Abstract
Messenger RNA export factors are recruited to genes in a transcription-dependent manner. To ascertain the mechanism of this process, we show that RNA polymerase II transcription is sufficient to recruit the Saccharomyces cerevisiae hnRNP protein Npl3 to a gene independent of RNA sequence. In contrast, the cotranscriptional recruitment of the RNA-binding protein Yra1 is dependent on pre-mRNA processing. Yra1 associates with introns of intron-containing genes in a splicing-dependent manner. Conversely, Yra1 recruitment to genes without introns is not dependent on splicing. Finally, 3′-end formation is required for Yra1 recruitment to genes regardless of intron status.
Keywords: Yra1, Npl3, mRNA export, transcription, splicing, chromatin IP
In eukaryotes, mRNAs are transcribed and processed in the nucleus and exported to the cytoplasm. During transcription, mRNAs are bound by RNA-binding proteins that function in pre-mRNA processing steps such as capping, polyadenylation/cleavage, and splicing (for review, see Bentley 2002; Proudfoot et al. 2002). In addition, transcription promotes the recruitment of mRNA export factors to genes (Lei et al. 2001). The cotranscriptional recruitment of pre-mRNA processing and mRNA export factors has been suggested to be a mechanism for efficient production of fully processed and export-competent ribonucleoparticles (RNPs).
In Saccharomyces cerevisiae, Npl3 is an abundant protein that serves as a prototype for hnRNP function in mRNA export. Identified based on its ability to UV-cross-link to poly(A)+ RNA (Wilson et al. 1994), Npl3 is one of the major RNA-binding proteins in yeast and may function to ensure structural integrity of the RNP. Mutation of NPL3 results in accumulation of mRNA in the nucleus (Singleton et al. 1995; Lee et al. 1996). Furthermore, Npl3 shuttles between the nucleus and the cytoplasm, and its export is dependent on ongoing transcription by RNA polymerase II (Pol II; Lee et al. 1996). Npl3 interacts physically with Pol II and genetically with TATA-binding protein (TBP) to promote mRNA export (Lei et al. 2001). For Npl3, cotranscriptional recruitment begins at an early stage of transcription, either at initiation or soon after elongation begins. Finally, Npl3 cotranscriptional recruitment is specific for Pol II-transcribed genes.
Nuclear export of mRNA also depends on the function of the RNA-binding protein Yra1. Acting at a later stage of mRNA export than Npl3, Yra1 bridges the interaction between the RNP and the soluble mRNA export receptor heterodimer Mex67/Mtr2 located primarily at the nuclear pore complex (NPC; Santos-Rosa et al. 1998; Sträßer and Hurt 2000). The human homolog of Yra1, Aly, is a component of the exon–exon junction complex (EJC) that is deposited on mRNAs as a result of splicing (Le Hir et al. 2000). Although there is not yet evidence of the existence of an EJC in yeast, both Aly and the nonsense-mediated decay factor Upf3 associate with the EJC and are conserved throughout eukaryotes (Le Hir et al. 2001). Aly promotes the export of both spliced and unspliced mRNAs (Zhou et al. 2000; Rodrigues et al. 2001), and interaction of Aly with the splicing factor UAP56 has been suggested to be a mechanism of recruitment of Aly to spliced mRNAs (Luo et al. 2001). Similarly, Yra1 interacts with the UAP56 yeast homolog Sub2 (Sträßer and Hurt 2001), which is also involved in splicing (Kistler and Guthrie 2001; Libri et al. 2001; Zhang and Green 2001). In turn, Sub2 and its human homolog recently have been shown to physically interact with the conserved THO complex of transcription elongation factors, which are required for proper mRNA export (Sträßer et al. 2002). In fact, Aly was originally isolated as a transcriptional coactivator (Bruhn et al. 1997).
