Abstract
Reconstruction of a gene with its introns removed results in reduced levels of cytoplasmic mRNA. This is partly explained by introns promoting the export of mRNA through coupling splicing to nuclear export processes. However, we show here that splicing signals can have a direct role in enhancing gene transcription. Removal of promoter proximal splice signals from a mammalian gene or the excision of introns from two different yeast genes results in a marked reduction in levels of nascent transcription, based on both nuclear run-on and direct image analysis. This further establishes that mRNA processing and transcription are tightly coupled mechanisms.
Keywords: Introns, splice sites, transcription, gene expression
The sequencing of eukaryotic genomes has revealed the widespread presence of introns within protein encoding genes. Whilst the principal role of introns may be to generate increased protein diversity by alternative splicing (Smith and Valcarcel 2000; Graveley 2001), they may also enhance gene expression. Since the initial discovery of introns in eukaryotic genes some 20 years ago, there have been numerous reports of inefficient expression of recombinant genes when their introns are removed (e.g., Brinster et al. 1988; Palmiter et al. 1991; Lacy-Hulbert et al. 2001). However, it remains general practice to place cDNA derivatives of genes in expression vectors, especially in gene therapy, where some gene delivery vectors often tolerate only moderately sized recombinant gene inserts (Somia and Verma 2000). Invariably, levels of expression from such cDNA constructs are low, indicating that introns within a gene facilitate full levels of gene expression.
One way in which introns may increase gene expression is through the enhancing effect of splicing on mRNA export from the nucleus to cytoplasm (Reed and Hurt 2002). The splicing reaction itself is crucial for the deposition of proteins around the splice junctions, which mark the mRNA as being ready for export. Splicing of terminal introns has also been shown to increase the efficiency of polyadenylation (Niwa et al. 1990; Vagner et al. 2000), which in turn promotes higher mRNA levels, because the poly(A) tail is crucial for mRNA export, stability, and mRNA translation. As well as enhancing more efficient nuclear export and polyadenylation, early results using transgenic mice indicated a link between introns and transcriptional efficiency. Gene constructs with nonnative introns near their 5′ ends produced more transcripts, and at least some of this effect occurred at the transcriptional level (Brinster et al. 1988; Palmiter et al. 1991)
The results presented here extend these early studies by showing that the presence and promoter proximity of a donor site significantly enhances nascent transcription levels in an HIV-1 minigene construct. Furthermore, in a more physiological context, two Saccharomyces cerevisiae genes, DYN2 and ASC1, are both shown to require promoter proximal introns for full transcriptional output. Taken together with recent in vitro data showing that splicing can enhance transcription (Fong and Zhou 2001; Tian 2001; Kwek et al. 2002), a picture emerges that closely associates these two fundamental stages in the expression of a gene.
Results
Spacing the 5′ splice site away from the site of initiation reduces steady-state RNA levels
We previously demonstrated that 5′LTR polyadenylation in HIV-1 is blocked by the presence of the downstream positioned major splice donor site (Ashe et al. 1995, 1997b, 2000). As part of these studies (Ashe et al. 2000), spacer sequences positioned between the promoter and poly(A) site were introduced into minigene constructs to investigate the effect of promoter proximity on the 5′ poly(A) site use. RNAse protection analysis revealed a minor effect of promoter proximity on 5′ polyadenylation, but it also showed that spacing caused a marked decrease in mRNA levels. The fact that mRNA levels decreased in a manner proportionate to the size of the spacer suggests the presence of a position-dependent cis element required for efficient minigene transcription.
