Abstract
Transcriptional activation of the human U1 snRNA genes is dependent on a noncanonical octamer element contained within an upstream enhancer. The U1 octamer only weakly recruits the Oct-1 POU domain, although recruitment is stimulated by a peptide containing the Oct-1-binding domain of SNAP190. Structural analysis of the Oct-1 POU domain/U1 octamer/SNAP190 peptide complex revealed that SNAP190 makes extensive protein contacts with the Oct-1 POU-specific domain and with the DNA phosphate backbone within the enhancer. Although SNAP190 and OCA-B both interact with the Oct-1 POU domain through the same Oct-1 interface, a single nucleotide within the U1 octamer ablates OCA-B recruitment without compromising activator recruitment by SNAP190.
Supplemental material is available at http://www.genesdev.org.
Keywords: Transcription, Oct-1, SNAPc, X-ray crystallography
In humans, the U1 snRNA multigene family maintains a total level of ∼106 transcripts per cell. These levels of U1 RNA suggest that preinitiation complex formation and polymerase recruitment to these genes are highly efficient (Hernandez 1992; for review, see Hernandez 2001; Lobo and Hernandez 1994). Activated transcription of most human snRNA genes depends on a distal sequence element (DSE) that serves as an enhancer of transcription. Typically, the DSE contains an octamer element that can recruit the transcriptional activator protein Oct-1, which recognizes the DSE via its POU DNA-binding domain. The POU domain is a bipartite DNA-binding motif consisting of a POU-specific region (POUS) and a POU homeodomain (POUHD) separated by a flexible linker (Herr et al. 1988). Both subdomains are important for stable sequence-specific DNA binding to cognate octamer elements and for transcriptional activation (for review, see Herr and Cleary 1995). Oct-1 displays an unusual potential for gene activation, activating transcription of certain immunoglobulin genes in lymphocytes in a tissue-specific pattern and transcription of snRNA genes ubiquitously (Tanaka et al. 1992; Das et al. 1995).
Oct-1 also contains an activation domain, but surprisingly, the Oct-1 POU DNA-binding domain is sufficient to maintain robust activation of snRNA gene transcription (Murphy et al. 1992; Mittal et al. 1996; Murphy 1997). During a search for potential targets for the Oct-1 POU domain, the SNAP190 subunit of the snRNA activating protein complex (SNAPC) was isolated (Wong et al. 1998). SNAPC, also known as PTF, is a general transcription factor that binds to the proximal sequence element (PSE) contained within the core promoters of human snRNA genes (Murphy et al. 1992; Sadowski et al. 1993). Further analysis demonstrated that Oct-1 cooperates with SNAPC to increase promoter recognition by SNAPC and identified the region within SNAP190 from amino acids 800 to 930 to be required for interactions with the Oct-1 POU domain (Ford et al. 1998; Mittal et al. 1999). Mutational data confirm that direct protein contacts between the Oct-1 POU domain and general transcription factor SNAP190 are required for transcriptional activation by the Oct-1 POU domain (Ford et al. 1998; Mittal et al. 1999).
The region of SNAP190 directly targeted by Oct-1 exhibits homology with the B-cell specific coactivator OCA-B (Luo et al. 1992; Gstaiger et al. 1995; Strubin et al. 1995; Ford et al. 1998). Furthermore, mutations in Oct-1 that abolish activation of human snRNA transcription and interaction with SNAP190 also block Oct-1 synergism with OCA-B (Babb et al. 1997; Ford et al. 1998; Mittal et al. 1999), suggesting that Oct-1 may use similar mechanisms to activate transcription of mRNA-type genes via the coactivator OCA-B and of snRNA-type genes via the general transcription factor SNAP190. The OCA-B interaction was revealed in the crystal structure of the Oct-1 POU domain bound to a high-affinity H2B octamer element in a complex with a peptide from OCA-B (Chasman et al. 1999). In this structure OCA-B binds to the major groove within the octamer element and interacts extensively with the POUS and POUHD domains, which contact the major groove on opposite faces of the DNA helix. Together OCA-B and the Oct-1 POU domain encircle the high-affinity H2B octamer element to form a stable ternary complex (Chasman et al. 1999). In order to compare the general transcription factor SNAP190 function to that of the OCA-B coactivator as a target for activation by Oct-1, the structure of a complex of the Oct-1 POU domain, a peptide containing the Oct-1-interacting region of SNAP190, and the human U1 octamer sequence was determined to 2.4 Å. It represents the first structure of a transcriptional activator in a complex with its general transcription factor target.