Less well understood is how 3′-end formation of mRNA contributes to its nuclear export. Disruption of cleavage/polyadenylation by either cis- or trans-acting mutations results in nuclear accumulation of transcripts (Eckner et al. 1991; Long et al. 1995; Huang and Carmichael 1996; Brodsky and Silver 2000). Furthermore, these defects cause accumulation of transcripts at a focus that may be near the site of transcription (Hilleren et al. 2001; Jensen et al. 2001). One candidate for a mediator of 3′-end formation stimulated mRNA export is the hnRNP protein Hrp1, which is a component of the cleavage complex CF I (Kessler et al. 1997). Hrp1 interacts genetically with Npl3 and is required for nuclear export of mRNA (Henry et al. 1996; Brodsky and Silver 2000).
Messenger RNA export factors are recruited to genes in a transcription-dependent manner to increase the efficiency of mRNA export (Lei et al. 2001). However, the mechanism of this cotranscriptional recruitment is not known. Here we show that Pol II transcription is sufficient to cotranscriptionally recruit Npl3 to an RNA. In contrast, Yra1 associates with intron-containing genes preferentially at introns in a splicing-dependent manner. Moreover, the splicing factor Sub2 associates with genes in a pattern identical to that of Yra1. However, SUB2 is not required for Yra1 recruitment to genes without introns. Additionally, 3′-end formation is required for Yra1 recruitment to all genes tested regardless of intron status. We present a model for cotranscriptional pre-mRNA processing-dependent packaging of mRNAs for nuclear export.
Results and Discussion
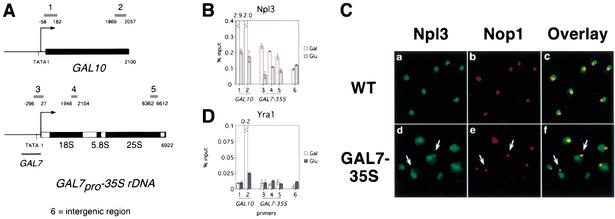
The specificity of Npl3 recruitment to mRNA could be achieved through interaction between Npl3 and RNA or by interaction of Npl3 with the Pol II transcription machinery. To test these two possibilities, we examined whether Npl3 can be recruited to a gene that is not normally transcribed by Pol II using a strain (YJV100) that expresses the 35S rDNA under the control of the inducible GAL7 Pol II promoter (GAL7pro–35S rDNA; Venema et al. 1995). This fusion gene is integrated into the rDNA locus in multiple copies, and the strain is deleted for RNA polymerase I (Δrpa135) so that all rRNA transcription is driven by Pol II. Cells were grown in galactose-containing media to stimulate transcription of galactose-dependent genes, including the Pol II-driven rRNA, or were transcriptionally repressed by addition of glucose for 1 h. We performed chromatin immunoprecipitations of Npl3 from chromatin of an average size of ∼200 bases using α-Npl3 antibodies. Quantitative PCR was performed using primer sets that span the galactose-dependent GAL10 gene, the GAL7pro–35S rDNA fusion gene, or an intergenic nontranscribed region to determine the amount of DNA associated with Npl3 (Fig. 1A).
Figure 1.
Pol II transcription is sufficient for Npl3 but not Yra1 cotranscriptional recruitment. (A) Diagram of GAL10 and GAL7pro–35S rDNA. Primer sets 1 and 2 span the 5′ and 3′ coding sequence of GAL10, respectively. Primer set 3 is specific for the GAL7pro–35S rDNA fusion. Primer sets 4 and 5 are directed to the 18S and 25S rDNA, respectively, recognizing the fusion gene as well as endogenous rDNA. (For GAL10, ATG = +1; for GAL7pro–35S rDNA, 5′ ETS = +1.) Primer set 6 spans a nontranscribed intergenic region. (B) Quantitation of Npl3-associated DNA using primer sets indicated in A. Raw values are expressed as a percentage of input for cells grown at 30°C in media containing 1% raffinose/1% galactose (white) and repressed for 1 h in 2% glucose (gray). Error bars are shown for a single experiment. Numerical values are shown above broken bars, which extend beyond the upper limit of the graph. (C) Colocalization of Npl3 and Nop1 in wild-type (WT, a–c) and GAL7pro–35S rDNA-expressing (GAL7–35S, d–f) cells. Indirect immunofluorescence with polyclonal antibodies to Npl3 (a,d), monoclonal antibody to Nop1 (b,e), and overlay (c,f) are shown. Arrows point to the region of colocalization in GAL7pro–35S rDNA-expressing cells. (D) Quantitation of Yra1–myc-associated DNA using primer sets indicated in A.