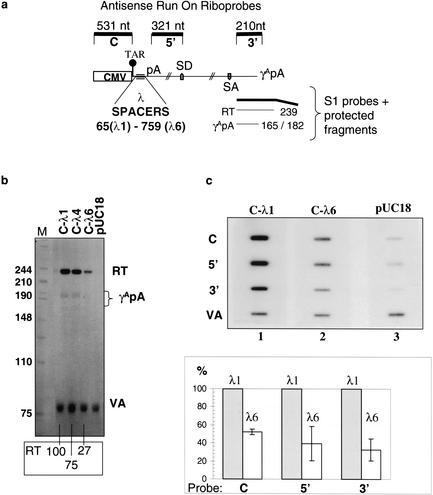
To investigate this phenomenon in more detail, we carried out further transcription analyses on three of these spacer constructs using both steady-state and nascent RNA analysis (nuclear run-on technique, NRO). The HIV-1 minigene constructs employed had a CMV promoter replacing the HIV-1 U3 promoter and contained a minimal Aγ-globin poly(A) signal marking the end of the minigene (Fig. 1a). The position of three spacer sequences (derived from phage lambda DNA; λ1, λ4, and λ6), which were chosen to lack any potential splice donor sequences, are also indicated (Fig. 1a). The minigene spacer constructs show a partial activation of the poly(A) site as well as a stepwise reduction in cytoplasmic RNA levels using a 5′-positioned RNA probe and RNAse protection analysis (Ashe et al. 2000). To further analyze these minigene transcripts, we performed a 3′-end S1 analysis on total RNA from transfected HeLa cells (Fig. 1b). The strong RT bands indicate that the γ-globin poly(A) signal works inefficiently, with the majority of transcripts reading into the plasmid vector. However, it is apparent that there is a stepwise reduction in the levels of these transcripts from λ1 to λ4 and then to λ6, compared to the expression of the cotransfected adenovirus VAI gene (VA). We suspect that the Aγ-globin poly(A) signal is inefficient due to the lack of downstream elements, recently demonstrated to be required in addition to the poly(A) signal for termination of transcription by RNA polymerase II (Pol II) in globin genes (Dye and Proudfoot 2001).
Figure 1.
The position of the 5′ splice site in HIV-1 minigenes influences the levels of steady-state RNA and nascent transcription. (a) Diagram of HIV-1 minigene. CMV promoter and TAR sequence are indicated. Poly(A) sites from HIV-1 5′LTR and at end of minigene (pA), major splice donor site (SD), splice acceptor sites (SA). Position and size of λ spacers are indicated. S1 probe and resulting protected fragments are indicated. Length and position of the antisense nuclear run-on riboprobes relative to the minigene are shown. (b) S1 nuclease analysis of total RNA. Transcripts polyadenylated at the γA globin poly(A) site (γApA), transcripts reading through the γApA (RT), and the VAI contransfection control (VA). Size markers are in bp; % levels of RT signal to C-λ1 are given. (c) Typical NRO analysis of Hela cells transfected with either C-λ1 or C-λ6 together with the cotransfection control VAI construct, showing the spacing effect on nascent transcription. pUC18-negative control: HeLa cells transfected with VAI containing plasmid and pUC18. Antisense riboprobes (C, 5′, 3′, and VA) are indicated at the left. VA-normalized quantitation of six independent transfections (three for the C probe) relative to the values obtained with the C-λ1 (gray column) construct over the corresponding probes is presented in the graph. All signals are corrected for the background hybridization shown in the pUC18-negative control. White column shows transcription levels obtained with λ6.
Insertion of spacer sequences affects the levels of nascent transcription
We next carried out a nascent transcription analysis on two of the minigene constructs, λ1 and λ6, in transfected HeLa cell nuclei. Transcription levels were assessed by NRO with three, single-strand, antisense RNA probes (C, 5′, and 3′) covering the minigene sequence as indicated (Fig. 1a). Each HeLa cell transfection was cotransfected with the VAI-expressing plasmid, to control for transfection efficiency. NRO signals obtained for the HIV minigene spacer constructs decreased (relative to the VAI control signal) as the spacer size increased (data not shown), resulting in a significant reduction in nascent transcription with the C-λ6 construct (Fig. 1c). Quantitation of six independent VAI normalized transfections confirms that insertion of a 759-nucleotide spacer sequence reduces nascent transcription levels over the 5′ and 3′ probes by an average of 2.5-fold and 3-fold, respectively (Fig. 1c, bottom panel). The C probe results in an average twofold decrease (three independent transfections). Note that the C probe detects nascent transcription that reads around the plasmid because Pol II termination occurs inefficiently in this minigene construct. In summary, these results demonstrate that some feature of these minigene spacer constructs has a direct effect on transcription levels from the CMV promoter. Either the lambda DNA inserts directly inhibit the promoter, or the altered position of sequences within the HIV-1 minigene affects transcription levels.