Results and Discussion
SNAP190 assists Oct-1 binding to the U1 octamer
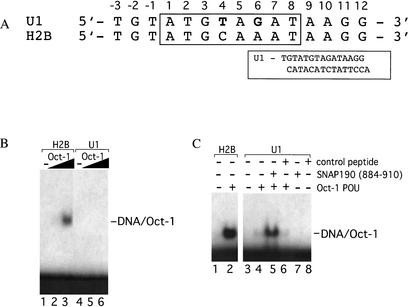
The U1 octamer element differs from the canonical H2B octamer sequence at position 4, where a T replaces C, and position 6, where a G replaces A (Fig. 1A). To assay the relative affinity of the Oct-1 POU domain for these two sequences, electrophoretic mobility shift assays were performed (Fig. 1B). As increasing amounts of Oct-1 POU domain (0.1 and 1 ng) were added to reactions containing a high-affinity H2B octamer element, efficient formation of a POU/octamer complex was observed (Fig. 1B, cf. lanes 2,3 and lane 1). In contrast, undetectable levels of complex formation were observed for reactions containing 0.1 or 1 ng of the Oct-1 POU domain and the human U1 octamer element probe (Fig. 1B, lanes 5,6, respectively). Overall, the relative affinity of the Oct-1 POU domain for the human U1 octamer is several orders of magnitude lower than for the H2B octamer element (Fig. 1B; data not shown). To determine whether SNAP190 can stimulate DNA binding by Oct-1, electrophoretic mobility shift assays were performed with the Oct-1 POU domain and a SNAP190 peptide containing amino acids 884–910. This peptide includes a region of SNAP190 that was previously shown to contact the Oct-1 POU domain during transcriptional activation by Oct-1 (Ford et al. 1998). As shown in Figure 1C, efficient complex formation is observed when 1 ng of the Oct-1 POU domain is incubated with the high-affinity H2B octamer element (Fig. 1C, lane 2). In contrast, weak complex formation was observed in reactions containing 30 ng of the Oct-1 POU domain and the U1 octamer element, as expected (Fig. 1C, lane 4; KD of ∼60 nM by complete titration and curve fitting analysis; data not shown). Interestingly, when the SNAP190 (884–910) peptide was also included in these reactions, a three- to fourfold enhancement (also calculated by titration and curve-fitting analysis; data not shown) of DNA binding by the Oct-1 POU domain was now observed (Fig. 1C, lane 5). The effect of the peptide was specific because comparable amounts of several nonspecific peptides had no effect on DNA binding by Oct-1 POU (Fig. 1C, lane 6; data not shown). The pIs of the nonspecific peptides ranged from 3.5 to 9.0. In addition, none of the peptides could bind DNA in the absence of the Oct-1 POU domain (Fig. 1C, lanes 7,8). Therefore, SNAP190 stimulates Oct-1 POU domain binding to the weak U1 octamer element.
Figure 1.
SNAP190 (884–910) assists Oct-1 POU DNA binding to the U1 octamer element. (A) The DNA sequences used in the gel shifts shown below. The octamer sequence is numbered 1–8, with the equivalent base on the opposite strand indicated by a prime. The two base pair changes in the U1 octamer relative to the H2B octamer sequence are shown in bold. (Inset) Complete U1 octamer oligonucleotide sequence used in crystallization. (B) Electrophoretic mobility shift assays were performed using 0.1 ng (lanes 2,5) or 1 ng (lanes 3,6) of human Oct-1 POU-domain protein with DNA probes containing a human histone H2B (lanes 1–3) or U1 snRNA (lanes 4–6) octamer element. Lanes 1 and 4 contain the probes alone. The position of the POU complex is indicated (DNA/Oct-1). (C) Electrophoretic mobility shift assays were performed using DNA probes containing a human histone H2B (lanes 1,2) or U1 snRNA octamer element (lanes 3–8) with 1 ng (lane 2) or 30 ng (lanes 4–6) of human Oct-1 POU-domain protein alone (lanes 2,4), with 10 μg of SNAP190 peptide (lane 5), or with an equimolar amount of a control peptide (lane 6). Lanes 7 and 8 contain the SNAP190 or control peptides alone, respectively. Lanes 1 and 3 contain probe DNA alone. The position of the POU complex is indicated (DNA/Oct-1).
