Abstract
p130 is a tumor suppressor of the pocket protein family whose expression is posttranscriptionally regulated and largely G0 restricted. The mechanism of down-regulation of p130 expression in proliferating cells was investigated. Our results indicate that the decline of p130 expression as G0 cells reenter the cell cycle is due to a decrease in protein stability. The enhancement of p130 turnover in late G1 and S phase compared with G0 and early G1 phase was dependent on Cdk4/6-specific phosphorylation of p130 on Serine 672, and independent of Cdk2 activity. The activity of the ubiquitin ligase complex Skp1–Cul1/Cdc53–F-box protein Skp2 (SCFSkp2) and the proteasome were necessary for p130 degradation. In vitro, recombinant Skp2 was able to bind hyperphosphorylated but not dephosphorylated p130. Furthermore, in vitro polyubiquitination of p130 by SCFSkp2 was specifically dependent on phosphorylation of p130 on Serine 672. Thus, like the Cdk inhibitor p27Kip1, p130 turnover is regulated by Cdk-dependent G1 phosphorylation leading to ubiquitin-dependent proteolysis.
Keywords: p130, Skp2, Cdk4, Cks1, ubiquitin, proteasome
p130 and its cognates, pRb and p107, constitute the “pocket protein” family. These proteins are key negative regulators of cell cycle progression and modulators of cellular differentiation processes in mammals. Although structurally related, their expression and biological activity are differentially regulated during the cell cycle, and their relevance to cell proliferation/differentiation tasks is also dependent on cell type and development stage (Garriga et al. 1998; Stiegler et al. 1998b; Lipinski and Jacks 1999). pRb serves as a critical regulator of the G1/S transition, because the functional inactivation of pRb is essential in most cell types for the progression beyond the G1/S boundary. The role of p107, which normally accumulates at the G1/S boundary, is not well understood. p130 appears to be involved in the maintenance of the G0 state, although it can also have a primary role in the control of G1/ S transition, depending on cell type (Grana et al. 1998; Hoshikawa et al. 1998; Mulligan and Jacks 1998; Stiegler and Giordano 1999). The antiproliferative function common to the three pocket proteins is transcription factor E2F (E2F)-dependent inhibition of transcription through the formation of suppressor complexes with E2F factors and histone deacetylase (HDAC) enzymes and/or switch/sucrose-nonfermenting chromatin remodeling (SWI/SNF) complexes (Trouche et al. 1997; Ferreira et al. 1998; Stiegler et al. 1998a; Takahashi et al. 2000; O'Connor et al. 2001; Singh et al. 2001; Rayman et al. 2002). Furthermore, p130 and p107 share the ability to bind to and directly inhibit the activity of Cdk2/cyclin E and A complexes (De Luca et al. 1997; Lacy and White 1997; Woo et al. 1997; Castano et al. 1998). Consistent with this function, cyclin E/Cdk2 activity is efficiently inhibited by p130 in serum-starved p27Kip1−/− p21Cip1−/− double knockout mouse ebryonal fibroblasts (MEFs; Coats et al. 1999).
Even though the ability to negatively control cell cycle progression is a common feature of all three members of the pocket protein family, only Rb meets all the requirements to be a bona fide tumor suppressor. Homozygous Rb mutations are very frequently found in tumors and reintroduction of a wild-type RB gene into RB-deficient tumor-derived cell lines often reverts the malignant phenotype. Most significantly, heterozygous RB null mutations in mice predispose to cancer (Weinberg 1995; Paggi et al. 1996; Mulligan and Jacks 1998). Targeted disruption of the gene for p130 or its cognate p107, on the other hand, gives rise to no evident phenotype. Yet it is clear that p130 has a role in cell cycle control in the context of development because mice of the mixed genotypes p130−/− p107−/− and p130−/− RB+/− exhibit severe developmental defects (Classon and Dyson 2001). In addition, the p130−/− genotype by itself confers developmental defects in the Balb/c background, which is mutated at the INK4a (p16) locus (LeCouter et al. 1998; Zhang et al. 1998). Although p130 disruption does not cause predisposition to cancer in mice, p130 is found to be mutated in some human cancers; furthermore, p130 levels inversely correlate with tumor grade (Baldi et al. 1996, 1997; Susini et al. 1998; Massaro-Giordano et al. 1999; Tanaka et al. 1999). Thus, whereas p130 functions appear to be limited to control of cell proliferation in the context of development and differentiation in the mouse, p130 has an additional, although not well-defined, role in suppression of cancer in humans (Yee et al. 1998; Classon and Dyson 2001; Paggi and Giordano 2001).
The expression of p107 and, to a lesser extent, pRb is transcriptionally regulated via E2F sites, consistent with their levels peaking at the G1/S boundary (Nevins 1998). pRb is also expressed at high levels in some terminally differentiated G0 cell types, by an E2F-independent transcriptional mechanism (Martelli et al. 1994). On the other hand, accumulation of p130 is posttranscriptionally regulated and largely restricted to G0. Although neither the p130 mRNA level nor its promoter activity fluctuate as a function of proliferation or cell cycle progression, p130 protein is abundant in G0 and early G1 and then progressively disappears as cells progress through the cell cycle (Richon et al. 1997; Grana et al. 1998; Nevins 1998; Smith et al. 1998; Fajas et al. 2000).
Exit from G0 and progression through G1 into S phase requires both the activity of Cdks and E2F-dependent transcription, both of which are regulated by pocket proteins (Grana and Reddy 1995; Helin 1998). The control of Cdk activity is carried out at multiple levels, one of which is binding to kinase inhibitors such as p130 (Morgan 1995; Arellano and Moreno 1997; Obaya and Sedivy 2002). E2F-dependent transcription is controlled by the modulation of the relative amounts of free and pocket protein-bound E2F factors, leading to either transcriptional activation or repression (Dyson 1998; Black and Azizkhan-Clifford 1999). Furthermore, the expression of E2F1–2–3 is transcriptionally repressed specifically by E2F4/p130 repressor complexes, which are the most abundant E2F species in G0 (Furukawa et al. 1999; Takahashi et al. 2000). The disappearance of such complexes occurs after the transition from G0 to G1 in cells reentering the cell cycle, following Cdk-dependent hyperphosphorylation of p130 and dissociation of the complexes (Smith et al. 1996). The activities of Cdk4/6 and Cdk2 appear to be redundant in this context (Cheng et al. 2000; Hansen et al. 2001). On the other hand, p130 binding to and inhibition of Cdk2 is not controlled by phosphorylation, but only by the relative abundance of p130, which progressively decreases as G0 cells reenter the cell cycle and approach S phase (Grana et al. 1998; Smith et al. 1998; Hansen et al. 2001). Therefore, the mechanism underlying p130 down-regulation is particularly relevant for p130 functions involving negative regulation of Cdk2 kinase activity.