Inactivation of the 5′ splice donor reduces the levels of nascent transcription
Three features of the minigene were tested to determine their potential effect on transcription levels; the major splice donor site, the TAR sequence, and the promoter proximal poly(A) signal (Figs. 1a, 2). First, the two constructs C-m1 and C-m2 containing altered splice donor sequences as indicated (Figs. 2a, 3a) were employed and subjected to nascent transcription analysis. Inactivation of the major splice donor site has a significant effect on transcription levels (Fig. 2a). Mutations affecting the splice donor sequences reduced nascent transcription levels by up to threefold compared to the levels obtained with a construct containing the wild-type (wt) donor sequence (Fig. 2a, cf. lanes 1 and 2,3). These results have been reproduced in multiple experiments. In contrast, no reduction in transcription levels was observed with the second set of constructs where either TAR (Δ-TAR) or the 5′ poly(A) site (Δ-pA) were deleted (Fig. 2b, lanes 1,3,5). However, mutation of the donor site combined with Δ-TAR or Δ-pA did produce the same reduction in nascent transcripts (Fig. 2b, lanes 2,6).
Figure 2.
NRO analysis of splice donor mutation, poly(A), and TAR deletion mutant HIV-1 minigene constructs. Quantitation adjacent to NRO signals indicates % nascent transcription levels for each probe relative to the corresponding wild-type SD constructs normalized to cotransfected VAI transcription and background pUC18 signals. Antisense riboprobes are indicated at the left of the panel. pUC18: HeLa cells transfected with VAI-containing plasmid and pUC18. (a) NRO showing the effect of splice donor mutation (C-m1 and C-m2) on nascent transcription. The mutation of the splice donor mutant C-m1 is indicated at the right of the panel. (b) NRO analysis of minigene wild-type (wt) and mutant splice donor constructs containing a deleted TAR sequence (ΔT) and wt and mutant splice donor constructs combined with the deletion of the 5′ poly(A) site (ΔpA).
Figure 3.
NRO analysis of C-wt versus C-m2 cotransfected with S-U1 or Sm2-U1 as indicated. (a) The sequences of the C-wt and C-m2 minigene donor sites and downstream sequence to which Sm2-U1 or S-U1 respectively bind are outlined. (b) NRO analysis of C-wt and C-m2 constructs cotransfected with indicated U1 expressing plasmids. cot: cotransfected. (c) VAI-normalized quantitation of five independent NRO's. Transcription levels obtained over the three riboprobes C, 5′, and 3′ with C-m2 cotransfected with S-U1 (open columns) are compared relative to values seen with C-m2 cotransfected with Sm2-U1 (shaded columns). Error bars are indicated. Note that expression levels measured from C-m2 cotransfected with Sm2-U1 are set to 100.
We also wanted to determine whether the reduced levels of nascent transcription observed with the donor site mutation construct (C-m1) could be directly attributed to disruption of U1 small nuclear RNA (snRNA) binding. We therefore employed C-m2, which contains the same donor site mutation as C-m1 as well as an additional G to A change that mutates an adjacent cryptic donor site. A modified hexamer sequence was also introduced further downstream to which a mutant U1 snRNA gene (Sm2-U1) can base pair (Fig. 3a; Ashe et al. 1997b). We then cotransfected the C-m2 construct with either the S-U1 or Sm2-U1 snRNA genes as well as C-wt with S-U1. The control S-U1 snRNA can only base pair with the wt HIV-1 sequence at the position indicated in Figure 3a. The C-m2 construct cotransfected with S-U1 gave reduced levels of nascent transcription (Fig. 3b, lanes 1,2) especially with the 3′ and C probes (compared to C-wt cotransfected with S-U1), consistent with the effect obtained by inactivation of the donor site. However, when C-m2 was cotransfected with Sm2-U1, partial rescue of nascent transcription levels was observed with the 3′ and C probes (Fig. 3b, lane 3). However, to rule out that the enhanced expression is due to an unspecific side effect of cotransfecting the Sm2-U1 plasmid, expression levels of C-wt and C-wt cotransfected with Sm2-U1 were compared, showing no detectible increase in NRO signal (Fig. 3b, lanes 5,6). Quantitation of five independent experiments shows that cotransfection of Sm2-U1 can increase the expression levels from C-m2 by up to twofold (Fig. 3c) compared to levels from C-m2 cotransfected with S-U1 (note that in these experiments, expression levels obtained with C-m2 cotransfected with Sm2-U1 were set to 100). These reproducible results indicate that recognition of the splice donor site by U1 snRNA plays a role in the transcriptional activation mechanism.
