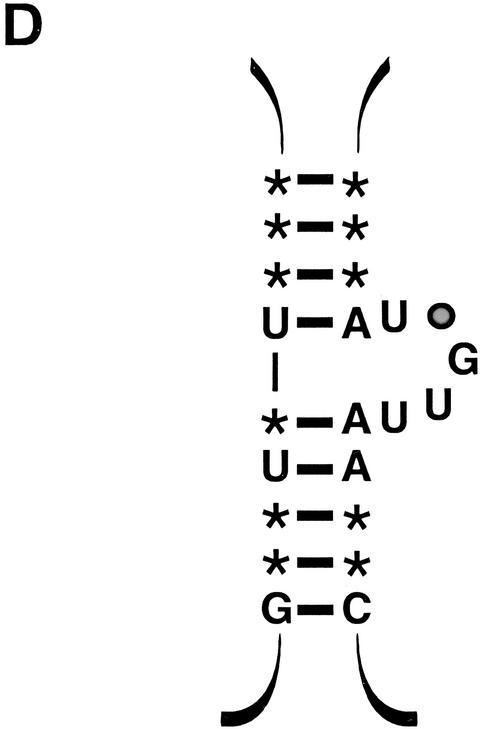Abstract
It is well established that the template for telomeric DNA synthesis is provided by the RNA subunit of telomerase; however, the additional functions provided by most of the rest of the RNA (>1000 nucleotides in budding yeast) are largely unknown. By alignment of telomerase RNAs of Saccharomyces cerevisiae and six Kluyveromyces species followed by mutagenesis of the S. cerevisiae RNA, we found a conserved region that is essential for telomere maintenance. Phylogenetic analysis and computer folding revealed that this region is conserved not only in primary nucleotide sequence but also in secondary structure. A common bulged-stem structure was predicted in all seven yeast species. Mutational analysis showed the structure to be essential for telomerase function. Suppression of bulged-stem mutant phenotypes by overexpression of Est1p and loss of co-immunoprecipitation of the mutant RNAs with Est1p indicated that this bulged stem is necessary for association of Est1p, a telomerase regulatory subunit. Est1p in yeast extract bound specifically to a small RNA containing the bulged stem, suggesting a direct interaction. We propose that this RNA structure links the enzymatic core of telomerase with Est1p, thereby allowing Est1p to recruit or activate telomerase at the telomere.
Keywords: Telomerase, RNA structure, TLC1, Est1p, yeast
RNA is an essential component of telomerase, the chromosome end-replicating enzyme (Greider and Blackburn 1989). Although a small portion of the RNA functions as the template for telomeric DNA synthesis, telomerase RNA subunits typically contain 150 to >1000 nucleotides in addition to their template. Some functions have been ascribed to these sequences. In one case, a long-range base-paired RNA structure has been shown to specify the template boundary (Tzfati et al. 2000). Other RNA structures or sequences are involved in the maturation and stability of the telomerase RNP (Mitchell et al. 1999; Seto et al. 1999; Teixeira et al. 2002), and a stem-loop of Saccharomyces cerevisiae telomerase RNA has been shown to have a genetic interaction with the DNA-repair protein Ku (Fig. 1A; Peterson et al. 2001). In addition, certain portions of telomerase RNAs have been implicated in the binding of core protein subunits (Fig. 1A; Licht and Collins 1999; Livengood et al. 2002), but it has proven difficult to identify specific structures of the type that are well known for RNA-protein interactions in other systems (for reviews, see Draper 1999; Leulliot and Varani 2001). In the present study, we describe the identification of an RNA bulged stem that is necessary for association of the telomerase regulatory protein Est1p with S. cerevisiae telomerase.
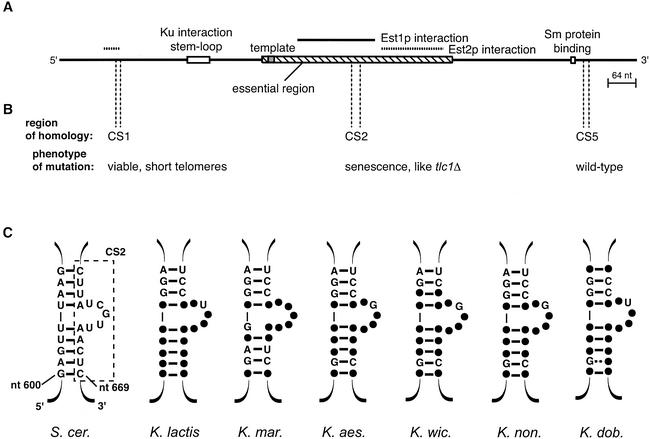
Figure 1.
Identification of a structural element conserved among budding yeast telomerase RNAs. (A) Location of previously characterized regions of the ∼1.3 kb TLC1 RNA. (B) Three regions of TLC1 RNA with homology with Kluyveromyces species telomerase RNAs were found by Clustal X alignment. The homologous regions corresponded to conserved sequences (CSs) 1, 2, and 5 as identified by alignment of the Kluyveromyces species RNAs. Mutant RNAs were generated in which one region is substituted with a block of unrelated sequence to test its importance in telomere maintenance. The effect of mutations on growth and telomere length is listed below the region tested. (C) CS2 (dashed box) was predicted by RNA mfold and X2s structure prediction programs to form one side of a bulged stem that is phylogenetically conserved among all the budding yeast examined (filled circles, identical nucleotides with S. cerevisiae). S. cer., Saccharomyces cerevisiae; K. lactis, Kluyveromyces lactis; K. mar., Kluyveromyces marxianus; K. aes., Kluyveromyces aestuarii; K. wic., Kluyveromyces wickerhamii; K. non., Kluyveromyces nonfermentans; and K. dob., Kluyveromyces dobzhanskii. (D) Phylogenetically conserved sequence in the bulged stem. Indicated nucleotides are absolutely conserved between the budding yeast examined; * indicates nucleotides that show co-variation; and grey circle indicates a position in the bulge where the sequence is not conserved.
In most organisms, an intact telomere containing a terminal tract of GT-rich repetitive sequence is critical for genome stability and cell proliferation. The action of telomerase replenishes telomeric sequence lost with each cycle of cell division, as the conventional DNA polymerases cannot replicate to the terminus of the chromosome. A specialized reverse transcriptase (TERT), in addition to the RNA subunit, comprises the catalytic core of telomerase. The conservation of telomerase is most apparent in TERT: All TERTs studied from evolutionarily diverse organisms have sequence similarity to previously defined reverse transcriptase motifs (Lingner et al. 1997b; Nakamura and Cech 1998). Another telomerase protein component, Est1p, is also essential for telomerase function in S. cerevisiae (Lundblad and Szostak 1989); Est1p either recruits telomerase to the telomere (Evans and Lundblad 1999) or activates telomerase that is already telomere-bound (Taggart et al. 2002). Est1p is also evolutionarily conserved, as homologs have been found by sequence comparison in Schizosaccharomyces pombe and human (H. Beernick and J. Cooper, pers. comm.; P. Reichenbach and J. Lingner, pers. comm.).