Structure of the SNAP190/Oct-1/U1 octamer complex
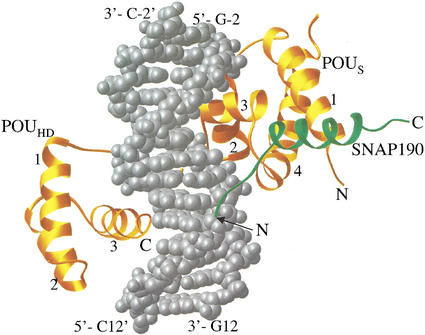
Recognition of regulatory elements by transcriptional activator proteins is presumed to be a prerequisite for subsequent recruitment of the general transcription machinery to gene promoters. Yet, the Oct-1 POU domain only weakly recognizes the human U1 octamer element, although binding is stimulated by SNAP190, suggesting that synergistic promoter recognition by these factors contributes to the activation of these genes. To understand the mechanism for SNAP190-mediated enhancement of DNA binding by the Oct-1 POU domain, X-ray structural analysis of a ternary complex of SNAP190, Oct-1 POU domain, and the human U1 octamer element was pursued. The ternary complex was formed using the 27-residue SNAP190 peptide encompassing amino acids 884–910, the Oct-1 POU domain containing both the POUS and POUHD DNA-binding modules, and a 14-bp DNA oligonucleotide containing the noncanonical U1 octamer sequence (see Fig. 1). The atomic structure of the Oct-1 POU domain/SNAP190/U1 octamer was solved at 2.4 Å resolution and represents the highest resolution structure of a monomeric POU domain complex to date. The relative orientation of the POUHD and POUS domains is similar to that seen in the structures of the Oct-1 POU/H2B octamer complex and the Oct-1/H2B octamer/OCA-B complex, with the two DNA-binding modules binding to the major groove on opposite sides of the DNA (Fig. 2). While the POUS interacts with the first four base pairs in the sequence (ATGT), the POUHD interacts with the last four base pairs (AGAT). As seen in all other structures of POU domain proteins, the linker peptide connecting the POUHD and POUS is disordered in this structure. Thus, interactions between SNAP190 and the Oct-1 POU domain barely perturb the overall structure of the POU domain (root mean square deviations, rmsd, for Cα atoms of the POU domains of the Oct-1 POU/H2B octamer and our Oct-1 POU/U1 octamer/SNAP190 complex are 0.6 Å).
Figure 2.
A Ribbons depiction of the SNAP190/Oct-1 POU/U1 octamer complex. Oct-1 POU is gold, SNAP190 (884–910) is green, and the U1 octamer DNA is silver and represented by a space-filling model (Carson 1991).
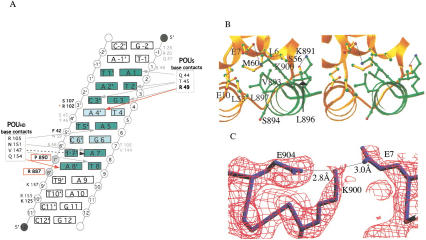
A more detailed characterization of the atomic interactions is described in Figure 3A and B, and in the Supplemental Material. In this structure, the ordered portion of the SNAP190 peptide begins at residue R887, which makes a salt bridge to the phosphate backbone of the DNA at position A8‘. An additional contact between SNAP190 P890 and the phosphate backbone at position T7‘ is also observed. Thus, DNA contacts by SNAP190 within the octamer element contribute to stable DNA binding by this complex. The SNAP190 chain then traverses the phosphate backbone and ends in a four-turn helix containing residues 892–906. This helix packs snugly against a surface of POUS that includes POUS helix 1, the loop between helices 3 and 4, and helix 4. The interactions between SNAP190 and Oct-1 are largely hydrophobic, with the core of the interaction defined by T892, V893, S894, L896, and L897 of SNAP190. The side chains of these residues make several hydrophobic contacts with the side-chain residues in the POUS domain, including L6, E7, E10, L53, and M60. Three hydrogen bonds are also made involving the main chains of SNAP190 and OCT-1 POUS: the SNAP190 V893 main-chain amide nitrogen to the POUS L55 main-chain carbonyl, the SNAP190 S894 main-chain amide nitrogen to the POUS L53 main-chain carbonyl, and the side-chain oxygen of T892 to the main-chain oxygen and nitrogen of L55. A perfect complement of shape is revealed in this interaction in which the side chains of the two peptides direct the main-chain atoms to make these hydrogen bonds. Notably, the only side-chain to side-chain hydrogen bond or salt bridge seen in our structure is between K900 of SNAP190 and E7 of the POUS domain (Fig. 3C). Interestingly, this interaction is sufficient to dictate activator-specific regulation of human snRNA gene transcription by POU-domain proteins and is critical for transcriptional activation of human snRNA genes by the Oct-1 POU domain (Mittal et al. 1996; Murphy 1997; Ford et al. 1998). Mutation of SNAP190 K900E completely abrogates Oct-1 binding. However, binding can be restored by the compensatory E7R mutation in Oct-1. This interaction is buttressed by a salt bridge between SNAP190 K900 and E904 that correctly positions K900 for this critical interaction with the Oct-1 activator. Importantly, this well-coordinated interaction helps to maintain the alignment of the critical hydrophobic interface between the Oct-1 POU domain and SNAP190 and explains, in part, the role of this single salt bridge in defining transcriptional specificity by POU-domain proteins.