Overall, these results demonstrate that in the HIV-1 minigene and by extrapolation in the HIV-1 provirus, the presence of the major splice donor site and most likely its interaction with U1 snRNA is required for full transcription levels. Although it has been shown in a number of studies that Pol II and in particular the hyperphosphorylated C-terminal domain (CTD) of its large subunit (Pol IIO) can enhance splicing (Hirose et al. 1999; Hirose and Manley 2000; Proudfoot et al. 2002), our present results show the reverse effect, that splice signals are capable of stimulating transcription.
Nuclear image analysis
We sought to confirm our RNA mapping data using a more direct image analysis of nuclear transcription. The minigene plasmids C-λ1 and C-λ6 on the one hand and C-wt and C-m1 on the other were transfected into HeLa cells. Following fixation of the transfected cells, they were subjected to RNA FISH analysis using minigene-specific oligonucleotide probes (Fig. 4). The top panel of Figure 4 shows a representative image of nuclei from C-wt and C-m1 transfections, and the graph below shows a statistical analysis of over 50 transfected nuclei for all four transfections, quantifying integrated transcript signals (x-axis) versus peak intensity (y-axis). Two results are apparent. First, as with NRO analysis, more nuclear transcript signal is evident for C-λ1 and C-wt. Secondly, the reduced efficiency constructs C-λ6 and C-m1 produce a more diffuse transcript pattern throughout the nucleus, in marked contrast to the focused transcript signals obtained with C-λ1 and C-wt. From these data we conclude that the RNA FISH confirms the NRO analysis showing the dependence of efficient transcription on a promoter proximal splice donor site.
Figure 4.
RNA FISH analysis of nascent transcript levels from HIV-1 minigenes. Representative images of HeLa cells transfected with C-wt and C-m1 are shown. In situ hybridization signals are superimposed on a phase image of the cells. For clarity the outlines of the nuclei are traced in white. Fifty cells chosen at random from each sample are plotted on the graphs below comparing integrated signal intensity per nucleus (x-axis) versus peak intensity (y-axis) for the two samples.
Removal of introns in the S. cerevisiae DYN2 and ASC1 genes reduces nascent RNA levels
We wished to determine the generality of transcriptional activation by promoter proximal splice sites in other eukaryotes. We therefore considered the case of Pol II transcribed genes in S. cerevisiae. Only a small minority (3.8%) of genes in this simple eukaryote possess introns. However, those that do normally contain a single intron positioned close to the gene’s promoter (Spingola et al. 1999) and are generally highly expressed (Ares et al. 1999; Lopez and Séraphin 1999). This genomic analysis is resonant with our results on the HIV-1 minigene showing that promoter proximal splice sites can enhance transcription. We therefore analyzed the effect of introns on transcription levels in DYN2, one of the few genes of S. cerevisiae to have two introns. This gene is nonessential and relatively highly expressed (Holstege et al. 1998). Furthermore, transcription analysis of DYN2 genes with separate deletions of either intron afforded a natural test on intron position with respect to the promoter.
A DYN2 deletion strain was transformed with plasmids containing a URA3 marker gene as well as either the wild-type DYN2 gene (wt) or DYN2 genes with intron 1 (A), 2 (B), or both (C) precisely removed (Fig. 5a). DYN2 mRNA levels were determined for each transformed strain using S1 nuclease digestion analysis of total RNA with both DYN2 and URA3 3′-end-labeled DNA probes (Fig. 5b, lanes 1,3–5). Control experiments were also performed with either RNA from the ΔDYN2 strain or no RNA (Fig. 5b, lanes 2,6). The URA3 gene allows relative quantitation of the levels of DYN2 mRNA for each strain. S1 analysis clearly shows a significant decrease of steady-state RNA levels to 39% when intron 1 is absent (Fig. 5b, cf. lanes 3 and 4). A similar reduction is observed when both introns are removed (Fig. 5b, lane 1).