Npl3 associates with the GAL7pro–35S rDNA fusion gene only when transcribed by Pol II. In cells grown in galactose, Npl3 associates strongly with the coding sequence of GAL10, and this association is reduced 10-fold when cells are glucose-repressed, similar to previous results (Fig. 1B, lanes 1,2; Lei et al. 2001). Association of Npl3 with the GAL7pro–35S rDNA is approximately twofold higher than the nontranscribed intergenic region in cells grown in galactose (Fig. 1B, lanes 3–6, white bars). Upon glucose repression, Npl3 association with the GAL7pro–35S rDNA is decreased to background levels (Fig. 1B, lanes 3–6, gray bars). Chromatin immunoprecipitations using an antibody against Pol II (8WG16) show a similar pattern of association with GAL10 and the GAL7pro–35S rDNA (data not shown). In wild-type cells, Npl3 does not associate with Pol I-transcribed 35S rDNA (data not shown). These results indicate that Pol II transcription is sufficient for Npl3 cotranscriptional recruitment independent of RNA sequence.
In cells expressing the GAL7pro–35S rDNA fusion gene, Npl3 is redistributed to the nucleolus. In wild-type cells, Npl3 localizes to a region of the nucleus that is adjacent to the crescent-shaped nucleolus, denoted by Nop1 staining, as previously described (Fig. 1C, panels a–c; Flach et al. 1994). When grown in galactose, cells expressing the GAL7pro-35S rDNA possess nucleoli that appear as a single focus that is adjacent but clearly distinct from the nucleoplasm as determined by comparison of Nop1 localization and DAPI staining (Fig. 1C, e; data not shown). In these cells, Npl3 staining is visible in the nucleoplasm as in wild type but also extends to a small adjacent region. In this area there is significant overlap of the Npl3 and Nop1 signals as indicated by orange to yellow signal in the overlay (Fig. 1C, d–f, arrows). Taken together, these results indicate that Pol II transcription is sufficient for Npl3 cotranscriptional recruitment.
Unlike Npl3, Yra1 is not recruited to the GAL7pro–35S rDNA. Epitope-tagged Yra1–myc was immunoprecipitated using α-myc antibody (9E11), and the amount of DNA associated was quantitated. In cells grown in galactose, Yra1–myc associates strongly with the 3′ but not the 5′ end of the GAL10 coding sequence compared with the nontranscribed intergenic region, and this association decreases >20-fold upon glucose repression, similar to previous results (Fig. 1D, lanes 1,2,6; Lei et al. 2001). In contrast, Yra1–myc association is not above background levels for the 5′, middle, or 3′ regions of the GAL7pro–35S rDNA even under transcriptionally repressing conditions (Fig. 1D, lanes 3–6). Therefore, Pol II transcription is not sufficient for Yra1 cotranscriptional recruitment. A possible explanation for the lack of Yra1 recruitment to this gene is that a specific binding site for Yra1 may not be present in the 35S rRNA. Alternately, because the 35S rRNA undergoes a series of processing steps that is entirely distinct from that of mRNA, some pre-mRNA processing steps may be a requirement for Yra1 recruitment.