Figure 3.
SNAP190 interacts with both the Oct-1 POUS domain and U1 octamer element. (A) Schematic representation of the protein/DNA contacts within the SNAP190/Oct-1/U1 octamer complex. POUS is numbered beginning with the N terminus of the domain, whereas POUHD begins at 101. The octamer bases are green. Lighter green highlights the differences between the U1 and H2B octamers. Red arrows indicate contacts not observed in the structure of Oct-1 POU alone. R49 (bold) makes a contact unique in SNAP190, whereas the Q154 contact is observed in both the OCA-B (Chasman et al. 1999) and SNAP190 structures. Gray arrows indicate contacts observed in all three Oct-1 POU-domain structures. The black dashed arrow indicates a base-specific contact shared only in the Oct-1 POU alone and SNAP190 structures. SNAP190/DNA phosphate backbone contacts (R887, P890) are in bold and boxed in red. S107 and F42 (bold) make phosphate backbone contacts that are observed in both the SNAP190 and OCA-B structures, whereas R102 (red asterisk) is seen only in the SNAP190 structure. The backbone contacts by K125 and K157 (dark gray type) are observed in the Oct-1/OCA-B/DNA and Oct-1/U1 DNA/SNAP190 structures. Amino acids within the POU domain that participate in phosphate backbone contacts observed in all three structures are shown in gray type. (B) A stereo view of the POUS (gold) interaction with SNAP190 (green). (C) A simulated annealing omit map contoured at 1.8 ς around SNAP190 K900, POUS E7, and SNAP190 E904. SNAP190 E904 buttresses K900, accurately positioning it to make a critical salt bridge with POUS E7 (Evans 1993).
Differential targeting of SNAP190 and OCA-B by Oct-1
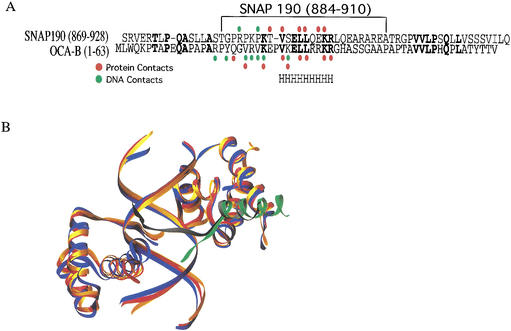
As shown in Figure 4A, the Oct-1-interacting regions of SNAP190 and OCA-B exhibit significant homology, especially in those regions that contact the POUS domain. In spite of the sequence conservation, there are important differences in the two interfaces. The N terminus of the OCA-B peptide makes several contacts with the POUHD domain, resulting in the DNA being surrounded by the Oct-1 POU/OCA-B peptide complex (Chasman et al. 1999). No similar interactions are observed between SNAP190 and POUHD in the SNAP190/Oct-1 POU/U1 octamer complex structure. These differences are also not surprising given that none of the residues in OCA-B that interact with the POUHD domain are conserved in the SNAP190 sequence. An interesting consequence of the interaction between OCA-B and the POUHD can be seen in Figure 4B. Alignment of the POUS domains from the Oct-1/H2B octamer (Klemm et al. 1994), Oct-1/H2B octamer/OCA-B (Chasman et al. 1999), and the Oct-1/U1 octamer/SNAP190 structures shows that although the Oct-1/H2B octamer and Oct-1/U1 octamer/SNAP190 structures are very similar, there is a significant change in the relative position of POUHD in the Oct-1/H2B octamer/OCA-B structure. In fact, the third helix of the POUHD has moved by >3 Å relative to the positions of either our structure or the Oct-1/H2B octamer structure. The motion combines a translation and rotation centered in the middle of the POUHD DNA-recognition helix. Both motions serve to pull POUHD helix 3 closer to the OCA-B peptide, allowing the interactions between OCA-B and helix 3 of the POUHD to occur. The rmsd between the Cα carbons