Figure 5.
The 5′ intron of DYN2 gene from S. cerevisiae affects transcription levels. (a) DYN2 and ASC1 gene maps showing the full-length constructs (wt) and DYN2 constructs lacking either intron 1 (A) or intron 2 (B) or both introns (C), and ASC1 construct lacking intron, Δ16. Numbers denote distance from start codon or chromosomal nucleotide coordinates. Position of transcription start sites (arrow) and tandem DYN2 poly(A) signals (pA) are indicated, as are positions of the DYN2 S1 protection and run-on probe. (b) S1 nuclease protection analysis of total RNA from DYN2 strains indicated (Δ, A, B, C, and wt denote ΔDYN2 strain and transformed with pA25, pB10, pC2, and pD16). DYN2 RT represents DYN2 transcripts that fail to polyadenylate at the DYN2 poly(A) site; DYN2 pA represents polyadenylated DYN2 transcripts. % DYN2 mRNA (polyadenylated and read-through transcripts) vs. pD16 strain are indicated in box below. (c) Run-on analysis of DYN2 strains Δ,A, B , C, and wt, and ASC1 strains Δ16 and wt. Quantitation as % of wt is indicated adjacent to each signal.
The removal of intron 2 alone also reduces steady-state RNA levels to 38% (Fig. 5b, lane 5). The decrease in RNA levels here is most likely due to inefficient 3′ end processing. Indeed, a slower migrating band appears in this mutant strain, clearly indicating that a substantial amount of transcripts fail to be correctly polyadenylated at the DYN2 poly(A) site and produce extended read-through RNAs (Fig. 5b, lane 5, DYN2 RT). The same read-through band also appears in mutant C where both introns are removed (Fig. 5b, lane 1). This failure to polyadenylate draws strong parallels with the mammalian system, where the coupling between last intron removal and efficient polyadenylation is well defined (Niwa et al. 1990; Dye and Proudfoot 1999). Therefore the reduction in steady-state RNA levels observed in mutant B and partially in C could be due to an inhibitory effect on polyadenylation and hence a reduction in the stability of the RNA. However, no such explanation can be given for the reduction in mRNA levels observed with the intron 1 deletion. Instead it appears that intron 1, as in the HIV minigene constructs tested above, enhances transcriptional levels.
To confirm these predictions, transcription run-on analysis was performed on these four DYN2 transformed yeast strains, using an oligonucleotide probe within exon 3 of the DYN2 gene, normalized to a probe for URA3 (Fig. 5c). Consistent with the steady-state analysis, it is clear that the intron 1 deletion has a greater effect than the intron 2 deletion on DYN2 nascent transcription levels (40% versus 77%; Fig. 5c, cf. A, B, and wt). As predicted, removal of intron 2 had only a minor effect on nascent transcription, supporting the model that its removal may be coupled with polyadenylation. Most importantly, removal of either intron 1 or introns 1and 2 (C) reduced levels of nascent transcription. To extend these results to a second intronic yeast gene, we also analyzed nascent transcription levels in the more highly expressed ASC1 gene, which contains a single intron (Fig. 5a). As with the DYN2 experiments, an ASC1 deletion strain was transformed with the URA3 plasmid containing ASC1 gene with or without its intron. Precise removal of the intron from the ACS1 gene reduces transcription levels to 38% of wild-type levels (Fig. 5c). This effect is close to the 40% reduction observed when the first intron of DYN2 is deleted. All of these yeast run-on analyses were reproduced in several independent experiments and together suggest that the presence of a promoter proximal intron activates transcription in yeast, consistent with our results in mammals.
Discussion
It is apparent that the requirement of a promoter proximal intron to enhance transcription levels is a general feature of eukaryotes. The data described in these studies on mammalian cells were obtained using a retroviral gene system, where it is likely that there is strong selection for maximal levels of gene expression. It is therefore significant that in retroviruses, the major splice donor is invariably placed near the beginning of the proviral promoter (Tang et al. 1999). Similarly, analysis of the human genome sequence reveals that the first exon of genes transcribed by Pol II are often relatively short, so that donor sites in these genes will be placed near the promoter (International Human Genome Sequencing Consortium 2001). Furthermore in S. cerevisiae, of the 250 genes containing introns, over 120 possess introns within 40 bp of the start codon (Ares et al. 1999), and many of these genes are highly expressed. These data correspond well with our results on DYN2, where intron 1 is placed 30 bp downstream of the start codon and acts to enhance transcription.