Previous studies have suggested that posttranscriptional regulation of p130 levels is at the level of proteasome-mediated protein turnover (Smith et al. 1996; Stubdal et al. 1996; Prince et al. 2002). Indeed, ubiquitin-dependent proteolysis by the proteasome is a common regulatory motif for a growing number of proteins involved in cell cycle control. Depending on either the substrate or the specific protein–ubiquitin ligase (or E3 enzyme) used, the signals triggering the polyubiquitination cascade can rely on different events such as (1) covalent or conformational modification of the target protein, (2) changes in its association with other proteins, or (3) direct activation of the relevant protein–ubiquitin ligase (Hershko and Ciechanover 1998; Peters 1998; Laney and Hochstrasser 1999; DeSalle and Pagano 2001; Yew 2001). A class of E3 enzymes, known as Skp1–Cul1/Cdc53–F-box protein (SCF) complexes, recognizes and polyubiquitinates substrates phosphorylated at specific sites. Roc1, Cul1, and Skp1 are the invariant core components of SCF complexes, and one of several F-box proteins is responsible for substrate recognition and specificity (Deshaies 1999; Krek 1998; Jackson et al. 2000). Specific SCF complexes polyubiquitinate phosphorylated IKBα, p27Kip1 and cyclin E, respectively, and target them to proteasome-mediated degradation (Clurman et al. 1996; Won and Reed 1996; Diehl et al. 1997; Vlach et al. 1997; Montagnoli et al. 1999; Winston et al. 1999; Koepp et al. 2001; Strohmaier et al. 2001). Correlation between p130 phosphorylation and down-regulation suggested that p130 might also be a target of SCF and subsequent proteasome-mediated degradation. In the present study, we show that this is indeed the case, with SCFSkp2 being the relevant protein–ubiquitin ligase. We show that recognition by SCFSkp2 is possible only after p130 is specifically phosphorylated at Ser 672 by Cdk4 and/or Cdk6. Cdk4 is already known to have a role in activation of Cdk2 by sequestration of the Cdk inhibitor p27 (Sherr and Roberts 1995, 1999; Cheng et al. 1998). Now we reveal an additional mechanism whereby Cdk4/6 activates Cdk2, triggering the degradation of the Cdk2 inhibitor p130.
Results
p130 is regulated through the cell cycle at the level of turnover
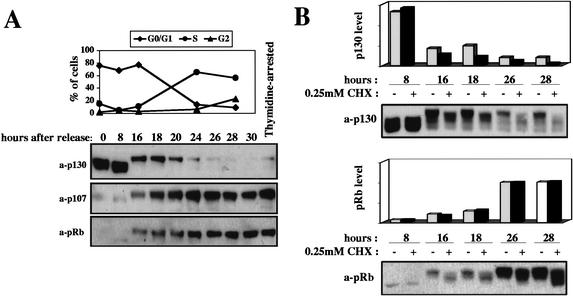
p130 levels are regulated in response to the proliferative state of cells. Serum-starved G0 human diploid fibroblasts contain high levels of p130 in a hypophosphorylated state (Fig. 1A). Subsequent to cell cycle entry in response to serum stimulation, mid-G1 cells exhibit an increase in p130 phosphorylation, which correlates with a decline in p130 steady state levels (Mayol et al. 1995; Grana et al. 1998; Fig. 1A.). Because p130 mRNA levels do not fluctuate through the cell cycle (Richon et al. 1997; Smith et al. 1998), we sought to determine whether p130 protein levels are regulated at the level of proteolysis. We first determined the stability of p130 at different time points after release from G0. Serum-starved human fibroblasts were released into the cell cycle by addition of serum, and duplicate samples were harvested at different time points after the release, ranging from early G1 to S/G2. For each time point, one sample was harvested immediately, and the other was cultured in the presence of a protein synthesis inhibitor, cyclohexemide, for two additional hours prior to harvest (Fig. 1B). Hyperphosphorylated p130 present from late G1 on was substantially less stable than the hypophosphorylated p130 associated with early G1. Although the related protein pRb was phosphorylated with similar cell cycle kinetics, no change in stability was observed (Fig. 1B). Thus, p130 levels are indeed regulated through controlled proteolysis, possibly in a phosphorylation-dependent manner.
Figure 1.
p130 expression and stability in human fibroblasts after release from G0 block. (A) G0-arrested human fibroblasts were released into the cell cycle and samples were harvested for Western blot analysis at the indicated time points after release. For each sample, the percentage of cells in the G0/G1, S, and G2 phases of the cell cycle was calculated by FACS analysis. The last sample on the right is from cells synchronized in early S phase by thymidine block. (B) G0-arrested human fibroblasts were released into the cell cycle and samples were harvested for Western blot analysis at the indicated time points after release. Samples treated with cyclohexemide (CHX) received the drug at the time at which the corresponding untreated samples were harvested, and were then harvested 2 h later.
p130 turnover is regulated by phoshorylation
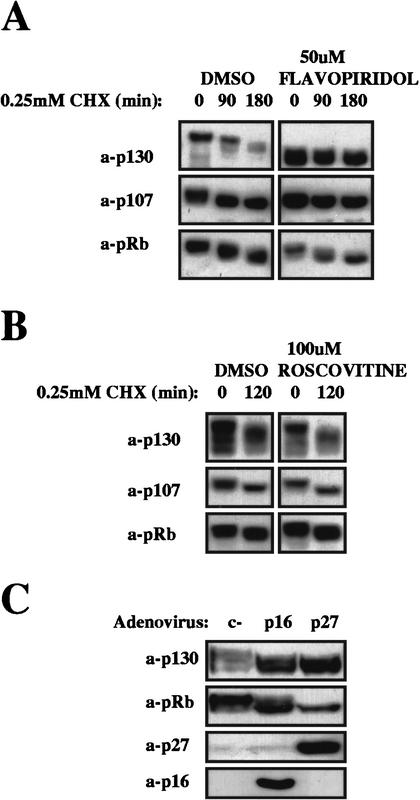
To establish a causal relationship between p130 hyperphosphorylation and its degradation, we investigated the effect of the protein kinase inhibitor flavopiridol. Flavopiridol targets Cdk2, Cdk4, and Cdk6, all of which can phosphorylate p130 (Hansen et al. 2001; D. Tedesco and S.I. Reed, unpubl.). To exclude cell cycle effects of the Cdk inhibitor, we first arrested cells in early S phase by treatment with thymidine, prior to treatment with the inhibitor. A similar approach was used with all subsequent manipulations that might affect the cell cycle distribution of cells under analysis. After the first 4 h of incubation with flavopiridol, cells were treated with cyclohexemide for various lengths of time before harvesting, and p130 stability was determined. Cdk inhibition by flavopiridol abolished both the hyperphosphorylation and the degradation of p130, as shown in Figure 2A. This confirms that p130 phosphorylation is potentially a signal for its degradation. Similar experiments were then carried out with the Cdk1/Cdk2-specific inhibitor roscovitine. Even though roscovitine can potentially affect Cdk4/6 activity indirectly, by inhibiting MAP kinases if administered in early G1, no effect on Cdk4/6 activity was detectable in thymidine-arrested cells, as judged by pocket proteins phosphorylation state (data not shown). After roscovitine treatment, p130 remained hyperphosphorylated and unstable, indicating that neither phosphorylation nor turnover depend on Cdk2 activity (Fig. 2B; Cdk1 is not active in S-phase cells). Furthermore, inhibition of Cdk4 and Cdk6 by adenovirus-mediated overexpression of the Cdk4/6 inhibitor p16INK4 in S-phase (thymidine)-arrested cells led to hypophosphorylation and accumulation of p130 similarly to p27, which when greatly overexpressed inhibits Cdk4/6 as well as Cdk2 (Fig. 2C). Thus, the response of p130 levels and stability to general and specific Cdk inhibitors suggests that Cdk4 and/or Cdk6, but not Cdk2 activity, is required for p130 degradation, possibly through site-specific phosphorylation.
Figure 2.
