Abstract
In eukaryotes, it is widely assumed that genes coding for proteins and structural RNAs do not overlap. Using a transposon-tagging strategy to globally analyze the Saccharomyces cerevisiae genome for expressed genes, we identified multiple insertions in an open reading frame that is contained fully within and transcribed antisense to the 25S rRNA gene in the nuclear rDNA repeat region on Chromosome XII. Expression of this gene, TAR1 (Transcript Antisense to Ribosomal RNA), can be detected at the RNA and protein levels, and the primary sequence of the corresponding 124-amino-acid protein is conserved in several yeast species. Tar1p was found to localize to mitochondria, and overexpression of the protein suppresses the respiration-deficient petite phenotype of a point mutation in mitochondrial RNA polymerase that affects mitochondrial gene expression and mtDNA stability. These findings indicate that coding information for protein and structural RNAs can overlap, raising issues regarding the coevolution of such complex genes, and also suggest that rDNA transcription and mitochondrial function are coordinately regulated in eukaryotic cells.
Keywords: rDNA, mitochondria, gene annotation, mitochondrial RNA polymerase, transposon
Present estimates as to gene number within fully sequenced eukaryotic genomes stem, in part, from computational algorithms designed to discriminate between coding and noncoding sequences. Often these algorithms use an explicit set of rules to make this distinction (Stein 2001); however, some genes do not conform to widely used rules regarding gene and genome architecture.
Stemming from a lack of evidence to the contrary, it has been assumed that protein-coding genes do not overlap sequences encoding ribosomal RNA (rRNA). In the budding yeast Saccharomyces cerevisiae, ∼140 rDNA genes (Goffeau et al. 1996) are arranged in a tandem array spanning >900 kb on Chromosome XII. As this locus was assumed to encode only rDNA, no protein-coding ORFs have been discovered. We report here a novel gene, TAR1 (Transcript Antisense to Ribosomal RNA), nested opposite to the 25S rRNA gene in the rDNA repeat region of Chromosome XII in yeast. By cellular fractionation and immunofluorescence analysis, we localize the Tar1p protein to the mitochondria. Consistent with a putative role as a mitochondrial protein, TAR1 expressed at low copy suppresses the petite phenotype of the R129D mutation in the mitochondrial RNA polymerase Rpo41p. In total, the discovery of TAR1 shatters the notion that structural RNAs and protein-coding sequences cannot overlap and holds significant implications in the evolution of these genes and the coordinate control of ribosome biogenesis and mitochondrial function.
Results and Discussion
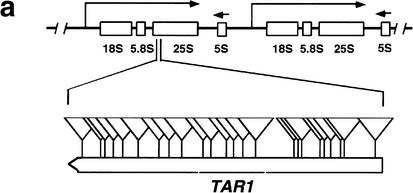
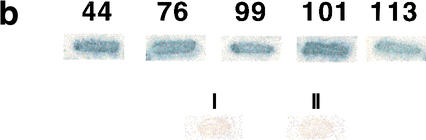
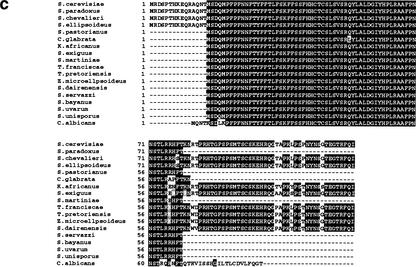
In the course of a large-scale analysis of the yeast genome by transposon tagging with a lacZ reporter that lacks both promoter sequences and an initiator ATG codon, we identified a number of transposon insertions in previously nonannotated open reading frames (NORFs; Ross-Macdonald et al. 1999; Kumar et al. 2002). In most cases, these insertions represent in-frame fusions of the NORF with the lacZ bacterial gene present in the transposon insertion construct. Intriguingly, we found ∼100 in-frame insertions in an NORF in the rDNA locus in S. cerevisiae Chromosome XII, a region not previously known to contain protein-coding sequences (Fig. 1a; Johnston et al. 1997). This NORF (hereafter called TAR1) is encoded on the opposite strand of that encoding the 25S rRNA gene (Fig. 1a; Philippsen et al. 1978; Petes 1979; Veldman et al. 1981). The TAR1 gene is 124 codons in length, encoding a protein with a predicted molecular mass of 14.34 kD. All of the insertion strains produce β-galactosidase activity as detected by filter assays (Fig. 1b), which is a strong indicator that TAR1 is transcribed and translated into protein. The high frequency with which the short TAR1 ORF was identified in the transposon screen (>50 times higher than the average yeast gene, size 1500 bp) suggests that many, if not all TAR1 units in the rDNA are transcribed. The Tar1p protein sequence is highly conserved in hemiascomycetous species (Fig. 1c; Souciet et al. 2000), but does not have any obvious motif or domain conserved in other proteins in the databases.