Previous explanations for both the paucity of introns in S. cerevisiae as well as the 5′ polarity of their position are often attributed to the effect of Ty transposition. Thus retroposition of spliced genes preferentially results in the loss of 3′ introns (Boeke et al. 1985). This could result from Ty-dependent reverse transcriptase incompletely copying genes and thereby allowing the retention of the 5′-positioned introns (Fink 1987). However, our results suggest that an additional evolutionary process may also select for genes with 5′-positioned introns. Thus higher level gene expression may be achieved by the transcriptional enhancement effect of 5′-positioned introns, which is consistent with a previous suggestion that intron-dependent interactions contribute to enhanced gene transcription (Spingola et al. 1999). The fact that only 3.8% of S. cerevisiae genes contain introns but produce 27% of mRNA argues that the stimulatory effect of introns is important.
Future work is needed to address the mechanism of this intron transcription enhancement function. We favor an RNA-mediated process in which the nascent transcript, associated with splicing factors which in turn associate with the Pol II CTD, directly regulate further transcriptional elongation and possibly indirectly regulate initiation. This could be envisioned to work through a mechanism whereby the initial Pol II elongation complex must transcribe up to an intron to allow efficient transcription through the rest of the gene. Alternatively, when the Pol II elongation complex reaches the intron, this may promote further release of initiation factors that could then allow more rapid recycling to new polymerase initiation events. In either scenario, intron transcriptional enhancement may provide a proofreading function to double-check that initiation has taken place on a real gene rather than an inappropriate cryptic promoter.
Several recent in vitro studies strongly support the above mechanisms. First, U1 snRNP can be coimmunoprecipitated with Pol II, and moreover, supplementing HeLa nuclear extract with RNA oligos containing splice donor-like sequences reduced transcription on a CMV promoter (Tian 2001). In addition, studies using an HIV-1-TAT-dependent in vitro system show a stimulation of transcriptional elongation by both splicing factors and splicing signals (Fong and Zhou 2001). Finally, U1 was recently demonstrated to act as a functional component of the general transcription factor TFIIH (Kwek et al. 2002). Furthermore, in transcription reinitiation assays performed with purified transcription components, the presence of a donor site, although it has little effect on the first round of transcription, strongly activates subsequent reinitiation of transcription (Kwek et al. 2002). This final result is highly resonant with our in vivo data and suggests that once the elongating Pol II complex reaches a promoter proximal donor site, transcription levels are enhanced.
It is well established that the poly(A) site of a gene is required to promote downstream transcription termination (Birse et al. 1998; Dye and Proudfoot 1999). The results described here now argue that at least for some classes of Pol II transcribed genes, the presence of promoter proximal introns may also facilitate more efficient transcription initiation or elongation. It would therefore appear that mRNA processing events control both the beginning and end of the transcription cycle.
Methods
DNA constructs, transfections, and steady-state RNA analysis
The construction of HIV-1 minigene plasmids is described in detail elsewhere (Ashe et al. 2000). The 5′ probe was generated by linearizing RP5′ (Ashe et al. 2000) with AflII. The 3′ probe was constructed by inserting the 210-bp EcoRI-XbaI fragment from the C-wt construct into pGem-4. C probe derives from insertion of the whole CMV enhancer/promoter sequence into pGEM-4. T7 RNA transcription was performed on EcoRI (3′ probe) or XbaI (C probe) linearized plasmids. Transfections were carried out using the SuperFect reagent from QIAGEN. Three micrograms of minigene plasmids for steady-state and 6 μg for nuclear run-on analysis were cotransfected into HeLa cells together with 1 μg plasmid containing the adenovirus VAI gene. Cells were harvested 24 h after transfection. S1-nuclease analysis was carried out as described (Furger et al. 2001) using the EcoRI linearized RP3′ end-filled with radiolabeled α32P-dATP as probe.