Effects of kinase inhibitors on p130 stability. Thymidine-arrested human fibroblasts were treated with either Flavopiridol (A) or Roscovitine (B). Protein-synthesis inhibitor cyclohexemide (CHX) was added after the first 4 h of treatment, and cells were harvested for Western blot analysis after the indicated time. DMSO, dimethyl sulfoxide. (C) Thymidine-arrested human fibroblasts were infected with the indicated recombinant adenoviruses, maintained under thymidine arrest, and harvested 18 h later for Western blot analysis.
p130 turnover depends on proteasome function
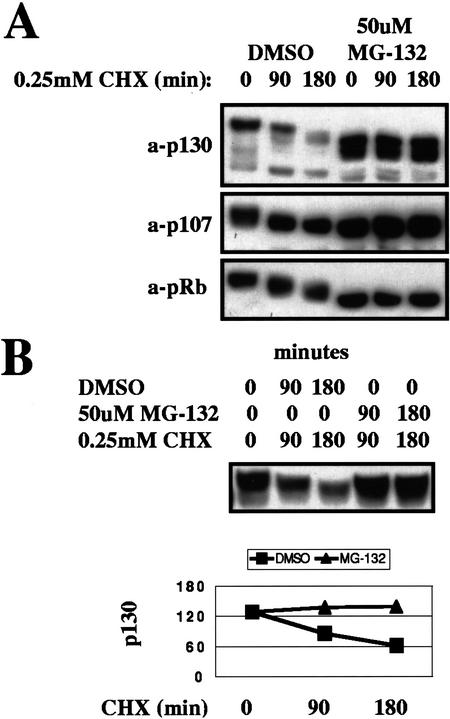
We next determined whether proteasomal function is required for p130 proteolysis. In an experiment analogous to those just described for Cdk inhibitors, S-phase-arrested human fibroblasts were pretreated with the proteasome-specific inhibitor MG-132, followed by treatment with cyclohexemide to measure the stability of p130 (Fig. 3A). Proteasome inhibition resulted in dramatic stabilization of p130, but also in an inhibition of p130 hyperphosphorylation. The latter phenomenon was likely attributable to the induced accumulation of the Cdk inhibitors p21 and p27 (data not shown), both substrates of the proteasome (Pagano et al. 1995; Sheaff et al. 2000). Therefore, it was not possible to determine whether p130 stabilization was a direct effect of proteasome inhibition or an indirect effect of inhibition of p130 phosphorylation. However, in a similar experiment in which both MG-132 and cyclohexemide were administered simultaneously so that kinase inhibitors could not accumulate, it was found that proteasome inhibition stabilized p130 without an effect on p130 phosphorylation (Fig. 3B).
Figure 3.
Effect of proteasome inhibition on p130 stability. (A) Thymidine-arrested human fibroblasts were treated with proteasome inhibitor MG-132. Protein-synthesis inhibitor cyclohexemide (CHX) was added after the first 4 h of treatment, and cells were harvested for Western blot analysis after the indicated time. DMSO, dimethyl sulfoxide. (B) Thymidine-arrested human fibroblasts were treated either with CHX alone, or with CHX and MG-132, and cells were harvested for Western blot analysis after the indicated time.
SCFSkp2 is required for the phosphorylation-dependent degradation of p130
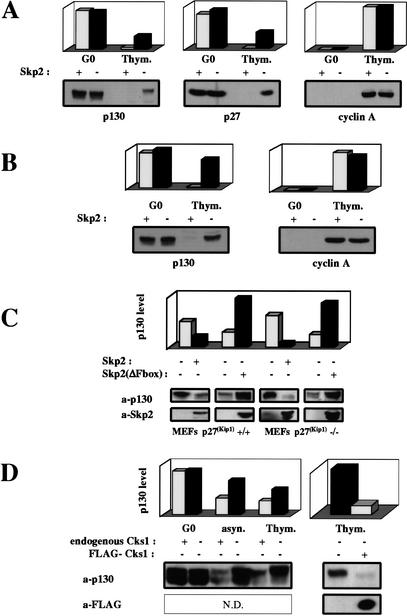
The cell cycle-dependent expression profile of p130 closely resembles that of the Cdk inhibitor p27, and the levels of both proteins are controlled through phosphorylation-triggered proteasome-mediated degradation. We investigated whether hyperphosphorylated p130 might be targeted to the proteasome by the SCFSkp2 protein–ubiquitin ligase, which is responsible for the polyubiquitination and subsequent degradation of p27 (Carrano et al. 1999). To this end, we compared the regulation of p130 in SKP2+/− and SKP2−/− mouse fibroblasts (Nakayama et al. 2000). In SKP2+/− and SKP2−/− fibroblasts arrested in G0, p130 was present at comparably high levels. In contrast, p130 levels in thymidine-arrested SKP2−/− cells were much higher than in corresponding SKP2+/− cells (Fig. 4A,B). To confirm that both SKP2−/− and SKP2+/− cells had exited G0, traversed G1, and similarly arrested in S phase, parallel immunoblots were performed for cyclin A, a marker for entry into S phase. As can be seen, cyclin A signals were comparable for both SKP2−/− and SKP2+/− fibroblasts (Fig. 4A,B). In Figure 4A, there is an apparent decrease in p130 levels as cells progress from G0 to S phase, suggesting that Skp2-independent mechanisms may also regulate p130 levels. However, a similar reduction was observed for p27, for which turnover is thought to be largely Skp2-dependent (Malek et al. 2001). Because loading of gels in Figure 4A was normalized to protein concentration, we hypothesized that the lower protein content of G0 cells compared with cycling cells might account for the apparent relative reduction in p130 levels in S-phase-arrested cells. Therefore another gel was run, in which loading was normalized to cell number (Fig. 4B). Using these assay conditions, it is clear that the amount of p130 per cell decreases minimally as SKP2−/− fibroblasts progress from G0 to S phase. Furthermore, in S-phase-arrested wild-type (wt) fibroblasts, adenovirus-mediated overexpression of wt Skp2 resulted in down-regulation of p130, whereas overexpression of dominant-negative, F-box-deleted Skp2 led to the accumulation of p130 in a hyperphosphorylated state (Fig. 4C). Similar results were obtained using p27Kip1−/− fibroblasts, thus ruling out the possibility that the effects of Skp2 expression on p130 levels might be mediated indirectly via changes in the level of p27Kip1, leading to changes in p130 phosphorylation (Fig. 4C).
Figure 4.
Effects of Skp2 and Cks1 on p130 expression. (A) G0-arrested and thymidine-arrested SKP2+/− and SKP2−/− primary MEFs were analyzed for p130, p27, and cyclin A levels by Western blot. Gel loading was normalized to protein concentration. Signals were quantitated by ImageQuant. (B) p130 and cyclin A levels were analyzed as in A, except that all loading was normalized to cell number. (C) Thymidine-arrested p27+/+ and p27−/− MEFs were infected with either empty, Skp2-expressing, or Skp2(ΔFbox)-expressing recombinant adenoviruses, maintained under thymidine arrest, and harvested 18 h after infection for Western blot analysis. (D) G0-arrested, asynchronous, and thymidine-arrested CKS1+/+ and CKS1−/− MEFs were analyzed for p130 expression by Western blot. Thymidine-arrested CKS1−/− MEFs were also infected with either empty or FLAG-tagged Cks1-expressing recombinant adenoviruses, maintained under thymidine arrest, and analyzed 18 h after infection. Loading was normalized to protein concentration.
Cks1 is required for SCFSkp2-mediated degradation of p130
Cks1 is en essential cofactor for Skp2-dependent degradation of phosphorylated p27 (Ganoth et al. 2001; Spruck et al. 2001), and thus might be required for ubiquitination of other SCFSkp2 substrates. In wt and CKS1−/− fibroblasts arrested in G0, p130 was present at comparably high levels (Fig. 4D). However, as with SKP2−/− fibroblasts, p130 levels in asynchronous and thymidine-arrested CKS1−/− cells were much higher than in corresponding wt cells, consistent with a role for Cks1 in p130 degradation. Transduction of CKS1−/− fibroblasts with a Cks1-expressing recombinant adenovirus restored p130 to wt levels, confirming that the defect in p130 regulation is due specifically to loss of Cks1 function (Fig. 4D; data not shown).