Unlike TERT, the RNA subunit has diverged greatly, as manifested by its variability in size and the paucity of overall primary sequence conservation between species. Phylogenetic analysis has been a powerful tool for the identification of conserved secondary structures in telomerase RNA. Analysis of 24 ciliate and 35 vertebrate RNAs has yielded a core secondary structure model that, despite extensive sequence divergence, appears to have been conserved between ciliates and vertebrates.
Structure determination of the yeast telomerase RNA has proved more elusive, primarily owing to the longer length of the molecule and to the limited number of sequences available. The only two yeast telomerase RNAs reported in the public database are ∼1300 bases in length, compared with the 150–200 nucleotides and 300–500 nucleotides in ciliates and vertebrates, respectively. To gain more insight into yeast telomerase RNA structure, we aligned the sequences of telomerase RNAs from S. cerevisiae (Singer and Gottschling 1994) and six Kluyveromyces species (Y. Tzfati and E. Blackburn, in prep.). We found a phylogenetically conserved bulged-stem structure that is essential for telomerase function in vivo, and provide genetic and biochemical evidence for the function of this bulged stem in interaction with the Est1 protein subunit. This bulged stem represents the first report of an RNA structural determinant for interaction with a yeast telomerase protein component. Because of the conservation of the Est1 protein component, we propose that a similar structural element will be present in telomerase RNAs from other organisms.
Results
Identification of a structural element conserved among budding yeast telomerase RNAs
To identify regions of budding yeast telomerase RNAs that are conserved in structure and/or function, we aligned the nucleotide sequence of the S. cerevisiae telomerase RNA (TLC1 RNA) with those of six Kluyveromyces species RNAs. Alignment, performed in Clustal X (1.64b), was facilitated by deletion of the highly variable and partially dispensable (Roy et al. 1998) sequence between the template and the 5′ element of the template boundary structure (corresponding to S. cerevisiae nucleotides 156–453 and Kluyveromyces lactis nucleotides 82–420). An alignment of the six Kluyveromyces species RNAs among themselves revealed seven regions of high conservation (Tzfati et al. 2000; Y. Tzfati and E. Blackburn, in prep.; see Materials and Methods for GenBank accession numbers).
Three of these conserved regions had limited sequence identity to the S. cerevisiae telomerase RNA (Fig. 1B). These sequences were targeted for block substitution (see Materials and Methods). Conserved sequence 1 (CS1) is located near the 5′ end of the RNA (encompassing nucleotides 125–135 in S. cerevisiae), within a region required for Est2p interaction (Livengood et al. 2002). Conserved sequence 2 (CS2; nucleotides 657–674) is centrally located within a region characterized as being essential for both cell viability and Est1p interaction (Livengood et al. 2002). Conserved sequence 5 (CS5) is found near the 3′ end of the RNA, from nucleotides 1178–1190. We substituted a block of unrelated sequence for each of these conserved regions. The CEN low-copy-number plasmids for expression of the tlc1 block mutants from the endogenous TLC1 promoter were individually transformed into a tlc1Δ, rad52− S. cerevisiae strain complemented with a wild-type TLC1 plasmid. Deletion of RAD52 prevents the secondary mode of survival of telomerase null yeast by recombination (Lundblad and Blackburn 1993). After loss of the complementing plasmid, the strains were assayed for growth and the ability to maintain telomeres. The CS5 block mutant allowed wild-type cell growth and telomere maintenance, whereas the CS1 mutant sustained cell growth with stably shorter telomeres. Mutation of CS2 had the most dramatic effect: a senescence phenotype, with loss of viability and telomeric DNA indistinguishable from those of a tlcΔ strain.
Because CS2 is essential for in vivo telomere maintenance, we explored its potential structure using the computer folding programs RNA mfold (Zuker et al. 1999) and X2s (Juan and Wilson 1999). The most energetically favorable structure predicted for this region in all the yeast telomerase RNAs is a bulged-stem element (Fig. 1C). The bulge-containing strand of the paired stem is contained within CS2, whereas its pairing partner is located more 5′ in the RNA, from S. cerevisiae nucleotides 600–609. Comparison of the predicted structures shows remarkable conservation, given the overall low sequence identity between the RNAs. The base-pairing in the stem is well supported by phylogenetic covariation (Fig. 1C). Interestingly, all but one of the changes found in the stem are transition mutations, which are more conserved in shape than transversions, further demonstrating the structure conservation of this element. In addition, a stretch of sequence, which comprises the 5 bulged nucleotides and 3 flanking paired nucleotides, is strictly conserved except for one position in the bulge (Fig. 1D). Together, this structural and primary sequence conservation is consistent with a significant role for this element in telomerase function.
The predicted bulged stem is essential for telomere maintenance in vivo
We constructed several mutants of S. cerevisiae telomerase RNA to test the predicted bulged-stem structure (Fig. 2A). These mutants were made in the context of the full-length wild-type RNA. The stem-disruption mutant contained three single-nucleotide changes to destabilize pairing in the stem. The stem-disruption compensatory mutant was designed to restore the pairing by mutation of the corresponding 3 nucleotides in the opposite strand. A series of mutants were also generated to test the bulge requirements: The bulge deletion eliminates all 5 bulged nucleotides; the bulge substitution replaces the 5-base wild-type sequence with a 5-base unrelated sequence; and the bulgeΔ2 and bulgeΔ4 mutants have deletions of 2 and 4 nucleotides, respectively. The U663A point mutant has a change in 1 of the invariant bulged nucleotides from an uracil to an adenine.
Figure 2.
The predicted bulged stem in telomerase RNA is essential for telomere maintenance. (A) RNA mutants generated to test the predicted bulged-stem secondary structure: stem-disruption mutant, 3 bp disrupted in the predicted stem; compensatory mutant, base-pairing potential restored in the stem; bulge-deletion mutant, deletion of the 5 bulged nucleotides; bulge-substitution mutant, the wild-type sequence of the bulged nucleotides replaced with an unrelated sequence; bulgeΔ2 mutant, 2 nucleotides deleted in the bulge; bulgeΔ4, 4 nucleotides deleted in the bulge; and U663A mutant, an invariant U in the bulge changed to an A. (B) Senescence phenotype of yeast expressing bulged-stem mutant RNAs. Tlc1Δ strains were transformed with a tlc1 RNA allele or an empty vector control and grown on selective medium for the indicated number of generations after loss of the complementing wild-type TLC1 plasmid. (C) Telomere length analysis of tlc1 bulged-stem mutants. After loss of the complementing TLC1 plasmid, a single colony was picked into liquid culture, diluted 200-fold, and then regrown to saturation. Four successive serial dilutions were performed, cell growth permitting. Genomic DNA was prepared from each dilution (corresponding to ∼14 generations), digested with XhoI to release the terminal telomeric fragment, and then analyzed by Southern blotting with probes to telomeric sequence and a region of chromosome IV. The chromosome IV signal served as an internal loading control as well as a marker for measuring relative telomere lengths.