ΔpA constructs: C-wt and C-m1 plasmids were digested with enzymes AflII and NarI end-filled and religated. ΔTAR is a deletion of +17 to 34 in the HIV-1 LTR sequence and its replacement with a BamHI linker deleting the upper stem and loop structure of TAR.
The U1 snRNA expression plasmids Sm2-U1 and S-U1 as well as the matching minigene plasmid C-m2 have all been described (Ashe et al. 1997a).
Yeast plasmids, strains, and probes
Nine hundred seventy-one base pairs of the S. cerevisiae DYN2 gene (Fig. 5a) and surrounding sequence were amplified by PCR and cloned into the EcoR1 site of the yeast shuttle vector pRS426 to generate pD16 (wt). Plasmids pA25 (A), pB10 (B), and pC2 (C) were constructed from pD16 (wt) through the deletion of the introns by PCR. Plasmids were transformed into S. cerevisiae BY4741 dyn2∷KanMX (Research Genetics, Invitrogen). The S. cerevisiae URA3 oligo and S1 antisense probes correspond to Chr V nucleotides 116167–116241 and 116898–116962, respectively. The DYN2 oligo and S1 probes are complementary to Chr IV nucleotides 1319510–1319436 and 1319512–131208, respectively. The S. cerevisiae ASC1 gene, including 769 bp of surrounding sequence (Fig. 5a), was amplified by PCR and cloned into pRS426, as above, to generate pASC9. The single intron was deleted by PCR to generate pD16 (Fig. 5a). Plasmids were transformed into S. cerevisiae BY4741 asc1∷KanMX (Research Genetics), and TRO analyses were performed using a probe corresponding to Chr XIII nucleotides 499457–499530.
Nuclear run-on analysis
Nuclei isolation and nuclear run-on analysis (NRO) were carried out as described (Ashe et al. 1997a). Subsequently RNA from the nuclei was isolated using the “hot phenol” method (Furger et al. 2001). Filters were prepared by adding 300–500 ng RNA probes per slot in R-loop buffer (Ashe et al. 1995) and denatured for 3′ at >85°C. Hybridization was carried out at 50°C–52°C overnight. Prehybridization was carried out at the same temperature for at least 3 h in the presence of 50 μg tRNA. Filters were washed in 1× SSPE 0.1% SDS at room temperature.
Transcription run-on analysis of the yeast strains was carried out as described (Birse et al. 1998).
RNA fluorescent in situ hybridization (FISH)
HeLa cells were grown directly on cover slips and transfected with the test plasmids and a cotransfection control plasmid using Effectene (QIAGEN). The cells were fixed 24 h after transfection in 4% formaldehyde/PBS for 15 min. The cells were permeabilized in 0.5% Triton X-100 for 4 min at 4°C. The cells were hybridized overnight in a mix containing 50% formamide, 2×SSC, 10% dextran sulphate, 50 ng/μL oligo dT(20), 1 μg/μL tRNA, and 5 ng/μL labeled oligo probe. Three oligo probes positioned downstream of the major splice donor site were employed. The oligo probes were labeled for 6 h at 37°C using terminal deoxynucleotidyl transferase (Promega) in the manufacturer's buffer with 125 pM dTTP and 70 pM digoxigenin-dUTP (Roche). Posthybridization washes were 30 min in each of 50% formamide in 2×SSC, 2×SSC, and 1×SSC. After blocking in 4×SSC and 4% BSA, rhodamine antidigoxigenin antibody was applied in 4×SSC and 1% BSA. Coverslips were washed in 4×SSC with 0.2% Tween 20 and then mounted in ProLong Antifade (Molecular Probes).
A Bio-Rad MCR 1024 confocal laser scanning microscope controlled with Lasersharp imaging software with constant settings for all samples was used. Images were acquired through a 60× PlanApo oil-immersion objective (NA 1.4) and an iris aperture of 1.0 Airy disk with the microscope in low-signal/low-scan mode. The laser power was set to the lowest power at which the brightest signals in the brightest sample fully utilized the 8-bit dynamic range of the detectors. The images were Kalman-filtered. Integrated signal intensities for each nucleus were measured using Image J software. Background fluorescence was measured in untransfected cells and subtracted.
