Abstract
To elucidate an outline of the mechanism of eukaryotic translation initiation, 48S complex formation was analyzed on defined mRNAs in reactions reconstituted in vitro from fully purified translation components. We found that a ribosomal 40S subunit, eukaryotic initiation factor (eIF) 3, and the eIF2 ternary complex form a 43S complex that can bind to the 5′-end of an unstructured 5′-untranslated region (5′-UTR) and in the presence of eIF1 scan along it and locate the initiation codon without a requirement for adenosine triphosphate (ATP) or factors (eIF4A, eIF4B, eIF4F) associated with ATP hydrolysis. Scanning on unstructured 5′-UTRs was enhanced by ATP, eIFs 4A and 4B, and the central domain of the eIF4G subunit of eIF4F. Their omission increased the dependence of scanning on eIFs 1 and 1A. Ribosomal movement on 5′-UTRs containing even weak secondary structures required ATP and RNA helicases. eIF4F was essential for scanning, and eIFs 4A and 4B were insufficient to promote this process in the absence of eIF4F. We report that in addition to its function in scanning, eIF1 also plays a principal role in initiation codon selection. In the absence of eIF1, 43S complexes could no longer discriminate between cognate and noncognate initiation codons or sense the nucleotide context of initiation codons and were able to assemble 48S complexes on 5′-proximal AUG triplets located only 1, 2, and 4 nt from the 5′-end of mRNA.
Keywords: eIF1, mRNA, ribosome, scanning, translation
The scanning model for translation initiation postulates a three-step mechanism by which eukaryotic ribosomes select the initiation codon in mRNA (Kozak 1978). A 43S complex comprising a 40S ribosomal subunit in association with initiator tRNA and eukaryotic initiation factors (eIFs) binds to the capped 5′ end of an mRNA (step 1) and scans downstream along the 5′-untranslated region (5′-UTR; step 2) until it encounters the first AUG triplet and stops (step 3). The resulting 48S complex is joined by a 60S subunit to form an 80S ribosome, and polypeptide synthesis begins. This model is supported by a substantial body of data derived from analysis of the effects of mRNA structure on the choice of initiation codon and the efficiency of initiation (Kozak 1991a), of factors required for cap-dependent attachment of ribosomes to mRNA (Gingras et al. 1999), and from genetic analysis of initiation codon selection (Donahue 2000). Nevertheless, many fundamental details about the mechanism of scanning are not known.
Initiator tRNA (Met-tRNAiMet), eIF2, and guanosine 5′-triphosphate (GTP) form a ternary complex, and its binding to a 40S subunit to form a 43S complex is stabilized by eIF1A and eIF3 (Dever 2002). Cellular mRNAs have a 5′-terminal m7G cap that enhances initiation through interaction with the cap-binding complex eIF4F, which promotes step 1, the 5′-end-dependent attachment of 43S complexes to mRNA (Banerjee 1980; Gingras et al. 1999). eIF4F consists of the cap-binding protein eIF4E, an ATPase/RNA helicase (eIF4A), and eIF4G, a large modular protein. eIF4E binds to the N-terminal third of eIF4G, and eIF4A binds to two sites in its central and C-terminal thirds; the central third also binds to eIF3 and RNA (Lamphear et al. 1995; Imataka and Sonenberg 1997; Lomakin et al. 2000). eIF4A cycles through the eIF4F complex (Yoder-Hill et al. 1993), and dominant negative mutant forms of eIF4A trap eIF4F in an inactive state and thus inhibit translation (Pause et al. 1994; Svitkin et al. 2001). The requirement for an active eIF4A subunit is consistent with ATP-dependent restructuring of the cap-proximal region of mRNA by eIF4F, which may provide a site of attachment for the 43S complex (Ray et al. 1985). It has been suggested that interactions between mRNA, eIF4G, and eIF3 (a component of the 43S complex) promote ribosomal attachment to mRNA (Carberry and Goss 1991; Lamphear et al. 1995). eIF4G613–1090 and eIF4A in place of eIF4F were sufficient to mediate a low level of end-dependent initiation in an in vitro reconstituted reaction (Morino et al. 2000). The conclusion that eIF4E and both terminal thirds of eIF4G play noncritical functions in promoting 5′-end-dependent attachment of ribosomes to mRNA is supported by observations that truncated forms of eIF4G lacking an eIF4E-binding site stimulated the translation of uncapped mRNAs in rabbit reticulocyte lysate (RRL; de Gregorio et al. 1998) and restored translation of capped mRNAs in RRL depleted of eIF4F (Ali et al. 2001). It is consistent with the finding that initiation by end-dependent scanning occurs with reduced efficiency but is not abolished by removal of the 5′-terminal cap from mRNAs (Banerjee 1980; Gunnery et al. 1997).
Ribosomal scanning (step 2) has not been observed or directly assayed, so almost nothing is known about the mechanism of this process. It is not known if the 40S subunit is intrinsically capable of movement on a 5′-UTR (by linear diffusion or transient dissociation-reassociation) or if it can only move along RNA by helicase-mediated translocation, and, if so, whether translocation is directly linked to RNA unwinding. Scanning 40S subunits with associated factors can unwind hairpins in the 5′-UTR that have a free energy of less than ∼−50 to −60 kcal/mole (Pelletier and Sonenberg 1985; Kozak 1986b). Scanning is reportedly ATP-dependent (Kozak 1980b), but it is not known whether this reflects a requirement for ATP hydrolysis for movement of the 40S subunit itself or only for RNA unwinding. eIF4B, eIF4G, and eIF4H all enhance the weak, nonprocessive ATP-dependent helicase activity of eIF4A (Rozen et al. 1990; Rogers et al. 2001), but it is not known whether any of these factors are associated with scanning ribosomes and whether any are required for ribosomal movement.
The third step in the scanning model is for the ribosomal complex to recognize the first AUG triplet that it encounters as the initiation codon and to arrest at it, forming a stable 48S complex. The critical interaction in ribosomal recognition of the initiator region is base-pairing between the initiation codon and the anticodon of initiator tRNA in the scanning ribosomal complex (Cigan et al. 1988). Genetic suppressor analyses in yeast of mutations that permit initiation at a UUG triplet have determined that eIF1, eIF2, and eIF5 influence start site selection (Donahue 2000). There are additional mRNA determinants of initiation codon recognition, and the original scanning model has been modified to account for two circumstances in which ribosomes bypass the first AUG triplet in an mRNA by “leaky scanning” (Kozak 1989). The first is if the context of the first AUG triplet deviates from the optimal/consensus sequence GCC(A/G)CCAUGG (in which the initiation codon is underlined, and mutations of the residues in bold have the greatest effects; Kozak 1986a, 1987a,b). Ribosomes can also initiate translation at the second AUG in an mRNA if the first AUG is located very close to the cap and do so progressively more frequently as the distance from the cap to the first AUG codon is decreased (Sedman et al. 1990; Kozak 1991b). The reason why 5′-proximal AUG triplets cannot promote efficient initiation is unclear. 5′-Terminal nucleotides may be unable to form stable codon–anticodon interactions when they enter the mRNA binding cleft of the 40S subunit, or the anticodon of initiator tRNA may be unable to inspect these terminal nucleotides. Leaky scanning can be partially suppressed by secondary structure downstream from the initiation codon that arrests the scanning ribosome at the suboptimal AUG triplet (Kozak 1990).
We recently reconstituted end-dependent initiation in vitro from purified translation components to be able to investigate the mechanism of this process (Pestova et al. 1998a, 2000). Here we report that a 43S complex comprising a 40S subunit, eIF3, and the eIF2 ternary complex can bind to the 5′-end of an unstructured 5′-UTR and in the presence of eIF1 scan along it and locate the initiation codon without a requirement for ATP or factors associated with ATP hydrolysis (eIF4A, eIF4B, eIF4F). However, in the absence of eIF4A, eIF4B, eIF4F, and ATP, 43S complexes could not move through even weak secondary structures located internally in otherwise unstructured 5′-UTRs. Although 43S complexes retained some capacity to scan along mRNA in the absence of eIF1, they could no longer discriminate between cognate and noncognate initiation codons or sense the nucleotide context of initiation codons and were able to assemble 48S complexes on 5′-proximal AUG triplets located 1, 2, and 4 nt downstream from the 5′-end.
Results
Ribosomal attachment and scanning on an unstructured 5′-UTR can occur independently of ATP hydrolysis
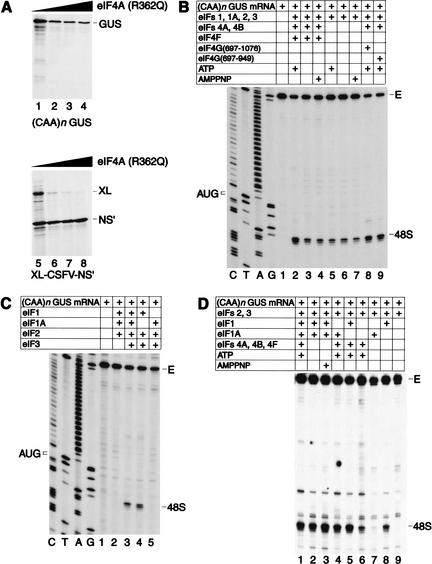
To investigate the ATP requirement for ribosomal attachment and scanning, we initially used uncapped in vitro transcribed (CAA)n-GUS mRNA, which comprises a 5′-UTR with the sequence 5′ GCAAGAA-(CAA)19-CACCAUGG (in which the initiation codon is underlined), and the β-glucuronidase (GUS) coding region (Wilson et al. 1990). This 5′-UTR does not contain any triplets that could act as noncognate initiation codons (Peabody 1989), and enzymatic probing indicates that it has a wholly single-stranded conformation (Tzareva et al. 1994). Translation of this mRNA in RRL was partially resistant to inhibition by R362Q negative trans-dominant mutant eIF4A, retaining ∼30% activity at concentrations that completely inhibited end-dependent translation of the first cistron of bicistronic XL-CSFV-NS‘ mRNA (Fig. 1A). This mutant had no effect on NS‘ translation mediated by the classical swine fever virus (CSFV) internal ribosomal entry site (IRES), as reported previously (Pestova et al. 1998b). Initiation on (CAA)n-GUS mRNA therefore does not show an absolute requirement for eIF4F, which in this experiment is inactivated by R362Q mutant eIF4A.