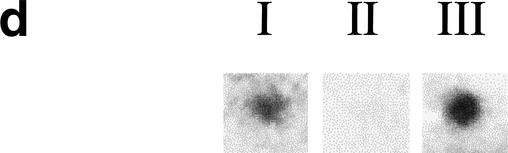
Figure 1.
(a) Diagram of two S. cerevisiae rDNA repeat units and minitransposon insertions in the TAR1 gene. The rDNA is transcribed from two start sites, one giving rise to the 37S transcript (subsequently processed into 18S, 5.8S, and 25S transcripts) and the other to the 5S transcript (indicated by boxes). The 25S rDNA region containing an open reading frame (TAR1) is enlarged in the bottom of the diagram; the TAR1 ORF is oriented in the antisense direction relative to the 25S rDNA; the 23 in-frame insertions of lacZ in TAR1 are indicated by triangles. (b) β-Galactosidase filter assays of five different insertion strains in TAR1. The numbers indicate the codon position of the transposon insertion in each insertion strain. Strains I and II carry insertions in regions of the 25S rDNA outside TAR1. (c) Protein sequence comparison of TAR1 homologs in different species of yeast. The indicated S. cerevisiae Tar1p sequence is translated from an ORF spanning base pairs 454697–455071 of Chromosome XII; for simplicity, coordinates within rDNA genes are provided for only the first repeat unit of the yeast rDNA locus. The predicted amino acid sequences were compared using the program CLUSTALW. Black shading indicates identity, whereas gray shading indicates similarity. (d) Dot-blot analysis of TAR1 expression. Biotinylated poly(A) RNA was hybridized against an array of 60-base oligonucleotides spotted onto nylon membrane; relevant signals are shown here. In lane I, labeled RNA was hybridized to an oligonucleotide complementary to the TAR1 transcript. Lane II indicates a lack of observable binding to an oligonucleotide derived from a noncoding region of yeast Chromosome XV. In lane III, labeled RNA is bound to an oligonucleotide complementary to PUT4, a known gene thought to encode a putative proline-specific permease.
To determine if the TAR1 gene is transcribed as RNA, we analyzed TAR1 expression levels by means of strand-specific, dot-blot analysis. Labeled poly(A) RNA was hybridized to a 60-base oligonucleotide complementary to the TAR1 coding sequence; identical assays were performed with oligonucleotides derived from known genes and noncoding segments of the yeast genome. As shown in Figure 1d, significant levels of TAR1 RNA transcript are observed relative to signals generated with oligonucleotides derived from noncoding regions of the yeast genome.
To determine if the TAR1 ORF is normally expressed as a protein, we tagged the C terminus of Tar1p with 13 copies of the myc epitope or with three copies of the haemagglutinin (HA) epitope (Tar1p–13myc and Tar1–3HA, respectively). As shown in Figure 2A (lane 2), two bands were detected by immunoblot analysis of total protein extracts probed with anti-myc antibodies. The protein with the slower mobility had an apparent molecular weight of ∼41 kD, and the protein of higher mobility had an apparent molecular weight ∼35 kD. The molecular weights of these proteins are similar to the predicted sizes of protein products derived from the first and second methionines of TAR1, respectively (Fig. 1c). Two bands were also detected when the TAR1 gene was tagged with three copies of the HA epitope (data not shown). Based on these findings and the fact that strains containing lacZ fusions throughout the TAR1 ORF express β-galactosidase activity (Fig. 1b), we conclude that TAR1 is a protein-coding gene encoded on the antisense strand of the 25S rRNA gene.