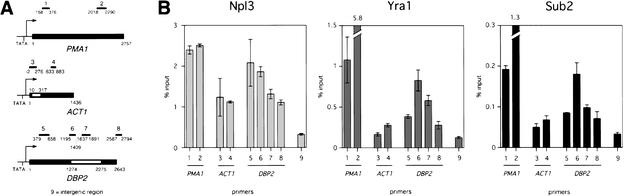
To test whether splicing affects mRNA export factor cotranscriptional recruitment, we first tested whether recruitment is affected by the presence of an intron. We performed chromatin immunoprecipitations using as reporters three genes with different intron statuses (Fig. 2A). PMA1 is a gene without an intron, and ACT1, like the majority of intron-containing genes in S. cerevisiae, contains a single intron that is situated close to the transcription initiation site, resulting in a short 5′ exon. For DBP2 and a handful of intron-containing genes, a single intron is located in the middle of the gene to produce a large 5′ exon of >1 kb. We predicted that if an EJC deposited by splicing exists in S. cerevisiae, it would be able to form on DBP2 but not ACT1 because a short 5′ exon is not capable of supporting EJC formation (Le Hir et al. 2001). As shown previously, Yra1–myc preferentially associates with the 3′ end of the intronless PMA1 gene more than fivefold compared with its 5′ end (Fig. 2B, center, lanes 1,2; Lei et al. 2001). Likewise, Yra1–myc association with ACT1 is higher at the 3′ end; however, the difference between the 3′ and 5′ ends is less than twofold, and overall levels of ACT1 association are considerably closer to background than PMA1 (Fig. 2B, center, lanes 1–4,9). In contrast, Yra1–myc association is biased clearly to the intron of DBP2, peaking at the intron at least twofold higher than the first and second exons, suggesting the existence of an EJC (Fig. 2B, center, lanes 5–9). It remains a possibility that an EJC or equivalent in S. cerevisiae is deposited cotranscriptionally on the ACT1 transcript but may be indetectable by this assay because of the proximity of the intron to the transcription start site. Notably, the characteristic 3′ bias of Yra1 is absent on DBP2, indicating that Yra1 associates differently with genes depending on intron status.
Figure 2.
Effects of intron status on mRNA export factor cotranscriptional recruitment. (A) Diagram of PMA1, ACT1, and DBP2. Primer sets 1 and 2 span the 5′ and 3′ coding sequence, respectively, of PMA1. Primer sets 3 and 4 span the intron and second exon, respectively, of ACT1. Primer sets 5, 6, 7, and 8 span the first exon, 5′-exon–intron junction, intron, and second exon, respectively of DBP2 (ATG = +1). Primer set 9 spans a nontranscribed intergenic region. (B) Quantitation of Npl3- (left), Yra1–myc- (center), and Sub2–HA-associated (right) DNA using primer sets indicated in A. Raw values are expressed as a percentage of input for cells grown in YPD at 30°C. Error bars are shown for a single experiment. Numerical values are shown above broken bars, which extend beyond the upper limit of the graph.
Sub2 is cotranscriptionally recruited in a manner similar to Yra1 to genes with and without introns. Sub2–HA was immunoprecipitated from chromatin using an α-HA antibody (12CA5), and the amount of DNA associated was quantitated. Consistent with the finding that Yra1 and Sub2 interact directly (Sträßer and Hurt 2001), the pattern of Sub2–HA association with PMA1, ACT1, and DBP2 is essentially identical to that of Yra1 (Fig. 2B, right, lanes 1–9). In addition, Sub2–HA is recruited strongly to the middle and 3′ end of the galactose-inducible gene GAL10 under transcriptionally active conditions comparable to Yra1, indicating that Sub2 recruitment to a gene is transcription-dependent (data not shown; Lei et al. 2001). Like Yra1, Sub2 is recruited to genes with and without introns, consistent with the result that SUB2 is required for the nuclear export of spliced and intronless mRNAs (Lei et al. 2001; Sträßer and Hurt 2001). In contrast to Yra1 and Sub2, Npl3 association with PMA1 and ACT1 is evenly distributed (Fig. 2B, left, lanes 1–4). For DBP2, Npl3 association is strongest at the 5′ end and tapers toward the 3′ end of the coding sequence similar to other genes tested (Fig. 2B, left, lanes 5–9; Lei et al. 2001). To reinforce our conclusions regarding intron status, we also tested the RPL30 and NOG2 genes, which display similar intron positions to ACT1 and DBP2, respectively, and found their behavior to be consistent (data not shown). Therefore, the presence and possibly the position of an intron affects Yra1 and Sub2 but not Npl3 cotranscriptional recruitment.