Figure 1.
Factor requirements for translation initiation on (CAA)n-GUS mRNA. (A) Partial resistance of (CAA)n-GUS mRNA translation to inhibition by R362Q mutant eukaryotic initiation factor (eIF) 4A. Uncapped (CAA)n-GUS (lanes 1–4) and XL-CSFV-NS‘ (lanes 5–8) mRNAs (0.2 μg) were translated in rabbit reticulocyte lysate (RRL; 15 μL) that had been preincubated with increasing amounts (0.3 μg, lanes 2,6; 1 μg, lanes 3,7; 2 μg, lanes 4,8) of mutant eIF4A at 30°C for 10 min, followed by addition of mRNA and further incubated at 30°C for 60 min. Lanes 1 and 5 did not contain added eIF4A. Samples were analyzed by SDS-PAGE and subsequent autoradiography. The positions of β-glucuronidase (GUS), XL, and NS‘ translation products are indicated to the right of appropriate panels. (B,C,D) Toeprint analysis of 48S complexes assembled on (CAA)n-GUS mRNA. Reaction mixtures contained 40S subunits, GMP-PNP, and either (B,C) aminoacylated total tRNA or (D) pure initiator tRNA in addition to translation components as indicated. Reaction mixture (C) did not contain adenosine triphosphate (ATP). Full-length cDNA is labeled E. cDNA products labeled “48S” terminated 15–17 nt downstream from the initiation codon of (CAA)n-GUS mRNA. The position of the initiation codon is shown to the left of the reference lanes, which show the (CAA)n-GUS sequence derived using the same primer as for toeprinting.
This experiment does not indicate whether initiation on (CAA)n-GUS mRNA is ATP-independent, because the free wild-type eIF4A in RRL might be used in the presence of mutant eIF4A. To investigate the ATP requirement, we used toeprinting to analyze 48S complex formation in a reconstituted in vitro system (Pestova et al. 1996). 48S complexes were formed on (CAA)n-GUS mRNA in the presence of ATP and eIFs 1, 1A, 2, 3, 4A, 4B, and 4F (Fig. 1); these components were then systematically omitted. Experiments were performed using either pure initiator tRNA (Fig. 1D) or unfractionated cytoplasmic tRNA that contains initiator and elongator tRNAs and a substantial amount of short RNA contaminants (“total tRNA”; Fig. 1B,C; Pestova and Hellen 2001). These contaminants may compete with input mRNA for RNA-binding factors such as eIF3 or eIF4F or for 43S complex. When total tRNA was used, 48S complex formation was reduced to 30% but not abolished on omission of eIFs 4A, 4B, and 4F with or without ATP or on inclusion of a nonhydrolyzable analog AMPPNP instead of ATP (Fig. 1B, lanes 3–7). eIF4F could be replaced by eIF4G697–1076 or eIF4G697–969 in reactions that contained eIFs 4A and 4B (Fig. 1B, lanes 2,8,9). In these conditions, eIFs 4A, 4B, and 4F therefore play an accessory role in promoting 48S complex formation on this unstructured 5′-UTR, and eIF4E and its cognate eIF4G-binding domain are not required for eIF4F’s contribution to this accessory role. 48S complex formation was not reduced in reactions containing pure Met-tRNAiMet either on omission of eIFs 4A, 4B, and 4F or omission of ATP or substitution of ATP by AMPPNP (Fig. 1D, lanes 2,3).
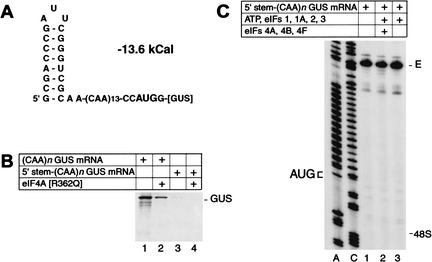
A control experiment confirmed that 48S complex formation on (CAA)n-GUS mRNA was not due to internal ribosomal entry. 5′-stem-(CAA)n-GUS mRNA, which has a stable hairpin (−13.6 kCal/mole) at its 5′ end (Fig. 2A), was almost inactive in translation in the presence or absence of R362Q mutant eIF4A (Fig. 2B) and did not support 48S complex formation in the reconstituted system (Fig. 2C). The inhibitory effect of this 5′-proximal hairpin on translation is consistent with previous reports of similar experiments (Kozak 1989). Taken together, these results indicate that 43S complexes containing eIFs 1, 1A, 2, and 3 are intrinsically able to recognize and bind to the 5′-end of an unstructured 5′-UTR and can scan to the initiation codon without ATP or factors associated with ATP hydrolysis (albeit at somewhat lower efficiency).
Figure 2.
Translation initiation on 5′-stem-(CAA)n-GUS mRNA. (A) Secondary structure of the 5′-UTR of 5′-stem-(CAA)n-GUS mRNA as predicted using mfold version 3.0 (Mathews et al. 1999). The theoretical standard free energy of the secondary structure is indicated. (B) Translation of (CAA)n-GUS and 5′-stem-(CAA)n-GUS mRNAs (0.2 μg) in RRL (15 μL) that had been preincubated without added mutant eIF4A (lanes 1,3) or with 1 μg R362Q mutant eIF4A (lanes 2,4) under conditions as described in the legend to Figure 1A. (C) Toeprint analysis of 48S complex formation on 5′-stem-(CAA)n-GUS mRNA in reaction mixtures that contained 40S subunits, GMP-PNP, and aminoacylated total tRNA in addition to translation components as indicated. Full-length cDNA is labeled E. The label “48S” indicates the expected position of toeprints caused by assembled 48S complexes on this mRNA. The position of the initiation codon is shown to the left of the two reference lanes, which show 5′-stem-(CAA)n-GUS sequence derived using the same primer as for toeprinting.
Omission of eIF1A did not affect 48S complex formation in reactions containing pure Met-tRNAiMet (Fig. 1D, lane 5). 48S complex formation was reduced on omission of eIF1 and was even more strongly reduced on omission of both eIFs 1 and 1A (Fig. 1D, lanes 4,6). Significantly, 48S complex formation was strongly reduced on omission of eIFs 4A, 4B, 4F, and 1A, almost abrogated on omission of eIFs 4A, 4B, 4F, and eIF1, and abolished when eIFs 4A, 4B, 4F, 1, and 1A were omitted (Fig. 1D, lanes 7–9). eIF3 was essential for 48S complex formation in all circumstances (Fig. 1C, lane 2; data not shown). These results indicate that eIFs 1 and 1A strongly increase the processivity of scanning 43S complexes on the (CAA)n 5′-UTR, and that their influence becomes crucial in the absence of eIFs 4A, 4B, and 4F.
The (CAA)n 5′-UTR is artificial, but we have found that 48S complex formation on natural mRNAs with similarly unstructured 5′-UTRs, such as alfalfa mosaic virus RNA-4, can also occur without ATP or factors associated with ATP hydrolysis (our unpublished data). This result is consistent with the partial resistance that this 5′-UTR confers to inhibition of translation by trans-dominant mutant eIF4A (Svitkin et al. 2001) and with earlier findings that translation of mRNAs with unstructured 5′-UTRs has a low requirement for the cap-binding complex eIF4F (Sonenberg et al. 1982; Gehrke et al. 1983).
Factor requirements for ribosomal scanning on structured 5′-UTRs
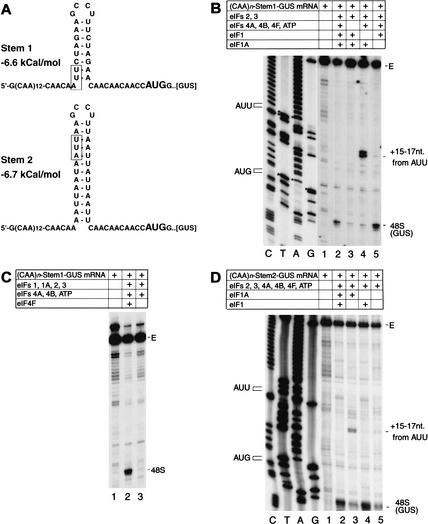
The observation that 43S complexes can bind to the end of the unstructured 5′-UTR of (CAA)n-GUS mRNA in the absence of ATP, eIF4A, 4B, and 4F allowed us to investigate the factor requirements for ribosomal scanning independently of the factor requirements for attachment of the 43S complex. In these experiments, we used derivatives of (CAA)n-GUS mRNA with the generic structure 5′ G-(CAA)14-STEM-CAACAACAACAAC CAUGG-[GUS] into which defined AU- or GC-rich hairpins (“STEM”) had been introduced into the 5′-UTR after 43 unstructured nucleotides, which we assumed would be sufficient for initial attachment of 43S complexes. AU-rich stem1 (−6.6 kcal/mole) contained 6 A-U base pairs and only 2 noncontiguous G-C base pairs, whereas stem2 (−6.7 kcal/mole) contained 13 A-U base pairs and no G-C base pairs (Fig. 3A). Formation of 48S complexes on (CAA)n-Stem1-GUS mRNA was very strongly reduced by omission of eIF4A, 4B, and 4F (Fig. 3B, lane 3), unlike in assembly reactions performed using the unstructured 5′-UTR of (CAA)n-GUS mRNA. Omission of only eIF4F had the same effect as omission of eIF4A, 4B, and 4F (Fig. 3C, lane 3). Identical results were obtained using (CAA)n-Stem2-GUS mRNA (data not shown). Replacement of total aminoacyl tRNA by purified natural or in vitro transcribed initiator tRNA did not lead to formation of 48S complexes on (CAA)n-Stem-GUS mRNAs in the absence of eIF4F (data not shown). These results indicate that 43S complexes are unable to scan through even weak AU-rich stems in the absence of eIFs 4A, 4B, and 4F. Moreover, eIF4F is required for scanning as well as for ribosomal attachment to mRNA. Although eIFs 4A and 4B may play independent roles in ribosomal scanning, these are not sufficient to promote scanning in the absence of eIF4F.