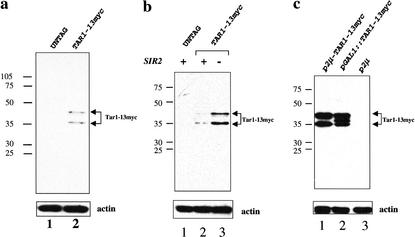
Figure 2.
Immunoblot analysis of Tar1p expression. Total protein extracts of logarithmically growing yeast cells carrying C-terminus 13myc-tagged TAR1 as a single copy in the chromosome (TAR1–13myc) or in different vectors (p2μ-TAR1–13myc, pGAL1∷TAR1–13myc) or untagged wild-type strains (untag) and a strain harboring an empty plasmid (p2μ) were prepared. The blots of separated proteins were probed with antibodies against the myc epitope. As a control for the protein amount loaded in each lane, the blots were stripped and probed with an antibody against actin. (a) Tar1p is detected as two bands (35 kD and 41 kD). (b) The amount of Tar1p in an SIR2 null strain (SIR2−) is increased relative to the wild-type strain (SIR2+). (c) Tar1p expression in strains carrying a multicopy vector containing TAR1 under the control of its own promoter (i.e., 500 bp of DNA upstream of the ATG, p2μ-TAR1–13myc) or a centromeric vector containing TAR1 under the control of the galactose-inducible GAL1 promoter (pGAL1∷TAR1–13myc). A third band is detected in the sample from a galactose-induced strain carrying pGAL1∷TAR1–13myc.
To learn more about the TAR1 protein we determined the subcellular localization of Tar1p, using indirect immunofluorescence and subcellular fractionation experiments. We detected a faint fluorescence signal in mitochondria of cells carrying a single chromosomal copy of the myc-tagged TAR1 gene. This mitochondrial localization was even more evident in cells that overexpressed Tar1p. A centromeric plasmid containing the 13Xmyc-tagged TAR1 gene under control of the inducible GAL1 promoter overproduces both forms of the protein when cells are grown on galactose (Fig. 2c, lane 2). Using indirect immunofluorescence of cells grown in medium containing galactose, we found that the tagged Tar1p localizes to the mitochondrial network (Fig. 3a, rows I,II); colocalization was observed with both mtDNA (DAPI staining) and proteins present in the outer mitochondrial membrane (αOM). Staining was not observed in cells grown in raffinose medium (data not shown) or in cells containing only the vector (Fig. 3a, row III). We also observed mitochondrial localization after overexpression of Tar1p tagged with three copies of the HA epitope (data not shown), in cells deleted for SIR2 (Fig. 3a, row IV; see below), and in cells carrying the TAR1 gene on a high-copy 2μ plasmid, under its endogenous promoter (Fig. 3a, row V).
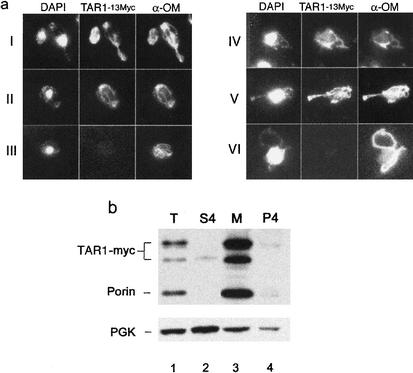
Figure 3.