of the POU domains of the Oct-1 POU/H2B octamer/OCA-B peptide structure and our Oct-1 POU/U1 octamer/SNAP190 peptide structure is 1.2 Å, significantly larger than the rmsd between our structure and the Oct-1 POU/H2B octamer structure (0.6 Å). This motion occurs without significant compensatory movement of the four bases of the octamer DNA bound by POUHD. Nevertheless, most of the interactions between the POUHD and the DNA bases are preserved, sometimes by changes in side-chain orientation. These interactions, however, do alter the trajectory of the flanking DNA on the POUHD side outside of the octamer sequence. In the case of the OCA-B ternary complex, the flanking DNA on the POUHD side has been pulled toward the domain as it has moved toward OCA-B. The consequences of flanking sequence movement for transcriptional activation are not known, but may influence the relative orientation of downstream components of the preinitiation complex at these promoters. These changes are apparently not due to crystal packing interactions as the deviation is seen in both molecules in the asymmetric unit, and no significant crystal packing interactions are seen in this region.
Figure 4.
SNAP190 and OCA-B use a similar interface for communication with the Oct-1 POU domain. (A) Sequence alignment of homologous regions of SNAP190 and OCA-B. H indicates the helical region that is common to both structures. (B) Overlay of the Oct-1/U1 octamer/SNAP190 (gold), Oct-1/H2B octamer (red), and the Oct-1/H2B octamer/OCA-B (dark blue) complex structures. The SNAP190 peptide is green, and OCA-B peptide is dark gray.
The structural divergence of the Oct-1 POUHD in the OCA-B- and SNAP190-containing structures is caused by significant differences in the N-terminal regions of these two peptides that affect their trajectories. The OCA-B peptide tracks across the minor groove of the DNA, making base-specific contacts with the A/T base pairs at positions 5 and 6 of the H2B octamer sequence. These interactions are mediated predominantly by V22 and V24 of OCA-B. In fact, OCA-B confers additional DNA-binding specificity to the complex because it will only bind to Oct-1/DNA complexes that have A/T base pairs at position 5 and 6 (Babb et al. 1997). Because the U1 octamer lacks the A/T base pair at position 6, it is not expected to have affinity for OCA-B. In contrast, those amino acids within OCA-B that make base-specific contacts are not conserved within SNAP190. Instead, SNAP190 tracks the phosphate backbone on only one side of the DNA, making two direct contacts with the DNA phosphate backbone (R887 and P890). In contrast to OCA-B, SNAP190 likely would not confer DNA specificity to the complex because its entire DNA interactions are with the phosphate backbone. Importantly, this critical base at position 6 in the U1 octamer reduces the affinity of OCA-B for DNA binding without compromising the ability of SNAP190 to assist Oct-1 activator binding to the U1 octamer element. Because the SNAP190 peptide is truncated at the N terminus, a different trajectory for a longer peptide or the full-length protein cannot be ruled out. Nevertheless, because none of the residues in either SNAP190 or OCA-B are conserved in this region, it is not surprising that the interface between SNAP190, the POUHD, and DNA is different.