Figure 3.
Factor requirements for 48S complex formation on derivatives of (CAA)n-GUS mRNA containing AU-rich hairpins in the 5′-UTR. (A) Secondary structures of the 5′-UTRs of (CAA)n-Stem1-GUS and (CAA)n-Stem2-GUS mRNA as predicted using mfold version 3.0 showing the initiation codons (bold). The theoretical standard free energies of the secondary structures are indicated. AUU triplets in the 5′-UTR of these mRNAs are boxed. (B,C,D) Toeprinting analysis of dependence on initiation factors and ATP of 48S complex formation on (CAA)n-Stem1-GUS and (CAA)n-Stem2-GUS mRNAs. Reaction mixtures contained 40S subunits, GMP-PNP, and aminoacylated total tRNA in addition to translation components as indicated. Full-length cDNA is labeled E. cDNA products labeled “48S” terminated 15–17 nt downstream from the initiation codon of (CAA)n-Stem1-GUS or (CAA)n-Stem2-GUS mRNA, as appropriate. cDNA products labeled “+15–17 nt from AUU” terminated 15–17 nt downstream from indicated AUU triplets in the 5′-UTRs of these mRNAs. The position of AUU triplets and initiation codons are shown to the left of appropriate reference lanes, which show cDNA sequences derived using the same primers as for toeprinting.
Introduction of GC-rich stems of increasing stability (−5.5 to −18.9 kCal/mole) at the same position progressively impaired but did not abolish GUS translation in RRL, but no 48S complexes were formed on these mRNAs in the reconstituted system (data not shown). This result indicates that additional factors may be present in RRL but missing from the reconstituted reaction that is required for scanning through stable GC-rich secondary structures, or that 40S subunits become less scanning-competent after purification through sucrose density gradients.
Aberrant initiation codon selection in the absence of eIF1
Insertion of stems 1 and 2 into the 5′-UTR of (CAA)n-GUS mRNA not only changed its secondary structure but also introduced sequences that contained potential near-cognate initiation codons (Fig. 3A). Omission of eIF1 from reactions very strongly reduced 48S complex formation at the GUS initiation codon in both (CAA)n-Stem1/Stem2-GUS mRNAs but resulted in the appearance of strong stops +15 to 17 nt from AUU triplets (Fig. 3B, lane 4, D, lane 3) at the 5′-base of stem 1 and near the top of stem 2 (Fig. 3A). The positions of these stops relative to these triplets indicated that scanning ribosomes had arrested on them. Omission of eIF1A from reactions did not strongly influence the efficiency of 48S complex formation on GUS initiation codons of these mRNAs but led to the appearance of very faint toeprints at positions +15 to 17 nt from the AUU triplets (Fig. 3B, lane 5, D, lane 4). These results indicate that in the absence of eIF1, 43S complexes can scan along a 5′-UTR but cannot recognize and reject mismatches between potential initiation codons and the anticodon of initiator tRNA.
Requirements for eIF4E and the m7G cap for 5′-end-dependent ribosomal recruitment
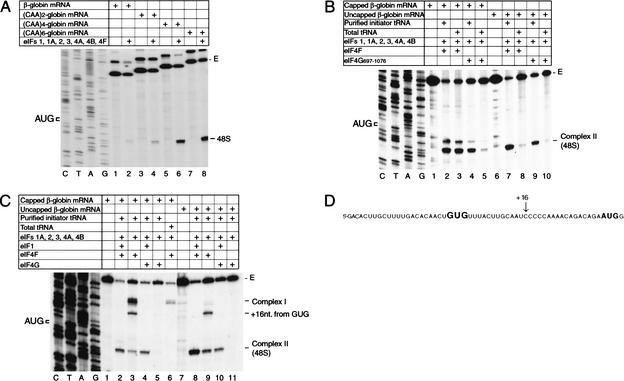
Unlike on native capped β-globin mRNA (Pestova et al. 1998a), 48S complex formation on uncapped β-globin mRNA transcripts is extremely inefficient in reactions containing total tRNA and either eIF4F or eIF4A and eIF4G697–1076 (Fig. 4A, lane 2, B, lanes 8,10). This is likely to be due to the low affinity of eIF4F and eIF4A/eIF4G697–1076 for the uncapped 5′-end of this mRNA. Enzymatic probing (Pavlakis et al. 1980; Lockhard et al. 1986) indicates that the 5′-end of the β-globin 5′-UTR is sequestered in secondary structure that may impair its recognition by eIF4G. Addition of increasing numbers of unstructured nucleotides to the 5′-end of uncapped β-globin mRNAs (2, 4, or 6 CAA triplets) progressively enhanced 48S complex formation (in reactions that contained total tRNA) up to the level of 48S complex formation on native capped β-globin mRNA (Fig. 4A). Thus, in these reactions containing total tRNA, eIF4F (and most likely its eIF4G subunit) does not efficiently interact with the partially sequestered 5′-end of a 5′-UTR in the absence of the 5′-terminal m7G cap but can interact with an uncapped 5′-end if it is unstructured.
Figure 4.
48S complex formation on β-globin mRNA and β-globin mRNA derivatives. (A–C) Toeprinting analysis of 48S complex formation on natural 100% capped or in vitro transcribed uncapped β-globin mRNA and uncapped derivatives thereof containing progressively longer unstructured 5′-terminal sequences, as indicated, in reaction mixtures that contained 40S subunits, GMP-PNP, ATP, aminoacylated pure initiator tRNA or total tRNA, and translation components as indicated. The label “48S” indicates the position of toeprints caused by 48S complexes assembled on these mRNAs in panel A; the same complex is indicated as complex II (48S) in panels B and C. Complex I in panel C terminated 21–24 nt from the 5′ end; a complex in this panel that terminated 16 nt from a GUG triplet in the 5′-UTR shown in panel D is designated “+16 nt from GUG.” Full-length cDNA is labeled E, and the position of the initiation codon is shown to the left of the β-globin sequence in reference lanes in panels A–C. (D) Sequence of the 5′-UTR of β-globin mRNA showing the AUG initiation codon and a GUG triplet in the 5′-UTR in bold, as well as the position (+16) of the leading edge of 48S complexes assembled at the GUG triplet.
However, uncapped β-globin mRNA lacking this additional unstructured 5′-end is translated in nuclease-treated RRL only 3 to 5 times less well than native capped β-globin mRNA (data not shown). This observation indicated that the low yield of 48S complexes in assembly reactions may be due to the presence of RNA contaminants in “total tRNA” that could compete with input β-globin mRNA for RNA-binding factors. Assembly reactions were therefore repeated using pure initiator tRNA to minimize competition. 48S complexes formed equally well on native capped β-globin mRNA when reactions contained eIF4F using either total tRNA or pure initiator tRNA (Fig. 4B, lanes 2,3). However, when eIF4A and eIF4G697–1076 replaced eIF4F, 48S complexes assembled with comparable efficiency only if reactions contained pure initiator tRNA and formed very inefficiently if total tRNA was used (Fig. 4B, lanes 4,5). 48S complexes assembled efficiently on uncapped β-globin mRNAs in reactions containing eIF4F or eIF4A/4G697–1076 only if the reactions also contained pure initiator tRNA and formed very inefficiently if total tRNA was included (Fig. 4B, lanes 7–10). These results indicate that 48S complexes can assemble efficiently on capped globin mRNA in the presence of eIF4A/4G697–1076 and on uncapped β-globin mRNA in the presence of either eIF4F or eIF4A/4G697–1076 only in the absence of competitor RNAs, and that competition is tolerated only when eIF4F is used with capped β-globin mRNA. This likely reflects the fact that the affinities of eIF4F for uncapped globin mRNA and of the eIF4G/4A complex for capped or uncapped globin mRNAs are lower than the affinity of eIF4F for capped globin mRNA.