The effect of the bulged-stem mutants on telomere maintenance was assayed by growth on plates and telomere-length analysis. The complementing TLC1 on a plasmid in a tlc1Δ, rad52− strain was replaced by each bulged-stem mutant, or by a control plasmid, using plasmid shuffle. The cells were assayed for their ability to continue to grow over several successive restreaks after loss of the complementing TLC1 plasmid.
The stem-disruption and bulge-deletion mutants exhibited a telomerase null phenotype (Fig. 2B; data not shown). By ∼75 generations, these mutant strains had ceased cell division, a rate of senescence comparable to a tlc1Δ strain. In comparison, the stem-disruption compensatory mutant complemented a telomerase null strain as efficiently as did the wild-type RNA. The bulgeΔ4 mutant had an intermediate senescence phenotype, with cell division ceasing after ∼100 generations rather than 75. The bulge-substitution mutant had an even more subtle phenotype: Growth was maintained for at least 10 restreaks, but the colonies were heterogeneous in size (data not shown). The bulgeΔ2 and U663A mutants had no discernible growth defect (data not shown).
The mutant strains were also subjected to telomere-length analysis, a more sensitive assay for in vivo telomerase function (Fig. 2C). Liquid cultures were inoculated with a single colony from the first restreak of the strains from Figure 2B. As each culture approached saturation, it was diluted 200-fold for four successive serial dilutions, or until the culture could not be regenerated. Each saturated culture (representing ∼14 generations) was harvested and genomic DNA prepared from the cells. The DNA was digested with XhoI to release a class of telomere-containing restriction fragments and was analyzed by Southern blotting and hybridization with telomeric and chromosome IV probes. (The chromosome IV nontelomeric fragment provides an internal length standard and a control for DNA loading.) In concordance with the growth assay, the telomeres of the stem-disruption and the bulge-deletion mutants shortened over time at a rate similar to that of a tlc1Δ strain. In contrast, the stem-disruption compensatory mutant had wild-type length telomeres. The bulgeΔ2 and bulgeΔ4 mutants had stably short telomeres, an intermediate telomere length phenotype, as did the bulge-substitution mutant. The U663A point mutant did not exhibit any telomere shortening (Fig. 2C).
These results validate the bulged-stem structure predicted by computer folding and comparative sequence analysis of budding yeast telomerase RNAs. We conclude from the senescence assay and the telomere length analysis that this conserved structure is essential for telomere maintenance in vivo.
Overexpression of telomerase protein Est1p but not Est2p suppresses the bulged-stem mutant phenotype
S. cerevisiae telomerase contains at least three protein subunits required for in vivo telomerase function, Est1p, Est2p, and Est3p (Lendvay et al. 1996). We predicted that if the bulged-stem mutants were weakened in interaction with one of these protein subunits, then overexpression of the protein might rescue the RNA mutant phenotype. To test this hypothesis, a 2-μm high-copy-number plasmid containing the ADH promoter for expression of Est1p, Est2p, Est3p, or, as a control, no protein, was transformed into tlc1Δ yeast expressing wild-type TLC1 or the bulged-stem mutants, or containing the control empty vector. The strains were assayed for senescence by successive restreaking on selective medium (Fig. 3A). Overexpression of Est1p, Est2p, Est3p, and empty vector had no effect on the growth of strains expressing wild-type TLC1 RNA or the stem-disruption compensatory mutant (data not shown for Est3p). In contrast, overexpression of Est1p restored wild-type growth to the stem-disruption, bulge-deletion, and bulgeΔ4 RNA mutants, but not to the empty vector control. This overexpression suppression effect was specific to Est1p, because growth was not restored by the empty control vector or by overexpression of Est2p (Fig. 3A) or Est3p (data not shown).
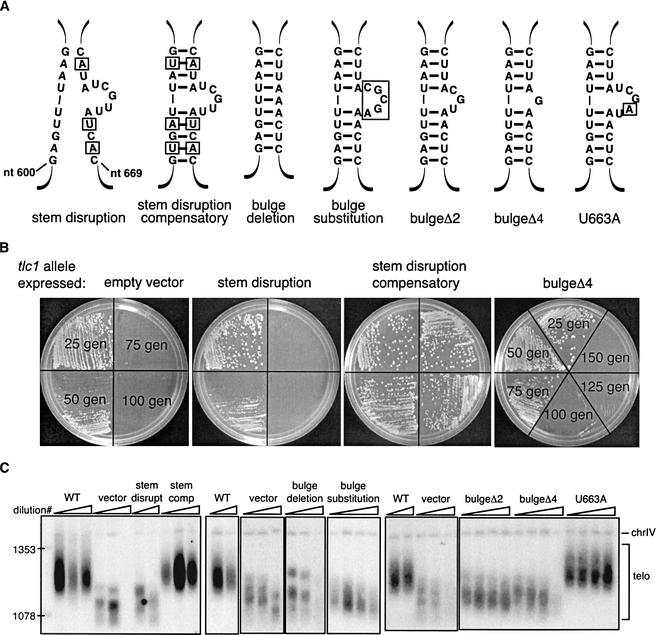
Figure 3.
Overexpression of Est1p but not Est2p rescues the senescence phenotype of bulged-stem mutants. (A) Senescence phenotype of yeast overexpressing a telomerase protein component. Yeast strains expressing the indicated tlc1 allele or an empty vector control were transformed with a 2-μm plasmid in which protein expression is driven by the ADH promoter. The RNA allele expressed is indicated above and below the plate. The protein overexpressed is indicated in the plate schematic on the left. The cell growth shown represents ∼80 generations after loss of the complementing TLC1 plasmid, or ∼75 generations after transformation with the overexpression plasmid. (B) Telomere-length analysis of strains shown in A. The experiment was performed as described for Figure 2C. Three successive serial dilutions were performed. Only dilution no. 3 is shown for the wild-type TLC1 strain and dilutions nos. 2 and 3 for the stem-disruption compensatory mutant. *, telomere maintenance by recombination; arrowheads, examples of amplified telomere-associated Y‘ fragments.
To examine more closely the effect of Est1p overexpression on telomerase function, we analyzed the telomere lengths in the bulged-stem mutant strains (Fig. 3B). Liquid cultures were inoculated with a single colony from the strains shown in Figure 3A. The cultures were serially diluted three times, for a total of ∼42 generations (as described for Fig. 2C). Telomeres of the tlc1Δ strain, carrying empty vector, were short and unaffected by overexpression of Est1p, Est2p, or the empty vector. Because the strain is RAD52+, the cells began to maintain telomeres by the recombination mode for survival. The resulting telomere rearrangements were apparent by the second dilution, as seen by a heterogenous distribution of telomeric hybridization and amplification of the telomere-associated Y‘ elements (Fig. 3B, lanes labeled by an asterisk, amplified Y‘ elements labeled with arrowheads; Lundblad and Blackburn 1993; Teng and Zakian 1999). Despite overexpression of Est2p or the vector, the telomeres of the stem-disruption and bulge-deletion mutants were clearly still short and began to recombine by the second dilution. In contrast, overexpression of Est1p suppressed the recombination mode of telomere maintenance in the stem-disruption mutant, presumably because the cells were able to maintain telomeres above the functional threshold, even though they were shorter than wild-type. Telomeres in the bulge-deletion mutant were restored to near-wild-type lengths by overexpression of Est1p. The bulgeΔ4 mutant, which maintained stably shorter telomeres with the empty vector or overexpression of Est2p, had restored wild-type length telomeres on Est1p overexpression.