Tar1p mitochondrial localization. (a) Micrographs of cells showing indirect immunolocalization of TAR1–13myc to the mitochondria using anti-myc antibody. The mitochondria were visualized by using an antibody against proteins present in the outer mitochondrial membrane (α-OM). The nucleus and mtDNA were visualized by staining the cells with DAPI. (Rows I,II) Two examples of galactose-induced cells carrying a centromeric vector containing TAR1 under the control of the galactose-inducible GAL1 promoter (pGAL1∷TAR1–13myc). (Row III) A galactose-induced cell containing an empty pGAL1 vector as a control. (Row IV) An sir2 cell harboring a 13myc-tagged TAR1 as a single copy in the chromosome. (Row V) A cell carrying a multicopy vector containing TAR1 under the control of its own promoter (p2μ-TAR1–13myc). (Row VI) A cell containing a multicopy (p2μ) empty vector as a control. (b) Fractionation of total cell extract by differential centrifugation. The samples derived from cells carrying a 13 myc-tagged TAR1 in the chromosome were prepared and analyzed by SDS-PAGE and immunoblotting. The blots were probed with an anti-myc antibody and anti-porin (mitochondrial marker) antibodies. The blot was stripped and probed again with an antibody against phosphoglycerokinase (PGK). T, total extract (lane 1); M, medium-speed mitochondrial-enriched pellet (lane 3). The supernatant of the medium-speed centrifugation was subjected to an ultracentrifugation, giving rise to a high-speed supernatant (S4; lane 2) and a high-speed pellet (P4; lane 4).
The subcellular localization of Tar1p was also examined using subcellular fractionation experiments. Total extracts of yeast cells expressing myc-tagged Tar1p from a single-copy chromosomal allele were prepared; mitochondria and other organelles were fractionated by differential centrifugation, and protein from the different fractions was analyzed by immunoblotting. Myc-tagged Tar1p cofractionated with porin, a mitochondrial protein, whereas the cytosolic protein, PGK, preferentially cofractionated with the soluble fraction (Fig. 3b, lanes 2,3). Altogether, these results demonstrate that Tar1p is a mitochondrial protein.
In an independent series of experiments, we also identified TAR1 in a random genetic screen for genes that when overexpressed suppress the phenotype of a point mutation that affects the mitochondrial RNA polymerase (Rpo41p; Greenleaf et al. 1986; Masters et al. 1987). This mutation changes arginine to aspartic acid at amino acid position 129 (R129D) in the N-terminal domain of the enzyme and causes a mitochondrial petite phenotype (impaired growth on YPG medium) at both 30°C and 37°C (Fig. 4; Rodeheffer et al. 2001). Initially, two separate low-copy centromeric plasmids, each containing a partial rDNA repeat unit (missing the 5S rRNA gene and large portions of the 18S and 25S rRNA genes), were found to suppress the rpo41-R129D mutation (Fig. 4a). Because the entire TAR1 ORF is present in both plasmids and Tar1p is localized to mitochondria, we predicted that TAR1 might be responsible for this multicopy suppressor activity. To test this idea, we transformed the R129D RNA polymerase mutant strain (rpo41-R129D) with a plasmid bearing TAR1 under control of the galactose-inducible GAL1 promoter. Tar1p expressed from this plasmid suppressed the rpo41-R129D growth defect manifested in glycerol medium at 30°C. Likewise, overexpression of TAR1 by the addition of galactose (0.2%) partially rescued the severe growth phenotype of the rpo41-R129D mutant at 37°C (Fig. 4b). These data indicate that the TAR1 gene encodes a mitochondrial protein that can rescue a defect in mitochondrial gene expression.
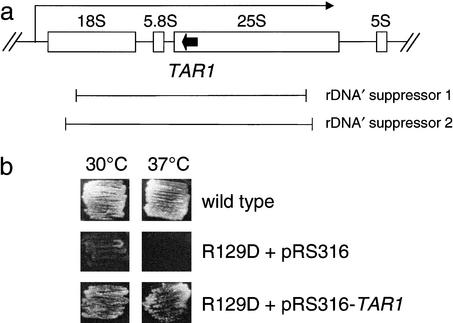
Figure 4.