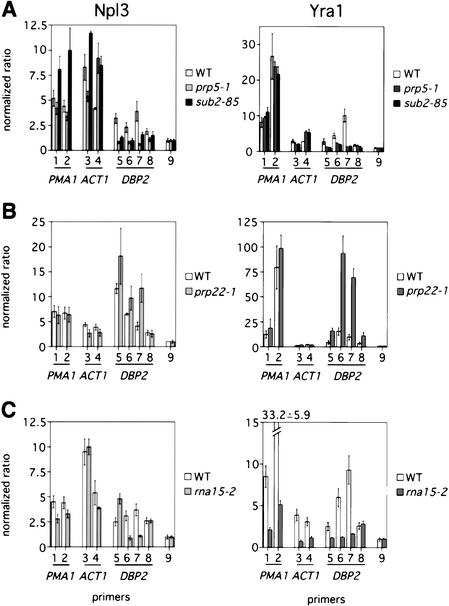
To determine whether splicing is required for mRNA export factor recruitment to genes, we examined cotranscriptional recruitment of Npl3 and Yra1 in the trans-acting splicing mutants prp5-1 and sub2-85. Mutations in both genes cause defects in early steps of spliceosome formation (Lustig et al. 1986; Kistler and Guthrie 2001; Zhang and Green 2001). Compared with wild type, prp5-1 mutants display normal recruitment of Npl3 to PMA1 and ACT1, although association with the second exon of ACT1 is increased by twofold (Fig. 3A, left, lanes 1–4, white and gray bars). However, Npl3 does not associate with DBP2 significantly above background in prp5-1 cells (Fig. 3A, left, lanes 5–9, white and gray bars). Similarly, in sub2-85 cells, Npl3 is not recruited to DBP2 but is increased up to 1.5-fold at PMA1 and ACT1 (Fig. 3A, left, lanes 1–9, white and black bars), showing that the requirement for splicing is specific for DBP2.
Figure 3.
Mutants in splicing and 3′-end formation affect mRNA export factor cotranscriptional recruitment. (A) Quantitation of Npl3- (left) and Yra1–myc-associated (right) DNA using primer sets indicated in Figure 2A from wild-type (white), prp5-1 (gray), and sub2-85 (black) cells. Values are normalized to the intergenic region, which is arbitrarily set to 1. Error is shown for a single experiment. Cells were grown at 25°C and shifted to 37°C for 30 min. (B) Quantitation of Npl3- (left) and Yra1–myc-associated (right) DNA using the primer sets indicated in Figure 2A from wild-type (white) and prp22-1 (gray) cells. Values are normalized to the intergenic region. Cells were grown at 25°C and shifted to 37°C for 2 h. (C) Quantitation of Npl3- (left) and Yra1–myc-associated (right) DNA using the primer sets indicated in Figure 2A from wild-type (white) and rna15-2 (gray) cells. Values are normalized to the intergenic region. Cells were grown at 25°C and shifted to 37°C for 30 min.
Splicing is also required for Yra1 recruitment to DBP2 but not PMA1 or ACT1. In comparison with wild type, Yra1 is recruited normally to PMA1 and ACT1 in the prp5-1 and sub2-85 mutant strains (Fig. 3A, right, lanes 1–4). In contrast, Yra1 does not associate with DBP2 above background levels in either mutant strain (Fig. 3A, right, lanes 5–9). Because sub2-85 and prp5-1 mutants behave very similarly in our cotranscriptional recruitment assays, we conclude that the function of SUB2 in recruitment of export factors is the same as that of a splicing factor. Furthermore, SUB2 is required for export of unspliced mRNAs (Sträßer and Hurt 2001) but does not affect mRNA export factor cotranscriptional recruitment to genes without introns; therefore, Sub2 must play a role in mRNA export that does not involve Yra1 recruitment.