In the strains carrying the wild-type and stem-disruption compensatory mutant TLC1 RNAs, Est1p overexpression caused slight telomere elongation (Fig. 3B). However, this elongation was not nearly as dramatic as that seen in the bulge mutants on Est1p overexpression, suggesting that the telomere lengthening in the bulge-deletion and bulgeΔ4 mutants was specific to Est1p overexpression rather than a general effect. Importantly, overexpression of Est1p appeared to obviate the need for the recombination pathway for survival in the stem-disruption and bulge-deletion mutants but not in the tlc1Δ strain. Nevertheless, we cannot exclude the possibility that Est1p overexpression causes generalized telomere elongation, and that such elongation is less dramatic in the wild-type strain than in the mutants because of tighter length regulation when telomeres are full-length. We therefore further addressed the allele-specificity of the Est1p overexpression by examining its effect on an unrelated RNA mutant, an Sm site mutant (tlc1-Sm2C4C; Seto et al. 1999). The short telomeres of this mutant were not affected by overexpression of Est1p (data not shown), demonstrating the allele specificity of the Est1p suppression for the bulged-stem RNA mutants. These data support the hypothesis that the bulged-stem structure in telomerase RNA interacts either directly or indirectly with the essential telomerase protein subunit Est1p.
An intact bulged stem is required for co-immunoprecipitation of telomerase RNA with the telomerase subunit Est1p but not Est2p
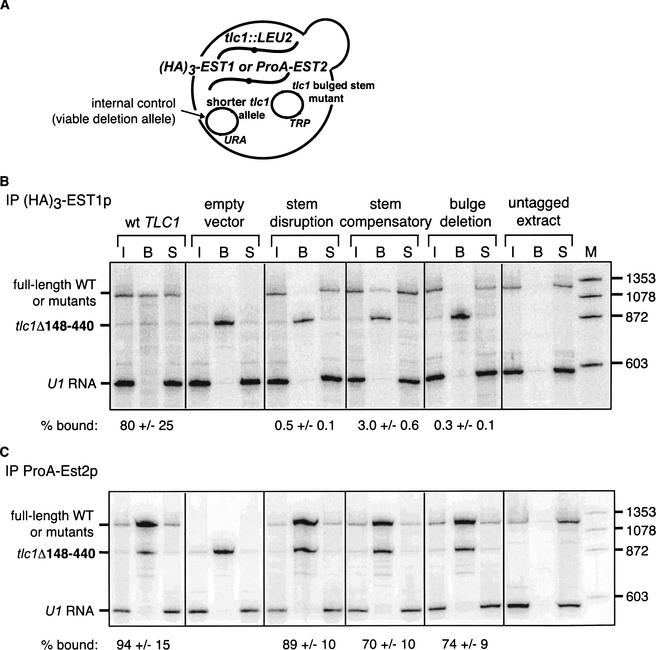
To more directly test the role of the bulged stem in Est1p interaction, we performed a series of co-immunoprecipitation experiments. Various mutants, as well as wild-type TLC1 and empty vector controls, were transformed into tlc1Δ strains with an epitope tag integrated into the endogenous locus of either EST1 (HA3-EST1) or EST2 (ProA-EST2; Fig. 4A). The strains were complemented with plasmid tlc1Δ148–440, encoding a telomerase RNA that co-immunoprecipitates with Est1p and Est2p (Livengood et al. 2002) but is electrophoretically distinct from the full-length RNA and therefore serves as an internal control for co-immunoprecipitations. (HA)3-Est1p or ProA-Est2p was immunoprecipitated from cell extract, and co-immunoprecipitated RNAs were analyzed by Northern blot hybridization. The extent of co-immunoprecipitation of each mutant tlc1 allele, or wild-type TLC1, was compared with that of the internal control tlc1Δ148-440 RNA (see legend for Fig. 4B).
Figure 4.
Bulged-stem mutants show reduced co-immunoprecipitation with Est1p but not Est2p. (A) Diagram of yeast strain used for co-immunoprecipitation assays. A tlc1Δ yeast strain carrying either (HA)3-EST1 or ProA-EST2 at their respective chromosomal loci is transformed with a tlc1 deletion allele (tlc1Δ148-440) that sustains cell growth and a full-length tlc1 bulged-stem mutant allele. (B) Northern analysis of RNAs that co-immunoprecipitate with (HA)3-Est1p. Extract was prepared from yeast strains expressing the indicated tlc1 allele or an empty vector control, or from an untagged wild-type strain. Extract was incubated with α-HA beads and RNA that co-immunoprecipitated on the beads was analyzed by Northern blot and probed for TLC1 and U1 RNAs. Lanes are as follows: M, radiolabeled φx174 DNA HaeIII fragments; I, input RNA; B, RNA bound to α-HA beads; and S, RNA in the supernatant of the immunoprecipitations. Approximately 2.5% of the total input and supernatant RNAs and 25% of bound RNA were loaded. Quantitation of bound RNAs was calculated as follows: [Bound(mutant)/Input(mutant)]/[Bound(Δ148-440)/Input(Δ148-440)] × 100. The mean and range of three independent experiments are reported. (C) Northern analysis of RNAs that co-immunoprecipitate with ProA-Est2p. Extracts from C were incubated with IgG beads to immunoprecipitate ProA-Est2p. Co-immunoprecipitated RNAs were analyzed by Northern blot and two independent experiments were quantitated, as described in B.
Wild-type TLC1 RNA co-immunoprecipitated with either Est1p (Fig. 4B) or Est2p (Fig. 4C) to a similar extent as did the internal control. U1 snRNA, which is not associated with the telomerase complex, was not co-immunoprecipitated with either Est1p or Est2p, confirming that the immunoprecipitation is specific. In three independent experiments, no co-immunoprecipitation of the stem-disruption or bulge-deletion mutant with Est1p was detected (the limit of detection is ∼0.5% of the wild-type level), whereas the compensatory mutant was co-immunoprecipitated to a limited extent (∼3% when normalized to the internal control; Fig. 4B). In contrast, the stem-disruption, stem-disruption compensatory, and bulge-deletion mutants co-immunoprecipitated with ProA-Est2p to a similar extent as wild-type TLC1 RNA when normalized to the internal control (Fig. 4C). Therefore, mutations in the bulged stem specifically affect the co-immunoprecipitation of TLC1 RNA with Est1p, but not with Est2p. These results suggest that an intact bulged stem is required for interaction with Est1p, rather than for stability, folding or subcellular localization of the RNA.