Skp2 binds to and targets phosphorylated but not unphosphorylated p130
Because p130 degradation was shown to be phosphorylation-dependent, in vitro binding assays were carried out to determine whether Skp2 could bind hyperphosphorylated, but not dephosphorylated p130, as is the case for other SCF substrates such as p27Kip1 (Carrano et al. 1999). Thymidine-arrested CKS1−/− fibroblasts were exploited as a source of native hyperphosphorylated p130 (see Fig. 4D). Anti-p130 immunoprecipitates were then challenged with purified recombinant glutathione-S transferase (GST)–Skp2/Skp1/Cks1 complexes. Figures 5A and 5B show that Skp2 bound untreated, but not protein phosphatase-treated, p130 immunoprecipitates. Recombinant adenoviruses were then used to show that the effect of Skp2 on p130 turnover was phosphorylation dependent. U2OS cells were chosen for these experiments because Skp2 activity was found to be limiting and p130, therefore, intrinsically stable in these cells (data not shown). Adenoviral transduction of wt Skp2 dramatically down-regulated endogenous p130, whereas neither the F-box-deleted Skp2 nor the Cdk4/6 inhibitor p16 caused accumulation of the already stable p130 (Fig. 6A). However, when Skp2 was expressed in conjunction with p16, the down-regulation of p130 was substantially reduced (Fig. 6A), indicating that in vivo, p130 requires phosphorylation by Cdk4/6 in order to be targeted by Skp2.
Figure 5.
In vitro Skp2 binding to hyperphosphorylated p130. (A) Nonimmune, anti-p130, and anti-p27Kip1 immunoprecipitates were immobilized on beads from thymidine-arrested CKS1−/− MEFs, and probed for binding to soluble GST–Skp2/Skp1/Cks1 complexes. After binding, beads were extensively washed and retained proteins were analyzed by Western blot. IP, immunoprecipitating antibody; NI, nonimmune serum. (B) Nonimmune and anti-p130 immunoprecipitates prepared as in A were either treated or not treated with λ protein phosphatase, and then assayed for binding, as in A.
Figure 6.
Phosphorylation requirements for Skp2 binding and destabilization of p130. (A) Thymidine-arrested U2OS cells were infected with either empty, Skp2-expressing, Skp2(ΔFbox)-expressing, or p16INK4-expressing recombinant adenoviruses, maintained under thymidine arrest, and harvested 18 h after infection for Western blot analysis. (B) Thymidine-arrested U2OS cells expressing the indicated HA-tagged recombinant alleles of p130 were infected with either empty or Skp2-expressing recombinant adenoviruses, maintained under thymidine arrest, and harvested 18 h after infection for Western blot analysis. (C) Pulse-chase analysis of p130 decay in thymidine-arrested U2OS cells expressing either β-galactosidase, HA-tagged wt p130, or HA-tagged S672A p130 after infection with either empty or Skp2-expressing recombinant adenoviruses.
Phosphorylation of Ser 672 is essential for Skp2 binding and p130 degradation
In order to confirm that the Skp2-induced degradation of p130 depends on site-specific phosphorylation of p130, p130 phosphorylation site mutants were analyzed for their susceptibility to Skp2 overexpression. Because p130 degradation is blocked by the inhibition of Cdk4/6, but not of Cdk2, at least one of the relevant phosphorylation sites in p130 has to be Cdk4/6 specific. By means of retroviral transduction, we obtained U2OS-derived populations expressing either hemagglutinin epitope (HA)-tagged p130 wt, HA-tagged p130 lacking the three previously identified specific Cdk4/6 phosphorylation sites (p130ΔCdk4; Hansen et al. 2001), or HA-tagged p130 lacking all Cdk phosphorylation sites (p130PM19A; Hansen et al. 2001). The effect of Skp2 activity was tested by adenoviral transduction of Skp2 in thymidine-arrested cultures. The Western blot in Figure 6B shows that mutation of the three Cdk4/6-specific phosphorylation sites sufficed to render p130 insensitive to Skp2 overexpression. To further refine the phosphorylation requirements for p130 degradation, we constructed single-residue mutants corresponding to individual Cdk4/6-specific phosphorylation sites and analyzed as described earlier. Of the three sites (T401, S672, and S1035), only the C-terminal two were considered in this analysis, because the N-terminal site is not conserved. It was found that a single substitution, S672A, rendered p130 as refractory to Skp2 activity as the triple mutant, whereas S1035A had no effect (data not shown), indicating that phosphorylation of Ser 672 is specifically required for p130 degradation (Fig. 6B). To ensure that the differences observed in steady-state level unambiguously reflect changes in turnover rate, we measured the p130 turnover directly by pulse-chase analysis. U2OS cells expressing either HA-wt-p130 or HA-S672A-p130 were transduced with either Skp2 or control adenoviruses, and the decay of the HA-tagged proteins was analyzed. As illustrated in Figure 6C, the results of two independent pulse-chase experiments confirmed that Skp2 overexpression induced a significant shortening of the p130 half-life that depended on the ability of p130 to be phosphorylated on Ser 672.
p130 is a target of ubiquitination by SCFSkp2
In vitro assays were performed to determine whether the effect of Skp2 on the stability of p130 is mediated by polyubiquitination. Soluble FLAG-tagged SCFSkp2 complexes were immunoaffinity purified from transfected 293T cells, and assayed for the ability to attach polyubiquitin chains to p130 molecules. Native endogenous p130 was purified from CKS1−/− fibroblasts, and HA-tagged wt and S672A p130 were prepared from retrovirally transduced U2OS cells. In vitro ubiquitination reactions were performed as previously described (Spruck et al. 2001; Strohmaier et al. 2001), and the reaction products analyzed by SDS-PAGE and Western blot. As illustrated in Figure 7A, SCFSkp2 complexes were capable of polyubiquitinating wt p130 but not p130 S672A, confirming that p130 phosphorylated on Ser 672 is a direct ubiquitination target of SCFSkp2. On the other hand, SCF complexes containing a different F-box protein, hCdc4, were incapable of ubiquitinating p130, although they were capable of ubiquitinating cyclin E (Fig. 7B). To eliminate the possibility that p130 was being monoubiquitinated at multiple sites rather than polyubiquitinated, reactions were performed using methylated ubiquitin rather than ubiquitin. Methylated ubiquitin can form monoubiquitin conjugates but not polyubiquitin conjugates. In vitro reactions using methylated ubiquitin produced a distinct monoubiquitinated p130 band, but no more slowly migrating species were observed (Fig. 7C). Therefore, the slowly migrating species observed in Figure 7A must correspond to polyubiquitin conjugates of p130.
Figure 7.
In vitro ubiquitination of p130 by SCFSkp2. (A) Anti-p130 immunoprecipitates were immobilized on beads from thymidine-arrested CKS1−/− MEFs, or thymidine-arrested U2OS cells expressing either wt or S672A HA-tagged p130. Ubiquitination reactions by soluble immunopurified SCFFLAG-Skp2 were performed, and polyubiquitination of the immobilized immunoprecipitates was assayed by Western blot analysis. (B) Anti-cyclin E and anti-p130 immunoprecipitates were immobilized on beads from thymidine-arrested U2OS cells. Ubiquitination reactions by soluble immunopurified SCFFLAG-hCdc4 were performed, and polyubiquitination of the immobilized immunoprecipitates was assayed by Western blot analysis. (C) Anti-p130 immunoprecipitates were immobilized on beads from thymidine-arrested U2OS cells. Ubiquitination reactions by soluble immunopurified SCFFLAG-Skp2 were performed in the presence of either native ubiquitin (Ub) or methylated ubiquitin (mUb), and mono/polyubiquitination of the immobilized immunoprecipitates was assayed by Western blot analysis.