To further test the hypothesis that splicing may be required for export factor recruitment, we performed the converse experiment of slowing mRNA release from the spliceosome and monitoring the association of export factors with genes. In comparison with wild type, Npl3 recruitment to PMA1 and ACT1 is unchanged in prp22-1 cells, which are defective at a late step of splicing (Fig. 3B, left, lanes 1–4; Company et al. 1991). However, Npl3 association with the intron of DBP2 is increased 1.5-fold to twofold in prp22-1 cells compared with wild type (Fig. 3B, left, lanes 5–9). Similarly, Yra1 recruitment to PMA1 and ACT1 is not altered (Fig. 3B, right, lanes 1–4). In sharp contrast, Yra1 recruitment is increased approximately threefold over the first and second exons of DBP2 and is enriched more than sixfold over the DBP2 intron (Fig. 3B, right, lanes 5–9). Importantly, Pol II association with DBP2 is not increased in the prp22-1 mutant, indicating that export factor accumulation at the gene is not simply a result of increased transcription (data not shown). Therefore, slowed kinetics of release of mRNA from the spliceosome causes accumulation of mRNA export factors at the site of DBP2 transcription. Our results show that cotranscriptional splicing promotes the recruitment of export factors to at least some intron-containing genes, perhaps dependent on intron position.
Because 3′-end formation is generally required for mRNA export (Eckner et al. 1991; Long et al. 1995; Huang and Carmichael 1996; Brodsky and Silver 2000), we reasoned that there may be a requirement for cleavage/polyadenylation in the cotranscriptional recruitment of mRNA export factors. Therefore, we examined the effects of export factor recruitment in the 3′-cleavage mutant, rna15-2. In comparison with wild type, Npl3 recruitment to PMA1 and ACT1 is unchanged in rna15-2 cells (Fig. 3C, left, lanes 1–4). Despite changes in the profile of Npl3 recruitment, Npl3 is cotranscriptionally recruited to DBP2 (Fig. 3C, left, lanes 5–9). In contrast, overall levels of Yra1 recruitment to PMA1, ACT1, and DBP2 are reduced dramatically in the rna15-2 mutant (Fig. 3C, right, lanes 1–9). In addition to mRNA export defects, 3′-end formation mutants display an apparent accumulation of mRNA that may be near the site of transcription (Hilleren et al. 2001; Jensen et al. 2001). Although the nature of the accumulated RNA is unknown, given the results presented here, we predict that Yra1 may not be bound to this pool of RNA, leaving it export-incompetent. These results show that 3′-end formation is required for Yra1 but not Npl3 recruitment to all genes regardless of intron status.
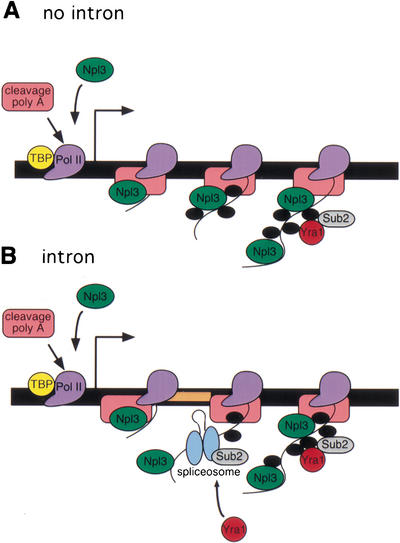
Here we show that pre-mRNA processing stimulates the cotranscriptional recruitment of mRNA export factors to promote mRNA export. Specifically, Yra1 is preferentially recruited to certain intron-containing genes dependent on splicing and generally recruited to all genes dependent on 3′-end formation. Taken together, our results suggest a model of cotranscriptional pre-mRNA processing stimulated packaging of mRNAs for export (Fig. 4). Although our findings indicate a requirement for both splicing and polyadenylation, given the extensive interplay among the various pre-mRNA processing steps, either splicing or polyadenylation could play a predominant role in this process. In contrast to Yra1, Npl3 can be recruited to a gene through interaction with the transcription machinery independent of the sequence of the RNA, indicating that specific pre-mRNA processing events are not absolutely required for Npl3 recruitment. More broadly, these results suggest that the transcription machinery can dictate the specificity of protein–RNA interactions.
Figure 4.