Specific interaction between the bulged stem and Est1p is independent of the full-length RNA
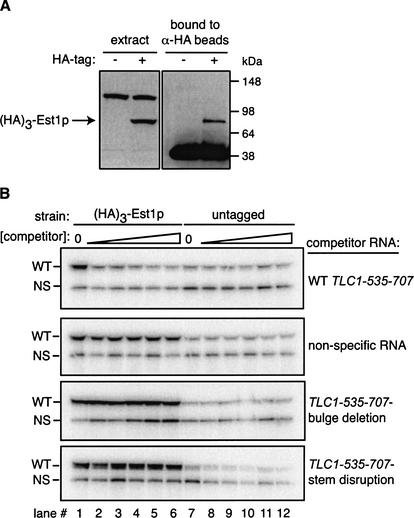
The telomerase RNA has been proposed to serve as a scaffold for the binding of telomerase protein subunits (Livengood et al. 2002). To address the possibility that the interaction between Est1p and the bulged-stem structure is mediated by another RNA-bound protein or another region of the RNA, we developed an in vitro assay for the binding of a fragment of TLC1 to Est1p. Extract was made from a tlc1Δ strain overexpressing (HA)3-Est1p. On a Western blot probed with α-HA antibodies, (HA)3-Est1p can clearly be detected directly from yeast extract and after immunoprecipitation by α-HA beads (Fig. 5A). TLC1 nucleotides 535–707, the region previously characterized to be essential for Est1p co-immunoprecipitation (Fig. 1A; Livengood et al. 2002), was in vitro transcribed from a bacteriophage T7 promoter. The P4-P6 domain of a group I intron from Dunaliella salina was also prepared as a nonspecific control RNA. Both radiolabeled RNAs were added simultaneously to the extract, then (HA)3-Est1p was immunoprecipitated on α-HA beads. Analysis of the RNA bound to beads showed that TLC1-535-707 bound (HA)3-Est1p at least fivefold over the background binding (Fig. 5B, cf. lanes 1 and 7).
Figure 5.
A small fragment of yeast telomerase RNA is sufficient for co-immunoprecipitation with Est1p. (A) Overexpression of (HA)3-Est1p in a tlc1Δ strain. Extract was prepared from a wild-type yeast strain or a tlc1Δ strain overexpressing (HA)3-Est1p from an ADH promoter on a 2-μm plasmid. The extract was analyzed by Western blot to the HA epitope tag (left panel) or by immunoprecipitation with α-HA beads followed by α-HA Western to protein bound to the beads (right panel). The upper band in the extract is a yeast protein recognized nonspecifically by the HA antibody. The lower band that is bound to the α-HA beads is the heavy-chain of the antibody on the beads. (B) In vitro binding of TLC1-535-707 to (HA)3-Est1p and competition by bulged-stem mutants. In vitro transcribed TLC1-535-707 and Dunaliella group I P4-P6 intron RNA were incubated with a wild-type (untagged) or tlc1Δ-extract overexpressing (HA)3-Est1p. Cold competitor RNA was added concurrently with the radiolabeled RNA at concentrations 1-, 2-, 4-, 8-, 16-, and 32-fold over that of the wild-type fragment. (HA)3-Est1p was immunoprecipitated with α-HA beads, and the co-immunoprecipitated RNAs were analyzed by denaturing SDS-gel electrophoresis and PhosphorImager analysis. The nonspecific binding of the Dunaliella group I intron served as an internal control for loading. WT, radiolabeled wild-type TLC1-535-707; NS, radiolabeled nonspecific RNA.
To determine the sequence specificity of the TLC1-535-707 binding to (HA)3-Est1p, competition for binding by TLC1-535-707 and the nonspecific RNA was assayed. Nonradiolabeled RNA was titrated in at concentrations 1-, 2-, 8-, 16-, and 32-fold over the radiolabeled RNA concentration (Fig. 5B, lanes 2–6 for the tagged extract, lanes 8–12 for the untagged). The wild-type TLC1 fragment competed very efficiently for the binding of the radiolabeled TLC1-535-707: A onefold excess of RNA reduced the binding by 66%, and an eightfold excess of RNA reduced the binding to near background levels (Fig. 5B). In contrast, the nonspecific RNA did not compete for TLC1 binding to (HA)3-Est1p. Similarly, the stem-disruption and bulge-deletion mutants, which did not form stable complexes with (HA)3-Est1p in vivo (Fig. 4A), also failed to compete efficiently for the wild-type binding signal (Fig. 5B, panels 3,4). As with the nonspecific RNA, at most a twofold reduction in binding was seen with these RNAs. Interestingly, the stem-disruption compensatory mutant also did not compete with the wild-type fragment for binding (data not shown), a result consistent with our finding that this mutant was partially impaired in co-immunoprecipitation with Est1p in vivo (Fig. 4A).
The in vitro binding results indicate that a small fragment of S. cerevisiae telomerase RNA is competent to bind to (HA)3-Est1p in a yeast extract in the absence of the full-length RNA. This binding is specific to TLC1 RNA, as a nonspecific RNA could not compete for the binding signal. Mutants that contain a disruption in the stem or deletion of the bulge did not compete for binding as well as wild-type, confirming that an intact bulged-stem structure is critical for the interaction between the telomerase RNA and Est1p.
Discussion
The newly identified bulged stem in budding yeast telomerase RNA provides a structural basis for the interaction of the core telomerase complex with Est1p. Est1p is not required for core telomerase in vitro enzymatic activity (Cohn and Blackburn 1995; Lingner et al. 1997a). However, Est1p is essential for telomerase action at the telomere (Evans and Lundblad 1999; Taggart et al. 2002); proposed functions are recruitment of the telomerase core to the telomere or activation of telomere-bound telomerase. Our results suggest that one molecular interaction necessary for Est1p to execute its in vivo function involves its binding to the bulged-stem RNA structure.
An essential bulged stem in budding yeast telomerase RNA
The phylogenetically conserved bulged-stem structure was tested by examination of the effect of disruption of the base-pairing in the stem or deletion of the bulged nucleotides on in vivo telomerase function. Both mutants had a tlc1Δ phenotype, whereas a compensatory mutant designed to reestablish the pairing in the stem exhibited wild-type growth and telomere lengths, suggesting that the structure of the stem is more important than its primary sequence. The presence of bulged nucleotides also appears to be more crucial than their sequence: Replacement of the 5 bulged nucleotides with a block of unrelated sequence yielded a telomerase in yeast that can sustain long-term growth, whereas deletion of 4 of the 5 nucleotides resulted in the more severe phenotype of delayed senescence.
The 44–77 nucleotides that lie between the sequences that create the bulged stem in various budding yeast are predicted to form interrupted stem-loop structures. However, no obvious conserved structure was seen in this intermediate region in our analysis. The sequence 3′ of the bulged stem appears to be dispensable for telomerase function, as a mutant with nucleotides 676–681 changed to unrelated sequence had no growth defect and wild-type length telomeres (data not shown). These results suggest that the bulged stem defines a structure of the RNA critical for telomerase function. A more detailed mutational study or phylogenetic comparison with more yeast RNA sequences may reveal additional elements involved in Est1p interaction.