The m7G cap-eIF4F interaction stabilizes ribosomal complexes assembled at the 5′-end of native β-globin mRNA
48S complexes do not form in in vitro reconstituted reactions on capped β-globin mRNA if eIF1A and/or eIF1 are omitted and instead an aberrant ribosomal “complex I” forms near the mRNA's 5′-end (Pestova et al. 1998a). The ability to use pure initiator tRNA to assemble 48S complexes on capped or uncapped mRNA with either eIF4F or eIF4G/4A enabled us to investigate formation of complex I in greater detail. Omission of eIF1 from reactions that included total tRNA and capped β-globin mRNA led to formation of complex I without formation of 48S complexes (Fig. 4C, lane 6), as described previously (Pestova et al. 1998a). However, substitution of total tRNA by pure initiator tRNA in an otherwise identical reaction mixture led to formation of complex I, of a small amount of 48S complex, and of an additional complex that mapped +16 nt downstream from a GUG triplet in the β-globin 5′-UTR (Fig. 4C, lane 3, D). No ribosomal complexes were detected by toeprinting on capped β-globin mRNA in reactions that lacked eIF1 and that contained eIF4A/eIF4G697–1076 in place of eIF4F (Fig. 4C, lane 5). In reactions in which β-globin mRNA was not capped, no ribosomal complexes formed in reactions that contained eIF4A/4G but lacked eIF1 (Fig. 4C, lane 11), and no complex I was formed in reactions that contained eIF4F but lacked eIF1 (Fig. 4C, lane 9). However, small amounts of 48S complex and of the “+16 nt” complex formed on a GUG triplet were formed in the last of these reactions.
These results indicate that formation of complex I is dependent on the eIF4F-cap interaction. Formation of 48S complexes and of the “+16 nt” complex in the absence of eIF1 occurred on uncapped and capped mRNAs only when pure initiator tRNA was used. It is likely that the eIF4F used in these experiments is not homogenous, and that the eIF4E subunit had dissociated from some eIF4F molecules. The resulting eIF4A/4G complexes can mediate initiation on capped mRNA without interacting with the m7G cap, so that some initiation events that occur in this reaction mixture will resemble those mediated by eIF4F on uncapped mRNA. Contaminating RNA present in total tRNA competes with globin mRNA for these eIF4A/4G complexes so that the only initiation events that occur are the result of the higher-affinity interaction of eIF4F with the capped mRNA (leading to complex I formation if eIF1 is absent). It may be significant that unlike with eIF4F, no ribosomal complexes were formed in reactions that contained eIF4A/4G697–1076 instead of eIF4F and that lacked eIF1. This indicates that ribosomal attachment and probably subsequent scanning are more dependent on eIF1 in this circumstance, which may be due to the integrity of eIF4G or the presence of eIF4E in the eIF4F heterotrimer. Formation of ribosomal complexes on GUG triplets in the absence of eIF1 reflected the inability of 43S complexes to recognize and reject mismatches between potential initiation codons and the anticodon of initiator tRNA in the absence of this factor noted previously.
Factor requirements for discrimination of initiation codon context
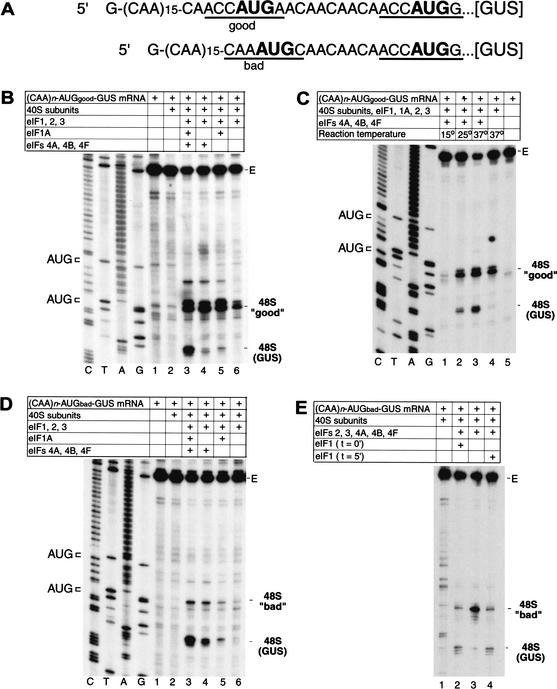
A significant aspect of the scanning process that is almost wholly uncharacterized is the mechanistic basis for the importance of sequence context in initiation codon recognition. The ability to assemble 48S complexes on (CAA)n-GUS mRNA in the absence of some factors allowed us to investigate which factors are responsible for the discrimination of initiation codon context. Additional AUG triplets in either “good” (accAUGa) or “bad” (caaAUGc) context were introduced upstream of the GUS initiation codon of (CAA)n-GUS mRNA (Fig. 5A), and 48S complex formation on the resulting (CAA)n-AUGgood-GUS and (CAA)n-AUGbad-GUS mRNAs was investigated.
Figure 5.
Factor dependence of initiation codon selection on mRNAs containing tandem initiation codons differing in sequence context. (A) Sequences of the 5′-UTRs of (CAA)n-AUGgood-GUS and (CAA)n-AUGbad-GUS mRNAs, showing the AUG initiation codons in bold. Context residues from −3 to +4 positions for each initiation codon are underlined; the A of the AUG codon is designated as +1. (B–E) Toeprinting analysis of 48S complex formation on (CAA)n-AUGgood-GUS and (CAA)n-AUGbad-GUS mRNAs, as indicated, in reaction mixtures containing 40S subunits, GMP-PNP, ATP, aminoacylated total tRNA, and translation components as indicated and incubated at 37°C (B,D,E) or at temperatures as indicated (C). eIF1 was included at the beginning of each assembly reaction except where indicated in panel E. Full-length cDNA is labeled E in panels B–E. The label “48S (GUS)” indicates the position of toeprints caused by 48S complexes assembled on the GUS initiation codon; the labels 48S “good” and 48S “bad” indicate 48 complexes assembled at upstream initiation codons in these mRNAs that are in either good or bad context. The position of these initiation codons are shown to the left of appropriate reference lanes (C,T,A,G) in panels B–D, which show cDNA sequences derived using the same primer as for toeprinting.
Approximately 60% of scanning ribosomes stopped at the new “good” context AUG triplet of (CAA)nAUGgood-GUS mRNA and the remainder scanned to the GUS initiation codon (Fig. 5B, lane 3). The propensity of ribosomes to bypass the first AUG triplet in an mRNA even if it has favorable context has been noted previously for 5′-UTRs with reduced secondary structure (Kozak 1980a). Approximately 90% of ribosomes scanned past the new “bad” context AUG triplet in (CAA)n-AUGbad-GUS mRNA to the GUS initiation codon (Fig. 5D, lane 3). This calculation was made with an assumption that the short distance between the two AUGs (13 nt for (CAA)n-AUGgood-GUS and 11 nt for (CAA)n-AUGbad-GUS mRNAs) does not allow these codons to be simultaneously occupied by 40S subunits. eIFs 1, 1A, 2, 3, 4A, 4B, and 4F are therefore sufficient to enable scanning ribosomes to discriminate between good and bad context for initiation codons. Omission of eIF1A alone or simultaneous omission of eIFs 4A, 4B, and 4F or eIFs 1A, 4A, 4B, and 4F from reactions significantly reduced 48S complex formation on the GUS initiation codon of (CAA)n-AUGgood-GUS mRNA (Fig. 5B, lanes 4–6), indicating that omission of these factors made scanning ribosomes less processive and possibly allowing more time to establish codon–anticodon interaction on the first “good” context AUG codon. However, we cannot exclude the possibility that one or more of these factors (eIFs 4A, 4B, 4F, 1A) may have some direct influence on recognition of the context of initiation codons. Progressive reduction of the temperature of assembly reactions from 37°C to 15°C also reduced 48S complex formation on the second AUG of this mRNA (Fig. 5C, lanes 1–3).
Separate or simultaneous omission of eIFs 1A, 4A, 4B, and 4F neither increased the amount of 48S complex that formed on the upstream “bad” context AUG codon of (CAA)n-AUGbad-GUS mRNA nor significantly altered the ratio of 48S complexes assembled on the first and second AUG codons of this mRNA (Fig. 5D, lanes 4–6). These four factors are therefore not mainly responsible for the ability of 43S complexes to distinguish between “good” and “bad” contexts. Omission of eIFs 4A, 4B, and 4F had an overall stronger inhibitory effect on 48S complex formation on both of these mRNAs [in particular, on (CAA)n-AUGbad-GUS mRNA] compared with 48S complex formation on (CAA)n-GUS mRNA. This fact can be explained by the possible existence of weak secondary structures in mutated 5′-UTRs.
However, omission of eIF1 from reactions very strongly increased 48S complex formation on the upstream “bad” context AUG codon of (CAA)n-AUGbad-GUS mRNA (Fig. 5E, lanes 2,3). Delayed addition of eIF1 after 5′-incubation to 48S complexes that had been preassembled on this codon in the absence of eIF1 led to dissociation of these complexes, so that the distribution of 48S complexes between the first and second AUG codons of this mRNA in this reaction resembled exactly the distribution of 48S complexes that was observed when eIF1 was present in reactions from the beginning (Fig. 5E, lane 4). This result shows that in addition to enabling ribosomal complexes to reject codon–anticodon mismatches (Fig. 3B,D), eIF1 also plays the principal role in enabling 43S complexes to distinguish between initiation codons in “good” and “bad” contexts.