Packaging model for coordinated transcription, splicing, 3′-end formation, and mRNA export. (A) In the absence of an intron, Npl3 (green) and 3′-end formation factors (pink) are recruited by the transcription machinery to genes at an early stage of transcription (Dantonel et al. 1997; Lei et al. 2001; Licatalosi et al. 2002). Shown are TATA-binding protein (TBP, yellow) and RNA polymerase II (Pol II, purple). Npl3 is transferred to the growing RNA and is joined by pre-mRNA processing and export factors (black). Sub2 (gray) and Yra1 (red) are recruited to the RNA at a later stage of transcription, dependent on the presence or activity of 3′-end formation factors and perhaps other RNA-binding proteins. (B) When an intron (peach) is present sufficiently far from the site of transcription initiation, the spliceosome (blue) including Sub2 is recruited to the site of transcription and preferentially recruits Yra1. Yra1 recruitment is dependent on splicing, 3′-end formation, and perhaps the presence of other RNA-binding proteins.
Conversely, we find that Pol II transcription is not sufficient for Yra1 recruitment. It has been proposed recently that the inessential THO complex of transcription elongation factors can recruit Sub2 and, by association, Yra1 to genes (Sträßer et al. 2002). Because it is present in substoichiometric amounts compared with Pol II, the THO complex may be required for only a subset of genes or under certain growth conditions. Interestingly, it has been shown that splicing factors can stimulate transcription elongation, suggesting that splicing can cause a transcriptional feedback (Fong and Zhou 2001). Therefore, we examined Pol II recruitment in all chromatin immunoprecipitation experiments but did not find a strong correlation of export factor and Pol II recruitment (data not shown). Hence, a clear transcriptional feedback could not be detected in these experiments. Furthermore, the ability to separate export factor association from that of Pol II in a mutant that affects pre-mRNA processing (Fig. 3B, DBP2) indicates that the chromatin immunoprecipitation assay is capable of detecting RNA-dependent protein association with genes.
Nuclear export of mRNA serves as a regulatory step to ensure that only properly processed mRNAs are translated (for review, see Lei and Silver 2002). It has been shown that polyadenylation is required for mRNA export, but the mechanism remains unknown. Furthermore, microinjection studies have shown that mRNAs that undergo splicing are exported more efficiently than unspliced mRNAs (Luo and Reed 1999). Subsequent studies showed that a stimulatory effect of splicing on mRNA export is not observed for all spliced mRNAs (Le Hir et al. 2001; Rodrigues et al. 2001). Although the work of Luo and Reed (1999) is provocative, these studies bypass transcription, offering a simplified view of mRNA export. Our studies go beyond these findings and provide evidence that splicing and 3′-end formation enhance mRNA export by stimulating the recruitment of mRNA export factors to the RNA as it is being transcribed.
Materials and methods
Chromatin immunoprecipitations were performed as described previously (Lei et al. 2001). Each experiment was performed at least twice. Quantitation and error are shown for a single experiment consisting of at least three independent measurements of a single immunoprecipitation extrapolated from a standard curve of at least five dilutions of input known to be in the linear range of PCR. For 18S and 25S rDNA primers, 18–20 cycles of PCR were performed, but for all others, 26–30 cycles were performed. For normalized experiments in Figure 3, the propagated error of the ratio of the percentage input determined for a given primer set to that of the intergenic region is shown for a single experiment. Western blotting was performed to confirm consistent protein levels and IP efficiency in each strain. Wild-type (PSY2408), prp5-1 (PSY 2692), sub2-85 (PSY2670), prp22-1 (PSY 2562), and rna15-2 (PSY2822) strains deleted for the genomic copy of YRA1 and expressing pNOPMYCA1L-YRA1 (Lei et al. 2001) were used for mutant analysis. Sub2–HA (PSY2673) was created by C-terminal integration of three copies of the HA epitope followed by the kanMX6 marker into PSY580 using a method previously described (Longtine et al. 1998). Wild-type (PSY2408) or Sub2–HA were used for steady-state analysis. Control chromatin immunoprecipitations using 12CA5 (α-HA) and 9E11 (α-myc) in untagged strains yielded signal <0.007% input. For YJV100 experiments, a plasmid encoding Yra1–myc (pPS2701) was used and was produced by subcloning an SacII/XhoI fragment of pNOPMYCA1L-YRA1 into pRS313 (Sikorski and Hieter 1989). Indirect immunofluorescence was performed as described previously (Krebber et al. 1999) using mouse monoclonal α-Nop1 at 1:2500 and polyclonal α-Npl3 at 1:5000 and rhodamine-donkey α-mouse and FITC-donkey α-rabbit, respectively. Wild-type (PSY580) or YJV100 cells were grown in YP containing 1% raffinose/1% galactose at 30°C.