Genetic interaction of the bulged stem with the essential telomerase protein Est1
In addition to the telomerase RNA, the protein subunits Est1p, Est2p, and Est3p are essential for in vivo telomerase function in S. cerevisiae (Lendvay et al. 1996). An overexpression analysis revealed that overexpression of Est1p, but not of Est2p or Est3p, restored wild-type growth to the bulged-stem mutants. We hypothesize that the bulged-stem mutants are impaired for interaction with Est1p, and therefore, increased levels of Est1p compensate for the weakened affinity. The implication is that there could be at least one other element—RNA or protein—in addition to the bulged stem that interacts with Est1p in the telomerase complex.
Interestingly, although Est1p overexpression rescued wild-type growth in the stem-disruption mutant, telomeres were not restored to wild-type lengths. Thus, overexpression of Est1p in this mutant may allow maintenance of telomeres that, although still short, are not short enough to cause senescence. In contrast, telomeres of the bulge-deletion mutant were restored to wild-type by Est1p overproduction. These results reveal a subtle difference between the stem-disruption and bulge-deletion mutants. We suggest that perhaps the stem-disruption mutant is more impaired for Est1p interaction than is the bulge-deletion mutant, so that even overexpression of Est1p is not sufficient to restore full function to this mutant. Alternatively, the stem-disruption mutant may have a slightly more acute telomere phenotype that, despite being indiscernible by the telomere blot analysis, is too severe for overexpression of Est1p to restore wild-type length telomeres. Disruption of the stem is expected also to change the conformation of the bulged nucleotides; the simultaneous impairment of the two potential determinants for the Est1p interaction site—the stem and the bulge—may therefore account for the more severe phenotype. Furthermore, we cannot preclude the possibility that an additional determinant for function, independent of Est1p interaction, is impaired in the stem-disruption mutant and therefore would not be expected to be compensated by Est1p overexpression.
Interaction with Est1p is dependent on an intact bulged stem and independent of the full-length RNA
Co-immunoprecipitation of the stem-disruption and the bulge-deletion mutant RNAs with Est1p was undetectable, whereas these mutants were still competent for binding to Est2p at levels comparable to that of wild-type. Surprisingly, the stem-disruption compensatory mutant was also partially impaired for co-immunoprecipitation with Est1p, despite wild-type growth and telomere length. Although the compensatory mutant co-immunoprecipitated at least sixfold more efficiently than did the stem-disruption and bulge-deletion mutants, it was significantly below the levels of wild-type RNA co-immunoprecipitation. One possible explanation for this difference arises from the fact that RNA mfold structure prediction for this mutant revealed an alternative, more energetically favored conformation lacking the bulged stem. If the calculation is accurate, most molecules would be likely to form this alternative structure. We propose that the small portion of the population having the wild-type fold accounted for the low level of co-immunoprecipitation, but was sufficient to confer a wild-type phenotype. Alternatively, as co-immunoprecipitation in a cell-free system is a more stringent assay for protein-RNA interactions, it may not detect cooperative and/or additionally stabilizing interactions within the telomerase complex that provide complementation of function in vivo. Therefore, the sequence changes on each side of the stem in the compensatory mutant may lower its affinity for Est1p slightly, but not enough to give an in vivo phenotype.
Our data support the hypothesis that a critical determinant for Est1p interaction is an intact bulged stem. This interaction occurs independently of most of the RNA, as a TLC1-535-707 fragment is sufficient for co-immunoprecipitation of Est1p. Co-immunoprecipitation of the telomerase RNA and Est1p has been shown to be independent of Est2p (Zhou et al. 2000), so it is unlikely that Est2p mediates this interaction. Furthermore, co-immunoprecipitation of the RNA with Est3p is dependent on the presence of Est2p (Hughes et al. 2000); therefore, Est1p interaction with the RNA should be independent of Est3p as well. Importantly, we observed no effect on the bulged-stem mutant phenotype upon Est2p and Est3p overexpression. Taken together with these findings, our results support an interaction of the bulged stem with Est1p independent of the other known telomerase protein components and 85% of the RNA subunit. However, because our experiments have been conducted in whole cell extracts, we cannot completely exclude the possibility that other proteins affect the interaction between Est1p and the bulged stem.
How might Est1p interact with the bulged stem?
The RRM (RNA recognition motif) or ribonucleoprotein (RNP) domain is the best-characterized and most widespread RNA-binding motif (Perez-Canadillas and Varani 2001). Est1p has been reported to contain a region with weak sequence homology with the consensus RRM, and mutants in this region do not complement an est1Δ strain and no longer co-immunoprecipitate TLC1 RNA (Zhou et al. 2000). The function of this region is not entirely clear, however, because a subsequent report showed that Est1p containing a deletion in this region was still able to partially co-immunoprecipitate the RNA (Evans and Lundblad 2002).
In the absence of additional functional or structural data about Est1p, we can only speculate as to its mode of recognition of the bulged-stem structure. Precedents are provided by well-characterized RNA-binding proteins that recognize single-stranded nucleotides in the context of secondary structure (Leulliot and Varani 2001), such as the HIV-1 Tat-TAR interaction. The TAR RNA is comprised of 3 bulged nucleotides situated within a helical stem, not unlike the bulged stem we describe for TLC1 RNA. Structural studies revealed that the ability of the helix to bend at the bulged nucleotides is critical for protein binding. The RRM-containing U1A protein binds to the U1 snRNA and to its own pre-mRNA (Oubridge et al. 1994; Allain et al. 1996) by recognizing 6 unpaired nucleotides in a sequence-specific manner (Hall 1994). The high sequence conservation of the bulged nucleotides of the TLC1 RNA bulged stem and the importance of an intact stem suggest that Est1p could recognize this structure, at least in part, in a similar manner.
Other known structural features in telomerase RNA
We report here the first defined structure in yeast telomerase RNA that is essential for in vivo telomerase function and is involved in interaction with a telomerase protein component. In S. cerevisiae, the only other known structural element in telomerase RNA involved in protein interaction is a stem-loop that functions through the Ku pathway (Peterson et al. 2001). A deletion analysis of K. lactis RNA identified two telomerase RNA regions essential for assembly into the telomerase complex (Roy et al. 1998). Tetrahymena thermophila telomerase RNA contains a 22-nucleotide region that is essential for tTERT binding; although this region is predicted to fold into a stem-loop, mutagenesis suggested that the determinants for binding were RNA sequence and sequence positioning rather than secondary structure (Licht and Collins 1999). A structural element important for tTERT interaction also specifies one template boundary (Lai et al. 2002). A portion of the human telomerase RNA has been shown to contain important sequences for hTERT binding, including a proposed stem-loop (Mitchell and Collins 2000). Mutagenesis of an analogous structure in the mouse RNA confirmed that this stem-loop is an important determinant for mTERT binding (Chen et al. 2002).
In addition to binding telomerase-specific proteins, telomerase RNAs also interact with factors important for their biogenesis and stability (Seto et al. 1999; Aigner et al. 2000; Mitchell and Collins 2000). Telomerase RNAs from different classes of organisms use different proteins for their biogenesis. Despite these differences, however, there is likely a conserved structural core within the telomerase RNAs, as has already been observed between ciliates and vertebrates (Chen et al. 2002). It remains to be seen whether the yeast RNAs will also display a homologous core structure.