Suppression of a mitochondrial RNA polymerase N-terminal-domain point mutation by a partial nuclear rDNA repeat containing TAR1. (a) Two independent plasmids harboring a portion of a single nuclear rDNA repeat (denoted rDNA‘ suppressors 1 and 2) were isolated as described (Bryan et al. 2002) through their ability to rescue the mitochondrial petite growth phenotype of the rpo41-R129D point mutation in mtRNA polymerase (Rpo41p) at 35°C. The genomic DNA insert contained in each plasmid is delineated in brackets below a diagram of a 9.1-kb rDNA repeat unit. The 18S, 5.8S, 25S, and 5S rRNA genes are depicted as boxes with the direction of transcription of the 35S rRNA precursor shown as a bent arrow. The TAR1 ORF embedded in the 25S rRNA is depicted as a solid black arrow, with the direction of the arrow indicating that it is transcribed antisense to the 25S rRNA gene. (b) Tar1p expressed from a GAL1 promoter suppresses the rpo41-R129D mutant phenotype. A plasmid that expresses TAR1 from the GAL1 promoter (pRS316-TAR1) was introduced into the rpo41-R129D strain (labeled R129D) as was the empty vector (pRS316) as a control. Under glycerol growth conditions (YPG medium + 0.2% galactose), the GAL1 promoter is derepressed, allowing elevated expression of Tar1p from the plasmid. At both 30°C (left column) and 37°C (right column) on YPG, the rpo41-R129C strain exhibits a severe growth defect compared with the wild-type strain, GS122. Expression of Tar1p from the GAL1 promoter (+pRS316-TAR1) substantially rescues this petite phenotype at both temperatures.
TAR1 is readily expressed when the coding sequence plus 500 bp upstream of the ATG are inserted into a high-copy plasmid (Fig. 2c; Fig. 3, row V), suggesting that it forms its own transcription unit in the rDNA. Some genes inserted into the rDNA locus are repressed by the silencing activity of Sir2p (Smith and Boeke 1997). To determine if TAR1 expression is affected by the activity of Sir2p, we compared the protein levels of the single copy, chromosomally encoded myc-tagged Tar1p in an SIR2 null strain with that of a wild-type (SIR2+) strain. A 2.5-fold increase in the amount of Tar1p–13myc was observed in the SIR2 deletion strain (Fig. 2b, lane 3). In addition, we readily detected Tar1p–13myc by indirect immunofluorescence in an SIR2 null strain carrying a single copy of the construct in the chromosome, whereas the protein was difficult to detect in SIR2 wild-type cells (Fig. 3a, row IV).
Our results demonstrate for the first time that the rDNA locus contains overlapping genes; the 25S rRNA is produced from one strand (as part of a larger 35S rRNA precursor), and Tar1p coding information is located on the opposite strand. Although the rRNA is transcribed by Pol I, it is likely that TAR1 is transcribed by Pol II because (1) the RNA is present in polyA+ RNA; (2) the RNA encodes protein; and (3) expression is elevated in sir2 mutant strains—all features normally ascribed to RNA polymerase II-transcribed messages. Additional experiments will be necessary to directly test this hypothesis.
After our paper was submitted, Kermekchiev and Ivanova (2001) reported the identification of a protein, ribin, whose cDNA sequence is derived from sequence complementary to the large rRNA in mouse, and from a different region than the Tar1 protein. The ribin cDNA sequence was not identical to the antisense strand of the rRNA, and the 3′ untranslated region does not contain rDNA sequences; therefore, the ribin transcript may not be encoded at the rDNA locus. The ribin protein is involved in rRNA transcription (presumably in the nucleolus), rather than mitochondrial function, raising the possibility that both rRNA complementary sequences are involved in gene expression, albeit in different cellular compartments.
The presence of overlapping genes in the rRNA locus, one encoding a protein and the other encoding a structural (and probably catalytic) RNA, raises interesting issues regarding the origin and conservation of such genes. It is interesting to speculate that the rRNA gene evolved first in the RNA world, and that the advent of protein-coding RNAs arose from the exact same gene, albeit it on the other strand (Joyce and Orgel 1993). Regardless of the mechanism, these genes are expected to be tightly linked in their evolution; presumably it will be difficult to mutate one gene without affecting the function of the other, and thus the sequence of this region is expected to be highly conserved. Such constraints might be reflected in the observation that the TAR1 ORF is highly conserved among very diverse yeast species.