Acknowledgments
We thank C. Guthrie, E. Hurt, F. Lacroute, and H. Raué for providing strains. We are indebted to M. Ares, C. Stern, and M. Yu for critical reading of the manuscript and members of the Silver laboratory for discussion. E.P.L. was supported by grants from the NCI and the Ryan Foundation. This work was supported by grants from the NIH to P.A.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pamela_silver@dfci.harvard.edu; FAX (617) 632-5103.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1032902.
References
- Bentley D. The mRNA assembly line: Transcription and processing machines in the same factory. Curr Opin Cell Biol. 2002;14:336–342. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- Brodsky AS, Silver PA. Pre-mRNA processing factors are required for nuclear export. RNA. 2000;6:1737–1749. doi: 10.1017/s1355838200001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhn L, Munnerlyn A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes & Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- Eckner R, Ellmeier W, Birnstiel ML. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 1991;10:3513–3522. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach J, Bossie M, Vogel J, Corbett A, Jinks T, Willins DA, Silver PA. A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- Henry M, Borland CZ, Bossie M, Silver PA. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics. 1996;142:103–115. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- Huang Y, Carmichael GC. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Patricio K, McCarthy T, Rosbash M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol Cell. 2001;7:887–898. doi: 10.1016/s1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- Kessler MM, Henry MF, Shen E, Zhao J, Gross S, Silver PA, Moore CL. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes & Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler AL, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes & Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebber H, Taura T, Lee MS, Silver PA. Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export. Genes & Dev. 1999;13:1994–2004. doi: 10.1101/gad.13.15.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes & Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Silver PA. Protein and RNA export from the nucleus. Dev Cell. 2002;2:261–272. doi: 10.1016/s1534-5807(02)00134-x. [DOI] [PubMed] [Google Scholar]
- Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes & Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Graziani N, Saguez C, Boulay J. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes & Dev. 2001;15:36–41. doi: 10.1101/gad.852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Long RM, Elliott DJ, Stutz F, Rosbash M, Singer RH. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA. 1995;1:1071–1078. [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Luo MJ, Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc Natl Acad Sci. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- Lustig AJ, Lin RJ, Abelson J. The yeast RNA gene products are essential for mRNA splicing in vitro. Cell. 1986;47:953–963. doi: 10.1016/0092-8674(86)90810-x. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Rodrigues JP, Rode M, Gatfield D, Blencowe B, Carmo-Fonseca M, Izaurralde E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc Natl Acad Sci. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Panté N, Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Chen S, Hitomi M, Kumagai C, Tartakoff AM. A yeast protein that bidirectionally affects nucleocytoplasmic transport. J Cell Sci. 1995;108:265–272. doi: 10.1242/jcs.108.1.265. [DOI] [PubMed] [Google Scholar]
- Sträßer K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- Sträßer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Venema J, Dirks-Mulder A, Faber AW, Raue HA. Development and application of an in vivo system to study yeast ribosomal RNA biogenesis and function. Yeast. 1995;11:145–156. doi: 10.1002/yea.320110206. [DOI] [PubMed] [Google Scholar]
- Wilson SM, Datar KV, Paddy MR, Swedlow JR, Swanson MS. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J Cell Biol. 1994;127:1173–1184. doi: 10.1083/jcb.127.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Green MR. Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes & Dev. 2001;15:30–35. doi: 10.1101/gad.851701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]