Discriminating role of eIF1 in 48S complex formation at 5′-proximal AUG codons
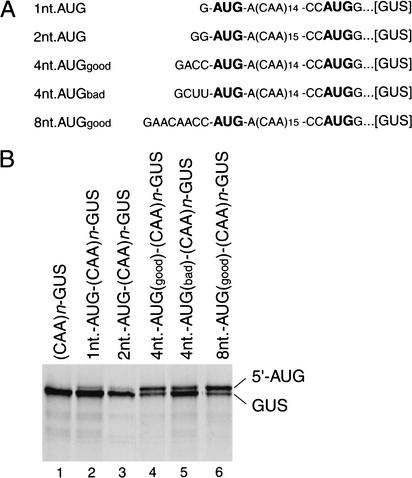
Ribosomes initiate translation inefficiently at the first AUG in an mRNA if it is located very close to the cap even if it has a favorable context (Sedman et al. 1990; Kozak 1991b). The molecular basis for discrimination against 5′-proximal AUG triplets is unknown. We investigated translation and 48S complex formation on derivatives of (CAA)n-GUS mRNA containing additional AUG codons located 1, 2, 4, or 8 nt from the 5′ terminus of the mRNA (Fig. 6A). Translation from AUG triplets located 1 or 2 nt from the 5′ terminus of the mRNA was very inefficient, so that almost all initiation events took place at the GUS initiation codon (Fig. 6B, lanes 2,3). The efficiency of translation from the novel upstream AUG codon increased significantly when its spacing from the 5′-end of the mRNA was increased to 4 nt, and initiation occurred much more often when it had the favorable context accAUGa than the unfavorable context cuuAUGa (Fig. 6B, lanes 4,5). Initiation at the 5′-proximal AUG triplet increased further when its separation from the 5′-end was increased from 4 to 8 nt (Fig. 6B, lane 6).
Figure 6.
Initiation of translation from 5′-proximal initiation codons. (A) Sequences of the 5′-UTRs of 1nt-AUG-(CAA)n-GUS, 2nt-AUG-(CAA)n-GUS, 4nt-AUGgood-(CAA)n-GUS, 4nt-AUGbad-(CAA)n-GUS, and 8nt-AUGgood-(CAA)n-GUS mRNAs, showing the AUG initiation codons in bold. (B) Products of translation in RRL of these mRNAs and of (CAA)n-GUS mRNA, as indicated. Translation was performed under standard conditions. 5′AUG and GUS indicate the products of translation that were initiated at upstream 5′-proximal initiation codons and at the GUS initiation codon, respectively.
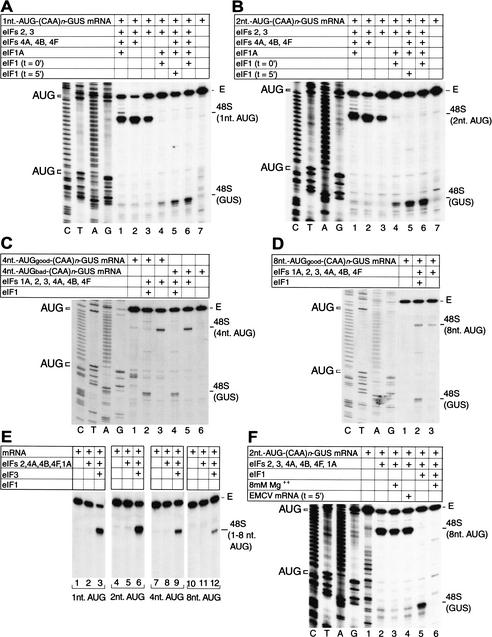
48S complexes did not form on the AUG triplets 1 and 2 nt downstream from the 5′-end of mRNA in the presence of eIFs 1, 1A, 2, 3, 4A, 4B, and 4F, although formation of 48S complexes on the GUS AUG codon was very efficient in these reactions (Fig. 7A, lane 6, B, lane 6). However, when eIF1 was omitted from reactions, 48S complexes formed very efficiently on these two 5′-proximal AUG triplets but did not form at all on the GUS AUG codon (Fig. 7A, lane 1, B, lane 1). These 5′-proximal 48S complexes could be formed on omission of eIFs 1 and 1A, and even in the presence of only eIF2 and eIF3, in all instances without 48S complex formation on the GUS AUG codon (Fig. 7A, lanes 2,3, B, lanes 2,3). Omission of eIF1A in addition to eIF1 even slightly increased 48S complex formation on 5′-proximal AUG triplets (Fig. 7A, lane 2, B, lane 2). 48S complex formation on these 5′-proximal AUGs depended completely on eIF3 (Fig. 7E). These 5′-proximal 48S complexes could be formed at high Mg++ concentration, in contrast with scanning-dependent assembly of 48S complexes on the GUS initiation codon (Fig. 7F, cf. lanes 3 and 6) and the β-globin initiation codon (Pestova et al. 1998a). We have previously noted similar resistance to high Mg++ concentrations for the formation of complex I on β-globin mRNA and for 48S complex formation on encephalomyocarditis virus (EMCV) and CSFV IRESs (Pestova et al. 1998a), none of which involve scanning. However, unlike complex I on β-globin mRNA, 48S complexes formed on the 5′-proximal AUGs of (CAA)n-GUS mRNAs were stable and resistant to delayed addition of competitor EMCV RNA (Fig. 7F, lane 4).
Figure 7.
Factor dependence of initiation codon selection on derivatives of (CAA)n-GUS mRNA containing 5′-proximal initiation codons. (A–F) Toeprinting analysis of 48S complex formation on mRNAs containing 5′-proximal initiation codons, as indicated, in reaction mixtures containing 40S subunits, GMP-PNP, ATP, aminoacylated total tRNA, and translation components as indicated. eIF1 was included at the beginning of each assembly reaction except where indicated in panels A and B. Encephalomyocarditis virus (EMCV) competitor mRNA (5 μg) was added to a reaction after 5 min of incubation where indicated on panel F. MgAc (8 mM) was added to reactions where indicated on panel F. Full-length cDNA is labeled E. The label “48S GUS” indicates the position of toeprints caused by 48S complexes assembled on the GUS initiation codon; other labels such as 48S (1 nt AUG) indicate 48 complexes assembled at the indicated 5′-proximal initiation codon in these mRNAs. The positions of initiation codons are shown to the left of appropriate reference lanes (C,T,A,G) in panels A–D and F, which also show cDNA sequences derived using the same primer as for toeprinting.
The delayed addition of eIF1 to 48S complexes assembled in its absence on 5′-proximal AUG triplets separated from the 5′-end of mRNA by 1 or 2 nt also led to the complete dissociation of these complexes and to the formation of 48S complexes exclusively on the GUS initiation codon (Fig. 7A, lane 5, B, lane 5).
In the absence of eIF1, 48S complexes also assembled efficiently on AUG triplets 4 nt downstream from the 5′-end whether they had “good” or “bad” context and did not form at all on the GUS AUG codon (Fig. 7C, lanes 3,5). However, this situation was completely reversed on inclusion of eIF1 in assembly reactions: 48S complexes formed on the GUS AUG codon, did not assemble at all on the “bad” context AUG codon, and assembled extremely inefficiently on the “good” context AUG codon (Fig. 7C, lanes 2,4). 48S complexes were formed on an AUG triplet in “good” context located 8 nt downstream from the 5′-end of (CAA)n-GUS mRNA both in the presence and in the absence of eIF1; 48S complexes also formed on the GUS AUG codon in the presence of eIF1 (Fig. 7D, lanes 2,3).
Taken together, these results indicate that the anticodon loop of initiator tRNA in a 43S complex is able to inspect mRNA from the very first nucleotide and to establish codon–anticodon base-pairing interactions with an AUG triplet separated by just 1 nt from the 5′-end of an mRNA. However, this mRNA is most likely not correctly fixed in the mRNA binding cleft of the 40S subunit, and the presence of eIF1 allows 43S complexes to sense that it is incorrect. Eight 5′-proximal nucleotides are sufficient for proper fixation of the initiation codon and flanking sequences in 48S complexes, which provide resistance to eIF1-induced destabilization.
Subunit joining protects 48S complexes assembled on 5′-proximal AUG codons from eIF1-induced destabilization
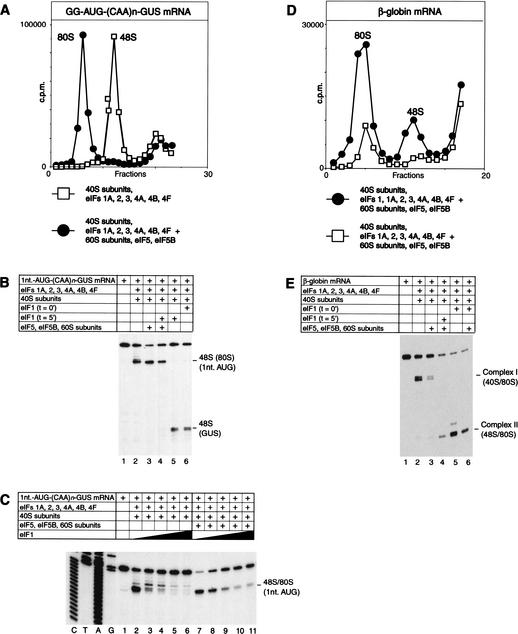
48S complexes assembled on 5′-proximal initiation codons in the presence of eIFs 1A, 2, 3, 4A, 4B, and 4F were stable in conditions of sucrose density gradient centrifugation and joined extremely efficiently with 60S subunits to form 80S ribosomes in a reaction mediated by eIFs 5 and 5B (Fig. 8A). It has been suggested that eIF1 can act on ribosomes during the elongation cycle, because mutations in eIF1 have been reported to increase ribosomal frame-shifting (Cui et al. 1998). The possibility that eIF1 could destabilize 80S complexes assembled on a 5′-proximal AUG was assayed by toeprinting analysis. 48S complexes and 80S complexes yield an identical pattern of toeprints unless translocation of the latter occurs (Anthony and Merrick 1992). Preincubation of 48S complexes assembled on an AUG triplet only 1 nt from the 5′-end of (CAA)n-GUS mRNA with 60S subunits, eIF5, and eIF5B did not change the position of toeprints but made ribosomal complexes resistant to destabilization by delayed addition of eIF1 (Fig. 8B, lanes 3,4). In a direct competition experiment, translation components required for 48S complex formation on this AUG triplet were incubated with increasing amounts of eIF1 in the presence or absence of 60S subunits, eIF5, and eIF5B. At the same concentrations of eIF1, more ribosomal complexes were formed at the 5′-proximal AUG triplet in the presence of 60S subunits, eIF5, and eIF5B than in their absence (Fig. 8C, cf. lanes 2–6 and 7–11). This experiment confirmed that subunit joining to 48S complexes assembled on 5′-proximal AUGs occurred in competition with eIF1-induced destabilization of these complexes. Protection by the 60S subunit of complexes assembled on 5′-proximal AUGs could explain the apparent paradox that in the presence of eIF1, formation of stable 48S complexes at 5′-proximal initiation codons did not occur in the in vitro reconstituted system, whereas initiation occurred weakly at these codons in RRL.