Discussion
Both cell cycle inhibitory functions of p130 are antagonized by phosphorylation-dependent mechanisms
The ability of p130 to confer a sustained G1 block when ectopically expressed has been ascribed to both phosphorylation-sensitive and phosphorylation-insensitive functions (Hansen et al. 2001). Here we show that, ultimately, all cell cycle inhibitory functions of p130 are phosphorylation-sensitive in that the steady-state level of p130 itself is regulated by phosphorylation-dependent proteolysis.
Our initial observation of a strong correlation between p130 hyperphosphorylation and instability prompted us to investigate the possibility of a causal relationship between the two phenomena. Using the kinase inhibitors flavopiridol and roscovitine, both active against Cdk2, but only the former active against Cdk4/6, we could show that Cdk2 activity was not necessary for triggering p130 degradation, whereas Cdk4/6 was. Additional experiments using adenovirus-mediated overexpression of the Cdk4/6 specific inhibitor p16 confirmed the requirement for Cdk4/6 in the regulation of p130 degradation. A caveat concerning the use of p16 as a Cdk4/6 specific inhibitor is the ability of INK4 family inhibitors to displace p27 from Cdk4/6 to Cdk2, thereby leading to inhibition of both Cdk4/6 and Cdk2 (Sherr and Roberts 1995; Cheng et al. 1998). However, an indirect inhibitory effect of p16 on Cdk2 activity requires a large pool of p27 to be released from Cdk4/6 containing complexes, which is the case in early or mid-G1 cells but not in thymidine-arrested S-phase cells used in our study. Thus, as cells emerge from G0 and enter the cell cycle with concomitant accumulation of D-type cyclins and formation of active cyclin D/Cdk4/6 complexes, p130 is simultaneously dissociated from E2F4 relieving transcriptional repression of E2F-dependent genes and targeted for proteasome-mediated degradation, relieving inhibition of cyclin E/Cdk2 complexes. In this manner, p130 serves as a link between positive proliferative signals shown to promote accumulation of D-type cyclins and the core cell cycle machinery driven by cyclin E/Cdk2 and E2F-dependent transcription.
The relationship between p130 and p27
The observation that Cdk4/6 activity is the functional trigger for p130 degradation and, as a consequence, the release of Cdk2 inhibition, suggests a parallelism between p130 and p27 in the regulation of Cdk2 activity. Both are Cdk2 inhibitors targeted for ubiquitin-dependent proteasome-mediated degradation at the G1/S boundary by Cdk-dependent phosphorylation. However, there are also key differences. First, p27 down-regulation in cells emerging from G0 begins early in G1 prior to p27 phosphorylation. At least in some cell types, this is likely to occur through a cytoplasmic mechanism that is dependent on polyubiquitination and proteasome-mediated degradation but independent of Skp2 (Hara et al. 2001). Second, Cdk2 inhibition by p27 is relieved not only by degradation, but also by the equilibration of p27 away from Cdk2 to cyclin D/Cdk4/6 complexes as these accumulate (Sherr and Roberts 1999). In cells exiting from G0, it is therefore conceivable that the release from p27-mediated inhibition of Cdk2 is achieved more rapidly than p130-mediated inhibition, because p27 degradation begins in early G1 and only the physical accumulation of cyclin D/Cdk4/6 complexes is needed to titrate p27 away from Cdk2. On the contrary, p130 is stable in early G1 and cannot be sequestered by Cdk4/6, and it is only through phosphorylation by Cdk4/6 and subsequent degradation of p130 that Cdk2 is released from inhibition. Although it is likely that the increase of p130 turnover following hyperphosphorylation is a rapid event, phosphorylation of the bulk of p130 that has accumulated in G0 cells appears to be rate limiting and is not complete until cells are in late G1 (Mayol et al. 1995; D. Tedesco and S.I. Reed, unpubl.). Furthermore, because the half-life of hyperphosphorylated p130 is not dramatically short (at least 1 h), p130 expression does not drop to minimal levels until cells are at the G1/S boundary. In light of these observations and considerations, the roles of p27 and p130 in regulation of Cdk2 activity, although overlapping, are probably not completely redundant. We suggest a sequential inhibition, first primarily by p27, and then when p27 pools have been reequilibrated to Cdk4/6 complexes, by p130. The resistance of p130 to rapid inactivation by titration would promote the accumulation of inactive cyclin E/Cdk2/p130 complexes until a threshold level of cyclin D/Cdk4/6 activity triggers p130 degradation and concomitant rapid activation of Cdk2. However, the ability of Skp2 to target both cell cycle inhibitors may explain its potency, when overexpressed, in driving quiescent cells into S phase (Sutterluty et al. 1999).
p27 and p130 are both targets of SCFSkp2
Many cell cycle regulatory proteins are targeted for proteasome-mediated degradation by polyubiquitination following site-specific phosphorylation and recognition by specific SCF ubiquitin ligases (DeSalle and Pagano 2001; Yew 2001). The degradation of the Cdk inhibitor p27 depends on polyubiquitination by SCFSkp2 following phosphorylation on Thr187 (Carrano et al. 1999). SCF-mediated ubiquitination of p27 depends on a physical interaction between Skp2 and the small Cdk interacting protein, Cks1 (Ganoth et al. 2001; Spruck et al. 2001). Consistent with this requirement, in both SKP2−/− and CKS1−/− cells, p27 is stable even when phosphorylated, and as a consequence, is maintained at a relatively high level even at the G1/S transition, where it is normally down-regulated. p130 exhibits a similar pattern of hyperaccumulation in SKP2−/− and CKS1−/− fibroblasts. In principle, p130 accumulation under these circumstances could be an indirect effect of p27 accumulation and concomitant inhibition of p130 phosphorylation. However, the fact the p130 accumulates in its hyperphosphorylated forms renders this mechanism unlikely, and prompted us to perform the series of experiments that led to the conclusion that Skp2 is the F-box protein responsible for the ubiquitination and degradation of p130. Thus, p27 and p130 are regulated in a similar fashion, the primary difference being the specific Cdk activity conferring affinity for Skp2 and hence triggering degradation: cyclin D/Cdk4/6 for p130 and cyclin E/Cdk2 for p27. Although it is not surprising that two proteins with partially redundant functions are regulated by parallel mechanisms, the apparent lack of homology between p130 and p27 in the regions surrounding the relevant phosphorylation sites raises some interesting questions concerning the nature of the Skp2 “degron,” the immediate protein structure actually recognized by Skp2. It has recently been reported that another F-box protein, Cdc4, recognizes a specific, although somewhat degenerate, consensus, surrounding the substrate phosphorylation site, designated the Cdc4 degron (Nash et al. 2001; Strohmaier et al. 2001). In contrast, it seems, based on sequence comparisons of p27 and p130, that a consensus degron for Skp2 might not exist, raising the possibility that Skp2 might interact with different substrates through different subregions in its large leucine-rich repeat domain (Kobe and Kajava 2001). In this context, the fact that Skp2 requires Cks1 for ubiquitination of both p27 and p130 suggests a general Skp2 activation function for Cks1.