Conservation of the bulged stem in higher eukaryotes
Est1p homologs have been identified in S. pombe and human (H. Beernick and J. Cooper, pers. comm.; P. Reichenbach and J. Lingner, pers. comm.), both of which are evolutionarily distant from the budding yeast. If the function of these homologs has been conserved, then perhaps the bulged-stem RNA structure has also been conserved through evolution. The current secondary structure predictions for ciliate and vertebrate telomerase RNAs do not reveal an obvious counterpart to the budding yeast bulged stem we have identified. Further higher-order structural analyses may be necessary to determine whether such a bulged stem exists in the vertebrate RNAs.
Materials and methods
Sequence alignment and secondary structure prediction
Secondary structure predictions for the region flanking CS2, corresponding to nucleotides 550–750 in S. cerevisiae, were obtained using the RNA mfold (Zuker et al. 1999) and X2s RNA structure prediction programs (Juan and Wilson 1999; RNA mfold version 2.3 with the folding temperature at 30°C, found at http://www.bioinfo.rpi.edu/∼zukerm). GenBank accession nos. are AY151277, AY151278, AY151279, AY151280, and AY151281, for the Kluyveromyces aestuarii, Kluyveromyces dobzhanskii, Kluyveromyces marxianus, Kluyveromyces nonfermentans, and Kluyveromyces wickerhamii telomerase RNAs, respectively.
Plasmid construction
pBM125-135 (CS1 block mutant), pBM657 (CS2 block mutant), pBM1178 (CS5 block mutant), p3PM658 (3-nucliotide stem-disruption mutant), p3PM658C (3-nucleotide stem compensatory), pDel660 (bulge deletion), pBM550 (bulge substitution), pBulge-3 (bulgeΔ2), pBulge-1 (bulgeΔ4), and pT663A (U663A point mutant) were constructed by introducing the appropriate mutated sequence into pSD107 (TRP, CEN, TLC1 with endogenous promoter and terminator sequences; Seto et al. 1999). Mutations were introduced by ligation of PCR fragments generated with primers containing the mutant sequence. The CS1 block mutant substituted the sequence of nucleotides 125–135 with 5′-ACGGCATGCGT-3′; the CS2 block mutant substituted the sequence of nucleotides 657–674 with 5′-AATCGCGATT GAGACCTT-3′; and the CS5 block mutant substituted nucleotides 1178–1190 with 5′-TCTAGATAAAAAA-3′. All mutant clones were sequenced over the region subjected to PCR amplification. Plamid ptlc1Δ148–440 was created in a previously characterized deletion library (Livengood et al. 2002). The plasmid pVL369 (URA3, 2 μm, EST2 with ADH promoter and terminator sequences) was obtained from V. Lundblad (Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX). For overexpression of untagged Est1p, the XmaI-SpeI fragment of pRS423 containing EST1 with ADH promoter and terminator sequences (K. Friedman, in prep.) was cloned into the same sites in pRS426 (URA3, 2 μm). For overexpression of (HA)3-Est1p, pVL1008 [TRP, 2 μm, (HA)3-EST1 with ADH promoter and terminator sequences] was obtained from V. Lundblad (Evans and Lundblad 2002).
Strain construction and manipulations
TCy43 (MATa ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 VR∷ADE2-TEL adh4∷URA3-TEL Δtlc1∷LEU2 rad52∷LEU2 pTLC1-LYS2-CEN; Seto et al. 1999), was transformed with pSD107, pAS500 (TRP, CEN, tlc1ΔNcoI-NsiI; Seto et al. 1999), pBM125-135, pBM657, pBM1178, p3PM658, p3PM658C, pDel660, pBM660, pBulge-3, pBulge-1, or pT663A. Single colonies were then picked to α-aminoadipate plates to counter-select pTLC1-LYS2-CEN. After growth on α-aminoadipate, a single colony was picked and streaked onto medium lacking tryptophan. Transformations were performed with lithium acetate as described previously (Rose et al. 1990). Replacement of TLC1 from nucleotide 128–1140 in the strain AVL78 (MATα leu2 trp1 ura3-52 prb prc pep4-3; Lingner et al. 1997a,b) with the S. pombe LEU2 gene was achieved by one-step gene replacement as described (Baudin et al. 1993). After growth on medium lacking leucine, AVL78-tlc1∷LEU2 was maintained by transformation with the complementing pRS316/TLC1 (URA, CEN, TLC1; Livengood et al. 2002). This strain was then transformed with pSD107, pAS500, p3PM658, p3PM658C, pDel660, and pBulge-1. Single colonies were grown on 5-fluoroorotic acid plates to counter-select for pRS316/TLC1. From 5-FOA, single colonies were picked for transformation with URA3-labeled plasmids for EST1, EST2, or EST3 overexpression, or with pRS426. Strains (HA)3-Est1–tlc1∷LEU2 and ProA-Est2–tlc1∷LEU2 have been described previously (Livengood et al. 2002).
Senescence assay and telomere length analysis
After loss of the complementing wild-type TLC1 plasmid, single colonies carrying pSD107, pAS500, pBM125-135, pBM657, pBM1178, p3PM658, p3PM658C, pDel660, pBM660, pBulge-3, pBulge-1, or pT663A were successively restreaked on medium lacking tryptophan to assay for senescence. In addition, for telomere-length analysis, single colonies were picked and grown in 5 mL selective media. The culture was grown to saturation, then diluted 200-fold to inoculate a fresh 5 mL culture. After 24 h of growth, the culture was rediluted 200-fold. Cells from the saturated culture were harvested, and DNA was isolated using the DNA-Pure Yeast Genomic Kit (CPG Inc.). Half of the DNA isolated was digested with XhoI, resolved on a 1.1% agarose gel, transferred to Hybond-N+ (Amersham Pharmacia) by osmoblotting, and hybridized at 55°C to telomeric and chromosome IV DNA fragments random-primed with High Prime reaction mixture (Roche) and [α32P]dCTP.
RNA co-immunoprecipitation with Est1p or Est2p from extracts
Co-immunoprecipitation experiments were performed as described previously (Livengood et al. 2002).