The function of Tar1p is not known. However, the N-terminal domain of mtRNA polymerase is involved in mtDNA stability (Wang and Shadel 1999) and interacts with Nam1p (Rodeheffer et al. 2001) and Sls1p (Bryan et al. 2002), two proteins involved in posttranscriptional gene expression in mitochondria. The presence of Tar1p in mitochondria and its ability to suppress the petite phenotype of a point mutation in this domain suggest that this protein increases the efficiency of oxidative phosphorylation directly or indirectly, by influencing mtDNA stability or mitochondrial gene expression at the posttranscriptional level. The fact that this gene is not highly expressed during vegetative growth, but is elevated in a sir2 null mutant strain, might indicate that Tar1p expression and rDNA transcription are inversely regulated. This type of regulation could provide a means to coordinate rDNA transcription and mitochondrial function in response to changing cellular needs or energy demands, for example, under different types of growth condition, during mitosis, or in aging cells. Additional studies are likely to further elucidate the function of Tar1p and its role in coordinating nuclear and mitochondrial functions.
Materials and methods
Strains and plasmid construction
Growth media and genetic manipulation were as described (Guthrie and Fink 1991). All strains are derivatives of the diploid strain Y270, a descendant of S228C, except for the sir2 and its isogenic wild-type strains, which are derived from the deletion collection available from Research Genetics (Winzeler et al. 1999). The epitope-tagged strains were grown at 30°C in YPAD medium (liquid media supplemented with adenine; Sherman et al. 1986). Plasmid-bearing strains were grown in synthetic complete medium. Cells bearing the GAL1-promoter plasmids were induced by adding galactose to the liquid medium to a final concentration of 2%.
The strain containing TAR1–13myc in the chromosome was constructed using the PCR epitope-tagging method of Longtine et al. (1998). The 13-myc epitope followed by the ADH1 terminator was integrated at the C terminus of the coding region (before the termination codon). Oligonucleotides BW1 (5′-CCAAATTACAACTCGGGCACCGAAGGTAC CAGATTTCAAATTCGGATCCCCGGGTTAATTAA-3′) and BW2 (5′-CCGGGATTGCCTTAGTAACGGCGAGTGAAGCGGCAAAAGCGA ATTCGAGCTCGTTTAAAC-3′) were used. Correct integration of the myc tag was confirmed by PCR analysis. The tagged constructs appear to be present at single or low copy in the rDNA as determined by PCR analysis.
p101, a CEN–URA3 plasmid containing TAR1 tagged with 13 copies of the myc epitope at the C terminus under the control of the inducible GAL1 promoter and ADH1 terminator (pGAL1∷TAR1–13MYC) was constructed as follows. A C-terminal fusion between TAR1 and 13 myc was made by in vivo homologous recombination between two PCR fragments (A + B) and a linearized vector. The first fragment (A), containing the TAR1 gene, was PCR-amplified from plasmid pRDN-wt-U (Chernoff et al. 1994; a gift from J. Warner, Albert Einstein College of Medicine) using oligonucleotide CC1 (5′-GTCAAGGAGAAAAAACCCCGGATC CAAGCTTATGCGAGATTCCCCTACC-3′, where the italicized portion is complementary to the TAR1 5′-ORF sequence, and the underscored portion is complementary to the vector pGAL1) and oligonucleotide CJ (5′-TTAATTAACCCGGGGATCCGAATTTGAAATCTGGTA CC-3′, where the italicized portion is complementary to the TAR1 C-terminal codons, and the underscored portion is complementary to the reading frame for the myc epitope in plasmid pFA6a-13myc-His3MX6 (Longtine et al. 1998). The second fragment (B), containing the 13 myc epitopes followed by the ADH1 terminator, was PCR-amplified from plasmid pFA6a-13myc-His3MX6 using oligonucleotide CI (5′-GGTAC CAGATTTCAAATTCGGATCCCCGGGTTAATTAA-3′, complementary to oligonucleotide CJ, see above) and oligonucleotide CE (5′-AA CAAAAGCTGGGTACCGGGCCCCCCCTCGAGGAGGAGGGAAGC TAAACAGATC-3′, where the italicized portion is complementary to the ADH1 terminator sequence, and the underscored portion is complementary to the vector pGAL1). The vector pGAL1 (a kind gift from Susana Vidan) was constructed by subcloning an EcoRI–SalI fragment containing the divergent GAL1,2 promoter from plasmid pBM272 (Johnston and Davis 1984) into an EcoRI–SalI-digested pRS316 (Sikorski and Hieter 1989). The final construct was obtained after yeast cotransformation of PCR fragments A and B and an XhoI–HindIII-cut pGAL1.