Cooperative promoter recognition and activation of human U1 transcription
Promoter recognition by activator proteins is key to the ability of these factors to modulate transcriptional activity. One important question is what makes the U1 octamer, which contains two base changes (positions 4 and 6) relative to the high-affinity H2B octamer, a poor binding site for Oct-1 POU. At first glance, interactions involving R49 from the Oct-1 POUS domain could play a critical role. Oct-1 POU R49 is the only residue that makes base-specific interactions with position 4 in the major groove of the Oct-1 POU/H2B octamer structure and is critical for DNA binding because the mutant R49A Oct-1 POU has very little affinity for any DNA sequence (Cleary and Herr 1995). Indeed, one of the two hydrogen bonds between R49 and G4‘ (now A4‘ in the U1 octamer) is lost in the SNAP190-containing structure. However, R49 compensates by moving significantly to make a much tighter hydrogen bond with the O6 of G at position 3 than it does in the Oct-1/H2B octamer complex structure (2.8 Å in our structure vs. 3.5 Å in the Oct-1 POU/H2B octamer structure). R49 also makes an additional hydrogen bond with N7 of G3 as well. Therefore, R49 is capable of making critical but flexible interactions with DNA in this complex, consistent with experimental evidence presented by Cleary and Herr (1995) and seen in the Pit1/DNA complex structure (Jacobson et al. 1997). The U1 octamer also contains a base change at position 6 from A to G, which changes the Oct-1 POU/DNA interface. In both the OCA-B-containing and original Oct-1/H2B octamer structures, Oct-1 R102 makes contacts with both the sugar and bases of the 5′, 6‘, 7, and 8 positions. In the Oct-1/U1 octamer/SNAP190 structure, these interactions are prevented from occurring because the trajectory of Oct-1 R102 is altered through a steric collision with the amine of G6. Given the above arguments, the base change in the 6 position probably has the more devastating effect on Oct-1 POU/DNA binding, although both base changes may contribute to the reduced affinity for the U1 octamer. Importantly, the nucleotide at position 6 that reduces Oct-1 binding is also critical for defining OCA-B affinity. The stimulation of Oct-1 binding to the U1 octamer by SNAP190 is probably caused by several factors, the most direct being that it bridges the complex by interacting with both POUS and the DNA. It also may further stabilize POUS by its extensive interface with this domain.
The weakened ability of Oct-1 to recognize the U1 octamer is interesting because abrogation of DNA binding plays a key role in the transcriptional specificity for snRNA gene regulation. For example, both SNAPc and TBP bind DNA poorly alone but can act together to stimulate promoter recognition by both factors at a human U6 promoter (Mittal et al. 1996). Oct-1 can activate U1 transcription, in part, by interacting directly with the SNAP190 subunit of SNAPC to overcome this intrinsic inhibition of DNA binding by SNAPC (Ford et al. 1998; Mittal et al. 1999). In a further twist to this theme, we show here that Oct-1 DNA binding is inhibited at the U1 octamer and that this inhibition can be relieved by interactions between Oct-1 and SNAP190. Therefore, at least at the U1 promoter, all proteins that interact directly with DNA (SNAPC, TBP, and Oct-1) are inhibited for this binding in the absence of at least one of the other proteins or protein complexes that also interact with DNA. Given that many SNAPC-dependent genes are highly expressed, it may be important to keep SNAPC from binding at functionless DNA sites. Binding of the full complex requires a series of synergistic interactions between all three factors that prevents these proteins from interacting at this or other promoters or DNA sequences in the absence of their functional partners.
Materials and methods
Crystallization
Preparation of Oct-1 POU (284–439) and DNA was described elsewhere (Aurora and Herr 1992; DeWees and Geiger 1999). The SNAP190 peptide (884–910) was purchased from the W.M. Keck Facility (HHMI, Yale University) and purified over a C18 column (Vydac). The three components were combined in a 1:1.2:3 ratio of Oct-1 POU, DNA, SNAP190 peptide, respectively, with a complex concentration of 5 mg/mL. The final complex was in 10 mM HEPES (pH 7.9) and 10 mM DTT.
The Oct-1 POU/SNAP190 peptide/U1 octamer complex was crystallized using the hanging-drop vapor diffusion method. Crystals were grown by mixing equal volumes of the complex-containing solution with a solution containing 20% isopropanol, 20% PEG 4000, 0.1 M Na Citrate (pH 5.6), and equilibrating over the latter. The crystals grew in 3 wk to maximum dimensions of 0.7 mm × 0.05 mm × 0.025 mm. The crystals were cryoprotected with 30% MPD in the precipitating agent solution. The Oct-1 POU/SNAP190 peptide/U1 octamer complex crystallizes in the P1 space group with unit cell dimensions of a = 36.43, b = 54.97, c = 77.61 Å, α = 94.93°, β = 99.59°, γ = 109.25°, with two complexes in the asymmetric unit (52% solvent). X-Ray diffraction data to a resolution of 2.3 Å were collected at the Structural Biology Center ID19 beamline at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). Diffraction data were indexed and integrated using HKL2000 and SCALEPACK (Otwinowski 1993). X-Ray diffraction data were 94.1% complete with an Rmerge of 0.096 for 24,916 unique reflections from a total of 110,313 measured reflections. Data collection parameters and refinement statistics are listed in Table 1.