Figure 8.
Differences between 48S complexes assembled on cap-proximal initiation codons and “complex I” aberrantly assembled on natural capped globin mRNA in subunit joining activities and in protection by 60S subunits from eIF1-induced destabilization. (A,D) Sucrose density gradient centrifugation of 48S and 80S complexes assembled on [32P]-labeled (A) GG-AUG-(CAA)n-GUS mRNA and (D) globin mRNA from purified translation components as indicated. Sedimentation was from right to left. (B,E) Effect of inclusion of eIF1 at indicated times to assembly reactions that contained (B) G-AUG-(CAA)n-GUS mRNA and (E) β-globin mRNA, GTP, aminoacylated tRNA, and other translation components as indicated. (C) Effect of inclusion of increasing amounts (0.05 μg, lanes 3,8; 0.15 μg, lanes 4,9; 0.45 μg, lanes 5,10; 1 μg, lanes 6,11) of eIF1 in assembly reactions that contained G-AUG-(CAA)n-GUS mRNA, GTP, aminoacylated tRNA, and other translation components as indicated. Lanes 2 and 7 did not contain added eIF1.
Because 48S complexes assembled on 5′-proximal AUGs could be protected from eIF1-induced destabilization by joining to a 60S subunit, we wished to determine whether or not the aberrant complex I assembled on β-globin mRNA could similarly be protected. Complex I was preassembled on globin mRNA under standard conditions (Pestova et al. 1998a) and then incubated with 60S subunits, eIF5, and eIF5B. Complex I did not form 80S ribosomes efficiently (Fig. 8D) in conditions under which 80S ribosomes readily formed on the correct β-globin initiation codon (Fig. 8D). As a result, addition of 60S subunits, eIF5, and eIF5B to reaction mixtures did not protect complex I from destabilization by eIF1 (Fig. 8E, lane 4). These results and the observation that 48S complexes assembled on 5′-proximal AUGs are intrinsically stable (Fig. 7F, lane 4) whereas complex I does not resist challenge by a competitor RNA (Pestova et al. 1998a) indicate that these complexes have completely different natures. 48S complexes at 5′-proximal AUGs are assembled correctly, whereas complex I is assembled aberrantly.
Discussion
The mechanism of ribosomal scanning has remained obscure because of a lack of appropriate experimental systems to investigate many of its aspects. Here we have used a series of mRNA templates with defined structures and an initiation system reconstituted in vitro from individual purified components to investigate the factor requirements for 5′-end-dependent ribosomal attachment to mRNA, ribosomal scanning on the 5′-UTR, and initiation codon recognition.
Ribosomal attachment
We found that 48S complex formation on unstructured 5′-UTRs is 5′-end-dependent and can occur in the absence of ATP or factors eIF4A, 4B, and 4F, which are associated with ATP hydrolysis. However, in such conditions, 48S complex formation required the presence of eIFs 1 and 1A. The initiation AUG codon on (CAA)n-GUS mRNA is separated from the 5′-end by 68 nt, and 48S complex formation on this mRNA should involve scanning. Therefore, the requirements for eIF1, and to a smaller extent for eIF1A, for 48S complex formation on (CAA)n-GUS mRNA may reflect their requirement for scanning rather than for attachment of 43S complexes to the 5′-end of mRNA. The fact that 48S complexes could be assembled on 5′-proximal AUGs of derivatives of (CAA)n-GUS mRNA containing additional AUG codons located 1 or 2 nt from the 5′ terminus of the mRNA in the absence of eIF4A, eIF4B, eIF4F, and eIF1 and eIF1A, allowed us to conclude that 43S complexes comprising only a 40S subunit, the eIF2 ternary complex, and eIF3 are capable of ATP-independent, 5′-end-dependent attachment to mRNA. The component of the 43S complex that is responsible for the specificity of its interaction with the 5′-end of mRNA is not known. eIF3 is thought to bind at a site at the 5′-end of capped mRNAs after the mRNA has been “prepared” by eIF4F (Carberry and Goss 1991), so eIF3 may be able to bind directly to the 5′-end of an mRNA if it is already appropriately unstructured. In our experiments, 5′-proximal AUGs were followed by at least 42 unstructured nucleotides. Additional experiments are required to determine the exact number of unstructured nucleotides that is sufficient for attachment of 43S complexes to the 5′-end of mRNA independently of eIF4A, eIF4B, eIF4F, and ATP.
Attachment of 43S complexes to a structured 5′-UTR requires the participation of eIFs 4A, 4B, and 4F. Analysis of ribosomal attachment to capped and uncapped versions of the same mRNA promoted by either eIF4F or eIFs 4A and 4G697–1076 (which lacks the eIF4E-binding site) with or without competition (total tRNA versus purified initiator tRNA) indicated that the cap-eIF4E interaction primarily functions to enhance binding of eIF4F to the 5′-end of an mRNA. The fact that in the absence of competition we were able to assemble 48S complexes on capped mRNA using eIF4A and a truncated form of eIF4G (amino acid 697–1076) is in complete agreement with a recent report that a truncated form of eIF4G can support translation of capped mRNA (Ali et al. 2001). Increasing the number of unstructured nucleotides at the 5′-end of a structured 5′-UTR enhanced its ability to support 48S complex formation in the absence of a cap in conditions of competition (Fig. 4A), consistent with previous reports (Kozak 1980a). Because 43S complexes alone are able to bind to the 5′-end of an unstructured 5′-UTR and the number of unstructured 5′-terminal nucleotides required for binding independently of eIF4A, eIF4B, eIF4F, and ATP is unknown, it is difficult to say exactly which process is most influenced by the presence of increasing numbers of unstructured 5′-terminal nucleotides. It could be binding of eIF4A/4G, binding of 43S complex by itself, or both of these processes. However, we favor the first possibility. It is also worth noting that complex I (Pestova et al. 1998a) can be assembled only on capped β-globin mRNA and only in the presence of eIF4F but not eIF4A/4G. This result emphasizes the role of the cap-eIF4E interaction in stabilizing ribosomal complexes assembled on the 5′-end of mRNA when ribosomal scanning does not occur because of the absence of eIF1. What is even more interesting is that in the absence of the cap-eIF4E interaction, 43S complexes were able to scan the 5′-UTR even in the absence of eIF1 (Fig. 4C, lane 9). This result could be explained by the possibility that attachment of 43S complexes to the 5′-end of mRNA occurs differently with and without the cap-eIF4E interaction. It is also possible that for processive scanning, the initial cap-eIF4E interaction has to be broken and this does not happen in the absence of eIF1. At present we have no experimental data to support any of these hypotheses.
Ribosomal scanning
Although 43S complexes containing only eIF2 and eIF3 can bind to mRNA without eIF1 and form 48S complexes on 5′-proximal AUG triplets only 1–2 nt downstream from the 5′-end (Fig. 7A,B), these complexes are not capable of scanning in its absence (Fig. 2D). Results presented here indicate that a 43S complex minimally comprising a 40S subunit, eIF1, the eIF2 ternary complex, and eIF3 is intrinsically capable of scanning without any requirement for ATP hydrolysis or for factors associated with ATP hydrolysis that could act as ribosomal translocases. This minimal scanning complex requires that the 5′-UTR is essentially unstructured. Its ability to scan is enhanced by eIFs 1A, 4A, 4B, and 4F. In the presence of eIFs 4A, 4B, and 4F, scanning becomes less dependent on eIFs 1 and 1A. eIFs 1, 1A, 4A, 4B, and 4F all contribute to the processivity of scanning, although most likely by different mechanisms. Although eIFs 4A, 4B, and 4F can restructure mRNA, eIFs 1 and 1A being part of 43S complexes may influence the structure of the mRNA binding cleft of the 40S subunit and/or the position of initiator tRNA in these complexes. eIFs 4A, 4B, 4F, and ATP became essential if even very weak hairpins were introduced internally in the 5′-UTR (Fig. 3). All these data indicate that although 43S complexes are intrinsically capable of movement on unstructured 5′-UTRs by a mechanism of linear diffusion or transient dissociation-reassociation, in normal circumstances their movement on structured 5′-UTRs requires RNA helicases.