Acknowledgments
We thank the members of N.J.P.'s lab for discussion and Shona Murphy and Sasha Akoulitchev for advice on the manuscript. We also thank Joan Monks for technical help. This work was supported by the UK Medical Research Council and The Wellcome Trust.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL nicholas.proudfoot@path.ox.ac.uk; FAX 01-865-275-556.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.983602.
References
- Ares M, Grate L, Pauling MH. A handful of intron-containing genes produces the lion's share of yeast mRNA. RNA. 1999;5:1138–1139. doi: 10.1017/s1355838299991379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes & Dev. 1997a;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe MP, Griffin P, James W, Proudfoot NJ. Poly(A) site selection in the HIV-1 provirus: Inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes & Dev. 1995;9:3008–3025. doi: 10.1101/gad.9.23.3008. [DOI] [PubMed] [Google Scholar]
- Ashe MP, Pearson L, Proudfoot NJ. The HIV-1 5' LTR poly(A) site is inactivated by U1 SnRNA interaction with the downstream major splice donor site. EMBO J. 1997b;16:5752–5763. doi: 10.1093/emboj/16.18.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe MP, Furger A, Proudfoot NJ. Stem-loop 1 of the U1 snRNP plays a critical role in the suppression of HIV-1 polyadenylation. RNA. 2000;6:170–177. doi: 10.1017/s1355838200991957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Garfinkel DJ, Styles CA, Fink JR. TY elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye MJ, Proudfoot NJ. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell. 1999;3:371–378. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- ————— Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell. 2001;105:669–681. doi: 10.1016/s0092-8674(01)00372-5. [DOI] [PubMed] [Google Scholar]
- Fink G. Pseudogenes in yeast. Cell. 1987;49:5–6. doi: 10.1016/0092-8674(87)90746-x. [DOI] [PubMed] [Google Scholar]
- Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- Furger A, Monks J, Proudfoot NJ. The retroviruses human immunodeficiency virus type 1 and Moloney murine leukemia virus adopt radically different strategies to regulate promoter-proximal polyadenylation. J Virol. 2001;75:11735–11746. doi: 10.1128/JVI.75.23.11735-11746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes & Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes & Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FCP, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuit of a Eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Kwek, K.Y., Murphy, S., Furger, A., Kimura, H., Thomas, B., O'Gorman, W., Proudfoot, N.J., and Akoulitchev, A. 2002. U1 snRNA associates with general transcription factor IIH and regulates transcriptional initiation. Nature Struct. Biol. (In press). [DOI] [PubMed]
- Lacy-Hulbert A, Thomas R, Li XP, Lilley CE, Coffin RS, Roes J. Interruption of coding sequences by heterologous introns can enhance the functional expression of recombinant genes. Gene Ther. 2001;8:649–653. doi: 10.1038/sj.gt.3301440. [DOI] [PubMed] [Google Scholar]
- Lopez PJ, Séraphin B. Genomic-scale quantitative analysis of yeast pre-mRNA splicing: Implications for splice-site recognition. RNA. 1999;5:1135–1137. doi: 10.1017/s135583829999091x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Rose SD, Berget SM. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes & Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- Palmiter RD, Sandgren EP, Avarbock MR, Allen DD, Brinster RL. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci. 1991;88:478–482. doi: 10.1073/pnas.88.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N, Furger A, Dye M. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Reed R, Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–533. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- Smith CW, Valcarcel J. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- Somia N, Verma IM. Gene therapy: Trials and tribulations. Nat Rev Genet. 2000;1:91–99. doi: 10.1038/35038533. [DOI] [PubMed] [Google Scholar]
- Spingola M, Grate L, Haussler D, Ares MA. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Kuhen KL, Wong-Staal F. Lentivirus replication and regulation. Annu Rev Genet. 1999;33:133–170. doi: 10.1146/annurev.genet.33.1.133. [DOI] [PubMed] [Google Scholar]
- Tian H. RNA ligands generated against complex nuclear targets indicate a role for U1 snRNP in co-ordinating transcription and RNA splicing. FEBS Lett. 2001;509:282–286. doi: 10.1016/s0014-5793(01)03188-x. [DOI] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3'-end processing and splicing. Genes & Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]