Table 1.
Data collection and refinement statistics
| Wavelength (Å) | 0.979 |
| Resolution range (Å) | 50.0–2.4 (2.48–2.38) |
| Reflections recorded (unique) | 110,313 (24,916) |
| Completeness (%) | 94.1 (92.5) |
| I/ς(I) | 12.7 (3.7) |
| R (I)merge (%)a | 9.6 (30.9) |
| R-factor (2.4 Å)b | 22.8 |
| R-Free (2.4 Å) | 29.4 |
| rmsd for bonds | 0.02 Å |
| rmsd for angles | 2.6° |
| % of residues in allowed Ramachandran regions | 100% (2% in generously allowed) |
Rmerge = Σ Σi/Ii − <I>|/Σi |I|, where Ii is an individual intensity measurement and <I> is the average intensity for this reflection, with summation over all data.
R-factor = Σ∥Fo| − |FC∥/Σ|FO|.
Values in parentheses refer to statistics for the last shell.
Structure determination and refinement
The structure was solved by molecular replacement using AMoRe (Navaza 1994), and the Oct-1 POU/H2B octamer structure (Klemm et al. 1994) was used as a search model with only the four central base pairs of DNA included in the initial search model. After an initial cycle of refinement, an Rfree of 42% and an R-factor of 33% was obtained excluding both peptides and most of the DNA. Clear electron density for both peptides and the DNA was observed in the map and built using TURBO FRODO. Then 244 water molecules were added and the model was refined with CNS (Brunger et al. 1998). The linker region (residues 368–379 Oct-1 numbering) is disordered in both molecules of the structure. The two DNA overhangs are also disordered. Final refinement statistics are tabulated in Table 1.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays were performed as described (Ford and Hernandez 1997) except 0.1 μg BSA was substituted for fetal calf serum. For assays with peptides, 50 mM sodium citrate buffer (pH 5.6) was substituted for HEPES buffer, and the reactions were separated on 5% polyacrylamide gels 19:1 (acrylamide:bis-acrylamide) rather than 39:1. The control peptide sequence was MSEPDPQELGAEC. The DNA sequences used were the U1 and H2B oligonucleotides shown in Figure 1A. The reaction volume was 20 μL with the molar concentrations of the protein, DNA, and peptides being 2.7 nM or 81 nM, 0.15 nM, and 0.16 mM, respectively.
Acknowledgments
We thank Ronguang Zhang at the SBC 1D19 beamline at the Advanced Photon Source for assistance in data collection, Winship Herr for the POU domain expression vector, and Brandon LaMere for technical assistance. This work was supported by grants from the National Science Foundation and Michigan Life Sciences Corridor Initiative to J.G. and from the American Cancer Society and National Institutes of Health to R.W.H. We also gratefully acknowledge the Michigan State University Foundation for their support of the Gene Expression in Development and Disease Initiative (to R.W.H., J.H.G. and S.H.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL geiger@cem.msu.edu; FAX (517) 353-1793.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1021002.