One of the factors that enhanced ribosomal scanning through weak hairpins is eIF4F, and significantly, eIF4A and eIF4B did not promote scanning except in its presence. This result provides the first evidence that eIF4F plays a role in scanning, as well as in ribosomal recruitment to mRNA, and indicates that eIF4A and eIF4B are not able to support scanning in the absence of eIF4F. A significant unresolved question is whether the unwinding of secondary structure in the 5′-UTR and ribosomal scanning are coupled processes and, if so, which components of the eIF4F complex are associated with the scanning ribosome. Is the eIF4E-cap interaction broken as the ribosome begins to scan or does eIF4F remain attached to the cap throughout the scanning process so that it is only displaced with other factors during subunit joining? Data presented here indicate that the eIF4E-cap interaction is not broken immediately after ribosomal attachment (Fig. 4C). If eIF4F remains associated with the ribosome after it begins scanning, cycling of eIF4A through the eIF4F complex (Yoder-Hill et al. 1993) may provide a mechanism to couple RNA unwinding and scanning and ensure the processivity of both processes. RNA unwinding by either eIF4A or eIF4F alone is nonprocessive (Rogers et al. 2001). An advantage of simultaneous binding of eIF4F to the cap and to the recruited ribosome as it scans from the 5′-end to the initiation codon is that it would facilitate rebinding of this scanning ribosomal complex to the same mRNA if its dissociates prematurely. However, it would also have the disadvantage of binding only one 40S subunit to the 5′-UTR of an mRNA at all times.
Initiation codon selection
In the absence of eIF1, 43S complexes retained their ability to move along mRNA, although at a reduced level. However, in the absence of eIF1, scanning 43S complexes could not discriminate between cognate and noncognate initiation codons and arrested at AUU and GUG triplets. This result is consistent with earlier findings that some mutations in yeast eIF1 enhance initiation on a UUG triplet (Yoon and Donahue 1992).
A 40S subunit, eIF1, the eIF2 ternary complex, and eIF3 were sufficient for assembly of a scanning ribosomal complex that is able to discriminate between initiation codons in good and bad contexts. Of these three factors, eIF1 played the critical role in discrimination, and without it scanning complexes were arrested at good and bad context initiation codons with similar efficiency. Most remarkably, eIF1 was able to dissociate 48S complexes preassembled at an upstream initiation codon with bad context, resulting in formation of a stable complex at the next downstream codon in good context.
In the absence but not in the presence of eIF1, 48S complexes were efficiently assembled on 5′-proximal AUG triplets 1, 2, and 4 nt downstream from the 5′-end. However, the delayed addition of eIF1 to 48S complexes preassembled on these AUGs also resulted in dissociation of these complexes. Therefore, eIF1 plays the major role in initiation codon selection during the scanning process. It is also responsible for the dissociation of incorrectly assembled ribosomal complexes. The mechanism of dissociation of aberrant ribosomal complexes by eIF1 is unknown. After binding of eIF1 to such ribosomal complexes, they could either continue to scan along mRNA searching for the correct initiation codon or dissociate from mRNA. If the latter scenario is correct, it is also not clear which component of the complex (initiator tRNA, mRNA, initiation factors?) is released first, thereby triggering dissociation of the whole complex.
Functions and mechanism of action of eIF1
The data presented here provide significant new insights into the function of eIF1 in initiation. First, they confirm its importance for ribosomal scanning, in which eIF1 acts synergistically with eIF1A (Pestova et al. 1998a). This function is particularly important in the absence of eIFs 4A, 4B, and 4F, indicating complementary roles of eIFs 1/1A and eIFs 4A/4B/4F in promoting the processivity of scanning. We also found that eIF1 acts in several circumstances to maintain the fidelity of initiation codon selection. eIF1 plays the principal role in enabling 43S complexes to distinguish between initiation codons in “good” and “bad” contexts, it destabilizes ribosomal complexes assembled on 5′-proximal AUG codons when the 5′-UTR is <8 nt long, and omission of eIF1 from assembly reactions yields complexes that cannot recognize and reject mismatches between noncognate initiation codons and the anticodon of initiator tRNA. These activities of eIF1 are strikingly similar to activities of the prokaryotic initiation factor IF3, which like eIF1 discriminates against initiation at non-AUG codons (Hartz et al. 1990) and at 5′-terminal AUG codons in leaderless mRNAs (Tedin et al. 1999) and also promotes initiator tRNA selection (Hartz et al. 1989, 1990). Only the C-terminal domain of this factor (IF3-C) is required to fulfill these functions (Petrelli et al. 2001). Recent footprinting and directed hydroxyl radical cleavage analysis data place IF3-C on the interface surface of the platform of the 30S subunit in the vicinity of the ribosomal P site (Dallas and Noller 2001). However, the precise position of IF3-C on 30S subunit indicates an indirect role for this factor in promoting discrimination of at least initiator tRNA (Dallas and Noller 2001). On the other hand, binding of IF3 induces conformational changes in the decoding site of the ribosome causing rearrangement of mRNA on the 30S subunit (La Teana et al. 1995; Shapkina et al. 2000; Petrelli et al. 2001). Such ribosomal structural changes induced by IF3 could be responsible for the discriminatory role of the factor.
Recent studies have shown that prokaryotic IF1 is structurally related to eukaryotic eIF1A (Sette et al. 1997; Battiste et al. 2000) and that prokaryotic IF2 and eukaryotic eIF5B have related sequences and a common function in promoting subunit joining (Pestova et al. 2000). The results reported here extend these observations. It will be interesting to determine whether eIF1 and IF3-C use similar mechanisms to discriminate against initiation at noncognate sites.
Although it is possible that context nucleotides are directly inspected by eIF1 in a scanning complex, we propose an alternative model to account for the discriminatory role of eIF1 in initiation codon selection in which it acts indirectly by influencing the conformation of the 43S complex. According to this model, 43S complexes can exist in two conformations: “closed” (in the absence of eIF1) and “open” (in its presence). Potential conformational changes induced by eIF1 could cause changes in positions of mRNA, initiator tRNA, or both on the 40S subunit. In its “closed” scanning-incompetent conformation, the anticodon loop of initiator tRNA can readily establish partial base-pairing interactions with non-AUG triplets. In its “open” scanning-competent form, the anticodon loop of initiator tRNA can establish stable interaction only with cognate AUG triplets surrounded by proper nucleotide context. This model indicates that context residues may interact directly with an rRNA or protein component of the 40S subunit to ensure such fixation of mRNA in the ribosomal mRNA binding cleft, which helps to establish the codon–anticodon interaction. Identification of the position of eIF1 on the 40S subunit would allow one to discriminate between direct and indirect mechanisms of action of eIF1.
Materials and methods
Enzymes and reagents
DNA restriction endonucleases and modifying enzymes were from New England BioLabs. DNA oligonucleotides were from Invitrogen and MWG Biotech. Avian myeloblastosis virus reverse transcriptase (AMV-RT) and RRL for in vitro translation were purchased from Promega Corp. Native rabbit total tRNA was from Novagen. Unlabeled nucleoside triphosphates and RNasin RNase inhibitor were from Amersham Biosciences. Radiochemicals [35S]methionine (44 TBq/mmole), [32P]UTP (111TBq/mmole), [32P]pCp (110TBq/mmole), and [32P]dATP (220TBq/mmole) were purchased from Amersham Biosciences and ICN Radiochemicals.
Plasmids
(CAA)n-GUS (Wilson et al. 1990), XL-CSFV-NS‘ (Pestova et al. 1998b), and pBS-(β-globin) (Hellen et al. 1993) transcription vectors and vectors for expression of recombinant eIF1 and eIF1A (Pestova et al. 1998a), wild-type eIF4A (Pestova et al. 1996), [R362Q] mutant eIF4A (Pestova et al. 1998b), eIF4B (Pestova et al. 1996), eIF4GI697–969 and eIF4GI697–1076 (Lomakin et al. 2000), eIF5, and eIF5B (Pestova et al. 2000) have been described previously. Insertion and substitution mutants derived from (CAA)n-GUS and pBS-(β-globin) were generated using PCR. All mutations were confirmed by sequencing of the complete 5′-UTR and adjacent coding region.
In vitro translation
XL-CSFV IRES-NS‘, (CAA)n-GUS and 5′-stem-(CAA)n-GUS mRNAs, and mRNAs of the (CAA)n-Stem-GUS and AUG-(CAA)n-GUS series were transcribed using T7 RNA polymerase and were translated in RRL under standard conditions in the presence of [35S]methionine and in the presence or absence of [R362Q] mutant eIF4A. Translation products were resolved by electrophoresis using 12% polyacrylamide gel. The efficiency of translation was determined by quantification of translation products using a Molecular Dynamics PhosphoImager.
Assembly and analysis of ribosomal complexes
Escherichia coli methionyl-tRNA synthetase from E. coli strain MRE 600 (American Type Culture Collection), ribosomal 40S and 60S subunits, and native and recombinant translation initiation factors were purified as described (Pestova et al. 1996, 1998a,b, 2000; Lomakin et al. 2000). Individual, fully aminoacylated rabbit initiator tRNA (Met-tRNAiMet) was prepared and purified exactly as described (Pestova and Hellen 2001). Ribosomal 48S complexes were assembled on (CAA)n-GUS, β-globin mRNAs, and their derivatives and were analyzed by primer extension using the primers 5′-CGCGCTTTCCCACCAACG-3′ (complementary to GUS nt 97–114) and 5′-GCATTTGCAGAG GACAGG-3′ (complementary to β-globin nt 177–194), as appropriate, and AMV-RT in the presence of [32P]dATP, essentially as described previously (Pestova et al. 1998a). Reaction mixtures (40 μL) containing 0.3 μg of mRNAs, 6 pmole total, or individual initiator aminoacylated tRNA, 5 pmole 40S subunits, 2 μg eIF2, 6 μg eIF3, 2 μg eIF4A, 1 μg eIF4B, 1 μg eIF4F, 0.3 μg eIF1, 0.3 μg eIF1A, 0.5 μg eIF5, 0.5 μg eIF5B, and 7 pmole 60S subunits (as indicated in figures) were incubated at 37°C for 7 min in a buffer containing 20 mM Tris at pH 7.5, 100 mM KAc, 2 mM DTT, 2.5 mM MgAc, 1 mM ATP, 0.4 mM GTP or GMPPNP, and 0.25 mM spermidine. cDNA products were analyzed by electrophoresis through 6% polyacrylamide sequencing gels. 80S ribosomal complexes were assembled using factors and conditions as described previously (Pestova et al. 2000). Native β-globin mRNA was 3′-end-labeled with [32P]pCp using T4 RNA ligase. [32P]-labeled GG-AUG-(CAA)n-GUS mRNA was transcribed in vitro in the presence of [32P]UTP. Reaction mixtures (120 μL) had the same buffer conditions and concentrations of translation components as those that were used for 48S complex formation. Ribosomal complexes were resolved by centrifugation through 10%–30% sucrose density gradients.