Materials and methods
Cell lines
hTERT-immortalized human fibroblasts were a kind gift of J.W. Shay (The University of Texas Southwestern Medical Center, Dallas, TX). CKS1+/+ and CKS1−/− MEF lines were previously described (Spruck et al. 2001). p27+/+ and p27−/− MEF lines were a kind gift of J. Roberts (Fred Hutchinson Cancer Research Center, Seattle, WA). Human osteosarcoma cell line U2OS is ATCC no. HTB-96. SKP2+/− and SKP2−/− MEFs were a kind gift of K.-I. and K. Nakayama (Kyushu University, Fukuoka, Japan).
Cell culture and synchronization
All cells were routinely cultured in 10% FBS DMEM. Synchronization in G0 was achieved by 48-h serum starvation of confluent cultures (0.1% FBS). Re-entry into the cell cycle of G0-arrested cells was achieved by trypsinization, sparse reseeding, and cultivation in 10% FBS DMEM. Synchronization in early S phase was achieved by restimulating G0-synchronized cells in the continuous presence of 2 mM thymidine for 24 h.
Immunological reagents and procedures
Immunoprecipitations and Western blots were performed as described (Harlow and Lane 1988), unless otherwise specified. Gel loading was normalized to protein concentration, unless otherwise specified. Signals were quantitated by ImageQuant. Antibodies used included the following: p130 (C-20, Santa Cruz), p130 (C-20-G, Santa Cruz), p107 (C-18, Santa Cruz), pRb (G3–245, PharMingen), cyclin A (H-432, Santa Cruz), GST (B-14, Santa Cruz), HA (12CA5 ascitis), HA (HA.11 monoclonal, BAbCO), SKP2 (GP45, Zymed), p27 (cat no. K25020, Transduction), p27 (C-19, Santa Cruz), p16 (cat no. 3501–1, Clontech), p21 (C-19, Santa Cruz), and FLAG (M2, Sigma).
Retroviral production and infections
Recombinant retroviruses were generated and stable cell lines expressing HA-tagged p130 alleles were obtained by retroviral infection and selection, as described (Morgenstern and Land 1990). Viral stocks were prepared as described, using the 293T-derived packaging cell line Phoenix-Ampho, and the retroviral plasmid vectors of the pBABE series (Morgenstern and Land 1990; Kinsella and Nolan 1996).
Adenoviral production and infection
Recombinant adenoviruses expressing either wt or ΔFbox Skp2 (Carrano et al. 1999) were generated as previously described (Bett et al. 1994; Strohmaier et al. 2001). The recombinant adenovirus for the expression of human p16 and human p27 were previously described (Niculescu et al. 1998; Schreiber et al. 1999). The recombinant adenovirus for the expression of human Cks1 was a kind gift of C. Spruck (The Scripps Research Institute, La Jolla, CA). Purified viral stocks were prepared by CsCl ultracentrifugation. U2OS cells and MEFs were infected at a multiplicity of infection of 400 and 1200 v.p./cell, respectively, by incubating thymidine-arrested cultures with CsCl-purified viral particles in media with no serum supplemented with 2 mM thymidine for 4 h. FBS to 10% final concentration was then added and cells were cultured in 2 mM thymidine 14 h further prior to harvesting.
In vitro binding assays
Baculovirus expressed GST–Skp2/6HIS–Skp1 complexes were purified on a Ni-NTA resin (QIAGEN) under nondenaturing conditions as previously described (Spruck et al. 2001). Cks1 was expressed in bacteria and purified as described (Bourne et al. 1996). Nonimmune, a-p130 and a-p27 immunoprecipitates immobilized on beads were prepared from CKS1−/− thymidine-arrested fibroblasts, and probed for binding to the Cks1/Skp2/Skp1 soluble complex. The binding reaction was carried out at +4°C for 3 h in a buffer containing 10 mM Na Pyrophosphate, 10 mM Na β-glycerophosphate, 10 mM MgCl2, 0.1% Tween-20, 5% glycerol, 1 mM DTT, 1% BSA, 2 μg/mL aprotinin, 2 μg/mL leupeptin, 2 μg/mL pepstatin, 1 mM PMSF, and 50 mM TrisHCl (pH 7.5). Pellets were then washed five times in such buffer, boiled in SDS-sample buffer and analyzed by Western blot with a monoclonal anti-GST antibody. In the λ-phosphatase experiment, phosphatase inhibitors were not included in the washing buffer, and samples were dephosphorylated at +30°C for 1 h following manufacturer's specifications (Calbiochem) before the binding reaction.
In vitro ubiquitination assays
Recombinant SCFSkp2 and SCFhCdc4 complexes were isolated from transfected 293T cells, as previously described (Strohmaier et al. 2001), and then eluted in ubiquitination buffer containing 3×-FLAG peptide (SIGMA) following manufacturer's specifications. Ubiquitination buffer contained 5mM NaF, 1 mM NaVO4, 1.5 mM MgCl2, 5 mM KCl, 1 mM DTT, 20 mM HEPES (pH 7.4), 2 μg/mL aprotinin, 2 μg/mL leupeptin, 2 μg/mL pepstatin, and 1 mM PMSF. Equal amounts of SCFSkp2 complex were added to immunopurified substrates immobilized on beads. Ubiquitination was carried out in a 30-μL volume at +30°C for 2 h with 2 μg either Bovine Ubiquitin (SIGMA) or Methyl Ubiquitin (Boston Biochem), 1.3 μg UbcH3/Cdc34 (Boston Biochem) and 0.65 μg E1 enzyme (Boston Biochem), 0.1 μg Cks1, 1 mM ATP, 20 mM creatine phosphate, and 5 μg creatine kinase. Reactions were terminated by washing the pellets three times in RIPA buffer and boiling for 3 min in SDS sample buffer. Eluted substrates were then analyzed by Western blot.
Pulse-Chase experiments
Pulse-chase experiments were performed on thymidine-arrested cultures. Cells were incubated with starvation medium (without methionine, 2 mM thymidine, no serum) for 15 min and then labeled (150 μCi/mL Redivue 35S-methionine, Amersham Pharmacia) for 45 min. Cells were then chased with normal medium supplemented with 10% FBS, 2 mM thymidine, and 100 mg/mL methionine. Cells were then washed twice in cold PBS and lysed in RIPA buffer. Immunoprecipitations were performed with anti-HA 12CA5 monoclonal antibody.
Acknowledgments
We acknowledge T. Herzinger, H. Strohmaier, and J. Bartek for contributions in the early stages of the project. We also thank K.-I. and K. Nakayama for SKP2−/− fibroblasts; J. Roberts for p27−/− fibroblasts; F.L. Graham, M. Schreiber, J.W. Shay, and M. Pagano for reagents; and C. Spruck and M. Henze for technical support and helpful discussions. D. Tedesco has been supported by a fellowship from the Consiglio Nazionale delle Ricerche, Italy, and a fellowship from the American Cancer Society. This work was supported by NIH grant CA78343 to S.I. Reed.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sreed@scripps.edu; FAX (858) 784-2781.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1011202.