p102, a 2μ–TRP1 plasmid containing TAR1 tagged with 13 copies of the myc epitope at the C terminus under the control of its own promoter (500 bp of the sequence upstream of the ATG) and ADH1 terminator (p2μ–TAR1–13MYC) was constructed using the same strategy as for p101 (pGAL1∷TAR1–13MYC). The first fragment (A), containing the TAR1 gene, was PCR-amplified from plasmid pRDN-wt-U using oligonucleotide CQ1 (5′-TCGACGGTATCGATAAGCTTGATATCGAATTCAAC TGATACGAGCTTCTG-3′, where the italicized portion is complementary to a sequence 500 bp upstream of the TAR1 start codon, and the underscored portion is complementary to the vector pRS424) and oligonucleotide CJ (see above). The second fragment (B), containing the 13 myc epitopes followed by the ADH1 terminator, was PCR-amplified from plasmid pFA6a-13myc-His3MX6 using oligonucleotide CI (5′-GG TACCAGATTTCAAATTCGGATCCCCGGGTTAATTAA-3′, complementary to oligonucleotide CJ, see above) and oligonucleotide CQ2 (5′-GCGGTGGCGGCCGCTCTAGAACTAGTGGATCCAGGCAAGCTA AACAGATC-3′, where the italicized portion is complementary to the ADH1 terminator sequence, and the underscored portion is complementary to the vector pRS424). The final construct was obtained after yeast cotransformation of PCR fragments A and B and an EcoRI–BamHI-cut pRS424 (Sikorski and Hieter 1989).
Indirect immunofluorescence
DAPI staining and immunofluorescence techniques were performed as described (Pringle et al. 1991). TAR1–13myc was detected by incubation with preabsorbed mouse monoclonal anti-myc primary antibody (9E10 from BABCO) diluted 1:400, followed by incubation with preabsorbed CY-3 conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch Laboratory, Inc.) diluted 1:600. Mitochondrial outer membrane proteins were detected by incubation with rabbit α-OM (gift from M. Smith and L. Pon, Columbia University) diluted 1:400, followed by incubation with FITC-conjugated goat anti-rabbit secondary antibodies diluted 1:600.
Immunoblot analysis and subcellular fractionation
Protein extracts were prepared from cells grown under YPAD rich medium (chromosome-tagged strain) or under selective conditions (plasmid-bearing strains) until mid-log phase. Cells were washed in cold water and lysed using glass beads in 250 μL of lysis buffer (50 mM Tris at pH 7.5, 100 mM NaCl, 10 mM EDTA, 0.1% Triton, 0.1% β-mercapthoethanol), 0.5 mM PMSF, and a Roche complete minimum protease inhibitor cocktail. The lysates were suspended in 5× Laemmli buffer, boiled 5 min before separating the proteins by SDS-polyacrylamide gel electrophoresis using a 12% minigel. The proteins were transferred onto Immobilon-P (Millipore) and probed with preabsorbed mouse monoclonal anti-myc primary antibody (9E10 from BABCO), which was detected using secondary goat anti-mouse horseradish peroxidase and ECL detection reagents (Amersham). The subcellular fractionation was performed essentially as described (Glick and Pon 1995).
RNA blot analysis
Poly(A) RNA isolated from vegetatively growing yeast (strain Y800) was directly labeled using biotinylated psoralen (Ambion). Biotinylated RNA was subsequently hybridized against a series of 60-bp oligonucleotides spotted onto a nylon membrane CAST microscope slide (Schleicher and Schuell). Oligonucleotides were selected from sequences complementary to the predicted TAR1 transcript, as well as from sequences encoding the known gene PUT4 and an intergenic region of yeast Chromosome XV (nt 702742–702801) predicted to be noncoding. The oligonucleotide designed from this noncoding region is as follows: 5′-GACGATGCGAGAAA GTCTAACATGATTAGATTGGCACCTGCACATTATATATTGCGT TTC-3′. No appreciable RNA hybridization to this sequence was detected; similar results have been observed for hundreds of other predicted yeast noncoding sequences assayed by this method (Kumar et al. 2002).