Acknowledgments
We thank T.M.A. Wilson for the (CAA)n-GUS plasmid, I.N. Shatsky for suggesting that we use (CAA)n-GUS mRNA, F. Dardel for pointing out the similarity in structures of eIF1 and IF3, and Christopher Hellen for helpful discussions and help in preparation of this manuscript. This work was supported by grant R01 GM59660 from the National Institute of General Medical Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tatyana.pestova@downstate.edu; FAX (718) 270-2656.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1020902.
References
- Ali IK, McKendrick L, Morley SJ, Jackson RJ. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 2001;20:4231–4244. doi: 10.1093/emboj/20.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony DD, Merrick WC. Analysis of 40S and 80S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem. 1992;267:1554–1562. [PubMed] [Google Scholar]
- Banerjee AK. 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev. 1980;44:175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battiste JL, Pestova TV, Hellen CUT, Wagner G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol Cell. 2000;5:109–119. doi: 10.1016/s1097-2765(00)80407-4. [DOI] [PubMed] [Google Scholar]
- Carberry SE, Goss DJ. Interaction of wheat germ protein synthesis initiation factors eIF-3, eIF-(iso)4F, and eIF-4F with mRNA analogues. Biochemistry. 1991;30:6977–6982. doi: 10.1021/bi00242a024. [DOI] [PubMed] [Google Scholar]
- Cigan AM, Feng L, Donahue TF. tRNAiMet functions in directing the scanning ribosome to the start site of translation. Science. 1988;242:93–97. doi: 10.1126/science.3051379. [DOI] [PubMed] [Google Scholar]
- Cui Y, Dinman JD, Kinzy TG, Peltz SW. The mof2/Sui1 protein is a general monitor of translational accuracy. Mol Cell Biol. 1998;18:1506–1516. doi: 10.1128/mcb.18.3.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- de Gregorio E, Preiss T, Hentze MW. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′-end-dependent. RNA. 1998;4:828–836. doi: 10.1017/s1355838298980372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Donahue TF. Genetic approaches to translation in Saccharomyces cerevisiae. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational control of gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 487–502. [Google Scholar]
- Gehrke L, Auron PE, Quigley GJ, Rich A, Sonenberg N. 5’-conformation of capped alfalfa mosaic virus ribonucleic acid 4 may reflect its independence of the cap structure or of cap-binding protein for efficient translation. Biochemistry. 1983;22:5157–5164. doi: 10.1021/bi00291a015. [DOI] [PubMed] [Google Scholar]
- Gingras A-C, Raught B, Sonenberg N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Gunnery S, Mälvali Ü, Mathews MB. Translation of an uncapped mRNA involves scanning. J Biol Chem. 1997;272:21642–21646. doi: 10.1074/jbc.272.34.21642. [DOI] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Gold L. Selection of initiator tRNA by Escherichia coli initiation factors. Genes & Dev. 1989;3:1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- Hartz D, Binkley J, Hollingworth T, Gold L. Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes & Dev. 1990;4:1790–1800. doi: 10.1101/gad.4.10.1790. [DOI] [PubMed] [Google Scholar]
- Hellen CUT, Witherell GW, Schmid M, Shin SH, Pestova TV, Gil A, Wimmer E. A cytoplasmic 57 kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Sonenberg N. Human eukaryotic translation initiation factor-4G (eIF4G) possesses two independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. How do eukaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978;15:1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- ————— Influence of mRNA secondary structure on binding and migration of 40S ribosomal subunits. Cell. 1980a;19:79–90. doi: 10.1016/0092-8674(80)90390-6. [DOI] [PubMed] [Google Scholar]
- ————— Role of ATP in binding and migration of 40S ribosomal subunits. Cell. 1980b;22:459–467. doi: 10.1016/0092-8674(80)90356-6. [DOI] [PubMed] [Google Scholar]
- ————— Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986a;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- ————— Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci. 1986b;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 1987a;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- ————— Analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987b;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The scanning model for translation: An update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991a;266:19867–19870. [PubMed] [Google Scholar]
- ————— A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1991b;1:111–115. [PMC free article] [PubMed] [Google Scholar]
- Lamphear B, Kirchenweger R, Skern T, Rhoads R. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- La Teana A, Gualerzi C, Brimacombe R. From stand-by to decoding site. Adjustment of the mRNA on the 30S ribosomal subunit under the influence of the initiation factors. RNA. 1995;1:722–782. [PMC free article] [PubMed] [Google Scholar]
- Lockhard RE, Currey K, Browner M, Lawrence C, Maizel J. Secondary structure model for mouse βMaj globin mRNA derived from enzymatic digestion data, comparative sequence and computer analysis. Nucleic Acids Res. 1986;14:5827–5841. doi: 10.1093/nar/14.14.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Hellen CUT, Pestova TV. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol Cell Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Morino S, Imataka H, Svitkin Y, Pestova TV, Sonenberg N. Eukaryotic translation initiation factor (eIF) 4E binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third of eIF4GI functions as a modulatory region. Mol Cell Biol. 2000;20:468–477. doi: 10.1128/mcb.20.2.468-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Méthot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlakis GN, Lockhard RE, Vamvakopoulos N, Rieser L, Rajbhandary UL, Vournakis JN. Secondary structure of mouse and rabbit α- and β-globin mRNAs: Differential accessibility of α and β initiator AUG codons towards nucleases. Cell. 1980;19:91–102. doi: 10.1016/0092-8674(80)90391-8. [DOI] [PubMed] [Google Scholar]
- Peabody DS. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT. Preparation and activity of synthetic unmodified mammalian tRNA iMet in initiation of translation in vitro. RNA. 2001;7:1496–1505. doi: 10.1017/s135583820101038x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CUT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998a;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CUT. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal initiation of translation of Hepatitis C virus and Classical swine fever virus RNAs. Genes & Dev. 1998b;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CUT. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- Petrelli D, LaTeana A, Garofalo C, Spurio R, Pon CL, Gualerzi CO. Translation initiation factor IF3: Two domains, five functions, one mechanism? EMBO J. 2001;20:4560–4569. doi: 10.1093/emboj/20.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray BK, Lawson TG, Kramer JC, Cladaras MH, Grifo JA, Abramson RD, Merrick WC, Thach RE. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985;260:7651–7658. [PubMed] [Google Scholar]
- Rogers GW, Jr, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. Bidirectional RNA helicase activity of eukaryotic initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedman SA, Gelembiuk GW, Mertz JE. Translation initiation at a downstream AUG occurs with increased efficiency when the upstream AUG is located very close to the 5′ cap. J Virol. 1990;64:453–457. doi: 10.1128/jvi.64.1.453-457.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette M, van Tilborg P, Spurio R, Kaptein R, Paci M, Gualerzi CO, Boelens R. The structure of translation initiation factor IF1 from E. coli contains an oligomer-binding motif. EMBO J. 1997;16:1436–1443. doi: 10.1093/emboj/16.6.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapkina TG, Dolan MA, Babin P, Wollenzien P. Initiation factor 3-induced structural changes in the 30S ribosomal subunit and in complexes containing tRNAfMet and mRNA. J Mol Biol. 2000;299:615–628. doi: 10.1006/jmbi.2000.3774. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Guertin D, Lee KAW. Capped mRNAs with reduced secondary structure can function in extracts from poliovirus-infected cells. Mol Cell Biol. 1982;2:1633–1638. doi: 10.1128/mcb.2.12.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedin K, Moll I, Grill S, Resch A, Graschopf A, Gualerzi CO, Blasi U. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol Microbiol. 1999;31:67–77. doi: 10.1046/j.1365-2958.1999.01147.x. [DOI] [PubMed] [Google Scholar]
- Tzareva NV, Makhno VI, Boni IV. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 1994;337:189–194. doi: 10.1016/0014-5793(94)80271-8. [DOI] [PubMed] [Google Scholar]
- Wilson TMA, Saunders K, Dowson-Day MJ, Sleat DE, Trachsel H, Mundry KW. In: Post-transcriptional control of gene expression. McCarthy JEG, Tuite MF, editors. 1990. pp. 261–275. . NATO ASI Series, vol. H49, Springer Verlag, Berlin/Heidelberg. [Google Scholar]
- Yoder-Hill J, Pause A, Sonenberg N, Merrick WC. The p46 subunit of eukaryotic initiation factor (eIF)-4F exchanges with eIF-4A. J Biol Chem. 1993;268:5566–5573. [PubMed] [Google Scholar]
- Yoon H, Donahue TF. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol Cell Biol. 1992;12:248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]