References
- Arellano M, Moreno S. Regulation of CDK/cyclin complexes during the cell cycle. Int J Biochem Cell Biol. 1997;29:559–573. doi: 10.1016/s1357-2725(96)00178-1. [DOI] [PubMed] [Google Scholar]
- Baldi A, Esposito V, De Luca A, Howard CM, Mazzarella G, Baldi F, Caputi M, Giordano A. Differential expression of the retinoblastoma gene family members pRb/p105, p107, and pRb2/p130 in lung cancer. Clin Cancer Res. 1996;2:1239–1245. [PubMed] [Google Scholar]
- Baldi A, Esposito V, De Luca A, Fu Y, Meoli I, Giordano GG, Caputi M, Baldi F, Giordano A. Differential expression of Rb2/p130 and p107 in normal human tissues and in primary lung cancer. Clin Cancer Res. 1997;3:1691–1697. [PubMed] [Google Scholar]
- Bett AJ, Haddara W, Prevec L, Graham FL. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci. 1994;91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black AR, Azizkhan-Clifford J. Regulation of E2F: A family of transcription factors involved in proliferation control. Gene. 1999;237:281–302. doi: 10.1016/s0378-1119(99)00305-4. [DOI] [PubMed] [Google Scholar]
- Bourne Y, Watson MH, Hickey MJ, Holmes W, Rocque W, Reed SI, Tainer JA. Crystal structure and mutational analysis of the human CDK2 kinase complex with cell cycle-regulatory protein CksHs1. Cell. 1996;84:863–874. doi: 10.1016/s0092-8674(00)81065-x. [DOI] [PubMed] [Google Scholar]
- Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Castano E, Kleyner Y, Dynlacht BD. Dual cyclin-binding domains are required for p107 to function as a kinase inhibitor. Mol Cell Biol. 1998;18:5380–5391. doi: 10.1128/mcb.18.9.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Sexl V, Sherr CJ, Roussel MF. Assembly of cyclin D-dependent kinase and titration of p27Kip1 regulated by mitogen-activated protein kinase kinase (MEK1) Proc Natl Acad Sci. 1998;95:1091–1096. doi: 10.1073/pnas.95.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Rossi F, Fang W, Mori T, Cobrinik D. Cdk2-dependent phosphorylation and functional inactivation of the pRB-related p130 protein in pRB(−), p16INK4A(+) tumor cells. J Biol Chem. 2000;275:30317–30325. doi: 10.1074/jbc.M005707200. [DOI] [PubMed] [Google Scholar]
- Classon M, Dyson N. p107 and p130: Versatile proteins with interesting pockets. Exp Cell Res. 2001;264:135–147. doi: 10.1006/excr.2000.5135. [DOI] [PubMed] [Google Scholar]
- Clurman BE, Sheaff RJ, Thress K, Groudine M, Roberts JM. Turnover of cyclin E by the ubiquitin–proteasome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes & Dev. 1996;10:1979–1990. doi: 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]
- Coats S, Whyte P, Fero ML, Lacy S, Chung G, Randel E, Firpo E, Roberts JM. A new pathway for mitogen-dependent Cdk2 regulation uncovered in p27(Kip1)-deficient cells. Curr Biol. 1999;9:163–173. doi: 10.1016/s0960-9822(99)80086-4. [DOI] [PubMed] [Google Scholar]
- De Luca A, MacLachlan TK, Bagella L, Dean C, Howard CM, Claudio PP, Baldi A, Khalili K, Giordano A. A unique domain of pRb2/p130 acts as an inhibitor of Cdk2 kinase activity. J Biol Chem. 1997;272:20971–20974. doi: 10.1074/jbc.272.34.20971. [DOI] [PubMed] [Google Scholar]
- DeSalle LM, Pagano M. Regulation of the G1 to S transition by the ubiquitin pathway. FEBS Lett. 2001;490:179–189. doi: 10.1016/s0014-5793(01)02121-4. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin–proteasome pathway. Genes & Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- Dyson N. The regulation of E2F by pRB-family proteins. Genes & Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Fajas L, Le Cam L, Polanowska J, Fabbrizio E, Servant N, Philips A, Carnac G, Sardet C. A CDE/CHR-like element mediates repression of transcription of the mouse RB2 (p130) gene. FEBS Lett. 2000;471:29–33. doi: 10.1016/s0014-5793(00)01363-6. [DOI] [PubMed] [Google Scholar]
- Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Natl Acad Sci. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y, Iwase S, Kikuchi J, Nakamura M, Yamada H, Matsuda M. Transcriptional repression of the E2F-1 gene by interferon-α is mediated through induction of E2F-4/pRB and E2F-4/p130 complexes. Oncogene. 1999;18:2003–2014. doi: 10.1038/sj.onc.1202500. [DOI] [PubMed] [Google Scholar]
- Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, Hershko A. The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat Cell Biol. 2001;3:321–324. doi: 10.1038/35060126. [DOI] [PubMed] [Google Scholar]
- Garriga J, Limon A, Mayol X, Rane SG, Albrecht JH, Reddy EP, Andres V, Grana X. Differential regulation of the retinoblastoma family of proteins during cell proliferation and differentiation. Biochem J. 1998;333:645–654. doi: 10.1042/bj3330645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grana X, Reddy EP. Cell cycle control in mammalian cells: Role of cyclins, cyclin dependent kinases (CDKs), growth suppressor genes and cyclin-dependent kinase inhibitors (CKIs) Oncogene. 1995;11:211–219. [PubMed] [Google Scholar]
- Grana X, Garriga J, Mayol X. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene. 1998;17:3365–3383. doi: 10.1038/sj.onc.1202575. [DOI] [PubMed] [Google Scholar]
- Hansen K, Farkas T, Lukas J, Holm K, Ronnstrand L, Bartek J. Phosphorylation-dependent and -independent functions of p130 cooperate to evoke a sustained G1 block. EMBO J. 2001;20:422–432. doi: 10.1093/emboj/20.3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Kamura T, Nakayama K, Oshikawa K, Hatakeyama S. Degradation of p27(Kip1) at the G(0)-G(1) transition mediated by a Skp2-independent ubiquitination pathway. J Biol Chem. 2001;276:48937–48943. doi: 10.1074/jbc.M107274200. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hoshikawa Y, Mori A, Amimoto K, Iwabe K, Hatakeyama M. Control of retinoblastoma protein-independent hematopoietic cell cycle by the pRB-related p130. Proc Natl Acad Sci. 1998;95:8574–8579. doi: 10.1073/pnas.95.15.8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD. The lore of the RINGs: Substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- Kinsella TM, Nolan GP. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- Krek W. Proteolysis and the G1-S transition: The SCF connection. Curr Opin Genet Dev. 1998;8:36–42. doi: 10.1016/s0959-437x(98)80059-2. [DOI] [PubMed] [Google Scholar]
- Lacy S, Whyte P. Identification of a p130 domain mediating interactions with cyclin A/cdk 2 and cyclin E/cdk 2 complexes. Oncogene. 1997;14:2395–2406. doi: 10.1038/sj.onc.1201085. [DOI] [PubMed] [Google Scholar]
- Laney JD, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- LeCouter JE, Kablar B, Whyte PF, Ying C, Rudnicki MA. Strain-dependent embryonic lethality in mice lacking the retinoblastoma-related p130 gene. Development (Suppl.) 1998;125:4669–4679. doi: 10.1242/dev.125.23.4669. [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Jacks T. The retinoblastoma gene family in differentiation and development. Oncogene. 1999;18:7873–7882. doi: 10.1038/sj.onc.1203244. [DOI] [PubMed] [Google Scholar]
- Malek NP, Sundberg H, McGrew S, Nakayama K, Kyriakides TR, Roberts JM, Kyriakidis TR. A mouse knock-in model exposes sequential proteolytic pathways that regulate p27Kip1 in G1 and S phase. Nature. 2001;413:323–327. doi: 10.1038/35095083. [DOI] [PubMed] [Google Scholar]
- Martelli F, Cenciarelli C, Santarelli G, Polikar B, Felsani A, Caruso M. MyoD induces retinoblastoma gene expression during myogenic differentiation. Oncogene. 1994;9:3579–3590. [PubMed] [Google Scholar]