Acknowledgments
We thank M. Smith and D. Gelperin for critical comments on the manuscript, and M. Smith and L. Pon for the anti-porin antibodies. This research was supported by grants from the NIH. to M.S. and G.S.S. P.S.R.C. was supported by an FAPESP (Sao Paulo, Brazil) postdoctoral fellowship and by a grant from the Burroughs Wellcome foundation. A.K. was supported by a postdoctoral fellowship from the American Cancer Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL michael.snyder@yale.edu; FAX (203) 432-6161.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1035002.
References
- Bryan AC, Rodeheffer MS, Wearn CM, Shadel GS. Sls1p is a membrane-bound regulator of transcription-coupled processes involved in Saccharomyces cerevisiae mitochondrial gene expression. Genetics. 2002;160:75–82. doi: 10.1093/genetics/160.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Vincent A, Liebman SW. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 1994;13:906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Pon LA. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Jonhston M, et al. Life with 6,000 genes. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Greenleaf AL, Kelly JL, Lehman IR. Yeast RPO41 gene product is required for transcription and maintenance of the mitochondrial genome. Proc Natl Acad Sci. 1986;83:3391–3394. doi: 10.1073/pnas.83.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. In: Abelson JNaSM., editor. Methods in enzymology. San Diego: Academic Press; 1991. pp. 1–933. [PubMed] [Google Scholar]
- Johnston M, Davis RW. Sequences that regulate the divergent GAL1–GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Hillier L, Riles L, Albermann K, Andre B, Ansorge W, Benes V, Bruckner M, Delius H, Dubois E, et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome XII. Nature. 1997;29:87–90. [PMC free article] [PubMed] [Google Scholar]
- Joyce GF, Orgel LE. Prospects for understanding the origin of the RNA world. In: Gesteland RFA, Atkins JF, editors. The RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1–25. [Google Scholar]
- Kermekchiev M, Ivanova L. Ribin, a protein encoded by a message complementary to rRNA, modulates ribosomal transcription and cell proliferation. Mol Cell Biol. 2001;21:8255–8263. doi: 10.1128/MCB.21.24.8255-8263.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Harrison PM, Cheung K-H, Lan N, Echols N, Bertone P, Miller P, Gerstein MB, Snyder M. An integrated approach for finding overlooked genes in yeast. Nat Biotech. 2002;20:58–63. doi: 10.1038/nbt0102-58. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Masters BS, Stohl LL, Clayton DA. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- Petes TD. Yeast ribosomal DNA genes are located on chromosome XII. Proc Natl Acad Sci. 1979;76:410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen P, Thomas M, Kramer RA, Davis RW. Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol. 1978;123:387–404. doi: 10.1016/0022-2836(78)90086-4. [DOI] [PubMed] [Google Scholar]
- Pringle J, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Rodeheffer MS, Boone BE, Bryan AC, Shadel GS. Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J Biol Chem. 2001;276:8616–8622. doi: 10.1074/jbc.M009901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-MacDonald P, Coelho PSR, Roemer T, Agarwal S, Kumar A, Jansen R, Cheung K-H, Sheehan A, Symoniatis D, Umansky L, et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink G, Hicks J. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes & Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Souciet J, Aigle M, Artiguenave F, Blandin G, Bolotin-Fukuhara M, Bon E, Brottier P, Casaregola S, de Montigny J, Dujon B, et al. Genomic exploration of the hemiascomycetous yeasts: 1. A set of yeast species for molecular evolution studies. FEBS Lett. 2000;487:3–12. doi: 10.1016/s0014-5793(00)02272-9. [DOI] [PubMed] [Google Scholar]
- Stein L. Genome annotation: From sequence to biology. Nat Rev Genet. 2001;2:493–503. doi: 10.1038/35080529. [DOI] [PubMed] [Google Scholar]
- Veldman GM, Klootwijk J, de Regt VC, Planta RJ, Branlant C, Krol A, Ebel JP. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981;9:6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shadel GS. Stability of the mitochondrial genome requires an amino-terminal domain of yeast mitochondrial RNA polymerase. Proc Natl Acad Sci. 1999;96:8046–8051. doi: 10.1073/pnas.96.14.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]