Western blotting and in vitro binding assay
AVL78, or AVL78-tlc1∷LEU2 carrying pVL1008, were grown to 0.8–1.5 OD600 at 30°C in YPD or media lacking tryptophan, respectively. Whole-cell extract was prepared by glass-bead lysis as previously described (Livengood et al. 2002), except with the addition of 1 mM PMSF and 10 U/mL RNasin (Promega), and insoluble material was sedimented by centrifugation at 8820g for 15 min followed by a high-speed spin at 100,000g for 1 h at 4°C. Total protein concentration of the S-100 fraction was determined by Bradford assay (Bio-Rad). Western blot analysis of (HA)3-Est1p was performed by separation of the S-100 fraction or α-HA-immunoprecipitated material on 4%–20% Novex Tris-Glycine gels (Invitrogen). Proteins were transferred to Hybond-P (Amersham Pharmacia) by wet transfer in 20 mM phosphate buffer (pH 6.8) at 0.5 A for 1.25 h. After blocking the membrane with 5% (w/v) reconstituted nonfat milk powder in PBS and 0.05% Tween-20, the blot was incubated with 1000-fold diluted HA.11 α-HA antibody (Covance) in 5% milk and PBS for 1 hr at room temperature or overnight at 4°C. Washes were performed with PBS and 0.05% Tween-20. The secondary antibody, goat anti-mouse IgG HRP-conjugated (Pierce), was incubated at a 2000-fold dilution for 1 h at room temperature. The blots were visualized using ECL solution and Hyperfilm-ECL (Amersham Pharmacia). Templates for in vitro RNA transcription of TLC1-535-707 were made by PCR amplification of pSD107, p3PM658, pDel660, or p3PM658C using a 5′ primer containing the T7 promoter and TLC1 sequence beginning with nucleotide 535 (5′-GGAATTCTAATACGACTCACTATAGGGATTTTTTTCT CGTTTTCTTATACCTAG-3′) and a 3′ primer ending in TLC1 nucleotide 707 (5′-CATGTTCCCTGACGTTCTTTTTCC-3′). RNA transcription and gel purification were performed as described (Sperger and Cech 2001) with 10 mg T7 RNA polymerase, 40 pmole DNA template, and 1 mM of each rNTP for 1.5 h at 37°C. Prior to gel-purification, transcription reactions were treated with 5 U RQ1 RNase-free DNase (Promega) for 15 min at 37°C to degrade the DNA template. Gel-purification was performed with 8% PAGE-8 M urea. In vitro transcribed Dunaliella group I intron P4-P6 RNA (158 nucleotides) was provided as a generous gift from E. Podell (Howard Hughes Medical Institute, University of Colorado at Boulder). For the RNA-protein binding reactions, RNAs were prefolded by incubation for 15 min at 50°C in 10 mM Tris-HCl (pH 7.5), 200 mM NaCl with cooling for 20 min at room temperature. Approximately 0.25 μg of S-100 fraction protein was added to the appropriate amount of cold competitor RNA. Immediately after addition of the cold competitor, 250,000 cpm of 5′ end-labeled wild-type TLC1-535-707 RNA and Dunaliella P4-P6 RNA were added. RNA-protein complexes were allowed to form for 20 min on ice. The total volume was then brought to 200 μL with extract buffer, and 5 μL of α-HA beads were added. After at least 2.5 h of rotation at 4°C, the beads were washed four times with extract buffer. RNAs bound to beads were removed by incubation in NuPAGE LDS sample buffer with 100 mM DTT (Invitrogen) for 2 min at 95°C. Samples were immediately loaded onto a NuPAGE 4%–12% Bis-Tris gel run in MOPS running buffer (Invitrogen). After electrophoresis, the gel was fixed by incubation in 25% isopropanol and 10% acetic acid for 15 min with shaking. After drying, the amount of radiolabeled RNA in the gel was determined by PhosphorImager analysis (Molecular Dynamics).
Acknowledgments
We are grateful to V. Lundblad, E. Podell, K. Friedman, and A. Zaug for gifts of strains, plamids, and reagents. We thank J. Cooper and J. Lingner for sharing unpublished results. A.G.S. thanks V. Lundblad, J. Goodrich and members of the Cech lab, particularly S. Aigner, P. Baumann, and A. Berglund for helpful discussions. This work was supported by National Institutes of Health grants GM28039 (to T.R.C.) and GM26259 (to E.H.B.), and a Human Frontiers Science Program Fellowship LT-415/96 (to Y.T.)
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL thomas.cech@colorado.edu; FAX (310) 215-8558.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1029302.
References
- Aigner S, Lingner J, Goodrich KJ, Grosshans CA, Shevchenko A, Mann M, Cech TR. Euplotes telomerase contains a La motif protein produced by apparent translational frameshifting. EMBO J. 2000;19:6230–6239. doi: 10.1093/emboj/19.22.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain FH, Gubser CC, Howe PW, Nagai K. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formation. Nature. 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-L, Opperman KK, Greider CW. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 2002;30:592–597. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M, Blackburn EH. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- Draper DE. Themes in RNA-protein recognition. J Mol Biol. 1999;293:255–270. doi: 10.1006/jmbi.1999.2991. [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- ———. 2002. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics (In press). [DOI] [PMC free article] [PubMed]
- Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Hall KB. Interaction of RNA hairpins with the human U1A N-terminal RNA binding domain. Biochemistry. 1994;33:10076–10088. doi: 10.1021/bi00199a035. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Evans SK, Weilbaecher RG, Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- Juan V, Wilson C. RNA secondary structure prediction based on free energy and phylogenetic analysis. J Mol Biol. 1999;289:935–947. doi: 10.1006/jmbi.1999.2801. [DOI] [PubMed] [Google Scholar]
- Lai CK, Miller MC, Collins K. Template boundary definition in Tetrahymena telomerase. Genes & Dev. 2002;16:514–520. doi: 10.1101/gad.962602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulliot N, Varani G. Current topics in RNA-protein recognition: Control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- Licht JD, Collins K. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes & Dev. 1999;13:1116–1125. doi: 10.1101/gad.13.9.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci. 1997a;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997b;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Livengood AJ, Zaug AJ, Cech TR. Essential regions of Saccharomyces cerevisiae telomerase RNA: Separate elements for Est1p and Est2p interaction. Mol Cell Biol. 2002;22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol Cell. 2000;6:361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Cech TR. Reversing time: Origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- Oubridge C, Ito N, Evans PR, Teo C-H, Nagai K. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- Perez-Canadillas J-M, Varani G. Recent advances in RNA-protein recognition. Curr Opin Struc Biol. 2001;11:53–58. doi: 10.1016/s0959-440x(00)00164-0. [DOI] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE. The function of the stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genetics. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. pp. 122–123. [Google Scholar]
- Roy J, Fulton TB, Blackburn EH. Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes & Dev. 1998;12:3286–3300. doi: 10.1101/gad.12.20.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Sperger JM, Cech TR. A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry. 2001;40:7005–7016. doi: 10.1021/bi0103359. [DOI] [PubMed] [Google Scholar]
- Taggart KP, Teng S-C, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Förstemann K, Gasser SM, Lingner J. Intracellular trafficking of yeast telomerase components. EMBO Rep. 2002;3:652–659. doi: 10.1093/embo-reports/kvf133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S-C, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfati Y, Fulton TB, Roy J, Blackburn EH. Template boundary in a yeast telomerase specified by RNA structure. Science. 2000;288:863–867. doi: 10.1126/science.288.5467.863. [DOI] [PubMed] [Google Scholar]
- Zhou J, Hidaka K, Futcher B. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol Cell Biol. 2000;20:1947–1955. doi: 10.1128/mcb.20.6.1947-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Mathews DH, Turner DH. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark BFC, editors. RNA Biochemistry and Biotechnology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]