- Massaro-Giordano M, Baldi G, De Luca A, Baldi A, Giordano A. Differential expression of the retinoblastoma gene family members in choroidal melanoma: Prognostic significance. Clin Cancer Res. 1999;5:1455–1458. [PubMed] [Google Scholar]
- Mayol X, Garriga J, Grana X. Cell cycle-dependent phosphorylation of the retinoblastoma-related protein p130. Oncogene. 1995;11:801–808. [PubMed] [Google Scholar]
- Montagnoli A, Fiore F, Eytan E, Carrano AC, Draetta GF, Hershko A, Pagano M. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes & Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- Morgenstern JP, Land H. Advanced mammalian gene transfer: High titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan G, Jacks T. The retinoblastoma gene family: Cousins with overlapping interests. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, Shirane M, Tsunematsu R, Tsukiyama T, Ishida N, et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 2000;19:2069–2081. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Nevins JR. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- Niculescu AB, III, Chen X, Smeets M, Hengst L, Prives C, Reed SI. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol Cell Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obaya AJ, Sedivy JM. Regulation of cyclin-Cdk activity in mammalian cells. Cell Mol Life Sci. 2002;59:126–142. doi: 10.1007/s00018-002-8410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor RJ, Schaley JE, Feeney G, Hearing P. The p107 tumor suppressor induces stable E2F DNA binding to repress target promoters. Oncogene. 2001;20:1882–1891. doi: 10.1038/sj.onc.1204278. [DOI] [PubMed] [Google Scholar]
- Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- Paggi MG, Giordano A. Who is the boss in the retinoblastoma family? The point of view of Rb2/p130, the little brother. Cancer Res. 2001;61:4651–4654. [PubMed] [Google Scholar]
- Paggi MG, Baldi A, Bonetto F, Giordano A. Retinoblastoma protein family in cell cycle and cancer: A review. J Cell Biochem. 1996;62:418–430. doi: 10.1002/(SICI)1097-4644(199609)62:3%3C418::AID-JCB12%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Peters JM. SCF and APC: The Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- Prince AM, May JS, Burton GR, Lyle RE, McGehee RE., Jr Proteasomal degradation of retinoblastoma-related p130 during adipocyte differentiation. Biochem Biophys Res Commun. 2002;290:1066–1071. doi: 10.1006/bbrc.2001.6291. [DOI] [PubMed] [Google Scholar]
- Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, te Riele H, Dynlacht BD. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes & Dev. 2002;16:933–947. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richon VM, Lyle RE, McGehee RE., Jr Regulation and expression of retinoblastoma proteins p107 and p130 during 3T3-L1 adipocyte differentiation. J Biol Chem. 1997;272:10117–10124. doi: 10.1074/jbc.272.15.10117. [DOI] [PubMed] [Google Scholar]
- Schreiber M, Muller WJ, Singh G, Graham FL. Comparison of the effectiveness of adenovirus vectors expressing cyclin kinase inhibitors p16INK4A, p18INK4C, p19INK4D, p21(WAF1/CIP1) and p27KIP1 in inducing cell cycle arrest, apoptosis and inhibition of tumorigenicity. Oncogene. 1999;18:1663–1676. doi: 10.1038/sj.onc.1202466. [DOI] [PubMed] [Google Scholar]
- Sheaff RJ, Singer JD, Swanger J, Smitherman M, Roberts JM, Clurman BE. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes & Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- ————— CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes & Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Singh P, Chan SW, Hong W. Retinoblastoma protein is functionally distinct from its homologues in affecting glucocorticoid receptor-mediated transcription and apoptosis. J Biol Chem. 2001;276:13762–13770. doi: 10.1074/jbc.M100137200. [DOI] [PubMed] [Google Scholar]
- Smith EJ, Leone G, DeGregori J, Jakoi L, Nevins JR. The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 cell state from a G1 cell state. Mol Cell Biol. 1996;16:6965–6976. doi: 10.1128/mcb.16.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EJ, Leone G, Nevins JR. Distinct mechanisms control the accumulation of the Rb-related p107 and p130 proteins during cell growth. Cell Growth Differ. 1998;9:297–303. [PubMed] [Google Scholar]
- Spruck C, Strohmaier H, Watson M, Smith AP, Ryan A, Krek TW, Reed SI. A CDK-independent function of mammalian Cks1: Targeting of SCF(Skp2) to the CDK inhibitor p27Kip1. Mol Cell. 2001;7:639–650. doi: 10.1016/s1097-2765(01)00210-6. [DOI] [PubMed] [Google Scholar]
- Stiegler P, Giordano A. Role of pRB2/p130 in cellular growth regulation. Anal Quant Cytol Histol. 1999;21:363–366. [PubMed] [Google Scholar]
- Stiegler P, De Luca A, Bagella L, Giordano A. The COOH-terminal region of pRb2/p130 binds to histone deacetylase 1 (HDAC1), enhancing transcriptional repression of the E2F-dependent cyclin A promoter. Cancer Res. 1998a;58:5049–5052. [PubMed] [Google Scholar]
- Stiegler P, Kasten M, Giordano A. The RB family of cell cycle regulatory factors. J Cell Biochem (Suppl.) 1998b;30–31:30–36. [PubMed] [Google Scholar]
- Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- Stubdal H, Zalvide J, DeCaprio JA. Simian virus 40 large T antigen alters the phosphorylation state of the RB-related proteins p130 and p107. J Virol. 1996;70:2781–2788. doi: 10.1128/jvi.70.5.2781-2788.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susini T, Baldi F, Howard CM, Baldi A, Taddei G, Massi D, Rapi S, Savino L, Massi G, Giordano A. Expression of the retinoblastoma-related gene Rb2/p130 correlates with clinical outcome in endometrial cancer. J Clin Oncol. 1998;16:1085–1093. doi: 10.1200/JCO.1998.16.3.1085. [DOI] [PubMed] [Google Scholar]
- Sutterluty H, Chatelain E, Marti A, Wirbelauer C, Senften M, Muller U, Krek W. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat Cell Biol. 1999;1:207–214. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: Distinct E2F proteins mediate activation and repression. Genes & Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Odajima T, Nakano T, Kimijima Y, Yamada S, Ogi K, Kohama G. Immunohistochemical investigation of new suppressor oncogene p130 in oral squamous cell carcinoma. Oral Oncol. 1999;35:321–325. doi: 10.1016/s1368-8375(98)00089-x. [DOI] [PubMed] [Google Scholar]
- Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlach J, Hennecke S, Amati B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes & Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won K-A, Reed SI. Activation of cyclin E/CDK2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. EMBO J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- Woo MS, Sanchez I, Dynlacht BD. p130 and p107 use a conserved domain to inhibit cellular cyclin-dependent kinase activity. Mol Cell Biol. 1997;17:3566–3579. doi: 10.1128/mcb.17.7.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee AS, Shih HH, Tevosian SG. New perspectives on retinoblastoma family functions in differentiation. Front Biosci. 1998;3:D532–D547. doi: 10.2741/a301. [DOI] [PubMed] [Google Scholar]
- Yew PR. Ubiquitin-mediated proteolysis of vertebrate G1– and S-phase regulators. J Cell Physiol. 2001;187:1–10. doi: 10.1002/1097-4652(2001)9999:9999<1::AID-JCP1049>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Zhang S, Ramsay ES, Mock BA. Cdkn2a, the cyclin-dependent kinase inhibitor encoding p16INK4a and p19ARF, is a candidate for the plasmacytoma susceptibility locus, Pctr1. Proc Natl Acad Sci. 1998;95:2429–2434. doi: 10.1073/pnas.95.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]