References
- Aurora R, Herr W. Segments of the POU domain influence one another’s DNA-binding specificity. Mol Cell Biol. 1992;12:455–467. doi: 10.1128/mcb.12.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babb R, Cleary MA, Herr W. OCA-B is a functional analog of VP16 but targets a separate surface of the Oct-1 POU domain. Mol Cell Biol. 1997;17:7295–7305. doi: 10.1128/mcb.17.12.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carson M. RIBBONS, a program for protein visualization. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- Chasman D, Cepek K, Sharp PA, Pabo CO. Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: Specific recognition of a protein–DNA interface. Genes & Dev. 1999;13:2650–2657. doi: 10.1101/gad.13.20.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary MA, Herr W. Mechanisms for flexibility in DNA sequence recognition and VP16-induced complex formation by the Oct-1 POU domain. Mol Cell Biol. 1995;15:2090–2100. doi: 10.1128/mcb.15.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G, Hinkley CS, Herr W. Basal promoter elements as a selective determinant of transcriptional activator function. Nature. 1995;374:657–660. doi: 10.1038/374657a0. [DOI] [PubMed] [Google Scholar]
- DeWees S, Geiger JH. Structural studies of the Msx-1 homeodomain–DNA complex I. Acta Crystallogr D. 1999;55:2039–2040. doi: 10.1107/s0907444999012081. [DOI] [PubMed] [Google Scholar]
- Evans SV. Setor—Hardware-lighted 3-dimensional solid model representations of macromolecules. J Mol Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- Ford E, Hernandez N. Characterization of a trimeric complex containing Oct-1, SNAPc, and DNA. J Biol Chem. 1997;272:16048–16055. doi: 10.1074/jbc.272.25.16048. [DOI] [PubMed] [Google Scholar]
- Ford E, Strubin M, Hernandez N. The Oct-1 POU domain activates snRNA gene transcription by contacting a region in the SNAPc largest subunit that bears sequence similarities to the Oct-1 coactivator OBF-1. Genes & Dev. 1998;12:3528–3540. doi: 10.1101/gad.12.22.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens CM. A B-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–362. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- Hernandez N. Transcription of vertebrate snRNA genes and related genes. In: McKnight S, Yamamoto K, editors. Transcriptional regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 281–313. [Google Scholar]
- ————— Small nuclear RNA genes: A model system to study fundamental mechanisms of transcription. J Biol Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- Herr W, Cleary MA. The POU domain: Versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes & Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- Herr W, Sturm RA, Clerc RG, Corcoran LM, Baltimore D, Sharp PA, Ingraham HA, Rosenfeld MG, Finney M, Ruvkun G, et al. The POU domain: A large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes & Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- Jacobson EM, Li P, Leon-del-Rio A, Rosenfeld MG, Aggarwal AK. Structure of Pit-1 POU domain bound to DNA as a dimer: Unexpected arrangement and flexibility. Genes & Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- Klemm JD, Rould MA, Aurora R, Herr W, Pabo CO. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- Lobo SM, Hernandez NT. Transcription of snRNA genes by RNA polymerases II and III. In: Conaway RC, Conaway JW, editors. Transcription: Mechanisms and regulation. New York, NY: Raven Press; 1994. pp. 127–159. [Google Scholar]
- Luo Y, Fujii H, Gerster T, Roeder RG. A novel B cell-derived coactivator potentiates the activation of immunoglobulin promoters by octamer-binding transcription factors. Cell. 1992;71:231–241. doi: 10.1016/0092-8674(92)90352-d. [DOI] [PubMed] [Google Scholar]
- Mittal V, Cleary MA, Herr W, Hernandez N. The Oct-1 POU-specific domain can stimulate small nuclear RNA gene transcription by stabilizing the basal transcription complex SNAPc. Mol Cell Biol. 1996;16:1955–1965. doi: 10.1128/mcb.16.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal V, Ma B, Hernandez N. SNAPc: A core promoter factor with a built-in DNA-binding damper that is deactivated by the Oct-1 POU domain. Genes & Dev. 1999;13:1807–1821. doi: 10.1101/gad.13.14.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S. Differential in vivo activation of the class II and class III snRNA genes by the POU-specific domain of Oct-1. Nucleic Acids Res. 1997;25:2068–2076. doi: 10.1093/nar/25.11.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S, Yoon JB, Gerster T, Roeder RG. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol Cell Biol. 1992;12:3247–3261. doi: 10.1128/mcb.12.7.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J. AMoRe, an automated program for molecular replacement. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- Otwinowski Z. Oscillation Data Reduction Program. In: Sawyer L, et al., editors. Data collection and processing. Daresbury, UK: SERC Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- Sadowski CL, Henry RW, Lobo SM, Hernandez N. Targeting TBP to a non-TATA box cis-regulatory element: A TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes & Dev. 1993;7:1535–1548. doi: 10.1101/gad.7.8.1535. [DOI] [PubMed] [Google Scholar]
- Strubin M, Newell JW, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Lai JS, Herr W. Promoter-selective activation domains in Oct-1 and Oct-2 direct differential activation of an snRNA and mRNA promoter. Cell. 1992;68:755–767. doi: 10.1016/0092-8674(92)90150-b. [DOI] [PubMed] [Google Scholar]
- Wong MW, Henry RW, Ma B, Kobayashi R, Klages N, Matthias P, Strubin M, Hernandez N. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol Cell Biol. 1998;18:368–377. doi: 10.1128/mcb.18.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]