apoptosis and scale formation. Science. 1996;272:738–741. doi: 10.1126/science.272.5262.738. [DOI] [PubMed] [Google Scholar]